Abstract
Fogo Selvagem (FS), the endemic form of pemphigus foliaceus, is mediated by pathogenic IgG4 autoantibodies against the amino-terminal extracellular cadherin domain of the desmosomal cadherin desmoglein 1 (Dsg1). Here we define the detailed epitopes of these pathogenic antibodies. Proteolytic footprinting showed that IgG4 from 95% of FS donor sera (19/20) recognized a 16-residue peptide (A129LNSMGQDLERPLELR144) from the EC1 domain of Dsg1 that overlaps the binding site for an adhesive-partner desmosomal cadherin molecule. Mutation of Dsg1 residues M133 and Q135 reduced the binding of FS IgG4 autoantibodies to Dsg1 by ~50%. Molecular modeling identified two nearby EC1 domain residues (Q82 and V83) likely to contribute to the epitope. Mutation of these residues completely abolished the binding of FS IgG4 to Dsg1. Bead aggregation assays showed that native binding interactions between Dsg1 and desmocollin 1 (Dsc1), which underlie desmosome structure, were abolished by Fab fragments of FS IgG4. These results further define the molecular mechanism by which FS IgG4 autoantibodies interfere with desmosome structure and lead to cell-cell detachment, the hallmark of this disease.
Keywords: cell adhesion molecule, desmosome, IgG4 autoantibody, skin autoimmunity
1. Introduction
Desmosomal cadherins, including desmogleins (Dsg1-4) and desmocollins (Dsc1-3), are desmosomal adhesion molecules that play a critical role in desmosome-mediated intercellular adhesions between epidermal keratinocytes [1]. Autoantibodies targeting Dsg1 and Dsg3 cause pemphigus foliaceus (PF) and pemphigus vulgaris (PV), a group of serious cutaneous blistering diseases [2]. Fogo Selvagem (FS), the endemic form of PF, is a cutaneous autoimmune blistering disease that exhibits geographic clustering in certain regions of Brazil [3, 4]. FS and PF are characterized by intraepidermal subcorneal blisters due to cell detachment, a process known as acantholysis [5]. It is well established that PF and FS patients possess IgG anti-epidermal autoantibodies, which correlate with disease-extent and activity [6, 7]. These autoantibodies bind specifically to Dsg1 [8] and do not bind other desmosomal cadherins, such as Dsg2, Dsg3, Dsg4 or Dsc1-3 [9–12]. Moreover, these autoantibodies are predominantly restricted to the IgG4 isotype [13], conformation- and calcium-dependent [14–17], carbohydrate-independent [17–19] and pathogenic [7, 13]. Both the whole IgG fractions and the F(ab)2 or monovalent Fab fragments from FS patients faithfully reproduce the human disease in neonatal micepassive transfer experiments [20, 21]. Additionally, the appearance of FS IgG4 anti-Dsg1 autoantibodies in individuals living in endemic areas of FS heralds the onset of disease by months or even years, i.e., these autoantibodies serve as disease predictors [22].
It was previously shown that the immunoreactive region of Dsg1 is located within the N-terminal EC domains of Dsg1 [23, 24]. Later, our group showed that IgG from FS patients preferentially binds to epitopes located on the EC1 and EC2 domains of Dsg1 [25]. However, these early studies were carried out using mainly domain-swapped segments of Dsg1 on Dsg3 backbones [24, 25]. Consequently, the fine specificity of the IgG4 anti-Dsg1 autoantibodies on Dsg1 had not been determined until now. Attempts to define Dsg1 epitopes may be relevant in understanding the molecular aspects of acantholysis and may provide new approaches to search for environmental antigens linked to FS.
In this investigation, we generated hybrid molecules encompassing the EC1 domain of Dsg1 grafted on three backbone (bb) carriers made of the EC2-EC5 domains of Dsg3 (Dsg1-EC1/Dsg3 bb), Dsg4 (Dsg1-EC1/Dsg4 bb) and desmocollin-1 (Dsc1) (Dsg1-EC1/Dsc1 bb). An additional hybrid was generated by inserting the EC1 domain of Dsg4 on a backbone of the EC2-EC5 domain of Dsg1 (Dsg4-EC1/Dsg1 bb). Using an enzyme-linked immunosorbent assay (ELISA), we showed that IgG4 from FS patients bound the Dsg1-EC1 in all three carriers; however, the hybrid of Dsg4-EC1 on the Dsg1 backbone was not reactive. Further, IgG4 autoantibodies bound specifically to a Dsg1-EC1/Dsg3 bb or Dsg1-EC1/Dsg4 bb Sepharose matrix and the eluted IgG4 autoantibodies were pathogenic when passively transferred into neonatal mice. We next tested purified FS IgG4 fractions from 20 FS patients by proteolytic footprinting [26–28] using recombinant Dsg1 ectodomain. Affinity-purified IgG4 from 19 out of 20 patients recognized a 16-amino acid peptide (A129LNSMGQDLERPLELR144); notably, residue A129 within the peptide lines the Trp 2 acceptor pocket, which is critical for adhesive binding. Site-directed mutagenesis of residues M133 and Q135 of the Dsg1-EC1 epitope decreased the binding of IgG4 anti-Dsg1 autoantibodies by approximately 50%. Additional mutations of residues Q82 and V83 of the Dsg1-EC1 epitope further decreased the binding of IgG4 autoantibodies to baseline levels. Most importantly, Fab fragments of FS IgG4 blocked heterophilic interactions of Dsg1 and Dsc1 in aggregation assays [29]. These findings further advance the understanding of the basic desmosomal cadherin interactions and how pathogenic autoantibodies from PF/FS patients specifically interfere with such interactions, possibly leading to cell detachment and blister formation.
2. Materials and methods
2.1. Sources of human FS sera
Well-characterized sera from 20 patients with classic FS, as defined by clinical, histological, immunological and epidemiological criteria [3–5], were available for epitope mapping studies. FS patients showed a widespread blistering eruption located on the trunk and extremities. Some patients showed facial and scalp lesions as well. Biopsies from these lesion showed typical subcorneal vesicles. Normal human sera from individuals living in endemic areas of FS in Brazil (n=30), and non-endemic areas of FS in Brazil (n=30) and the USA (n=30) were also tested. Sera of FS patients and controls were collected following IRB policies from the University of North Carolina at Chapel Hill (USA) and the University of São Paulo (Brazil). The sera are kept frozen at −30°C at the UNC Dermatology Research Laboratories.
2.2. Purification of IgG4 autoantibodies from FS sera
IgG4 autoantibodies were purified from the sera of FS patients by affinity chromatography using Capture Select® human IgG4 affinity matrix (BAC BV, Leiden, The Netherlands). The Capture Select® human IgG4 affinity matrix contains a 12kD llama antibody fragment that specifically recognizes human IgG4 without cross-reacting with other human IgG subclasses 1, 2 or 3. Briefly, FS serum was loaded onto the matrix and later washed with PBS, pH=7.4. The matrix was eluted with 0.1 M Glycine, pH=3.0. Bound and unbound fractions were tested by quantitative sandwich ELISA, developed in our laboratory using goat F(ab)2 anti-human IgG (Fab-specific) as the capture antibody, and monoclonal anti-human IgG or IgG subclass horseradish-peroxidase conjugates as detecting antibodies. Both fractions were dialyzed against PBS at pH=7.4, concentrated by ultrafiltration and stored at −20°C. The values of IgG and IgG subclasses were expressed as percentages from the total IgG loaded onto the matrix. The eluted fraction contained 97% IgG4. The unbound fraction contained 92% IgG1, with small amounts of IgG2 and IgG3. In addition, total IgG was purified by Protein G-affinity chromatography using HiTrap Protein G HP cartridge (GE Healthcare, Pittsburgh, PA, USA), from a normal individual living in endemic areas of FS. Normal human IgG4 kappa utilized as control was purchased from Sigma-Aldrich (Saint Louis, MO, USA).
2.3. Preparation of Fab fragments normal human and FS IgG4
4 mg of affinity purified IgG4 from a normal human and a FS patient (FS-1) were digested with papain according to PierceTM Fab Preparation Kit, Pierce Biotechnology(Rockford Illinois, USA), to generate IgG4 Fab fragments. The IgG4 Fab samples were concentrated to 1mg/ml using Amicon Ultra-0.5ml Centrifugal Filters Ultracel −3K (Merk Millipore Ltd Tullagreen Carrightwohill Co Cork, Ireland) and exchanged to PBS buffer at pH 7.5. The Fab fragments from normal human serum and FS-1 were tested by indirect IF against human skin cryosections, immunoblotting and ELISA using Fab and Fc specific sera. Additionally, the Fab fragments were tested by passive transfer experiments on the neonatal mouse model and the aggregation assay of bead coated with Dsg1 and Dsc1 as described by Harrison et al [29]
2.4. Plasmid constructs of Dsg1, Dsg3, Dsg4 and Dsc1
The ectodomain of human Dsg1 and human Dsg3 were already available in our laboratory [25]. Briefly, cDNA encoding the entire extracellular domain of Dsg1, including a COOH-terminal Histidine-tag, was subcloned into the baculovirus transfer vector pVL1393 (BD Biosciences, San Jose, CA, USA) and later sequenced to verify sequence integrity. The cDNA encoding the entire extracellular domain of human Dsg4 harboring an E-tag and His-tag was cloned into the pQE-TriSystem vector (QIAGEN, Inc., Chatsworth, CA, USA), as previously described [11]. Dsg4 cDNAs [11] and desmocollin-1 (Dsc1) cDNAs [12] were kindly provided by Dr. Masayuki Amagai (Keio University, Tokyo, Japan) and Dr. Takashi Hashimoto (Kurume University, Kurume, Japan), respectively.
2.5. Expression and purification of recombinant desmosomal proteins
Plasmids encoding the wild type ectodomains of desmosomal proteins and their mutants were co-transfected and expressed in a baculovirus expression system (BD BaculoGold, BD Biosciences, San Jose, CA, USA) using Sf9 insect cells media containing 5% fetal bovine serum(Invitrogen, Life Technologies Corp., Waltham, MA, USA), according to the manufacturer’s instructions. High-Five insect cells cultured in Express-Five serum-free medium (Life Technologies Corp.) were subsequently infected with baculovirus stocks to produce soluble recombinant antigens releasing in the culture supernatant. Next, antigens were isolated by affinity chromatography using Ni-NTA Superflow cartridges (QIAGEN, Chatsworth, CA), and subsequently dialyzed against Tris-buffered saline buffer containing 5 mM calcium (TBS-Ca2+) and stored at −80°C.
2.6. Generation of immunoreactive hybrid molecules by domain swapping
Three hybrid proteins were generated by introducing the EC1 domain of Dsg1 on three different backbones: the EC2 to EC5 domains of Dsg3, Dsg4 and Dsc1, as shown in Figure 2A. A fourth hybrid molecule was created by swapping the EC1 domain of Dsg1 for the EC1 domain of Dsg4 on a Dsg1 (EC2-5) backbone (Dsg4-EC1/Dsg1 bb) and used as a control. The Dsg1-EC1/Dsg3 bb construct has been previously described [25]. The Dsg1-EC1/Dsg4 bb construct was generated by grafting the EC1 domain of Dsg1 on the Dsg4 backbone (EC2-EC5) into a pQE-TriSystem vector harboring COOH-terminal E-Tag and His-tag, using specific primers: forward-5′-CAAAGGAGATATACCATGGACTGGAGTTTCTTCAGAG-3′ and reverse-5′-GTATACACTTTGCGATGAAAACACTGGAGGGTTGTC-3′ with NcoI/AccI cloning sites. The Dsg1-EC1/Dsc1 bb hybrid was generated using two pairs of specific primers. The first pair was used to generate the EC1 domain of Dsg1 including BglII/DralII insertion sites: forward-5′-GCCAGATCTCCTATAAATATGGACTGGAGTTTCTTCAG-3′ and reverse-5′-AGTCACTCTGTGTTCTGAAAACACTGGAGGGTTG-3′. The second pair was used to generate the EC2-EC5 domains of the Dsc1 backbone with DralII/XhoI sites: forward-5′-GAACACAGAGTGACTATCTTTACTG-3′ and reverse-5′-CCCCTCGAGTCTTCCAAGTATTACATTTG-3′. Both fragments were inserted into a receiving pEVmod vector harboring a COOH-terminal His-tag via BglII/XhoI to generate the Dsg1-EC1/Dsc1 bb harboring a Histidine tag [10]. Finally, the Dsg4-EC1/Dsg1 bb was digitally assembled from the reference sequences of human Dsg4 (uniprot.org, Q86SJ6) and human Dsg1 (uniprot.org, Q02413) with the additional sequences of the COOH-terminal His-tag and BamHI/NotI cloning sites. This DNA was synthesized and inserted into a pUC57 plasmid from GenScript Co. (Piscataway, NJ, USA) and later cloned into a pVL1939 vector. All four DNA constructs were expressed in insect cell culture and recombinant proteins were isolated as described above.
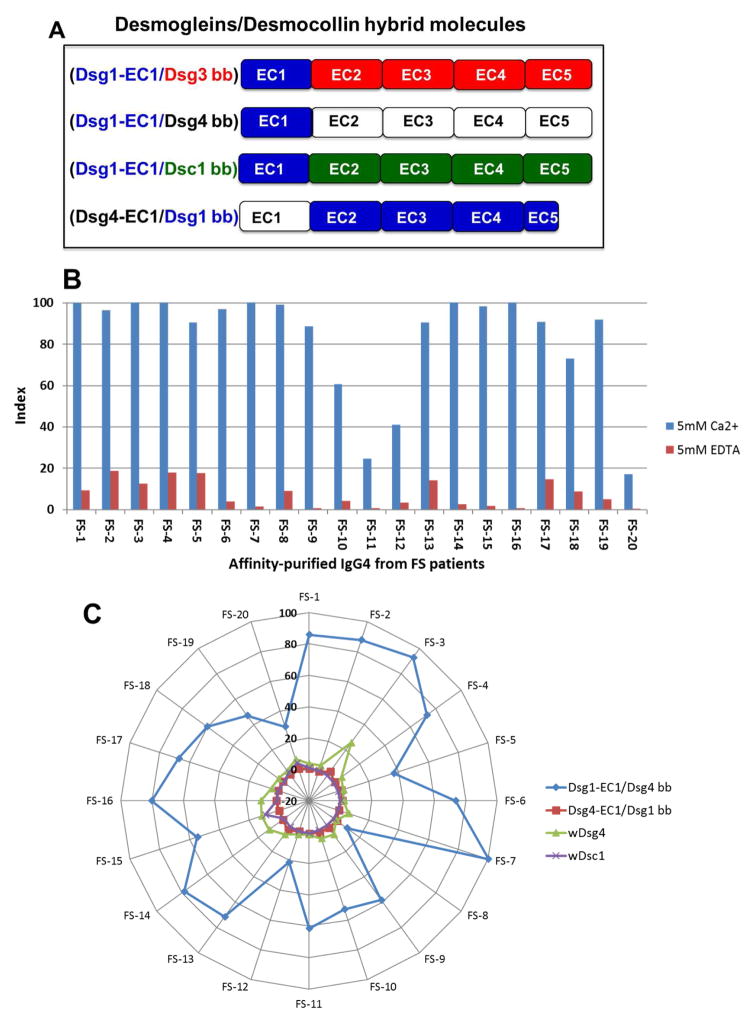
Figure 2. FS IgG4 autoantibodies mainly recognize Dsg1 EC1 domain.
(A) The EC1 domain of Dsg1 (blue) was grafted on three molecular backbones: Dsg3 (red domains), Dsg4 (white domains) and Dsc1 (green domains). We also grafted the EC1 domain of Dsg4 on a Dsg1 backbone (blue domains) as a control hybrid. These constructs were expressed in the baculovirus system and the recombinant proteins were used to test against FS sera. (B) Affinity-purified FS IgG4 on the llama anti-human IgG4 column was tested against chimeric Dsg1-EC1/Dsg3 bb in the presence of calcium and EDTA. A stock of IgG4 (0.5 mg/ml) of each patient at a dilution of 1:300 was tested on microtiter wells coated with 50 ng/well of Dsg1-EC1/Dsg3 bb. Peroxidase-labeled mouse anti-human IgG4 (Southern Biotech) at a dilution of 1:2,000 was used as an indicator. The results are expressed in index values. Similar results were observed when testing the FS IgG4 on plates coated with Dsg1-EC1/Dsg4 bb (data not shown). (C) Affinity-purified FS IgG4 on the llama anti-human IgG4 column was tested against chimeric Dsg1-EC1/Dsg4 bb (blue), Dsg4-EC1/Dsg1 bb (red), wDsg4 (green) and wDsc1 (purple) in the presence of calcium. A stock of IgG4 (0.5 mg/ml) of each patient at a dilution of 1:300 was tested on microtiter wells coated with 50 ng/well of the chimeric protein. Peroxidase-labeled mouse anti-human IgG4 (Southern Biotech) at the dilution of 1:2,000 was used as an indicator. The results are expressed in index values (Scale of -20 to 100 is shown in the figure).
2.7. Epitope mapping by epitope excision and mass spectrometry
Purified IgG4 from each of the 20 FS patients was immobilized on CNBr-activated Sepharose 4B (GE Healthcare, Pittsburgh, PA, USA) in compact reaction columns (CRC, USB Corporation, Cleveland, OH, USA) and incubated with the soluble ectodomain of human Dsg1. The Sepharose-IgG4-Dsg1 complex was then sequentially digested with sequencing grade chymotrypsin (Promega, Madison, WI, USA) for one hour at 37°C, immediately followed by digestion with sequencing-grade TPCK-treated trypsin (Worthington, Lakewood, NJ, USA) for 20 minutes at 37°C. Dsg1 peptides that remained bound to the human IgG4 after digestion were eluted with 0.1% Trifluoroacetic acid (Thermo Scientific, Waltham, MA, USA) and immediately analyzed on a 4800 Plus MALD-TOF/TOF-MS/MS in conjunction with Protein Pilot software (AB SCIEX, Foster City, CA, USA) [26–28]. The samples were spotted on a stainless-steel target with α-cyano-4-32hydroxycinnamic acid matrix (Sigma-Aldrich). The instrument used has a YAG laser with λ=355 nm. The potential difference between the source acceleration voltage and the collision cell was set at 2 kV. Calibration was done internally with self-digested TPCK-treated trypsin (same as above). Peak absorbances in MS spectra are not indicative of the abundance of peptide species due to differences in individual peptides’ ability to ionize. All analyses were done with Protein Pilot using an NCBI Mascot search.
2.8. Site-directed mutagenesis
Point mutations were carried out with a QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) following manufacturer’s instructions and using sets of primers containing the desired mutations. Thus, we generated a Dsg1 mutant named Dsg1-m1 [(M133R)(R135E)] by the replacing M133 and Q135 residues of Dsg1 with their Dsg4 counterparts (R133 and E135). We also generated a Dsg1-m2 [(M133R)(Q135E)-(Q82K)(V83I)] mutant, in which two additional amino acids, Q82 and V83 of Dsg1, were mutated by insertion of the corresponding residues of Dsg4.
2.9. Identification by molecular modeling of other residues relevant on the EC1-Dsg1 epitope
Finally, we used molecular modeling of the EC1 domain of Dsg1 to probe for additional components of the conformational epitope bound by FS IgG4 autoantibodies. Molecular modeling was performed at the R. L. Juliano Structural Bioinformatics Core Facility at the University of North Carolina at Chapel Hill. Structural templates were identified using the HHpred Fold Recognition Server [30, 31] and the extracellular domains of the human Dsg1 (hDsg1) and Dsc1 (hDsc1) homology models were built using the MODELLER software package [32]. The two structural templates used for modeling were: 1) PDB ID 3Q2W (mouse N-cadherin ectodomain), 2) PDB ID 3Q2V [mouse E-cadherin ecotodomain [33]], 3) PDB ID 5EQX (human Desmoglein-3), and 4) PDB ID 5IRY [human Desmocollin-1 (PUBMED: 27298358). The cadherin and desmocollin structural templates were in the homodimeric adhesive conformations. The structure of Dsg1 and Dsc1 were predicted in their heterodimeric adhesive conformation (dark gray and light gray monomers, respectively, (Figure 5).
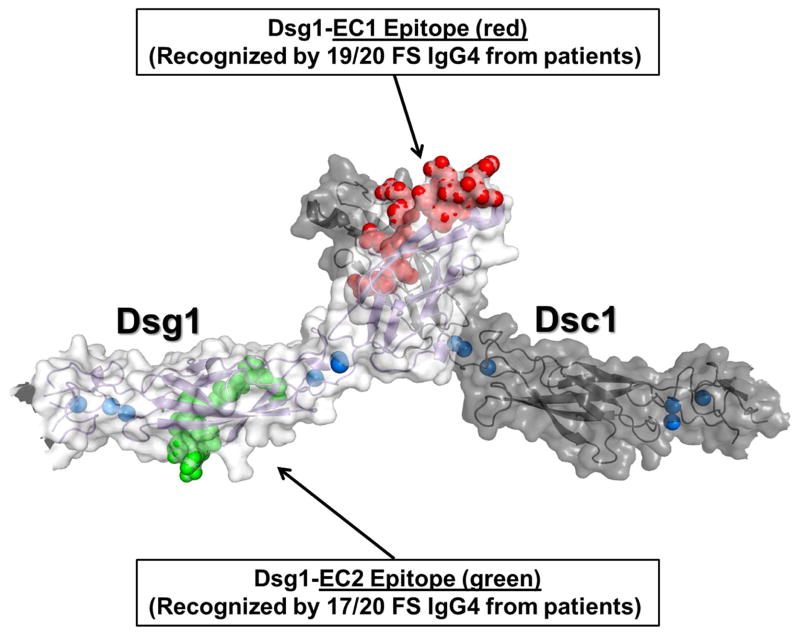
Figure 5. Homology model of trans heterodimer formed between EC1 domains of Dsg1 and Dsc1.
One Dsg1 monomer is colored light gray while Dsc1 is colored dark gray. Bound calcium ions are colored blue, with the EC1 and EC2 epitopes colored red and green, respectively.
2.10. Detection of IgG4 autoantibodies specific to desmosomal chimeric proteins, and the mutated proteins, by ELISA
Microtiter plates were coated with different amounts of each purified recombinant protein and normalized by anti-Histidine tag ELISA. Each normalized protein was then tested with different dilutions of the patient IgG4 fraction following a chessboard IgG4 ELISA protocol, as follows. ELISA plates were coated with each antigen in TBS buffer containing 5 mM of Ca2+(TBS-Ca2+), pH 7.5, at 4°C for overnight. After washing five times with TBS-Ca2+ buffer containing 0.05% Tween-20 (TBS-Ca2+/T-20), the plates were blocked with 1% BSA in TBS-Ca2+/T-20 at room temperature for one hour. For some experiments, we replaced calcium with 5 mM of EDTA in the buffer. Plates were washed and incubated with samples of diluted serum (1:300 from 0.5 mg/ml concentration) in duplicate for one hour at room temperature. Following the washes, plates were incubated with a 1:2,000 dilution of horseradish-peroxidase (HRP)–conjugated mouse anti-human IgG4 for one hour. The color development was achieved with the peroxidase substrate o-Phenylenediamine. Results were expressed as index values, calculated as previously reported [22]. The optimal amount of coating antigen and IgG4 dilution fraction was determined for each antigen tested.
2.11. Affinity purification of FS IgG4 autoantibodies specific to Dsg1-EC1/Dsg3 bb and Dsg1-EC1/Dsg4 bb chimeric proteins
FS IgG4 specific to the EC1 domain of Dsg1 was obtained by purifying the FS IgG4 using Ni-NTA Superflow matrix to immobilize the Dsg1-EC1/Dsg3 bb or Dsg1-EC1/Dsg4 bb protein. Briefly, 700 ml of the supernatant containing the chimeric protein from the High Five insect cell culture media were dialyzed against TBS-Ca2+, pH=7.5, with several changes. After the NaCl concentration was raised to 300 mM, 2 mM Imidazole were added and then the pH was adjusted to 8.0, the binding buffer condition. The supernatant was then cleared through the 0.45 micron filtration system (VWR, Radnor, PA, USA) and pumped onto the 5 ml Ni-NTA Superflow cartridge at a maximum flowrate of 1 ml per minute. After washing 20 column volumes with the binding buffer, and 10 column volumes with the binding buffer containing a higher concentration of Imidazole at 17 mM, the matrix-protein complex was removed from the cartridge and then divided into two portions of 2 ml and 3 ml. Each portion was transferred to a 15 ml column. The column containing the 3 ml of Ni-NTA Superflow matrix and protein complex was re-equilibrated with 20 column volumes of TBS-Ca2+, pH=7.5 and incubated with 10 ml of a FS IgG4 in TBS-Ca2+, pH=7.5, at the concentration of 0.59 mg per ml. The incubation was carried out by rotating at room temperature for three hours. After washing with TBS/Ca2+, pH 7.5 and high salt buffer (1.0M NaCl in TBS/Ca2+) the bound IgG4 was eluted with 4 M MgCl2 [25]. Each eluted fraction was determined by IgG4 ELISA to identify the existence of the IgG4 in the fractions. The identified fractions were pooled together, subjected to dialysis/ultrafiltration (Millipore, Billerica, MA, USA), using PBS, pH 7.5.
2.12. Passive transfer studies
Balb/c neonatal mice (24–36 hours old, body weight 1.4–1.8 g) were intradermally injected as previously described [7, 13, 20, 21], following IACUC UNC protocols with different quantities (contained in a volume of 100 μL) of either IgG4 or IgG1 purified fractions from FS sera. Commercially available human IgG4 (Sigma Aldrich, St Louis, MO, USA) was injected as a control. At 18–24 hours post-injection, the skin of the mice was evaluated for blisters. Skin biopsies were obtained for histology and direct immunofluorescence studies.
2.13. Bead aggregation and inhibition assays
Human Dsg1 EC1-4, Dsg3 EC1-5, and Dsc1 EC1-5 ectodomain fragments containing C-terminal biotinylated Avi-tags were purified from conditioned media of transfected human embryonic kidney (HEK) 293 cells as described previously [29]. Green or red fluorescent Neutravidin coated microspheres (Fluospheres, Life Technologies) were washed with binding buffer (BB) containing 10mM Tris-Cl pH 7.4, 150mM NaCl, 1mg/ml BSA and 0.05% Tween-20 and resuspended in BB containing 3mM CaCl2 and 5μM biotinylated Dsg1, Dsg3 or Dsc1. Samples were mixed by inversion for 45 minutes at 25°C to allow capture of biotinylated cadherin by neutravidin; unbound protein was then removed by washing with BB+3mM CaCl2. Coated beads were triturated to ensure monodispersity and mixed in equal quantities (approximately 5×107 beads total in a final volume of 10μl) in red/green combinations: Dsg1/Dsg1, Dsg3/Dsg3, Dsg1/Dsc1 or Dsg3/Dsc1. Mixtures were allowed to aggregate with inversion at 25°C for 1 hour and 5 μl samples were viewed at 4X magnification using a fluorescence microscope. For inhibition experiments, Dsg1 or Dsg3 coated beads were pre-incubated with purified IgG4 Fab fragments at concentrations of 0.5mg/ml (~10μM) to 0.03125mg/ml for 15 minutes prior to mixing with untreated Dsc1-coated beads for aggregation. Note that the antibodies remain present at half their initial concentration during aggregation. Representative images from duplicate experiments are shown [29].
3. RESULTS
3.1. Affinity-purified FS IgG4 autoantibodies to Dsg1 are pathogenic
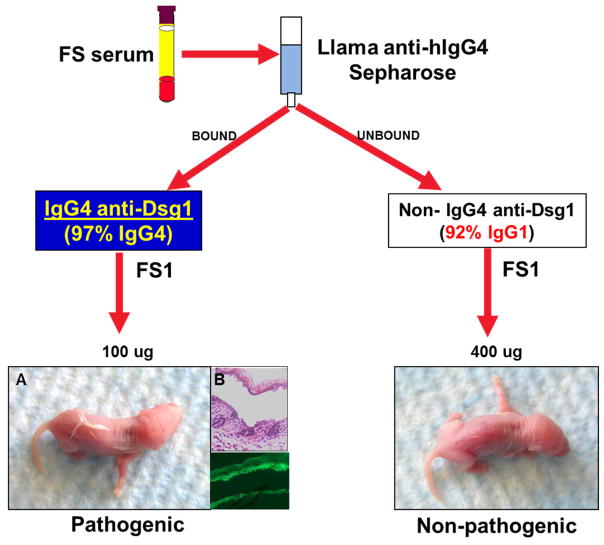
Since IgG4 autoantibodies are known to be involved in FS pathogenesis, we purified the IgG4 fractions from FS patients’ sera using a llama anti-human IgG4-Sepharose matrix. This bound fraction contained 97% IgG4, while the unbound fraction contained other IgG subclasses, of which IgG1 comprised 92%, as measured by quantitative IgG4 and IgG1 ELISA (data not shown). Both fractions stained the epidermal intercellular spaces when tested by indirect immunofluorescence (IF) (data not shown). Importantly, only the IgG4 fraction was pathogenic, inducing extensive blistering in neonatal mice, as determined by passive transfer experiments (Figure 1). Only 100 μg of the affinity-purified FS IgG4 was necessary to induce disease in mice. In contrast, 400 μg of the unbound IgG1 fraction was not pathogenic in the mouse model.
Figure 1. FS IgG4 autoantibodies are pathogenic.
The IgG4 fraction from FS1 serum (from patient #1 in the set of 20 tested, FS1 is used throughout the paper) was purified on a llama anti-human IgG4 Sepharose affinity column. The bound fraction contained 97% pure IgG4. The unbound fraction contained non-IgG4 subclasses with IgG1 representing the majority (92%). The IgG4 and non-IgG4 fractions were dialyzed, concentrated and injected subcutaneously into neonatal mice. A. Mice injected with FS1 IgG4 (left) developed extensive blistering, shown as fine wrinkling of the epidermis (induced by slight friction or pinching, the so-called Nikolsky’s sign). B. On histological examination large sheets of epidermis were separated from the naked dermis (x200) and direct immunofluorescence shows bound IgG4 to the roof of the split (x100). Mice receiving a higher dose of non-IgG4 fraction showed no disease (right).
We then obtained IgG4 anti-Dsg1-specific autoantibodies by affinity chromatography using the ectodomain of Dsg1 linked to agarose and the purified FS IgG4 fraction described above. The FS IgG4 anti-Dsg1 fractions were pathogenic in the mouse model, reproducing the classical features of FS in the epidermis of these animals (photo not shown). 10 μg of anti-Dsg1 IgG4 was sufficient to induce extensive skin blistering in these animals. These results strongly suggest that the epitope(s) recognized by FS IgG4 autoantibodies on Dsg1 are responsible for the blister formation in FS.
3.2. FS IgG4 autoantibodies recognize hybrid molecules bearing the EC1 domain of Dsg1 on different backbones (Dsg3, Dsg4 and Dsc1)
To determine which domains of Dsg1 are bound by FS IgG4 autoantibodies, we tested four recombinant hybrid proteins: (Dsg1-EC1/Dsg3 bb, Dsg1-EC1/Dsg4 bb, Dsg1-EC1/Dsc1 bb and Dsg4-EC1/Dsg1 bb) by ELISA (Figure 2A).
Grafting of Dsg1’s EC1 domain on an EC2-EC5 backbone of Dsg3, Dsg4 and Dsc1 (Figure 2A) made these hybrid molecules become reactive with affinity-purified IgG4 from FS patients. The reactivity of FS IgG4 autoantibodies with wild type Dsg1 (wDsg1) was the strongest. The reactivity was lower in the Dsg1-EC1/Dsg3 bb, Dsg1-EC1/Dsg4 bb and Dsg1-EC1/Dsc1 bb hybrids, as determined by ELISA (data not shown). We used the Dsg1-EC1/Dsg3 bb or Dsg1-EC1/Dsg4 bb hybrid to affinity purify Dsg1-EC1-specific FS IgG4 autoantibodies. These affinity-purified FS IgG4 autoantibodies were pathogenic, as shown by passive transfer experiments into neonatal mice (photo not shown). Figure 2B shows the reactivity of FS IgG4 autoantibodies derived from 20 patients to the Dsg1-EC1/Dsg3 bb hybrid, which was abolished when EDTA was incorporated in the ELISA reaction buffer. The same results were obtained with Dsg1-EC1/Dsg4 bb (data not shown), suggesting that the fractions of IgG4 purified by these hybrid proteins also bind to Ca2+-dependent conformational epitopes. Figure 2C shows the reactivity of FS IgG4 autoantibodies from 20 FS patients with Dsg1-EC1/Dsg4 bb, Dsg4-EC1/Dsg1 bb, wDsg4 and wDsc1 by ELISA, demonstrating the specificity of these autoantibodies for the EC1 domain of Dsg1.
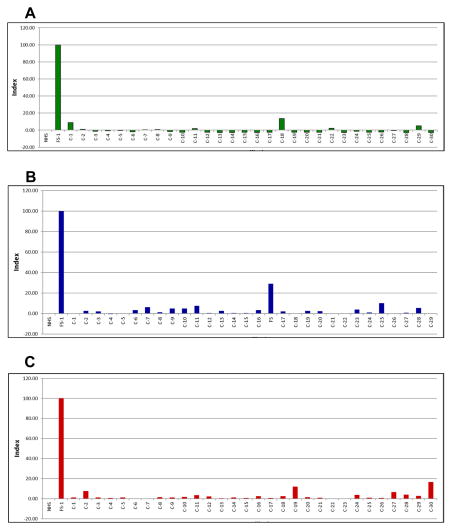
Finally, the sera of 30 healthy individuals living in endemic areas of FS in Brazil, 29 from non-endemic areas of FS (Brazil) and 30 from the US did not react with hybrid Dsg1-EC1/Dsg3 bb, as determined by ELISA (Figure 3), further confirming the Dsg1 EC1 specificity of the FS IgG4 antibodies.
Figure 3. Normal donors from endemic, non-endemic and US do not recognize Dsg1-EC1/Dsg3 bb.
The sera of three sets of normal individuals were tested by ELISA against Dsg1-EC1/Dsg3 bb coated plates. The sera were tested at 1:300 dilution and mouse anti-human IgG4 was used at the 1:2000 dilution. The plates were coated with 100 ng/well of Dsg1-Ec1/Dsg3 bb. The results are expressed in index values (scale of -20 to 100 is shown in the figure). Panel A shows the lack of reactivity of the sera of thirty normal individuals (C-1 to C-30) from the highly endemic area of Limao Verde, Brazil [4] compared with the reaction of FS-1 included as a positive control. Normal human serum (NHS) from the US served as a negative control. Panel B shows the lack of reactivity of the sera of twenty nine normal individuals (C1 to C-29) from non-endemic areas of Brazil (urban inhabitants of the cities of Sao Paulo and Belho Horizonte) compared with FS-1 and NHS. Donor C-17 was found later to be a case of FS undergoing therapy and in clinical remission. Panel C shows the lack of reactivity of the sera of thirty normal individuals (C-1 to C-30) from the UNC Blood Bank compared with FS-1 and NHS.
3.3. Identifying Dsg1 conformational epitopes that are recognized by IgG4 autoantibodies from 20 FS patients
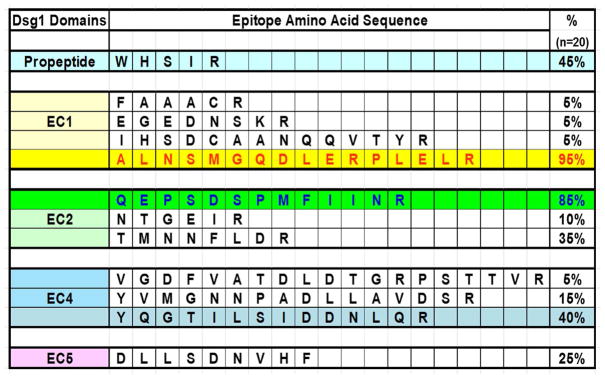
To determine the locations and sequences of the Dsg1 epitopes that are recognized by FS IgG4 autoantibodies, we employed epitope excision and mass spectrometry techniques [26–28]. From the purified IgG4 fractions of 20 FS patients, 19 (95%) recognized a 16-amino acid peptide located on the EC1 domain of Dsg1 (residues A129 – R144) (Figure 4). One of these residues (A129) at the N-terminal end of the peptide is known to line the Trp 2 acceptor pocket that functions in Dsg1 adhesion. The C-terminal end of the peptide is close to the Ca2+-binding linker between EC1 and EC2 [34]. A second epitope that comprises 13 residues, located on the EC2 domain of Dsg1 (residues Q201 – R213), was recognized by IgG4 fractions of 17/20 FS IgG4 (85%). Figure 5 shows the location of the two main epitopes recognized by FS IgG4 autoantibodies on a homology model of trans-dimeric structure of Dsg1 and Dsc1. Additional epitopes mapped to the propeptide sequence, and the EC4 and EC5 domains of Dsg1, but in lower frequencies (5–45%) (Figure 4).
Figure 4. Epitopes recognized by FS IgG4 autoantibodies.
Five Dsg1 peptides were bound by FS IgG4 from 20 patients: Propeptide (W41 to R45) (45%); EC1 (A129 to R144) (95%); EC2 (Q201 to R213) (85%); EC4 (Y460 to R473) (40%) and EC5 (D538 to F546) (25%). This investigation focused on the EC1 epitope.
To substantiate the epitope mapping results, we utilized the IgG4 fraction purified from healthy human serum. This fraction did not recognize any epitopes on Dsg1 by our epitope excision method. To confirm that the identified peptides were specific, we digested the recombinant Dsg1 with trypsin/chymotrypsin prior to incubation with FS IgG4-Sepharose. No relevant peptides were detected by MALDI-MS/MS in these digested fractions. These results suggest that the majority of identified peptides bound by FS IgG4 contain amino acids that are components of the epitopes.
Furthermore, the two major peptides identified within the EC1 and EC2 domains were synthesized alone or linked to biotin. The synthetic peptides were used to perform ELISAs and inhibition assays to determine if these linear epitopes bind FS IgG4 autoantibodies. None of the FS sera or purified IgG4 fractions recognized the synthetic peptides, as demonstrated by direct ELISA, or ELISAs on Streptavidin-coated microtiter plates (data not shown). These findings indicate that FS IgG4 autoantibodies bind conformational epitopes.
3.4. Point Mutations of the EC1 domain of Dsg1 further define the EC1 conformational epitope bound by FS IgG4 autoantibodies
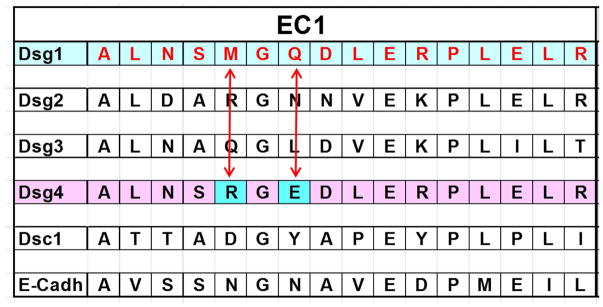
The 16 residues of the Dsg1-EC1 epitope obtained by epitope excision and mass spectrometry were compared with the analogous regions of other desmosomal cadherins, as shown in Figure 6. We found that the sequence of the Dsg1-EC1 epitope differs from its Dsg4 counterpart in only two residues: M133 and Q135 in Dsg1, which correspond to R133 and E135 in Dsg4. Knowing that the FS IgG4 does not bind to Dsg4 (Figure 2C), we used site directed mutagenesis to mutate the M133 and Q135 residues of Dsg1 to Dsg4 residues R133 and E135, respectively. The resultant mutant was named Dsg1-m1 (M133R)(Q135E) (Figure 7A).
Figure 6. The 16-residue alignment among different desmosomal proteins.
The 16-residue peptide bound by 19/20 FS IgG4 (A129 to R144) on Dsg1 was aligned with similar peptides from Dsg2, Dsg3, Dsg4, Dsc1 and E-Cadherin. The Dsg4 peptide shows two residues, R133 and E135 that are different from the Dsg1 counterpart. Human Dsg4 does not bind FS IgG or IgG4 autoantibodies.
Figure 7. Identification of key amino acid residues for FS IgG4 binding.
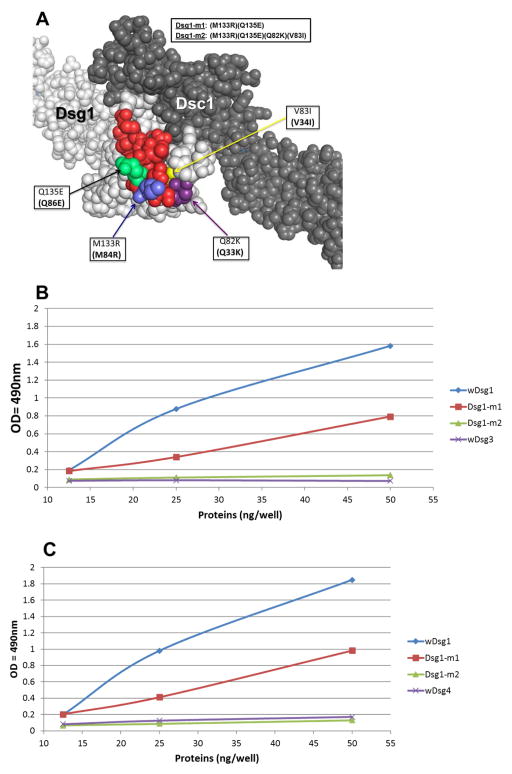
(A)The locations of the mutated residues (blue, lime green, yellow and dark purple) in the EC1 epitope (red) are shown on the homology model of the Dsg1/Dsc1 heterodimer (light (Dsg1) and dark (Dsc1) gray). (B) Affinity-purified FS1 IgG4 antibodies on the llama anti-human IgG4 and Dsg1-EC1/Dsg3 bb columns were tested against wDsg1 (blue), Dsg1-m1 (red), Dsg1-m2 (green) and wDsg3 (purple) by ELISA. A stock of IgG4 (0.3 mg/ml) from each patient at a dilution of 1:5,000 was tested on microtiter wells coated with different amounts of the recombinant protein (from 12.5 ng/well to 50 ng/well). Peroxidase-labeled mouse anti-human IgG4 at a dilution of 1:2,000 was used as an indicator. The results are expressed in OD490nm units. (C) Affinity-purified FS1 IgG4 antibodies on the llama anti-human IgG4 and Dsg1-EC1/Dsg4 bb columns were tested against wDsg1 (blue), Dsg1-m1 (red), Dsg1-m3 (green) and wDsg4 (purple) by ELISA. A stock of IgG4 (0.3 mg/ml) from each patient at a dilution of 1:5,000 was tested on microtiter wells coated with different amounts of the recombinant protein (from 12.5 ng/well to 50 ng/well). Peroxidase-labeled mouse anti-human IgG4 at a dilution of 1:2,000 was used as an indicator. The results are expressed in OD490nm units.
We determined that the Dsg1-EC1 epitope, as well as residues M133 and Q135, were located close to the acceptor pocket of Dsg1, and mutating these residues led to a decrease in autoantibody binding affinity of over 50% by ELISA. Two additional residues, Q82 and V83, which are in proximity to the hydrophobic pocket were considered as possible components of the EC1 epitope based on their location in the homology modeled structure (Figure 7A). These residues were therefore mutated to the amino acids found in Dsg4, K82 and I83 and the double mutant was named Dsg1-m2 [(M133R)(Q135E)-(Q82K)(V83I)].
FS IgG4 autoantibodies specific for the EC1 domain of Dsg1 were affinity purified using immobilized Dsg1-EC1/Dsg3 bb hybrid protein and then tested against wDsg1, wDsg3, Dsg1-m1 and Dsg1-m2 by ELISA (Figure 7B). The same FS IgG4 autoantibodies were affinity purified using Dsg1-EC1/Dsg4 bb hybrid protein and tested against wDsg1, wDsg4, Dsg1-m1 and Dsg1-m2 (Figure 7C). The reactivity of both sets of FS IgG4 autoantibodies was reduced by approximately 50% on plates coated with Dsg1-m1 protein compared to wDsg1. Further reduction to background levels was seen in plates coated with Dsg1-m2, demonstrating that residues Q82 and V83 are part of a larger conformational epitope that consists of amino acids from multiple discontinuous secondary structure elements. Consistent with our previous findings, wDsg3 and wDsg4 were not recognized by FS IgG4 autoantibodies.
Finally, another mutant was constructed by introducing the same amino acid substitutions of Dsg1-m2 into the hybrid Dsg1-EC1/Dsg4 molecule. Likewise, this mutant also has no reactivity with FS IgG4 autoantibodies, as determine by ELISA (data not shown).
3.5. FS IgG4 Fab inhibits the heterophilic interaction of Dsg1 and Dsc1
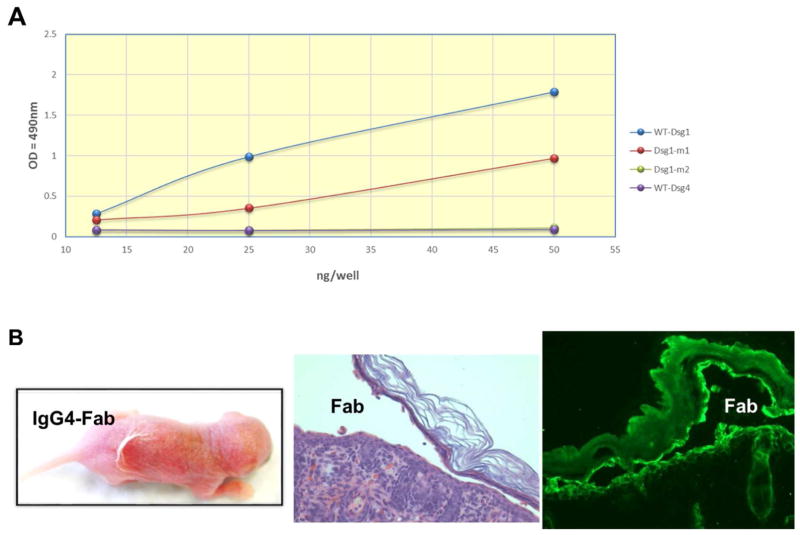
Since the major epitope of FS IgG4 identified in this work overlaps the Trp 2 acceptor pocket critical for Dsg1 adhesive interactions, we hypothesized that these autoantibodies could interfere with trans interactions between Dsg1 and its partner Dsc1 [29]. To test this hypothesis, we prepared Fab fragments from FS IgG4. Like the whole IgG4, Fab fragments from FS-1 reacted with wDsg1 but not with Dsg1-m2 by ELISA (Figure 8A) and induced skin disease in mice by passive transfer (Figure 8B).
Figure 8.
(A) Fab fragments from FS-1 IgG4 bind wDsg1(blue), bind partially to Dsg1-m1 (red) and do not bind Dsg1-m2 (green) or to wDsg4 (purple) that overlap in a single line. (B) Same Fab fragments from FS1 IgG4 are pathogenic when tested on the mouse model. The animals show skin blisters, which are subcorneal and the Fab fragments are bound to the surface of keratinocytes undergoing detachment at the site of injury by direct immunofluorescence (IF). Orignal magnification of histology and direct IF, x200.
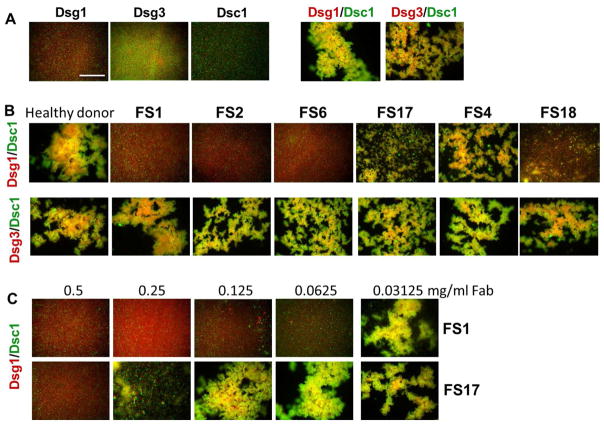
The Fab fragments from high titer (FS1, FS2 and FS6), mid titer (FS18) and low titer (FS17 and FS4) IgG4 fractions were coded and then tested blindly for their ability to inhibit aggregation of fluorescent beads coated with desmosomal cadherin adhesive pairs. Mixtures of beads coated separately with heterophilic pairs Dsg1/Dsc1 or Dsg3/Dsc1 result in robust aggregation in this assay due to heterophilic trans interactions, while beads coated with only the individual desmosomal cadherins remain monodisperse (Figure 9A and ref. 29). Remarkably, un-blinding of the inhibition results revealed that pre-incubation with Fab fragments from donors FS1, FS2, and FS6 completely inhibited the aggregation of beads coated with Dsg1 and Dsc1, whereas the normal control Fab fragments showed little effect (Figure 9B, top panel). Fab fragments from donors FS4, FS17, and FS18 showed intermediate levels of inhibition consistent with a lower titer of FS autoantibodies observed. The FS Fab fragments showed no effects on the aggregation of beads coated with Dsg3/Dsc1 confirming that the inhibitory effect was mediated via Fab binding specifically to Dsg1 (Figure 9B, center panels). Titration of the high titer FS1 and lower titer FS17 Fab fragments in the inhibition assay with Dsg1/Dsc1 showed dose-dependent inhibition with complete inhibition observed above 0.0625mg/ml Fab for FS1 and only at the highest concentration tested (0.5mg/ml) for FS17 (Figure 9C, lower panels).
Figure 9. Inhibition of desmosomal cadherin-mediated bead aggregation by FS antibodies.
(A) Co-aggregation of cadherin-coated red and green fluorescent beads mixed in homophilic (Dsg1/Dsg1, Dsg3/Dsg3, Dsc1/Dsc1) or heterophilic combinations (Dsg1/Dsc1 and Dsg3/Dsc1). (B) Inhibition experiments showing heterophilic aggregation of green Dsc1-coated beads with red Dsg1 (top panels) or Dsg3- coated beads (bottom panels) pre-incubated with purified IgG4 fractions from FS patients or from a healthy donor (see methods for details). (C) Dose-dependence of IgG4 Fab mediated inhibition of Dsg1/Dsc1 bead aggregation from donors FS 1 and FS17. Scale bar 0.5mm.
4. DISCUSSION
We have previously demonstrated that autoantibodies from FS patients with active disease bind to the EC1 and EC2 domains of Dsg1, whereas patients in clinical remission recognize EC5 [25]. In the current study, we further demonstrate that pathogenic FS IgG4 autoantibodies bind a conformational and Ca2+ dependent epitope located on the EC1 domain of Dsg1 inserted in hybrid molecules composed of the EC2 to EC5 domains of Dsg3, Dsg4 and Dsc1. We then applied epitope excision and mass spectrometry procedures [26–28] to further dissect the location of the Dsg1-EC1 conformational epitope. We found that affinity-purified FS IgG4 from 19 out of 20 patients recognize a 16-amino acid peptide located on the EC1 domain of Dsg1 (residues A129 – R144). Mutation of residues M133 and Q135 of this peptide reduce the binding of FS IgG4 autoantibodies by approximately 50% compared to binding to wDsg1, implying that these amino acids form part of the conformational epitope. Moreover, tridimensional atomic modeling of Dsg1 suggests that two of these residues (A129 and L130) on the N-terminus of the peptide overlap with the “RAL adhesive” site of Dsg1 and its C-terminus is separated by V145 from the Ca2+ binding sequence (R146 – N153) of EC1 [34]. The modeling information also suggested that two additional residues, Q82 and V83, were possible components of the conformational epitope. Mutation of these additional residues originated mutants that failed to bind FS IgG4 autoantibodies when tested by ELISA. Hence, M133, Q135, Q82 and V83 are residues that form part of the Dsg1-EC1 conformational epitope that are recognized by pathogenic FS IgG4 autoantibodies.
Sekiguchi et al. [24] generated a Dsg3 mutant by introducing three residues of Dsg1 (H25, C28 and A29, numbering from mature protein) to Dsg3. The resultant Dsg3 mutant became reactive with PF autoantibodies. The residues H74, C77 and A78 are near the Q82 and V83 residues identified in our study as crucial components of the Dsg1 epitope recognized by FS IgG4 autoantibodies. Our studies also identified other epitopes located on the EC2 domain (85%), propeptide (45%), and the EC4 and EC5 domains in lower frequencies. The Dsg1 EC2 epitope may possess unique immunological characteristics that needs to be addressed in detail, since in our initial studies FS IgG4 autoantibodies showed no or weak reactivity by ELISA with chimeras containing the Dsg1 EC2 domain grafted on Dsg3 or Dsg4 backbone. Hence, characterization of this Dsg1 EC2 epitope will be the subject of a subsequent communication. Previous studies have reported that non-pathogenic monoclonal antibodies derived from PF patients [35, 36] and endemic Tunisian PF [37] bind the precursor peptide of Dsg1. Such antibodies may develop as a consequence of self-immunization to intracellular keratinocyte antigens exposed during epidermal injury. The relevance of other epitopes recognized by FS IgG4 will be the subject of further studies. It is worth considering that the IgG autoimmune response to Dsg1 in FS patients may follow an epitope spreading mechanism, as suggested by Li et al. [25].
In the epidermis, four desmogleins (Dsg1, Dsg2, Dsg3 and Dsg4) and three desmocollins (Dsc1, Dsc2 and Dsc3) comprise the desmosomal cadherins that show differential expression patterns and play a critical role in epidermal integrity [1, 25]. The assembly and disassembly of desmosomes, the trafficking and turnover of desmosomal cadherins and their role in cell signaling have been thoroughly reviewed [38–41]. Mechanotransduction processes mediated by cadherins and likely desmosomal cadherins have been recently reported [42–45]. The implications of these later mechanisms in acantholysis remain unexplored. It is curious that the integrity of normal epidermis, (a highly specialized “biochemical Velcro” for single cell-cell adhesion [44] is maintained by cell adhesion molecules interacting with structural cytoskeletal proteins. The epidermis, at both the tissue and cellular level, remains flexible and resilient when exposed to mechanical forces. For example, single applications of moderate friction to the epidermis are harmless in healthy individuals; however, similar friction applied to the affected epidermis of FS patients removes superficial layers of epidermis due to the underlying acantholysis, a phenomenon known as Nikolsky’s sign [46].
Cytoplasmic regions of desmosomal cadherins are linked to cytokeratin intermediate filament networks via desmosomal plaque proteins. Their ectodomains interact with each other through trans and cis interactions, leading to junction assembly and cell-cell adhesion. For trans interactions, Trp 2 from a desmosomal cadherin from one cell surface is inserted into an “acceptor pocket” in an adhesive-partner desmocollin from an apposed cell. This pocket in part overlaps the RAL (desmogleins) or YAT (desmocollins) sequences [47, 48]. Mutations of Trp 2 or residues or the Trp 2 binding pocket impair the adhesive function of these molecules [49–53]. Previous studies from different laboratories have suggested either homophilic [54] or heterophilic interaction [55] between the ectodomains of desmogleins and desmocollins. A recent study by Harrison et al. [29] has demonstrated that strong trans interactions are formed between desmogleins and desmocollins, while desmocollin/desmocollin and desmoglein/desmoglein trans interactions are weak. These data suggest that heterophilic interaction between Dsg1 and Dsc1 may represent the dominant mode of adhesive mechanisms in the upper epidermis [29].
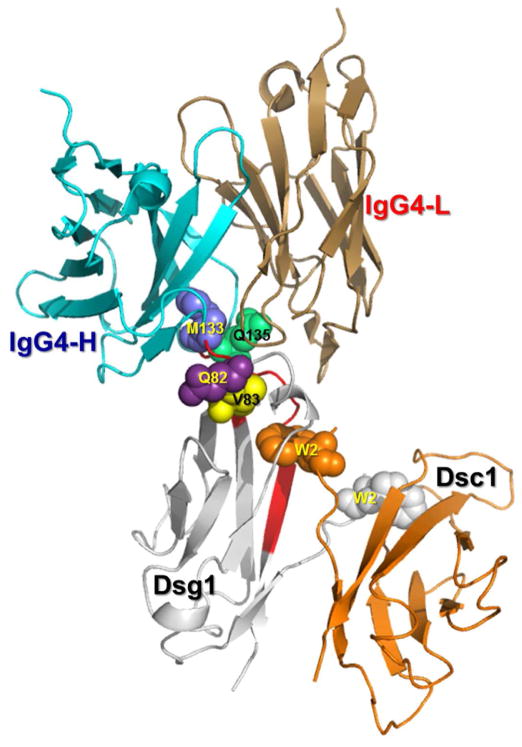
Binding of FS IgG4 autoantibodies to their epitope induces acantholysis and blistering in vivo, as demonstrated by passive transfer experiments in neonatal mice [7, 13, 20, 21]. It is envisioned that the IgG4/Dsg1 immune complexes are localized on the surface of keratinocytes facing the intermembranous spaces, known as epidermal intercellular spaces (ICS). The epidermal ICS was conceived as a dynamic “biological molecular mesh” formed by interacting surface transmembrane glycoproteins of cells sharing this compartment [56]. It was also suggested that external environmental factors and internal constituents, including antibodies, were likely to permeate the epidermal ICS by diffusion. Thus, pathogenic autoantibodies would diffuse out of the intravascular space to the dermis, pass the dermal/epidermal junction and reach the epidermal ICS where they bind their epitopes, triggering the detachment of epidermal cells. As demonstrated in this study, FS IgG4 autoantibodies bind an epitope in close proximity to the “acceptor pocket” of Dsg1, which impairs the adhesive interactions between Dsg1/Dsc1, thus leading to acantholysis. Additional data generated by Fab fragments from FS IgG4 that bind Dsg1, induces disease is neonatal mice by passive transfer and impairs the aggregation of beads coated with Dsg1 and Dsc1 further supports this hypothesis. Figure 10 shows the binding of IgG4 on the Dsg1/Dsc1 trans-interactive dimer. The antigen-binding site of the FS IgG4 autoantibody binds an epitope, where M133, Q135, Q82 and V83 residues are relevant components of the conformational epitope.
Figure 10. Modeled complex of Fab IgG4 on the modeled Dsg1/Dsc1 heterodimer showing the potential interaction of the FS IgG4 binding region with the Dsg1 conformational epitope.
Residues M133, Q135, Q82 and V83 (blue spheres) of Dsg1 are located close to the hydrophobic acceptor pocket, where the Trp2 (W2) of Dsc1 inserts. The EC1 domains of Dsg1 (magenta) and Dsc1 (pink) are shown as a strand swapped heterodimer, in which both Trp2 residues are exchanged between partnering molecules. It is hypothesized that the binding of FS IgG4 (cyan/sand) to its Dsg1 conformational epitope impairs the heterophilic interaction and adhesion of Dsg1 and Dsc1.
The amounts and the specificity of FS IgG4 autoantibodies reaching their target epitope in the epidermal ICS are known to be relevant in the induction of cell detachment. Also important in this detachment process might be the availability of the target epitope on Dsg1. Consequently, the synthesis, turnover and trafficking of these desmosomal cadherins likely influences the acantholysis process triggered by these autoantibodies. How binding of these autoantibodies to an epitope on the EC1 domain of Dsg1 triggers cell detachment is open to further investigation. It is known that this process is complement-independent [20, 21], but other mechanisms may be at work, such as internalization of the autoantibody/Dsg1 immune complexes and removal of the Dsg1 monomer/dimer from the interacting Dsc1, which would impair the adhesive function of these molecules [57–61]. Although theoretical, it is feasible that binding of autoantibodies to their target epitope may induce antigenic modulation in the keratinocyte, with a decrease of Dsg1 on the keratinocyte surface. Some investigators have proposed that pathogenic autoantibodies bind and impair the adhesive function of Dsg1 by steric hindrance [2], while others have shown that IgG from pemphigus patients, after binding their epitopes, triggers intracellular signaling [62–65]. It has also been proposed that acantholysis may result from a combination of both processes. In the cadherin field, investigators have used polyclonal and monoclonal antibodies against conformational epitopes located on the ectodomains of these molecules and showed that they induce cadherin conformational changes and allosteric activation of adhesion [66–68]. Our study suggests that FS IgG4 autoantibodies may impair the adhesive function of Dsg1 by steric hindrance.
Finally, defining relevant epitopes on Dsg1 would enable us to further investigate whether environmental antigens cross-react with FS autoantibodies that directly or indirectly contribute to the development of pathogenic IgG4 autoantibodies in FS. Our previous epidemiological studies suggest that exposure to hematophagous insect bites may be a risk factor of FS [3, 4, 69]. In addition, we have recently identified a Lutzomyia longipalpis salivary antigen (LJM11) that cross-reacts with FS IgG4 [70–72]. It remains unclear whether the LJM11 antigen mimics the pathogenic or non-pathogenic epitopes on Dsg1. If the epitope on LJM11 shares its conformational structure with non-pathogenic epitopes on Dsg1, this would suggest that LJM11 may serve as an antigen that activates naïve B cells, which would subsequently facilitate pathogenic autoantibody development via an epitope spreading mechanism [25]. Therefore, determining a disease-specific conformational epitope(s) on Dsg1 may also help to identify environmental trigger(s) of autoimmune response in FS.
5. Conclusions
Structural and biological studies have recently established that “trans” interactions of desmosomal desmogleins (Dsg) and desmocollins (Dsc) are heterophilic. Hence, the RAL of Dsg and the YAT of Dsc, located on the EC1 domain of these molecules constitute the acceptor pockets where the W2 of Dsg or Dsc respectively are inserted generating adhesive forces that bring about cell-cell adhesion [29]. Epitope mapping studies carried out with pathogenic IgG4 autoantibodies from fogo selvagem (FS) patients bind an epitope located on or around the RAL pocket of Dsg1. Mutation of four residues of the Dsg1 pocket abolish the binding of FS IgG4 autoantibodies. Additionally, the Fab fragments of FS IgG4 autoantibodies inhibit the heterophilic aggregation of Dsg1/Dsc1 in a dose dependent manner, but do not alter the aggregation of Dsg3/Dsc1. These studies strongly suggest that pathogenic FS IgG4 autoantibodies induce cell detachment and blisters in the epidermis of FS patients by inhibiting the interaction of Dsg1 and Dsc1 desmosomal cadherins. Steric hindrance and/or intracellular signaling or apoptosis are possible mechanisms under investigation.
Highlights.
Autoimmune Fogo Selvagem is mediated by pathogenic IgG4 anti-Dsg1 autoantibodies
Desmogleins (Dsg) and desmocollins (Dsc) TRANS interact to mediate epidermal adhesion
IgG4 anti-Dsg1 autoantibodies inhibit the trans interaction of Dsg1 and DsC1
Pathogenic IgG4 anti-Dsg1 autoantibodies bind an epitope on the RAL site of Dsg1
Pathogenic IgG4 anti-Dsg1 autoantibodies cause cell detachment and blisters
Acknowledgments
This work was supported by the National Institute of Health [RO1 AR32599, LA Diaz; P30 CA016086, BR Temple; R01 AR067315, Y Qian; R01 AR061372, N Li; AI07924 and AI40768, Z Liu; GM118584, L Shapiro]. We thank Dr. Ronald J. Falk for from the UNC Nephrology Division for sharing with us the epitope excision and mass spectrometry techniques for epitope mapping. We also acknowledge the important input of Dr. Jesus Valenzuela from the Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, Rockville, Maryland.
Abreviations
- FS
fogo selvage
- Dsg
desmoglein
- Dsc
desmocollin
- EC
extracellular
- PF
pemphigus foliaceus
- HEK
human embryonic kidney
Footnotes
Conflict of interest statement: The authors have declared that no conflict of interest exists.
Author contribution:
Designed the study: L.A.D, Z.L., B.T. and N.L.
Conducted experiments: F.E., A.R., P.P., O.J.H, J.B.
Data Analysis: L.A.D., N.L., J.V., Z.L., D.C., B.H., L.S. and Y.Q.
Wrote the manuscript: F.E., N.L., J.V., Y.Q., D.C., Z.L., J.B., O.J.H., B.H., L.S, and L.A.D.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harbor perspectives in biology. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amagai M, Stanley JR. Desmoglein as a target in skin disease and beyond. J Invest Dermatol. 2012;132:776–84. doi: 10.1038/jid.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz LA, Sampaio SA, Rivitti EA, Martins CR, Cunha PR, Lombardi C, et al. Endemic pemphigus foliaceus (Fogo Selvagem): II. Current and historic epidemiologic studies. J Invest Dermatol. 1989;92:4–12. doi: 10.1111/1523-1747.ep13070394. [DOI] [PubMed] [Google Scholar]
- 4.Hans-Filho G, dos Santos V, Katayama JH, Aoki V, Rivitti EA, Sampaio SA, et al. An active focus of high prevalence of fogo selvagem on an Amerindian reservation in Brazil. Cooperative Group on Fogo Selvagem Research. J Invest Dermatol. 1996;107:68–75. doi: 10.1111/1523-1747.ep12298213. [DOI] [PubMed] [Google Scholar]
- 5.Diaz L, Sampaio S, Rivitti E, Martins C, Cunha P, Lombardi C, et al. Endemic pemphigus foliaceus (fogo selvagem). I. Clinical features and immunopathology. J Am Acad Dermatol. 1989;20:657–69. doi: 10.1016/s0190-9622(89)70079-7. [DOI] [PubMed] [Google Scholar]
- 6.Beutner EH, Prigenzi LS, Hale W, Leme Cde A, Bier OG. Immunofluorescent studies of autoantibodies to intercellular areas of epithelia in Brazilian pemphigus foliaceus. Proc Soc Exp Biol Med. 1968;127:81–6. doi: 10.3181/00379727-127-32626. [DOI] [PubMed] [Google Scholar]
- 7.Roscoe J, Diaz L, Sampaio S, Castro R, Labib R, Takahashi Y, et al. Brazilian Pemphigus Foliaceus Autoantibodies Are Pathogenic to BALB/c Mice by Passive Transfer. J Invest Dermatol. 1985;85:538–41. doi: 10.1111/1523-1747.ep12277362. [DOI] [PubMed] [Google Scholar]
- 8.Stanley JR, Klaus-Kovtun V, Sampaio SA. Antigenic specificity of fogo selvagem autoantibodies is similar to North American pemphigus foliaceus and distinct from pemphigus vulgaris autoantibodies. J Invest Dermatol. 1986;87:197–201. doi: 10.1111/1523-1747.ep12695334. [DOI] [PubMed] [Google Scholar]
- 9.Flores G, Culton DA, Prisayanh P, Qaqish BF, James K, Maldonado M, et al. IgG autoantibody response against keratinocyte cadherins in endemic pemphigus foliaceus (fogo selvagem) J Invest Dermatol. 2012;132:2573–80. doi: 10.1038/jid.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hisamatsu Y, Amagai M, Garrod DR, Kanzaki T, Hashimoto T. The detection of IgG and IgA autoantibodies to desmocollins 1-3 by enzyme-linked immunosorbent assays using baculovirus-expressed proteins, in atypical pemphigus but not in typical pemphigus. Br J Dermatol. 2004;151:73–83. doi: 10.1111/j.1365-2133.2004.05995.x. [DOI] [PubMed] [Google Scholar]
- 11.Nagasaka T, Nishifuji K, Ota T, Whittock NV, Amagai M. Defining the pathogenic involvement of desmoglein 4 in pemphigus and staphylococcal scalded skin syndrome. J Clin Invest. 2004;114:1484–92. doi: 10.1172/JCI20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ota T, Amagai M, Watanabe M, Nishikawa T. No involvement of IgG autoantibodies against extracellular domains of desmoglein 2 in paraneoplastic pemphigus or inflammatory bowel diseases. J Dermatol Sci. 2003;32:137–41. doi: 10.1016/s0923-1811(03)00072-0. [DOI] [PubMed] [Google Scholar]
- 13.Rock B, Martins CR, Theofilopoulos AN, Balderas RS, Anhalt GJ, Labib RS, et al. The pathogenic effect of IgG4 autoantibodies in endemic pemphigus foliaceus (fogo selvagem) N Engl J Med. 1989;320:1463–9. doi: 10.1056/NEJM198906013202206. [DOI] [PubMed] [Google Scholar]
- 14.Eyre RW, Stanley JR. Human autoantibodies against a desmosomal protein complex with a calcium-sensitive epitope are characteristic of pemphigus foliaceus patients. J Exp Med. 1987;165:1719–24. doi: 10.1084/jem.165.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalczyk AP, Anderson JE, Borgwardt JE, Hashimoto T, Stanley JR, Green KJ. Pemphigus sera recognize conformationally sensitive epitopes in the amino-terminal region of desmoglein-1. J Invest Dermatol. 1995;105:147–52. doi: 10.1111/1523-1747.ep12316680. [DOI] [PubMed] [Google Scholar]
- 16.Labib RS, Rock B, Robledo MA, Anhalt GJ. The calcium-sensitive epitope of pemphigus foliaceus antigen is present on a murine tryptic fragment and constitutes a major antigenic region for human autoantibodies. J Invest Dermatol. 1991;96:144–7. doi: 10.1111/1523-1747.ep12515943. [DOI] [PubMed] [Google Scholar]
- 17.Olague-Alcala M, Diaz LA. The epitopes on bovine pemphigus foliaceus antigen are calcium dependent and located on the peptide backbone of this glycoprotein. Chron Dermat. 1993;2:189–209. [Google Scholar]
- 18.Amagai M, Ishii K, Hashimoto T, Gamou S, Shimizu N, Nishikawa T. Conformational epitopes of pemphigus antigens (Dsg1 and Dsg3) are calcium dependent and glycosylation independent. J Invest Dermatol. 1995;105:243–7. doi: 10.1111/1523-1747.ep12317587. [DOI] [PubMed] [Google Scholar]
- 19.Li N, Park M, Zhao M, Hilario-Vargas J, McInnes DM, Prisayanh PS, et al. The Thomsen-Friedenreich antigen-binding lectin jacalin interacts with desmoglein-1 and abrogates the pathogenicity of pemphigus foliaceus autoantibodies in vivo. J Invest Dermatol. 2010;130:2773–80. doi: 10.1038/jid.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espana A, Diaz LA, Mascaro JM, Jr, Giudice GJ, Fairley JA, Till GO, et al. Mechanisms of acantholysis in pemphigus foliaceus. Clin Immunol Immunopathol. 1997;85:83–9. doi: 10.1006/clin.1997.4407. [DOI] [PubMed] [Google Scholar]
- 21.Rock B, Labib R, Diaz L. Monovalent Fab′ immunoglobulin fragments from endemic pemphigus foliaceus autoantibodies reproduce the human disease in neonatal Balb/c mice. J Clin Invest. 1990;85:296–9. doi: 10.1172/JCI114426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qaqish BF, Prisayanh P, Qian Y, Andraca E, Li N, Aoki V, et al. Development of an IgG4-based predictor of endemic pemphigus foliaceus (fogo selvagem) J Invest Dermatol. 2009;129:110–8. doi: 10.1038/jid.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olague-Alcala M, Giudice GJ, Diaz LA. Pemphigus Foliaceus Sera Recognize an N-Terminal Fragment of Bovine Desmoglein 1. J Invest Dermatol. 1994;102:882–5. doi: 10.1111/1523-1747.ep12382794. [DOI] [PubMed] [Google Scholar]
- 24.Sekiguchi M, Futei Y, Fujii Y, Iwasaki T, Nishikawa T, Amagai M. Dominant autoimmune epitopes recognized by pemphigus antibodies map to the N-terminal adhesive region of desmogleins. J Immunol. 2001;167:5439–48. doi: 10.4049/jimmunol.167.9.5439. [DOI] [PubMed] [Google Scholar]
- 25.Li N, Aoki V, Hans-Filho G, Rivitti EA, Diaz LA. The role of intramolecular epitope spreading in the pathogenesis of endemic pemphigus foliaceus (fogo selvagem) J Exp Med. 2003;197:1501–10. doi: 10.1084/jem.20022031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bautz DJ, Preston GA, Lionaki S, Hewins P, Wolberg AS, Yang JJ, et al. Antibodies with dual reactivity to plasminogen and complementary PR3 in PR3-ANCA vasculitis. J Am Soc Nephrol. 2008;19:2421–9. doi: 10.1681/ASN.2008030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker CE, Tomer KB. MALDI/MS-based epitope mapping of antigens bound to immobilized antibodies. Mol Biotechnol. 2002;20:49–62. doi: 10.1385/MB:20:1:049. [DOI] [PubMed] [Google Scholar]
- 28.Roth AJ, Ooi JD, Hess JJ, van Timmeren MM, Berg EA, Poulton CE, et al. Epitope specificity determines pathogenicity and detectability in ANCA-associated vasculitis. J Clin Invest. 2013;123:1773–83. doi: 10.1172/JCI65292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison OJ, Brasch J, Lasso G, Katsamba PS, Ahlsen G, Honig B, et al. Structural basis of adhesive binding by desmocollins and desmogleins. Proc Natl Acad Sci U S A. 2016;113:7160–5. doi: 10.1073/pnas.1606272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alva V, Nam SZ, Soding J, Lupas AN. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016;44:W410–5. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Protein Sci. 2016;86:2 9 1–2 9 37. doi: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- 33.Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–56. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilles LA, Parry DA, Powers EE, Angst BD, Wagner RM, Green KJ. Structural analysis and expression of human desmoglein: a cadherin-like component of the desmosome. J Cell Sci. 1991;99(Pt 4):809–21. doi: 10.1242/jcs.99.4.809. [DOI] [PubMed] [Google Scholar]
- 35.Yamagami J, Kacir S, Ishii K, Payne AS, Siegel DL, Stanley JR. Antibodies to the desmoglein 1 precursor proprotein but not to the mature cell surface protein cloned from individuals without pemphigus. J Immunol. 2009;183:5615–21. doi: 10.4049/jimmunol.0901691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokouchi M, Saleh MA, Kuroda K, Hachiya T, Stanley JR, Amagai M, et al. Pathogenic epitopes of autoantibodies in pemphigus reside in the amino-terminal adhesive region of desmogleins which are unmasked by proteolytic processing of prosequence. J Invest Dermatol. 2009;129:2156–66. doi: 10.1038/jid.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toumi A, Saleh MA, Yamagami J, Abida O, Kallel M, Masmoudi A, et al. Autoimmune reactivity against precursor form of desmoglein 1 in healthy Tunisians in the area of endemic pemphigus foliaceus. J Dermatol Sci. 2013;70:19–25. doi: 10.1016/j.jdermsci.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broussard JA, Getsios S, Green KJ. Desmosome regulation and signaling in disease. Cell Tissue Res. 2015;360:501–12. doi: 10.1007/s00441-015-2136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cadwell CM, Su W, Kowalczyk AP. Cadherin tales: Regulation of cadherin function by endocytic membrane trafficking. Traffic. 2016;17:1262–71. doi: 10.1111/tra.12448. [DOI] [PubMed] [Google Scholar]
- 40.Celentano A, Mignogna MD, McCullough M, Cirillo N. Pathophysiology of the Desmo-Adhesome. J Cell Physiol. 2017;232:496–505. doi: 10.1002/jcp.25515. [DOI] [PubMed] [Google Scholar]
- 41.Nekrasova O, Green KJ. Desmosome assembly and dynamics. Trends Cell Biol. 2013;23:537–46. doi: 10.1016/j.tcb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bazellieres E, Conte V, Elosegui-Artola A, Serra-Picamal X, Bintanel-Morcillo M, Roca-Cusachs P, et al. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat Cell Biol. 2015;17:409–20. doi: 10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffman BD, Yap AS. Towards a Dynamic Understanding of Cadherin-Based Mechanobiology. Trends Cell Biol. 2015;25:803–14. doi: 10.1016/j.tcb.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Leckband DE, de Rooij J. Cadherin adhesion and mechanotransduction. Annu Rev Cell Dev Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 45.Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17:533–9. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 46.Goodman H. Nikolsky sign; page from notable contributors to the knowledge of dermatology. AMA Arch Derm Syphilol. 1953;68:334–5. doi: 10.1001/archderm.1953.01540090096013. [DOI] [PubMed] [Google Scholar]
- 47.Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, et al. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–37. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 49.Chitaev NA, Troyanovsky SM. Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J Cell Biol. 1998;142:837–46. doi: 10.1083/jcb.142.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitagawa M, Natori M, Murase S, Hirano S, Taketani S, Suzuki ST. Mutation analysis of cadherin-4 reveals amino acid residues of EC1 important for the structure and function. Biochem Biophys Res Commun. 2000;271:358–63. doi: 10.1006/bbrc.2000.2636. [DOI] [PubMed] [Google Scholar]
- 51.Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. Embo J. 1999;18:1738–47. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, et al. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148:579–90. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–63. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 54.Nie Z, Merritt A, Rouhi-Parkouhi M, Tabernero L, Garrod D. Membrane-impermeable cross-linking provides evidence for homophilic, isoform-specific binding of desmosomal cadherins in epithelial cells. J Biol Chem. 2011;286:2143–54. doi: 10.1074/jbc.M110.192245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chitaev NA, Troyanovsky SM. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell-cell adhesion. J Cell Biol. 1997;138:193–201. doi: 10.1083/jcb.138.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diaz LA. Molecular structure of the epidermal extracellular spaces. Int J Dermatol. 1979;18:434–42. doi: 10.1111/j.1365-4362.1979.tb01946.x. [DOI] [PubMed] [Google Scholar]
- 57.Calkins CC, Setzer SV, Jennings JM, Summers S, Tsunoda K, Amagai M, et al. Desmoglein endocytosis and desmosome disassembly are coordinated responses to pemphigus autoantibodies. J Biol Chem. 2006;281:7623–34. doi: 10.1074/jbc.M512447200. [DOI] [PubMed] [Google Scholar]
- 58.Cirillo N, Gombos F, Lanza A. Changes in desmoglein 1 expression and subcellular localization in cultured keratinocytes subjected to anti-desmoglein 1 pemphigus autoimmunity. J Cell Physiol. 2007;210:411–6. doi: 10.1002/jcp.20856. [DOI] [PubMed] [Google Scholar]
- 59.Lanza A, De Rosa A, Femiano F, Annese P, Ruocco E, Gombos F, et al. Internalization of non-clustered desmoglein 1 without depletion of desmoglein 1 from adhesion complexes in an experimental model of the autoimmune disease pemphigus foliaceus. Int J Immunopathol Pharmacol. 2007;20:355–61. doi: 10.1177/039463200702000216. [DOI] [PubMed] [Google Scholar]
- 60.Mao X, Choi EJ, Payne AS. Disruption of desmosome assembly by monovalent human pemphigus vulgaris monoclonal antibodies. J Invest Dermatol. 2009;129:908–18. doi: 10.1038/jid.2008.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel HP, Diaz LA, Anhalt GJ, Labib RS, Takahashi Y. Demonstration of pemphigus antibodies on the cell surface of murine epidermal cell monolayers and their internalization. J Invest Dermatol. 1984;83:409–15. doi: 10.1111/1523-1747.ep12273480. [DOI] [PubMed] [Google Scholar]
- 62.Berkowitz P, Chua M, Liu Z, Diaz LA, Rubenstein DS. Autoantibodies in the autoimmune disease pemphigus foliaceus induce blistering via p38 mitogen-activated protein kinase-dependent signaling in the skin. Am J Pathol. 2008;173:1628–36. doi: 10.2353/ajpath.2008.080391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li N, Zhao M, Wang J, Liu Z, Diaz LA. Involvement of the apoptotic mechanism in pemphigus foliaceus autoimmune injury of the skin. J Immunol. 2009;182:711–7. doi: 10.4049/jimmunol.182.1.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao X, Sano Y, Park JM, Payne AS. p38 MAPK activation is downstream of the loss of intercellular adhesion in pemphigus vulgaris. J Biol Chem. 2011;286:1283–91. doi: 10.1074/jbc.M110.172874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waschke J, Bruggeman P, Baumgartner W, Zillikens D, Drenckhahn D. Pemphigus foliaceus IgG causes dissociation of desmoglein 1-containing junctions without blocking desmoglein 1 transinteraction. J Clin Invest. 2005;115:3157–65. doi: 10.1172/JCI23475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison OJ, Corps EM, Berge T, Kilshaw PJ. The mechanism of cell adhesion by classical cadherins: the role of domain 1. J Cell Sci. 2005;118:711–21. doi: 10.1242/jcs.01665. [DOI] [PubMed] [Google Scholar]
- 67.Petrova YI, Spano MM, Gumbiner BM. Conformational epitopes at cadherin calcium-binding sites and p120-catenin phosphorylation regulate cell adhesion. Mol Biol Cell. 2012;23:2092–108. doi: 10.1091/mbc.E11-12-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shashikanth N, Petrova YI, Park S, Chekan J, Maiden S, Spano M, et al. Allosteric Regulation of E-Cadherin Adhesion. J Biol Chem. 2015;290:21749–61. doi: 10.1074/jbc.M115.657098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diaz LA, Arteaga LA, Hilario-Vargas J, Valenzuela JG, Li N, Warren S, et al. Anti-desmoglein-1 antibodies in onchocerciasis, leishmaniasis and Chagas disease suggest a possible etiological link to Fogo selvagem. J Invest Dermatol. 2004;123:1045–51. doi: 10.1111/j.0022-202X.2004.23438.x. [DOI] [PubMed] [Google Scholar]
- 70.Qian Y, Jeong JS, Abdeladhim M, Valenzuela JG, Aoki V, Hans-Filhio G, et al. IgE anti-LJM11 sand fly salivary antigen may herald the onset of fogo selvagem in endemic Brazilian regions. The Journal of investigative dermatology. 2015;135:913–5. doi: 10.1038/jid.2014.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qian Y, Jeong JS, Maldonado M, Valenzuela JG, Gomes R, Teixeira C, et al. Cutting Edge: Brazilian Pemphigus Foliaceus Anti-Desmoglein 1 Autoantibodies Cross-React with Sand Fly Salivary LJM11 Antigen. J Immunol. 2012;189:1535–9. doi: 10.4049/jimmunol.1200842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qian Y, Jeong JS, Ye J, Dang B, Abdeladhim M, Aoki V, et al. Overlapping IgG4 Responses to Self- and Environmental Antigens in Endemic Pemphigus Foliaceus. J Immunol. 2016;196:2041–50. doi: 10.4049/jimmunol.1502233. [DOI] [PMC free article] [PubMed] [Google Scholar]