Abstract
Far from being just “bugs in our guts,” the microbiota interacts with the body in previously unimagined ways. Research into the genome and the microbiome has revealed that the human body and the microbiota have a long-established but only recently recognized symbiotic relationship; homeostatic balance between them regulates body function. That balance is fragile, easily disturbed, and plays a fundamental role in human health—our very survival depends on the healthy functioning of these microorganisms. Increasing rates of cardiovascular, autoimmune, and inflammatory diseases, as well as epidemics in obesity and diabetes in recent decades are believed to be explained, in part, by unintended effects on the microbiota from vaccinations, poor diets, environmental chemicals, indiscriminate antibiotic use, and “germophobia.” Discovery and exploration of the brain-gut-microbiota axis have provided new insights into functional diseases of the gut, autoimmune and stress-related disorders, and the role of probiotics in treating certain affective disorders; it may even explain some aspects of autism. Research into dietary effects on the human gut microbiota led to its classification into three proposed enterotypes, but also revealed the surprising role of blood group antigens in shaping those populations. Blood group antigens have previously been associated with disease risks; their subsequent association with the microbiota may reveal mechanisms that lead to development of nutritional interventions and improved treatment modalities. Further exploration of associations between specific enteric microbes and specific metabolites will foster new dietary interventions, treatment modalities, and genetic therapies, and inevitably, their application in personalized healthcare strategies.
Introduction
The human intestinal microbiome has emerged as an important research frontier with profound implications for understanding disease pathogenesis. As technology has advanced, research has expanded from simply identifying these microorganisms, to understanding their functions and interactions within the body, to correlating these findings with human health and disease states. Genomics, transcriptomics, metagenomic sequencing, proteomics, and metabolomics technologies have profoundly transformed the field of microbiology just as the invention of the microscope transformed the science of biology.
The Human Microbiome Project (HMP), which investigated the structure, function, breadth, and diversity of the microbiome in healthy adults, found that there were substantial taxonomic variations in the composition of the microbial community at different anatomical locations in the same person (intra-individual), as well as substantial variations at the same anatomical site in different people (inter-individual).1 The eight anatomical sites chosen for taxonomic classification were the hair, skin, nostrils, oral cavity, esophagus, stomach, colon, and vagina.1 The intestinal microbiome is perhaps the most complex of the eight sites studied.
The term microbiota refers to the collection of eukaryotic microbes and viruses, as well as bacteriophages, archaea, and bacteria which live in the human gut, while the term microbiome refers to the genomes of the microbiota, both the microbial genes and gene products.1 Although the human microbiota is dominated by only 4 bacterial phyla (Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria) out of more than 50 known phyla,2 it has been estimated that the average human gut contains trillions of bacteria and archaea.3 This vast bacterial biomass contains many unique or minimally redundant bacterial genes,4 but because different bacterial species share functional traits, there is a high degree of functional redundancy.3
Background
One of the biggest limitations for researchers has been the inability to identify the vast array of intestinal microbiota using laboratory culturing methods,4 because it is extremely difficult to successfully maintain anaerobic culture conditions, which are required by the majority of intestinal microbes.5 This limitation has been largely eliminated by the speed, ease, and accuracy of gene sequencing.6 Using 16S ribosomal RNA and DNA, researchers have been able to quickly detect, identify, and classify most of the microbes found in the healthy human gut,6 although reference sequences are still unknown for about one-third of the metagenome.4
Culture-independent methods have their own limitations. Organisms considered to be of the same species based on 16S ribosomal RNA gene sequencing can have large differences in other DNA sequences, and often have different sets of gene clusters that regulate production of specialized metabolites.7 Further, even if microbial species membership and abundance remain constant, changes in available dietary or xenobiotic substrates can alter the expression of metabolic functions.7
Although genomic sequencing of intestinal microbes can verify their presence or absence in the gut, neither function nor biological activity can be inferred simply from their presence, because the intestinal ecosystem is complex, interdependent, and not fully understood.4, 8 In addition, the symbiotic relationship between the gut microbiome and the host results in extensive modulation of the metabolism and physiology of the interacting genomes, which therefore cannot be studied in isolation from each other.9
As with any new technology, there have been challenges to overcome, and new challenges identified. Gene sequencing and cataloging has been hampered by differences in donor recruitment, protocols, and methodologies; human DNA contamination of specimens; as well as errors and artifacts generated during data production and processing.6 These issues have been resolved for the most part, yet despite much effort, research using genomic techniques has not been able to establish a “core microbiome” common to all healthy people.1, 8, 10 Metagenomic sequencing categorized by gene function may ultimately prove more satisfactory for this purpose than whole-genome sequencing.4 From preliminary studies, it appears that each human being has a virtually unique microbiome,2 but despite individual variances in its taxonomy and microbial composition, the metabolic capabilities and functions carried out by the microbiota are fairly constant and remarkably stable.1, 4
Recent metagenomic sequencing of the human intestinal microbiome, in fecal samples from a cohort of 124 people in the MetaHIT project, has proposed the existence of a functional core or minimum human gut metagenome necessary for the normal functioning of the gut ecosystem; this functional core is composed of about 3364 non-redundant genes and between 1000 to 1150 bacterial species.3, 11 In the study population, each individual in the cohort harbored at least 160 of these bacterial species; although there was a high level of functional similarity between individuals, there was also a high inter-individual specificity of intestinal microbiota, with 38% of an individual's bacterial genes shared by 50% of the population, but only 9% shared by 80% of the population.3, 11
The Intestinal Microbiota
A robust, diverse intestinal microbiota is needed to develop and maintain the immune system,1, 2 regulate immunological function, as well as establish and maintain physiological homeostasis2, 12 The microbiota stimulates production of cross-reactive antibodies, primarily immunoglobulin A (IgA), which are secreted into the gut lumen, and profoundly influences development of lymphatic structures in the gastrointestinal (GI) tract and differentiation of lymphocytes.5 The presence of beneficial bacteria is critically important to prevent excessive colonization by pathogenic organisms, which occurs through competition for mucosal attachment sites, depletion of essential nutrients and/or oxygen, and by secretion of peroxides, antimicrobials, or bacteriocidins to inhibit other bacteria. 5, 13-15 This protective effect is known as ‘colonization resistance.’13
The intestinal microbiota has coevolved with its human host, and although within the lumen, it actually exists outside of the human body and functions independently from it but in close cooperation with it.1 However, the intestinal microbiota could be considered an organ within an organ,5 because it mediates various metabolic effects, directly influences epithelial cell proliferation, induces IgA production, stimulates lymphoid tissue development,3, 5, 16 and strongly influences the host's metabolic response to environmental factors, drugs, and disease.8 It is also intimately involved in digestion and metabolism of food, biosynthesis of vitamins and hormones that are absorbed by the host, as well as production of other compounds that the host cannot generate.2, 5, 15, 17 Intestinal bacteria produce highly specific metabolic products and critical growth factors, which support epithelial growth and metabolism, and induce genes in epithelial cells to produce digestive enzymes that are required for physiological digestive processes.5
Perhaps most importantly, the nutritional value of the food that is eaten—what can be absorbed and utilized from it—depends on the composition of the microbiota itself, which is directly affected by the type of long-term diet that is consumed.18 It should be noted as well that dietary components and xenobiotic agents are metabolized in different ways by different microbial communities.7 However, the process has layers of complexity: microbial metabolites may interfere with host metabolism of xenobiotics; diet-derived nutrients can regulate microbial metabolism of xenobiotics; and xenobiotics can modulate the expression and activity of the gut microbiome.7
Composition of the microbiota
The microbial community of the intestines is composed of both beneficial and pathogenic bacteria, and can include fungi and parasites, as well as viruses.19 Pathogenic microorganisms have the inherent ability to cross anatomical barriers, resist host defenses, and induce host responses.2 Infection with a pathogen sometimes causes disease, which is a function of the host/pathogen interaction,2 however, some people can be carriers of pathogenic microorganisms without ever developing symptoms.
Commensal and mutualistic bacteria are resident microorganisms that have, respectively, a benign or mutually beneficial relationship with their host; generally, these symbionts benefit the host by preventing colonization by pathogens, and the genes of these resident microorganisms are included in the gene pool of the host.2 However, if these normally harmless bacteria enter the body or overpower host defenses, they become pathogenic and the consequent opportunistic infection can lead to serious or fatal illness.5
We interact mostly with symbionts on a daily basis, which have not been associated with significant pathology.20 It has been proposed that the immune system is primarily shaped by symbionts, and that pro-inflammatory immunity may be a necessary component of establishing and maintaining a state of homeostasis between host and microbiota.20 Further, it has been suggested that the pathogenicity of microbes is defined by the context in which the microbe interacts with its host: mutualists can become pathogens if penetrating the wrong niche, and notorious pathogens can become mutualists when restricted to the right niche.20
Changes in composition
Changes in the composition of the intestinal microbiota have been associated with the development of diseases of the upper GI tract (ranging from gastritis, peptic ulcers, gastroesophageal reflux, and Barrett's esophagus, to stomach and esophageal cancers, and celiac disease) as well as the lower intestine (including inflammatory bowel disease [IBD], ulcerative colitis, Crohn's disease, and colorectal cancer).3, 4, 12 There appears to be a characteristic microbiota community associated with many of these diseases, indicated by both taxonomic and functional markers, some of which may precede the disease state.1, 2, 4 In addition, the immune status of the host can influence the composition of beneficial communities.21 Age-related changes in the microbiota can have harmful effects, including changes in the ratio between Firmicutes and Bacteroidetes species, progressive development of gastric atrophy, and cancer-related somatic mutagenesis.4
The composition of the microbiota has also been changed by the use of antibiotics, vaccines, improvements in healthcare, and changes in lifestyle that have lowered infant mortality rates and increased lifespans, but which have also resulted in reduced horizontal bacterial transfer and greatly increased selection against existing ancestral microorganisms.2, 4 It is thought that changes in human ecology, such as clean water, more frequent bathing, widespread use of antibiotics and antimicrobial soaps, smaller family size, less social crowding, more Cesarean births, and less breastfeeding may have had unintended effects on the composition of the intestinal microbiota, and therefore human physiology and disease risks.2
The microbiome is not inherited fully formed; it is acquired following birth via vertical or horizontal transfer.1, 4, 16 If born by vaginal delivery, the founding microbial population reflects the vaginal microbiome of the mother, being primarily Lactobacillus, Prevotella, or Sneathia species; if born by Cesarean section, it is similar to the skin microbiome of the mother, with primarily Staphylococcus, Propionibacterium, and Corynebacterium species.22 Lactobacilli 4 and Bifidobacteria23 are also contributed from mother's milk. Regardless of delivery mode, the initial bacterial colonies are undifferentiated in the newborn, with homogeneous distribution across the skin, nasopharyngeal, oral, and intestinal habitats.22 The initial colonization of the intestinal microbiota by pioneer colonizers prepares the baby's GI tract for subsequent additional bacterial population.4, 22 Bacterial diversity increases with age, achieving a configuration similar to adults by about three years of age.24
Several studies have documented that birth method influences fecal microbiota populations in infants.22, 24, 25 One study comparing babies born by Cesarean and vaginal delivery found that intestinal colonization was delayed and remained disturbed for six months or longer in those born by Cesarean.25 Lactobacillus and Bifidobacterium colonization normalized at 10 days and 30 days, respectively, but only 36% of Cesarean-delivered infants were colonized by Bacteroides fragilis at 6 months, versus 76% of vaginally-delivered infants (p=0.009); colonization by Clostridium perfringens at 30 days was 57% in babies delivered by Cesarean, versus 17% in vaginally-delivered infants (p=0.003).25
Colonization by B. fragilis may never normalize, due to interbacterial inhibition by one or more ecological competitors established in its absence.25 Reduced colonization may have unintended consequences, both because B. fragilis prevents colitis by supporting differentiation of T cells that express interleukin (IL)-10, which reduces inflammatory responses in the intestine, and because a capsular component of B. fragilis, polysaccharide A (PSA), plays a regulatory role in autoimmune processes in the central nervous system (CNS).13 In contrast, with vaginal birth, the primary means of intestinal colonization appears to be vertical transmission from the mother, with the founding microbial community of the baby principally composed of Lactobacilli from the vagina and mother's milk.4
The disappearing microbiota hypothesis postulates that loss of indigenous beneficial bacteria over time (for whatever reason) has contributed to the recent rapid increases in conditions such as type 2 diabetes mellitus (T2DM), obesity, and metabolic syndrome.2 The loss of keystone species can result in the extinction of secondary species as well, with a cascading downstream effect on the host and on co-colonizing species; functional redundancy can mask these effects in the short-term, but over time, the microbiota's ability to adapt and respond appropriately is compromised.4 The loss of Helicobacter pylori in particular is related to the increased incidence of allergies, asthma, and esophageal reflux, with concurrent decreases in rates of peptic ulcers and certain gastric cancers.2
A related concept, known as the hygiene hypothesis, suggests that reduced exposure to microbes and infectious diseases in childhood is directly linked to the increasing incidence in recent years of IBDs and chronic immune-related disorders.5 Exposure to foreign microorganisms in early childhood influences microbial composition and stimulates correct development of the immune system.5, 26 The use of antibiotics, especially in early infancy, may therefore be linked to the rising incidence of food and airborne allergies, other immune-related disorders, metabolic disorders, and autoimmune diseases.26
The Intestinal Barrier
Several mechanisms work together in the intestinal lumen to protect host integrity and prevent bacterial invasion. These include the mechanical barrier of the epithelium; tight junctions where cell-cell contact occurs; expression of mucins to form the mucus layer; secretion of chloride; production of defensins and cytokines; the protective presence of beneficial bacteria; and gut motility.5 The epithelium is a single layer of cells, composed of enterocytes, goblet cells, and enteroendocrine cells,27 which supports the largest mucosal surface of the human body.21
The primary function of the epithelium lining the GI tract is to maintain health and homeostasis.21 It does this by regulating digestion, absorption, and transport of vital nutrients and macromolecules, while simultaneously tolerating food antigens, avoiding overreaction and potentially destructive inflammatory responses to beneficial bacteria, and preventing invasion by pathogenic organisms.21, 26, 27 Maintaining homeostatic balance between tolerance and immunity is a regulatory challenge that involves delicate choreography, orchestrated by the epithelial cells, between the signals and responses of the adaptive and innate immune systems. Antigens activate both of these systems; any molecule that can be recognized by the immune system serves as an antigen,28 however, every antigen evokes a unique type and degree of response.
The innate immune system does not require prior exposure to specific antigens, but instead identifies broadly distributed antigens as “self,” distinguishes potentially harmful or foreign molecules and cells as “non-self,” and mounts an immediate defense against them.15, 28 The primary components of innate immune response include antimicrobial peptides and pattern recognition receptors (PRRs).15 Other components of the innate immune system include phagocytes, which ingest foreign antigens; natural killer cells, which kill virus-infected cells and some tumor cells; and leukocytes, which release inflammatory mediators.15, 28
Defensins are peptides that are expressed in mucosal epithelial cells and phagocytes, and are released into the intestinal lumen.15 In addition to acting as effector and regulatory molecules of the innate immune response, they induce the release of inflammatory mediators, regulate the complement system, and increase antigen-specific immune response by interacting with dendritic cells and T cells.15 Cytokines, complement, and acute phase proteins are molecular components that participate in both innate and adaptive immunity.28 Cytokines are not antigen-specific, but they influence the magnitude of inflammatory or immune responses and can act sequentially, synergistically, or antagonistically.29
Although the acquired or adaptive immune system takes longer to respond after initial exposure to an antigen, after it acquires antigen-specific memory, subsequent response to the same antigen is rapid.28, 30 Cell-mediated responses are derived from activation of certain T cell receptors, while humoral responses are derived from B cells, which secrete soluble antigen-specific antibodies.15, 28 The innate and adaptive components of the immune system also coordinate their efforts; for example, most microorganisms are killed after they are phagocytosed, but when necessary, phagocytes can be stimulated to produce more lytic enzymes and microbicidal products by T cell-derived cytokines such as interferon-γ (IFN-γ).28
PRRs are a class of proteins that respond to small molecular sequences that are consistently found on pathogens and other microbes, such as flagella, cilia, Gram-positive peptidoglycans, Gram-negative lipopolysaccharides (LPSs), oligosaccharides, and nucleic acids. 1, 15, 17, 21, 27, 28 Upon detection of these antigens or other bacterial ligands, toxins, bacterial DNA, or viral double-stranded RNA, PRRs on epithelial cells trigger signaling cascades that tailor the immunological memory of the adaptive immune system to recognize and respond to these molecular patterns in specific ways.1, 15, 17, 21, 27, 28 Activation of some signaling pathways leads to initiation of the adaptive immune response, including induction of T cells and maturation of dendritic cells, which confer a homeostatic response and tolerance to future contact, whereas activation of other pathways elicits a defensive response, involving activation of the innate immune system and release of antimicrobial peptides, inflammatory cytokines, chemokines, and macrophages.15, 26, 27, 31
The synergistic interaction between the microbiota, the epithelial barrier, and the immune system is complicated by the fact that all three respond to and are affected by dietary influences; this has been called the diet-microbiota-immune axis.32 Metabolites from epithelial cells directly regulate the functions of antigen-presenting cells (APCs) and lymphocytes;21 metabolites in the diet indirectly influence the function of the mucosal barrier;17 and the intestinal microbiota produce most of the metabolites found in blood plasma.4 Metabolic activities of the microbiota are monitored by the immune system, which is in turn modified by microbial signaling and mucosal absorption of metabolites and other dietary compounds.32
Establishing and preserving homeostasis
The preservation of homeostasis requires microbial sensing by epithelial cells; this in turn depends on the adequate presence and correct functioning of these receptors, which are critical for preventing the development of chronic intestinal inflammation, infectious colitis, and even cell apoptosis.31 These conditions can lead to increased permeability of the intestinal barrier and translocation of the microbiota across the epithelium.31 However, this damage is mitigated by intraepithelial lymphocytes, located between the epithelial cells in the intestinal barrier,15 which promote repair of injured epithelia and release antimicrobial peptides that prevent entrance of enteric bacteria.26
The tissue directly beneath the epithelium (on the basolateral side) is known as the lamina propria; it contains a variety of plasma cells, lymphocytes, stromal cells, and APCs.15, 27 It also produces both secretory IgA (sIgA), which is constantly transported across the intestinal epithelium into the gut lumen,33 and polymeric IgA, which promotes excretion of antigens present in the lamina propria and neutralizes pathogens intracellularly during transepithelial transport.34 The lamina propria is a key component of the innate immune system,35 which quickly responds to defend, support, or restore mucosal barrier integrity and function, and prevent uptake of pathogenic organisms and toxins.17
The apical side of the epithelial cells is protected by the glycocalyx, which consists of a high-density mixture of mucins (glycoproteins), glycolipids, and proteoglycans attached directly to the epithelial cell membrane,14, 36 as well as immunoglobulins, antimicrobial peptides, and electrolytes.36 The glycocalyx is negatively charged and repels most microorganisms that are also negatively charged.26 On the apical side of the glycocalyx, the negatively charged net-like polymer of mucins secreted by the enterocytes and goblet cells of the epithelium is heavily glycosylated and forms two layers of viscous and relatively impermeable extracellular mucus; the glycans in the inner layer are more densely packed than those in the outer layer, and their smaller pore size physically prevents bacterial penetration.14, 21
Mucins are composed of a variety of both membrane-bound mucins (MUC1, MUC3, MUC4, MUC12, MUC13, MUC16, and MUC17) and gel-forming mucins (MUC2, MUC5AC, MUC5B, and MUC6); the majority of the mucus in the GI tract from duodenum to colon is composed of the gel-forming mucin MUC2.36, 37 MUC6 is also secreted by Brunner's glands in the duodenum, while MUC5AC and MUC6 are secreted in the stomach.37 Membrane-bound mucins of the inner layer influence cell growth, metastasis of tumors, and immune system recognition, and may play a role in cell differentiation, but are primarily involved in cell signaling.38 Gel-forming secreted mucins of the outer layer provide lubrication as well as protection from digestive enzymes, gastric pH, free radicals, toxins, and carcinogens.37 This unregulated, baseline constitutive mucus secretion can be quickly augmented by receptor-mediated secretion, when stimulated by alcohol, mustard, other chemical irritants, prostaglandins, secretions from monocytes and macrophages, or bacterial toxins.37, 38
The mucus layers are the only point where there is direct contact between the host and the intestinal microbiota. Mucus keeps epithelial cells hydrated; provides a chemical, physical, and immunological barrier that protects the underlying epithelium against luminal microbes; and has hydrophobic qualities that also help fight enteric bacteria and regulate gut permeability.15, 37 Thus, defects that affect the function or differentiation of goblet cells and the production of mucus result in gut inflammatory disorders, dysfunction, and impaired resistance to infection.15, 31
Bacterial invasion is prevented by antimicrobial peptides that are produced in large quantities throughout the GI tract, and by antibodies and defensins that are usually bound to the mucins; despite attachment of segmented filamentous bacteria (SFB) to the epithelium, in a healthy individual the inner mucus layer is virtually sterile.5, 13, 14 SFB are host-friendly bacteria that penetrate the mucus layers and adhere tightly to the epithelium, which induces development of the adaptive immune system, including strong IgA responses, as well as differentiation and activation of T cells.13, 20, 31 SIgA coats beneficial microorganisms and anchors them to the mucus layer, which allows their colonization.26 SIgA antibodies are also believed to be involved in the progressive, controlled establishment of the newborn's microbiota.34
Antigen-specific sIgA antibodies attached to the mucus layer block specific binding sites on the bacterial cell wall, prevent direct contact with the epithelium by binding with pathogenic bacteria and toxins, and interfere with their adherence to epithelial receptors by stearic hindrance, all of which result in decreased colonization and faster removal through a process known as immune exclusion.13, 31, 33, 34, 39 Immune exclusion, which involves agglutination, entrapment in mucus, and/or clearance through peristalsis,34 may be the main and most important function of intestinal antibodies, because their ability to selectively interfere with specific bacterial adherence determines the composition of the resident microbiota.39
The term mucosal barrier refers to the lamina propria, epithelium, and glycocalyx, the mucus layers of the intestine, and the membrane-bound microbiota; the release of various chemical mediators and regulatory molecules drives multidirectional communication between the components of this local regulatory network.15 Homeostasis is possible—and is maintained—because barrier integrity is maintained while the intestinal epithelial cells closely regulate the activity of dendritic cells and the responses of macrophages and lymphocytes to prevent overreaction to resident microbiota.27
The relative percentage of mucus-secreting goblet cells increases from 4% in the duodenum to 16% in the descending colon; perhaps not surprisingly, the thickness of the mucus layers varies in each part of the GI tract,40 and is thickest in the highly colonized colonic segment of the distal gut.14, 15 There is very little data on the thickness of human mucus layers, and much disagreement as to which methodology gives the most accurate results, but the murine GI tract can be used as a representative model,37 although the layout of the intestinal tract is different.
An in vivo study in rats found that the firmly adherent, continuous inner mucus layer of the colon was 115 μm ± 51 μm, whereas the inner mucus layer of the small intestine was extremely thin (ranging from 15 μm ± 2 μm in the duodenum and jejunum to 29 μm ± 8 μm in the ileum), and was patchy and discontinuous.41 Similarly, the less dense outer mucus layer of the colon was 714 μm ± 109 μm, whereas the outer mucus layer of the duodenum, jejunum, and ileum was 154 μm ± 39 μm, 108 μm ± 5 μm, and 447 μm ± 47 μm, respectively.41 Extrapolating from the available data, it appears that human mucus layers should have similar relative proportions.37
The thinner outer mucus layer of the duodenum and jejunum, and the extremely thin, discontinuous inner layer are necessary for enterocytes to absorb nutrients, but this leaves the epithelium much more vulnerable to bacteria.41 In humans, direct bacterial contact is minimized because transit time through the small intestine is rapid, the mucus layers are continuously replenished, and antimicrobial peptides and lysozymes are expressed in much higher quantities that are concentrated closer to the epithelial cells in the small intestine than elsewhere in the GI tract.14 In addition to their absorptive and transport functions, enterocytes also develop immunological activity in self-defense; they not only express innate immune receptors and secrete several chemokines and cytokines, they also function as non-professional APCs.15 Additional protection is conferred by antibodies present in the glycocalyx that bind with antigens, as well as by pancreatic enzymes, which adsorb to the surface of the enterocytes and facilitate the breakdown of these antigen-antibody complexes; similarly, secretory antitoxins protect against the effects of toxic bacterial by-products.39
Although these defense strategies can limit exposure of the epithelium to gut microbes and prevent hypersensitive immune responses, controlled and regulated bacterial contact and direct sampling throughout the GI tract are required to maintain optimum gut immune functions.15, 19 Specialized regions of organized lymphoid tissues such as microfold (M) cells, Peyer's patches, and mesenteric lymph nodes play a critical role in direct antigen sampling of the gut; they are considered the inductive sites of the immune system that both induce and regulate immune responses.15, 19 Dendritic cells, which are found throughout the lamina propria and lymphoid tissue of the mucosa, also produce finger-like membrane extensions called dendrites that are extruded between epithelial cells to obtain direct antigen samples from the bacteria in the lumen.27, 42
Dendritic cells are the most important of the APCs in the innate immune system, and express up to 100 times more major histocompatibility complex (MHC) molecules than other APCs; those in the lymphoid tissue drive differentiation of naïve T cells and production of sIgA, whereas those in the lamina propria kill invading microorganisms and induce inflammatory responses to defensive signaling.27, 42 Among other factors, the type of microorganism encountered, the type and number of PRRs activated, and the local cytokine/chemokine environment influence the type of immune response.19, 42
Host-Microbial Interactions
Antigen binding to mature naïve B cells stimulates their differentiation into memory cells, with slow and limited production of antibodies; subsequent binding by the same antigen induces rapid proliferation and maturation of memory cells into plasma cells with immediate production of large amounts of antigen-specific antibodies.30 Although B cells can present antigens to T cells and secrete cytokines, their primary function is to develop into plasma cells that manufacture and secrete antibodies.30
T cell-dependent adaptive responses are typically triggered by MHC molecules expressed by APCs.43 Class I MHC molecules, which are expressed by all nucleated cells,43 present intracellular antigens such as those found on viruses to CD8 cytotoxic T cells.30 Class II MHC molecules, which are constitutively expressed by B cells, monocytes, macrophages, and dendritic cells, complex with peptides derived from extracellular antigens, which are then presented to CD4 helper T (TH) cells.30, 42
Microbial composition is an important factor determining the intricate balance between different T cell subsets in the GI tract; beneficial microbiota have been shown to drive local expansion of TH cell populations.19 There are three types of T cells: cytotoxic T cells, regulatory T cells (TREG), and TH cells.30 Cytotoxic T cells can secrete cytokines and are critical for eliminating intracellular pathogens, especially viruses, usually by inducing apoptosis of infected cells, whereas TREG cells either suppress the immune response by direct cell-to-cell contact or by secreting cytokines with immunosuppressive properties.30
TH cells are differentiated into three types, each of which secrete several cytokines.30 TH1 cells promote cell-mediated immunity against intracellular pathogens (e.g., viruses) by facilitating cytotoxic T cell and macrophage responses, whereas TH2 cells stimulate antibody production by B cells and direct responses to extracellular pathogens (e.g., parasites, bacteria).30 In contrast, TH17 cells promote tissue inflammation,30 and are also important mediators of intestinal immune defenses which protect against intestinal pathogens.13 TH17 cells are induced by the intestinal microbiota via SFB colonization of the epithelium, by phagocytosis of infected apoptotic cells by dendritic cells, and by direct flagellin-mediated stimulation of lamina propria dendritic cells, but are negatively regulated by inhibition of T cell proliferation following microbial recognition by intestinal epithelial cells.13
In addition, natural killer T (NKT) cells are a distinct subset of T cells in the adaptive immune system; when activated, NKT cells secrete IL-4 and IFN-γ, and are thought to regulate immune responses.30 NKT cells are discreet from natural killer cells, which are cytotoxic leukocytes in the innate immune system.30 Natural killer cells secrete various cytokines, influence the adaptive immune system by stimulating and inhibiting differentiation of TH1 cells and TH2 cells, respectively,30 and some subtypes appear to have mucosal barrier protective effects.26
Communication between the mucosal layer and the microbiota is bi-directional. Cross-talk is the term used to describe this ability to sense and appropriately respond to the microbial environment within the lumen; the release of cytokines, chemokines, and other signaling molecules, and the response by leukocytes and other immune cells, enables the body to mount a defensive response only when necessary.44 This dynamic interplay between the intestinal microbiota, epithelial cells, and gut-associated immune cells of the intestinal barrier maintains the crucial balance between immunity and inflammation in the gut.44
Different responses by the immune system to beneficial and pathogenic bacteria are evoked by the recognition of their different signature molecules; inflammatory and defensive responses occur in response to pathogen-associated molecular patterns (PAMPs), but these responses are not triggered by the microbe-associated molecular patterns (MAMPs) of commensal bacteria.44 The adequate presence and correct functioning of PRRs and signaling mechanisms in epithelial and monocytic cells are critical to prevent chronic stimulation by beneficial microorganisms of inappropriate and potentially destructive inflammatory responses, while simultaneously maintaining robust defensive responses against pathogenic organisms.19, 27, 44
These very different responses can initiate via the same pathway or receptor, depending on what activates it and where it is located, because the apical side of the epithelial cell interacts with the microbiota via the mucin layer, but the basolateral side interacts with immune cells in the lamina propria.15, 31 For example, if TLR-9 receptors on the apical side of epithelial cells are triggered, a tolerogenic response is induced, but when TLR-9 receptors on the basolateral side are triggered, the innate immune system is activated and expresses proinflammatory cytokines and chemokines.15, 27 Similarly, intrinsic signaling by nuclear factor kappa-B (NF-KB) prevents apoptosis of epithelial cells,40 yet when the NF-KB pathway is triggered by defensive signaling, it initiates a pro-inflammatory response, and if triggered by homeostatic signaling, it confers tolerance of resident bacteria, regulates TH cell responses, and prevents inflammation.13, 15, 19, 27
Surface-associated proteins, such as pili, flagella, and fimbria, not only provide motility for bacteria, but allow adhesion to glycosylated mucins or fibronectins in the extracellular matrix.36, 45 Once shed in the lumen, their monomeric subunits interact directly with epithelial and immune cells to induce β-defensins, tumor necrosis factor α (TNF-α), and IL-8 cytokines.45 Bacteria can also highjack certain signaling pathways and actively inhibit others.21 For example, both pathogenic and beneficial bacteria have evolved mechanisms to suppress or induce NF-KB activation, and inhibit or modulate its signaling pathway; recent evidence suggests that bacteria-derived secreted factors may be involved.19, 45, 46 The microbiome produces numerous such compounds that regulate expression of antimicrobial peptides26 and modulate intestinal barrier function,17 so alterations to the composition of the intestinal microbiota can affect mucosal barrier integrity and susceptibility to intestinal pathogens.13, 17, 44
Microbial Mechanisms and Effects
Commensal bacteria help to determine the composition of the resident microbiota by inhibiting pathogen colonization, and can thus have an anti-inflammatory effect, but they can also help to trigger inflammatory diseases and other gut disorders.45, 47 Likewise, pathogenic bacteria can have beneficial effects. For example, the polysaccharide chain and lipid tail of LPS are components of the bacterial outer cell membrane that identify it as “non-self” and there is evidence that trace amounts of LPSs bound to Gram-negative bacteria act beneficially to stimulate or “prime” the innate immune system without initiating a full-blown response.16 Free LPS in large quantities in body fluids (such as during sepsis, gastroenteritis, or altered bowel permeability) induces synthesis of prostaglandins and production of cytokines (IFNs, IL-1, IL-6, and TNF-α), activates complements, and stimulates macrophages and leukocytes.16
In contrast, when trace amounts of LPS are bound to very low-density or low-density lipoproteins, chylomicrons, bile salts, serum albumin, Ig, or LPS binding protein, the LPS remains biologically active and has a physiological effect rather than a pathological effect.16 Locally, it induces IgA and cytokine production in the gut-associated lymphoid tissue (GALT) and stimulates enterocyte differentiation, whereas systemically it results in highly restricted, controlled immunological responses in the reticuloendothelial cells of organs such as bone marrow, spleen, liver, and lungs.16 The beneficial effect of this physiological stimulation is that the immune system is maintained in a state of alertness, primed for an immediate response, but without toxic effects from high circulating levels of cytokines and without developing a tolerance for LPS.16
Most probiotics are derivatives of the intestinal bacteria and have beneficial effects on the host and most of the resident microbiota, with either inflammatory or anti-inflammatory capacity, which stimulate immune and non-immune cells, respectively.31, 45 Probiotics can preserve or restore intestinal epithelial integrity and homeostasis, contribute to increased production of antimicrobial substances, and compete with pathogenic bacteria for adhesion sites.35, 44, 46 Additional proposed mechanisms include down-modulating the immune response; stimulating innate immunity that protects against chronic inflammation; stimulating production of intestinal mucins and regeneration of epithelial cells; reducing permeability; and improving barrier tightness.15, 31, 35, 44, 46, 48 Specific strains can: prevent reduction in transepithelial resistance induced by hydrogen peroxide; down-regulate nitric oxide production by macrophages; promote cytoprotective responses; reduce bacterial translocation to the mesenteric lymph nodes from the gut; increase mucus production; maintain colonic mucosal barrier integrity; and regulate apoptosis.15, 46 One novel mechanism that has been observed in some intestinal bacteria is the production of antibacterial substances that, when cleaved by the pancreatic enzyme trypsin, are activated against Gram-positive pathogenic bacteria.44
Recent advances in technology and greater understanding of functional genomics have enabled researchers to better understand some of the processes by which bacterial colonization and survival occur, and the mechanisms by which cross-talk occurs between bacteria and host mucosal cells.36, 44-46, 48 Through technological advances, these very properties have been exploited for a variety of uses, from cosmetics, toiletries, paints, printer ink, and pesticides, to the medical field (including drug delivery, anticoagulants, plasma substitutes, wound dressings, tissue reengineering and replacement, and tumor inhibition) as well as functional foods such as probiotics.49 Digestive enzymes and low pH of the stomach are natural barriers to many bacteria, and the small intestine harbors an equally harsh environment that includes pancreatic enzymes, bile salts, and anti-microbial peptides such as defensins and cathelicidins.48 Probiotics help to prevent infection from surviving pathogens by stimulating production of these peptides and over-competing for colonization sites in the gut.44, 48
Bacteria have developed acid-induced survival mechanisms, such as strain-specific changes in sulfur amino acid metabolism; increased production of ATP synthases to reduce cytoplasmic H+ accumulation; increased production of ammonia and branched-chain amino acids, which capture excess H+; as well as changes in surface properties and fermentation capabilities that increase acid tolerance.48 Exposure to bile, which has a detergent-like effect on the lipid layer of the bacterial cell membrane, has resulted in strain-specific adaptive responses that include increased production of glycolytic enzymes and enzymes that synthesize fatty acids; expression of bile salt hydrolase to deconjugate bile salts; and increased activity of efflux pumps to reduce cytoplasmic accumulation of bile acids.48 Other adaptive responses to bile exposure include overexpression of chaperones and proteases to repair misfolding and prevent aggregation; induction of SOS response proteins to repair DNA damage; and production of exopolysaccharides.48 Exopolysaccharides will be discussed in more detail in the next section under the heading ‘Adhesins and Receptors.’
In addition to exopolysaccharides, some bacteria produce H2O2, some secrete bactericidins, and some secrete lactic acid and acetic acid, which lower the pH in the environment; these secretions inhibit growth and colonization of some pathogenic bacteria, whereas secretion of teichoic acid and lipoteichoic acid increases adherence by certain strains of Gram-positive bacteria.44, 50 Secreted molecules interact with dendritic cells to modulate their regulatory mechanisms and maintain immune tolerance for the bacteria.45 The cell wall associated proteins p40 and p75 also modulate dendritic cell response and serve as adhesion molecules, but in addition, they stimulate phosphorylation of epidermal growth factor receptors, and prevent epithelial injury and cytokine-induced apoptosis of epithelial cells.45, 46 In response to signals by probiotic and commensal bacteria, IgA antibody production and phagocytic activity are higher, and translocation of pathogens across the mucosal barrier is lower.44
Intestinal barrier dysfunction, which can include both increased intestinal permeability and inflammation, often involves dysfunction of tight junctions between epithelial cells.15 Pathogenic bacteria use flagella for motility, or produce mucus-degrading enzymes such as glycosidases to break down mucin oligosaccharides so they can digest mucus barriers to reach the host epithelial layer.36, 40 They have also developed strategies to gain access to the lamina propria, such as activating the inflammatory cascade, directly disrupting paracellular tight junctions, and inducing fluid and electrolyte secretions in response to toxin production.15 Vibrio cholera disrupts tight junctions through its cytotoxin hemagglutinin protease; Clostridium difficile produces two distinct exotoxins that cause increased paracellular permeability; Clostridium perfringens enterotoxin triggers massive permeability changes; and enteropathic Escherichia coli directly disrupts tight junctions and enhances intestinal permeability through selective dephosphorylation and phosphorylation.15
Bacterial survival strategies can make it difficult for the body to mount an effective immune defense, and also increase the possibility of autoimmune issues.49 Some pathogenic bacteria (e.g., E. coli) produce capsular polysaccharides known as glycosaminoglycans for molecular camouflage, and some of the most challenging pathogenic bacteria are difficult to treat because they form biofilms (e.g., Streptococcus pyogenes, which causes necrotizing fasciitis), whereas polysaccharides on some bacteria are very similar to those on human cells (e.g., Neisseria meningitidis).49
The immunogenic lipopolysaccharides produced by E. coli suppress macrophage growth and promote secretion of TNF-α, whereas commensal exopolysaccharides have the opposite effect, inhibiting secretion of TNF-α and promoting macrophage growth.45 Similarly, commensal and probiotic exopolysaccharides have been correlated with stimulation of Peyer's patch cells in the intestinal mucosa and increased natural killer cell activity. 45 They have also demonstrated an ability to inhibit biofilm formation, attenuate infection-related cytotoxic effects on enterocytes through their antioxidant and free-radical scavenging activity, and improve recovery time after viral infection.45
Antigens, Adhesins, and Receptors
In the human body, the immune system uses cell-surface molecules known as antigens to distinguish “self” from “not self.” Infection by pathogens occurs when they are able to evade or resist the body's immune response, express sufficient amounts of the appropriate adhesion molecules to bind with the host's receptors, establish residency in the tissue of choice, and produce toxins or other virulence factors in large enough amounts to cause destruction or disruption of normal cellular function.50, 51 Phagocytes have learned to recognize and respond to certain bacterial antigens, with phagocytosis being assisted by the actions of Toll-like receptors (TLRs) and scavenger receptors: TLRs can recognize and bind with LPS and lipopeptides, which upregulate scavenger receptors, whereas both TLRs and scavenger receptors bind with bacterial polysaccharides.52
In their turn, pathogenic bacteria have evolved several ways to avoid detection and elimination by the immune system: the IgA binding proteins that are found in hemolytic Streptococcus species and S. aureus (specifically M-proteins, β-antigen, and SSL), some of which also bind complement proteins, prevent phagocytosis and block the respiratory burst of neutrophils.53 The filamentous hemagglutinin (FHA) of B. pertussis upregulates complement receptor 3 (CR3) by binding with the phagocytic integrin signal transduction complex, and when it then binds with CR3, it is safely internalized within the macrophage; similarly, the FimH adhesin of E. coli binds to the CD48 receptor of macrophages, which results in internalization and survival within the phagocyte.52
After surviving macrophage engulfment, Salmonella enterica serovar Typhi and serovar Paratyphi are then spread through the blood and lymph systems of their human hosts.54 E. coli, N. meningitidis, and some Streptococcus species are among the pathogens that use molecular mimicry to evade detection by the host's immune system and protect themselves from phagocytosis; negatively-charged polysaccharides such as polysialic acid, heparosan, chondroitin, and hyaluronan not only repel phagocytes, but allow the bacteria to masquerade as “self,” which increases its virulence.55
Superantigens (SAgs) are an extremely potent bacterial defense against the immune system, which cause fever, shock, and often death; they stimulate mitogenesis of T-cells by first binding with MHC class II molecules and then binding to the variable β-chain region of T-cell receptors, leading to uncontrolled T-cell activation followed by systemic release of massive amounts of TNFα, IL-2, and other pro-inflammatory cytokines.56 The first bacterial SAgs identified were toxins from Gram-positive S. aureus and S. pyogenes, and there are now more than 40 known SAgs that have been identified with genomic sequencing, with some resulting from horizontal gene transfer between subgroups of streptococcal and staphylococcal species.56 SAgs have also been identified in Mycoplasma arthritidis and Yersinia pseudotuberculosis, which are Gram-negative bacteria.56 SAgs cause staphylococcal food poisoning and endotoxic shock, staphylococcal toxic shock syndrome, streptococcal toxic shock syndrome (which is lethal in up to 50% of cases), acute rheumatic fever, and Kawasaki disease; some studies have also found a possible SAg role in autoimmune diseases, such as multiple sclerosis (MS) and Type 1 diabetes mellitus (T1DM).56
Histo-Blood Group Antigens
Histo-blood group antigens (HBGAs) are specific surface-associated structures that have been classified into more than 30 blood group systems by researchers investigating the cause of life-threatening reactions to blood transfusions and rejection of tissue or organ transplants.57 The most well-known HBGAs are the ABO and Rh “blood type” antigens found on red blood cells (RBCs) and platelets, but HBGAs are found on the surface of most endothelial and epithelial cells, and in the body fluids and secretions of about 80% of those of European descent.57-59 Because blood group antigens are found throughout the body, they are considered histocompatibility antigens.57 To reflect the fact that these antigens are not just found on RBCs, the term “histo-blood group antigens” is used to specifically indicate that antigens from these blood group systems are also found on histological tissues.57, 59 Interested readers are referred to an extensive review of the associations between blood type biochemistry and human disease, which has been previously published.60
To better understand the microbiome, we need to consider the blood type antigens we were born with and their influence on the microbiota. These surface-associated markers not only serve as antigens, they also are a food source for intestinal bacteria.61, 62 In the intestine, the glycocalyx is constantly renewed, with an average turnover time of 6-12 hours, while the epithelial cells of the colon have a 24-hour turnover rate.14 Terminally differentiated goblet cells, enteroendocrine cells, and enterocytes reach the tip of the villi within 3-5 days, undergo apoptosis, and are extruded into the intestinal lumen.15, 40 Mucosal bacteria degrade the glycoproteins found in mucus, including the glycans on antigens.63 A small but intriguing study in Finland found a statistically significant correlation between the composition of the intestinal mucosal microbiota and the ABO blood type of the host.63 The overall mucosal microbial profile as well as the relative proportion of major bacterial groups in the mucosal layer were significantly different in the presence of the B antigen (from B and AB blood types).63
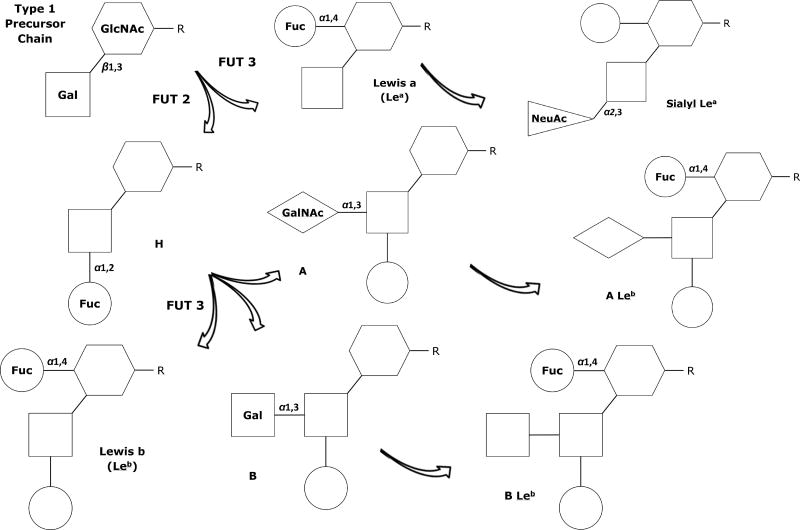
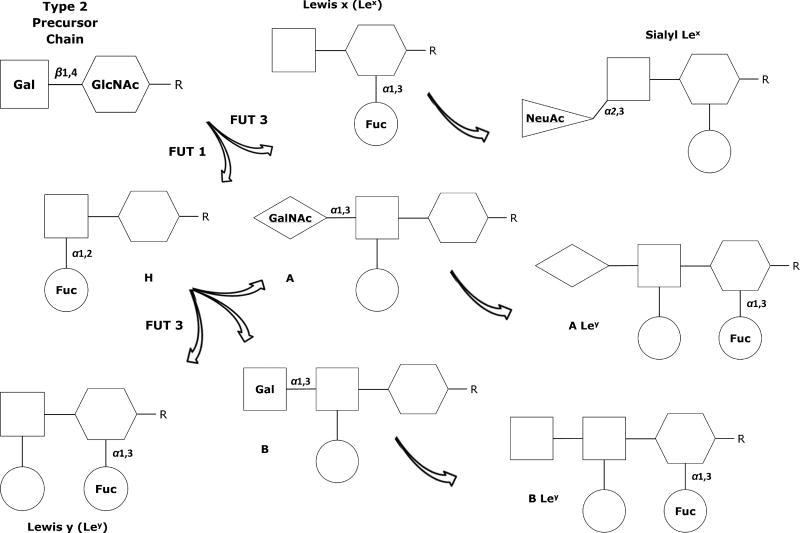
The HBGAs found on cell surfaces and in body fluids and secretions are not identical. There are two glycoside “precursor” chains or acceptor substrates with slightly different structures, known as Type 1 and Type 2 chains. Type 1 chains and their terminal antigens, shown in Figure 1, are detectable in sweat, breast milk, saliva, mucous, and other body fluids of individuals who are known as “secretors,” but these antigens are missing in the body fluids of about 20% of people of European descent, who are known as “non-secretors.”60, 64 Type 2 chains, depicted in Figure 2, are the basis for the addition of the ABO “blood type” antigens that serve as receptors on the surface of almost every cell and tissue in the body, including the endothelial cells of the GI tract.60, 64
Figure 1.
Type 1 precursor chain and associated antigen structures; the Lewisb antigen is found in the body fluids of about 80% of “secretors.”
Fuc – Fucose; FUT2 - α1,2-Fucosyltransferase enzymes expressed by Se genes; FUT3 - α1,3/4-Fucosyltransferase enzymes expressed by Le genes; Gal – Galactose; GalNAc - N-acetylgalactosamine; GlcNAc - N-acetylglucosamine; NeuAc - N-acetylneuraminic (sialic) acid
Figure 2.
Type 2 precursor chain and associated antigen structures, found on endothelial, epithelial, and red blood cells throughout the body. The H antigen is the acceptor molecule needed for attachment of the A, B, or AB terminal glycosides.
Fuc – Fucose; FUT1 - α1,2-Fucosyltransferase enzymes expressed by HH genes; FUT3 - α1,3/4-Fucosyltransferase enzymes expressed by Le genes; Gal – Galactose; GalNAc - N-acetylgalactosamine; GlcNAc - N-acetylglucosamine; NeuAc - N-acetylneuraminic (sialic) acid
Each person's “blood type” is determined by the combined expression of the HH gene and the ABO gene, which results in six possible allelic combinations and four phenotypes.57 Those with the H/H or H/h alleles have the fucosyltransferase enzyme FUT1 required for the acceptor substrate, but if they lack the glycosyltransferase enzymes which are required to attach either of the final glycosides, these individuals have OO antigens and type O blood.57, 60, 64 People with type A blood have either AO or AA antigens; those with type B blood have either BO or BB antigens; whereas those with type AB blood have alleles that express both enzymes and therefore have both the A and B antigens.60, 64, 65 Those with the so-called “Bombay” phenotype have the h/h allele, and because the FUT1 enzyme is non-functional, they do not carry the O, A, or B antigens.64
The Lewis (Le) and Secretor (Se) genes function independently from the HH and ABO loci, but play a significant role in antigen expression; they both express fucosyltransferase enzymes that attach a terminal fucose in specific ways to Type1 and Type 2 precursor chains and their derivative structures, as shown in Figures 1 and 2.60 Sialylation or sulfuration occurs when N-acetylneuraminic (sialic) acid is attached in α2-3 linkage to a β-D-galatose residue of an existing chain.57 Fucosyltransferase enzymes expressed by Le genes (FUT3) synthesize and release Lewis antigens from exocrine epithelial cells; those with Le/Le or Le/le alleles express Lewis antigens, while those with le/le do not.60, 66, 67 These antigens can be glycolipids or glycoproteins, found as membrane-bound antigens on the surface of cells and tissues or as oligosaccharides that freely circulate in blood and body fluids, and can be taken up by RBCs, platelets, and lymphocytes.60, 66, 67
The fucosyltransferase enzymes expressed by Se genes (FUT2) primarily interact with Type 1 precursor chains; about 80% of Caucasians are “secretors” with Se/Se or Se/se alleles, while those known as “non-secretors” have two non-functional copies (se/se) and the ABO antigens are not found in their saliva or other body fluids.60, 66, 67 By adulthood, the distal colon and rectum only express Lea and Lex antigens, regardless of secretor status, but the cells and secretions of the esophagus, stomach, and small intestine continue to strongly express the same HBGAs found in the fetal GI tract, with levels of HBGAs 10 to 12 times higher in the GI tract of secretors than in non-secretors.58
HBGAs are found on tissues derived from ectodermal, mesodermal, and endodermal origins, as well as RBCs and platelets; although ABO(H) and Lewis antigens may be detected on lymphocytes, they are acquired or adsorbed from the plasma.57 Regardless of secretor status, lymphocytes, granulocytes, and monocytes do not express ABO(H) and Lewis antigens that require FUT2 activity, however, they do have FUT3 activity and express Lex and sialyl-Lex antigens, which serve as ligands for selectins, a type of cell adhesion molecule found on endothelium, leucocytes, and platelets.57
Because HBGAs are “self” antigens, the adaptive immune system must be trained not to mount a response to these antigens. Central tolerance mechanisms include negative selection, in which MHC class I and class II molecules present endogenously-expressed antigens from peripheral tissues in medullary thymic epithelial cells, whereupon most but not all self-reactive T-cells are eliminated.68 Peripheral tolerance involves the presentation of self-peptide-MHC complexes by CD8+ dendritic cells to circulating T-cells in the lymph nodes, which subdues or eliminates the remaining self-reactive T-cells.68 Antigen proteins from parenchymal tissues can also be transported to draining lymph nodes, where MHC class I and class II molecules present them to circulating naïve T-cells.68
However, APCs are not required for induction of tolerance to antigens from intestinal epithelial cells.68 Instead, CD8+ dendritic cells present naïve T-cells with self-antigens from the intestinal mucosa (both epithelial cells and mucus) in the secondary GALT, and cortical stromal cells (which constitutively express many peripheral tissue antigens) present intestinal mucosa self-antigens in a similar manner in lymph nodes throughout the body.68 By activating and inducing deletional-tolerance in T-cells, lymph node stromal cells function like medullary thymic epithelial cells to directly eliminate self-reactive T-cells.68
In a similar manner, MHC class II molecules present microbiota-derived antigens from intestinal commensal bacteria to CD4+ T-cells in group 3 innate lymphoid cells (ILC3), thus inducing negative selection and deletion of activated commensal bacteria-specific T-cells.69 This process is very similar to negative selection of self-reactive T-cells by the medullary thymic epithelial cells and stromal cells, and results in control of adaptive immune responses which maintains intestinal homeostasis.69 Dysregulation of this process may be a factor in development of IBD. In tissue biopsies from pediatric Crohn's disease patients, ILC3 MHC class II levels were inversely correlated with colonic TH17 cell frequencies and commensal-bacteria specific serum IgG titers, indicating pro-inflammatory responses to commensal intestinal bacteria by the adaptive immune system.69
Dysregulation of mucosal immune function is thought to occur as a result of inadequate or abnormal initial colonization of the GI tract.62 The small intestine of the fetus has a thin, immature epithelium, with slow mucosal cell turnover and a limited amount of GALT cells in the lamina propria, whereas in the infant, the mature epithelium is thicker, with fast mucosal cell turnover and robust, diverse, and abundant GALT.62 Although the fetal intestinal mucosa is permeable, which allows exchange between fetal serum and amniotic fluid, intestinal membrane closure occurs during the first week after birth; intestinal epithelial growth and maturation is stimulated by human milk hormones and growth factors.70
The intestinal mucosal barrier continues to grow and develop during the first year of life, undergoing significant tissue remodeling and profound modifications of absorptive and digestive functions.70, 71 Immunological and morphological maturation of the mucosal barrier requires the presence of intestinal microbiota.23, 70, 71 For example, they induce expression of microbicidal proteins from Paneth cells as well as development of blood vessels in the villi.70 Maturation of the mucosal barrier also depends on crosstalk between the colonizing bacteria and the immature mucosal barrier of the newborn, which produces the initial functional innate and adaptive immune phenotype and establishes homeostasis in the GI tract.62 The maturation of the mucosal barrier is further strengthened with the introduction of oral feedings and solid foods.62, 71
The newborn is not entirely defenseless, but the function of all components of the innate immune system is weaker at birth than compared with later in life, and the adaptive immune system actively promotes self-tolerance.72 Macrophages and monocytes are immature, natural killer cells are hypo-responsive, and serum concentrations of complement are lower than in adults, with less biological activity.72 In the fetal intestine, T-cells and macrophages appear as early as the 11th week of gestation, and by the 16th week, Peyer's patches form; the mucin gene is fully expressed between the 23rd and 27th week of gestation; dendritic cells are seen throughout the gut; by about the 19th week, CD4 T-cells begin to predominate in Peyer's patches, but CD8 T-cells don’t appear until around the time of birth.73
During pregnancy, the maternal immune system is biased toward TH2 responses, allowing tolerance of the fetus; it is thought that fetal T-cells are from a separate hematopoietic lineage that is predisposed to a tolerogenic response to antigens encountered before birth.73 Intraepithelial lymphocytes are found which differ from naïve systemic lymphocytes and respond at a lower threshold.72, 73 The fetus receives passive immunity from transplacental maternal IgG, and at birth, as a result of sensitization to maternally derived antigens, the newborn displays immune competence with T-cells that are capable of antigen-specific responses; by the second week, the newborn produces cells containing IgA and IgM.73 In breast-fed babies, mother's milk contains sIgA, which binds with microbial antigens and is thought to protect the baby's developing immune system from its own intestinal microbiota.71
The tolerogenic bias in the newborn's immune system allows for the colonization of the intestinal microbiome without an overwhelming inflammatory response.73 In maturing epithelial cells of the intestine, surface TLR expression and downstream signaling is reduced and inhibitory factor kappa B is increased, which suppresses NF-KB activation.73 In addition, commensal bacteria downregulate expression of IL-17 and upregulate expression of IL-25 and IL-10, which results in anti-inflammatory effects; they also promote sealing of tight junctions and apical tightening, which maintains barrier integrity.73 This tolerogenic bias benefits the newborn in another way: although ABO(H) and Lewis antigens are expressed throughout the fetal GI tract,57, 59 in the first months of life, babies do not produce ABO antibodies.57 Fortunately, loss of self-reactive T-cells and adaptation to commensal bacteria and HBGAs occurs during the period before the baby's immune response loses the tolerogenic bias.
Because ABO(H) blood group antigens result in negative selection and deletion of self-reactive T-cells, the adaptive immune system does not produce antibodies to pathogenic bacteria that exhibit blood group antigens.61 Fortunately, the innate immune system contains various glycan-binding proteins, such as C-type lectins and galectins, which recognize a variety of cell-surface carbohydrate moieties and participate in innate immune responses to pathogens bearing blood group antigens.61 The galectins Gal-4 and Gal-8 are specifically reactive to blood group A and B antigens on pathogens, regardless of the individual's blood group antigen profile, but do not react to blood group A and B antigens if present on the individual's RBCs.61
Unlike other innate immune lectins, Gal-4 and Gal-8 do not require or activate complement, but directly kill bacteria by drastically disrupting membrane integrity and altering bacterial motility.61 Galectins are highly expressed in the intestinal mucosa, where the adaptive immune system has been trained to tolerate “self” antigens, thus providing direct protection via the innate immune system against bacteria masquerading as “self.”61 Galectins are able to bind with and kill bacteria expressing either α-Gal or β-Gal epitopes, indicating that their protection is not limited to bacteria expressing HBGAs; indeed, they function as pathogen recognition proteins for multiple populations of intestinal bacteria, and may play a previously-unrecognized role in modulating the intestinal microbiome.61
Some pathogens mimic HBGAs as a way of evading detection by the immune system. For example, E. coli O86 mimics the blood group B antigen and thus does not generate an immune response in blood group B people, but blood group A or O people generate high titers of blood group B antibodies when exposed to the bacteria.61 H. pylori expresses both Lea and Leb HBGAs on its cell surface,54 and the Leb antigen mediates attachment of H. pylori to gastric epithelial cells.43 Secretor status influences susceptibility to pathogens such as V. cholera, Campylobacter jejuni, H. pylori, Norwalk, and certain respiratory viruses; non-secretors are known to be more susceptible to Crohn's disease, primary sclerosing cholangitis, and other chronic inflammatory diseases.74 Although negatively charged surface-associated carbohydrate antigens, such as heparan sulfate and sialic acid, are recognized as receptors by most viruses, bacteria, and protozoans, some pathogens recognize and bind to HBGAs, which do not carry a charge.54
Adhesins and Receptors
Bacterial adhesion is a critical mechanism in the human gut, for without it, colonization and survival are unlikely, and bacterial virulence is impaired.36, 44 Exopolysaccharides serve a variety of functions for bacterial species, including slime and biofilm formation, establishment of colonies, as well as migration, adherence, and protection of individual microorganisms.45, 49 Exopolysaccharides can be made of homogeneous repeating monomers or heterogeneous polysaccharides, which affects the chemical nature of the polymers and the type of linkage bonds that are formed.49 The monomeric units found in homopolysaccharides include fructans, polygalactans, and α- and β-D-glucans, while the heteropolysaccharide units include L-rhamnose, D-glucose, D-galactose, and sometimes glucuronic acid (GlcA), N-acetylglucosamine (GlcNAc), and N-acetylgalactosamine (GalNAc).49
Exopolysaccharides that remain attached to the surface of the bacteria are known as capsules; these capsular polysaccharides are the antigenic basis for bacterial serotyping as well as antibody development leading to successful vaccines, and account for the diversity of diseases caused by bacteria of the same species.49 Exopolysaccharides can also be made in the external environment by bacterial enzymes attached to the external cell wall, or made within the bacteria and then secreted through the cell wall; some of these extracellular proteins are known as adhesins.46, 48, 49
In addition to polysaccharides, bacteria secrete extracellular proteins and small peptides which bind with receptors on host mucosal cells in the lumen, activate signaling pathways, and stimulate or suppress gene expression, with resulting physiological changes in the host cells.44-46, 75 As shown in Table 1, quite a few secretion systems have been identified, each with unique functional features and structural mechanisms. These secretion systems are used to transport extracellular proteins, peptides, toxins, and assembled adhesins across bacterial membranes, with different systems used by Gram positive and Gram-negative bacteria; it is quite common for a bacterial species to use multiple systems.52-54, 75-92
Table 1. Known secretion systems of Gram negative and Gram positive bacteria.
| Gram | Secretion System |
Membrane(s) Spanned |
Starting molecule |
Target Environment |
Some of the Known Bacteria Using Each System |
Secreted molecule(s) |
Examples | Ref |
|---|---|---|---|---|---|---|---|---|
| Neg, Pos | General Secretion Pathway | Bacterial inner | Preproteins containing signal peptide | Periplasm | Most | Unfolded proteins; lipoproteins | Sortase-assembled pili | 52, 75, 77, 82, 83, 91 |
| Pos | Accessory Secretion Pathway | Bacterial inner | Large serine-rich glycoproteins | Periplasm | Bacillus spp.; Streptococcus spp.; Staphylococcus spp.; Listeria monocytogenes; Mycobacterium tuberculosis; Clostridium difficile; Corynebacterium glutamicum | Virulence factors | FbpA; SodA; MnSOD; SRRPs; catalase-peroxidase; autolysin p60 | 52, 82 |
| Neg, Pos | Twin Arginine Translocase | Bacterial inner | Folded proteins with signal peptide | Periplasm | Escherichia coli; Bacillus subtilis; Streptomyces spp. | Folded proteins | Peroxidase; phosphodi-esterase | 82, 83 |
| Neg | Curli Biogenesis System | Bacterial outer | Unfolded soluble peptide | Extracellular space | Salmonella spp.; enteric E. coli | Insoluble amyloid fiber | Curli | 52, 89 |
| Neg | Chaperone/ Usher | Bacterial outer | Unfolded proteins | Extracellular space | E. coli spp.; Burkholderia spp.; Yersinia spp.; Salmonella enterica; Pseudomonas spp; Acinetobacter spp.; Bordatella spp,; Haemophilus spp.; Klebsiella spp.; Proteus mirabilis | B, γ-, κ-, and π-Fimbrial adhesins; non-fimbrial surface structures; Dra-blood group antigens | Type I pili; P pili; Afa/Dr | 54, 75, 77, 85, 86 |
| Neg | Alternate Chaperone/ Usher | Bacterial outer | Unfolded proteins | Extracellular space | Human enterotoxigenic E. coli; Yersinia pestis; Aeromonas hydrophila; Salmonella enterica serovar Typhi; Burkholderia spp.; | α-Fimbrial adhesins | Coli surface antigen 1 | 85, 86 |
| Neg | Type 2 | Bacterial inner and outer | Unfolded or folded proteins | Extracellular space | Klebsiella oxytoca; Klebsiella pneumoniae; Pseudomonas aeruginosa; Vibrio cholera; Neisseria gonorrhoeae; Neisseria meningitidis; E. coli spp.; Legionella pneumophila; Yersinia spp.; S. enterica serovar Typhi; Shewanella spp.; Chlamydia spp. | Hydrolytic enzymes; slime proteins; toxins; adhesins; cytochromes | Type IV pili; Pullulanase; pseudolysin; Cholera toxin | 54, 75, 77, 87, 88, 90, 92 |
| Neg | Type 5 | Bacterial outer | Unfolded proteins | Extracellular space | Neisseria gonorrhoeae; Neisseria meningitidis; Yersinia spp.; Bartonella henselae; S. enterica serovar Typhimurium; Helicobacter pylori; Bordatella pertussis | Virulence factors, adhesins, proteases | OMPs; Intimin; ATs; TAAs; TPS/FHA; IgA binding proteins | 53, 75, 77, 92 |
| Neg | Type 1 | Bacterial inner and outer | Preproteins containing signal peptide | Extracellular space | Uropathogenic E. coli; S. enterica | Large partially unfolded proteins | α-hemolysin; BAP family; SiiE | 75, 77 |
| Neg | Type 3 | Bacterial inner and outer, and target cell wall | Preproteins containing signal peptide | Target cell cytoplasm | S. enterica; Yersinia spp; Pseudomonas spp ; Shigella spp; enteropathogenic Escherichia spp | Partially unfolded proteins, virulence factors, effector molecules | Tir | 75, 77, 87 |
| Neg, Pos | Type 4 | Bacterial inner and outer, and target cell wall | Proteins containing signal peptide | Extracellular space; target cell cytoplasm | B. pertussis; Neisseria gonorrhea; Helicobacter pylori; Brucella spp; B. henselae; Campylobacter jejuni; Ricksettia typhi; Ricksettia felis; Ricksettia bellii; Legionella pneumophila; Enterococcus faecalis | Proteins, virulence factors, or complexes of single-stranded DNA and proteins | Pertussis toxin, CagA; RalF | 75, 77, 80, 81, 84, 87 |
| Neg | Type 6 | Bacterial inner and outer, and target cell wall | Proteins without signal peptides | Target cell cytoplasm or periplasm | Vibrio cholera; Aeromonas hydrophila; Pseudomonas aeruginosa; Burkholderia spp. | Effector proteins, antibacterial toxins | VrgG-1, Hcp, Tse1-3, Tle1-5 | 76, 78, 79 |
| Pos | Type 7 | Bacterial inner, peptidoglycan layer (and mycomembrane), cell capsule | Protein complexes containing C-terminal signal sequences | Extracellular space | M. tuberculosis; Mycobacterium leprae; L. monocytogenes; Corynebacterium diphtheriae; Norcardia spp.; Streptomyces spp.; Clostridium spp.; Staphylococcus aureus; Streptococcus agalactiae; B. subtilis; Bacillus anthracis | Protein complexes; virulence factors | ESAT-6; (EsxA) CFP-10 (EsxB); WXG100; PE/PPE proteins; Rv3881c (MTB48) | 77, 82 |
Each secretion system transports different types of molecules across the bacterial membrane(s), and some also enable translocation of effector proteins, single-stranded DNA, or toxins through the membrane of target cells.
Secretion Systems
The general secretion pathway (Sec) is used by both Gram-negative and Gram-positive bacteria; in Gram-negative bacteria, the Sec pathway is a mechanism composed of multi-subunit complexes, including an ATPase subunit, located in the bacterial inner membrane to transport unfolded proteins and lipoproteins across the inner membrane into the periplasm.82, 91 There are two separate processes, known as co-translational and post-translational, which result in proteins being translocated by this mechanism.75 In the co-translational process, a cytoplasmic signal recognition particle (SRP) binds to the signal peptide as it emerges from the ribosome, which temporarily suspends translation; the SRP guides the ribosome to the Sec mechanism and docks with it, which causes translation to resume and the protein emerges directly into the channel of the mechanism.75, 83, 91
In the post-translational pathway, cytoplasmic chaperone proteins prevent premature folding as they guide unfolded preproteins to the protein conducting channel of the mechanism for translocation, whereupon periplasmic signal peptidases cleave the N-terminal signal peptide from them to produce mature proteins.75, 83, 91 Lipoprotein precursor proteins are modified by the addition of diacylglycerol prior to translocation, which enables recognition and cleavage of their N-terminal signal peptide by a second type of periplasmic signal peptidases.82
In Gram-positive bacteria, the Sec pathway also transports preproteins across the cell membrane, where the N-terminal signal peptide is cleaved and the mature protein is released, unless the presence of a C-terminal sorting signal causes it to be retained.52, 91 Different sorting signals result in the retention of surface proteins that can either impede detection by the immune system, enable tissue adhesion, promote heme-iron uptake, or polymerize into pili.52, 91 The sorting signals are cleaved by membrane-anchored transpeptidases known as sortase enzymes; the housekeeping sortase of individual surface proteins catalyzes the transfer of the protein to pentaglycine cross-bridges that connect peptidoglycan strands in the cell wall, and in the case of pilus assembly, sequential transpeptidation reactions by pilin-specific sortases result in polymerization of pilin subunits until the completed pilus is transferred to the cell wall and anchored there by the housekeeping sortase.91
The accessory secretion pathway (Sec2) is a second independent Sec pathway found in primarily pathogenic Gram-positive bacteria; Sec2 exports virulence factors that are not translocated by Sec because they are highly glycosylated.82, 91 These large proteins are also anchored in the bacterial cell wall and facilitate pathogenic activities as diverse as platelet aggregation, endocarditis, and biofilm formation.52, 91 The twin arginine translocase (Tat) is a multi-subunit complex in the inner or cell membrane of Gram-negative and Gram-positive bacteria, respectively, that uses proton motive force to translocate certain precursor proteins when it recognizes a highly-conserved twin arginine consensus motif in signal peptides attached to them.82, 83 These are usually proteins that contain a cofactor and therefore must fold prior to translocation, but some rapidly-folding proteins without cofactors are also translocated through this pathway.82, 83
The Curli biogenesis system is a multi-subunit, outer membrane Sec-dependent mechanism that generates thin fimbriae that are unstructured soluble peptides until they get to the outer surface, where interaction with a nucleator subunit in the mechanism results in a conformational change that produces insoluble amyloid fibers.52, 89 Two unique features of this system are that it uses an entropy gradient that is diffusion-based and therefore does not require ATP for translocation of the subunit proteins,87 and that the amyloids it secretes self-polymerize when they contact the nucleator subunit.89 Similar to other systems, it uses periplasmic chaperones to prevent premature amyloid formation and to direct the peptide subunit to the secretion mechanism.87, 89 Of interest, closely related bacteria (e.g., E. coli and S. enterica) can “cross-seed” with their own unstructured soluble peptides to accelerate the other's amyloid polymerization.89
Gram-positive bacteria use the Type 4 secretion system (T4SS) or the Type 7 secretion system (T7SS) to export mature proteins.81, 82 In Gram-negative bacteria, mature proteins are secreted through the outer membrane via one of three additional systems: the chaperone/usher secretion systems, which transport fimbrial adhesins and pili; the Type 2 secretion system (T2SS), which secretes most bacterial enzymes and toxins; and the Type 5 secretion system (T5SS), which is the mechanism used by bacterial autotransporter proteins.75, 77 The Sec and T2SS pathways are ATP-dependent, but the T5SS and chaperone/usher mechanisms are not.75, 77 Regardless of the secretion system used, translocated proteins must fold properly and quickly into their native, active conformation to avoid degradation by proteases in the periplasmic or extracellular environment; the close proximity of metal ions such as calcium was shown to increase the speed with which protein folding occurred, and the release of folded proteins was significantly decreased when calcium was removed.93
The classical and alternate chaperone/usher secretion systems are located in the outer membrane and are mechanisms by which discrete fimbrial adhesin proteins are secreted.75, 85, 86 These fimbriae enable biofilm formation, host cell recognition, and cell-to-cell attachment, and contribute to pathogenicity.87 In both mechanisms, periplasmic proteins (the “chaperones”) bind with pilins to ensure proper folding, prevent premature assembly of the pilus in the periplasm, and guide the pilins to an integral protein (the “usher”) in the outer membrane, which translocates folded proteins and serves as a platform for pilus assembly,75, 85 via non-covalent polymerization of pilin subunits.85
However, the chaperone and usher proteins involved in the classical mechanism as well as the pilins that are assembled in the pilus itself are genetically distinct from those in the alternate mechanism.85, 86 A classification system86 has recently been proposed based on the genetic distinctions of the usher proteins and their associated chaperones and substrates (see reference 86 for a review of this system). In both secretion systems, the pilus has a rigid rod-like component anchored to the usher platform and a flexible structure at the tip; both components contribute to binding specificity.75, 87
The T2SS consists of a large multi-subunit ring-shaped complex in the outer bacterial membrane (the “secretin pore”) that is attached to an inner membrane secreton also consisting of multiple subunits, some of which extend into the cytoplasm of the bacteria.75, 90 A periplasmic pseudopilus participates in loading periplasmic proteins into the mechanism, and then pushes the proteins through the secretin pore channel and across the outer membrane; it is not clear whether the pseudopilus extends in a piston-like manner or turns in a manner reminiscent of Archimedes’ screw.87, 90
Many components that are similar to those of the T2SS are also found in the Type IV pilus assembly mechanism, in bacterial competence systems, and in the systems that construct archaeal pili and flagella.88, 90 Important distinctions are that the pili generated by Type IV assembly mechanisms are long and stabilized by covalent disulfide bonds which require energy from a retraction ATPase for their disassembly and retraction, whereas the pseudopilus of the T2SS is short and unstable, relying on electrostatic and hydrogen bonds, and does not retract.90
The T5SS is only found in the outer membrane and consists of a variety of autotransporter proteins, such as surface-attached coiled-coil oligomeric autotransporters, classical single-polypeptide autotransporters, and the two-partner system (TPS) of autotransporters, consisting of two proteins that are translated separately with each containing one of the components needed for protein secretion.75, 77 Autotransporters have a translocation domain or unit and a passenger domain or unit.75, 87 The translocated proteins are primarily virulence factors, but adhesins are also involved in biofilm formation and cell-to-cell adhesion.87
With TPS and classical autotransporters, the translocation component is a single protein that forms a β-barrel in the outer membrane and the passenger component is secreted through this pore; once in the extracellular space, it may remain attached or it may be cleaved and released.75, 77, 87 With oligomeric autotransporters, the β-barrel is formed by the coiled coiling of the C-terminal translocation units of a number of proteins which form an oligomeric complex in the outer membrane; this complex anchors the non-fimbrial adhesin proteins that are secreted through this pore.75 TPSs have been shown to bind to flagella on some enterohemorrhagic and enterotoxigenic E. Coli, however, the exact manner is unknown.52
The Type 1 secretion system (T1SS), Type 3 secretion system (T3SS), and T4SS are complexes that act as protein-translocation channels but, unlike the Sec-dependent pathways, they do not secrete periplasmic proteins,75, 77 with the exception of B. pertussis, which uses a T4SS to translocate the pertussis toxin from the periplasm.81 The T1SS is a heterotrimeric complex anchored in the inner membrane that forms a pore in the outer membrane, allowing secretion directly from the bacterial cytoplasm to the extracellular space, whereas the T3SS and T4SS are multi-subunit complexes that actually create a mechanism that spans both the bacterial membranes as well as the target cell membrane, so that secretion is directly into the cytoplasm of the target cell.75, 77 All three use ATP to provide the energy for translocation, but the T3SS transports proteins with N-terminal signal peptides,75, 77 whereas the T1SS and T4SS recognize C-terminal signal peptides attached to a variety of non-proteinaceous substrates, proteins, or protein-DNA complexes.77, 81 In the T1SS, this signal peptide is recognized by the ATP-binding cassette subunit anchored in its inner membrane.75, 81
The T3SS has evolved from and is similar in structure to flagellar systems, yet has the ability to deliver its translocator and effector proteins directly into the target cell; this capacity and the shape of the multi-subunit complex, which has a long cylinder with a needle-like extension at the tip, has resulted in the T3SS being called an “injectosome” or a “molecular needle.”75, 77 The inner membrane, multi-unit ring complex found at the base makes use of cognate chaperones to prevent proteins from folding before they are loaded into the cylinder.75, 87 It is thought that while the cylinder is being constructed, the loaded proteins are subunits and needle components needed to complete its length, and when its polymerization is complete, the same chaperones begin to load effector proteins for export.75, 87
The T4SS has similar DNA sequences that appear to have evolved from bacterial conjugation systems, and not surprisingly, has the ability to translocate single-stranded DNA-protein complexes as well as single proteins.75, 77, 81, 87 This process appears to involve the use of a surface protein structure, such as a filament, pilus, adhesin, or fibrous mesh,81, 87 and is used by both Gram-positive and Gram-negative bacteria.87 This secretion system allows bacteria to perform three very different functions: cell-to-cell DNA conjugation; export of DNA to the extracellular environment; and translocation of proteins from cell to cell.75, 81 The translocation process has been further subdivided into Type 4A and Type 4B: Type 4A has been shown to secrete effector proteins that induce inflammation and alter the target cell's cytoskeleton; Type 4B (more recently discovered) has been associated with secretion of the protein RalF, a guanosine nucleotide exchange factor protein that upregulates the target cell's ADP-ribosylation factor (Arf).75 To date, translocation of RalF has only been found in Legionella pneumophila and some Rickettsia infections, but in both cases, it alters normal cellular function.80, 84
After phagocytosis of L. pneumophila, membrane fusion between the phagosome and the lysosome is prevented, and the phagosome is positioned at the endoplasmic reticulum; the translocation of RalF recruits Arf1 to the surface of the phagosome and activates it, whereupon it assists in formation of an intracellular vacuole that the bacteria use for replication and survival.80, 84 RalF is also translocated by Ricksettia typhi, R. felis, and R. bellii, but with slight differences: Rickettsia RalF recruits Arf6 to the plasma membrane and increases the concentration of phosphatidylinositol bisphosphate, causing actin remodeling that induces phagocytosis and facilitates cellular invasion, following which Rickettsia species lyse the phagosome and replicate in the cytoplasm.84 R. typhi and R. felis remain co-located with the plasma membrane, but R. bellii co-locates with the perinuclear membrane.84
The Type 6 secretion system (T6SS) has a complex multi-subunit two-part mechanism consisting of a membrane-spanning component, with proteins that are similar to the Type 4B secretion system, and another cytoplasmic component that is perpendicular to the inner cell membrane.76, 78, 87 Reminiscent of the bacteriophage sheath, tube, and tail-spike assembly, the T6SS forms a needle-like cell-puncturing device and the extended tube of this sheath contracts as if it were spring-loaded to penetrate and eject its toxic contents through the outer membrane or cell wall of the target cell.76, 78, 87 In contrast to some of the other secretion systems, direct cell-to-cell contact is the trigger for the T6SS,76, 79 which is used by some of the most pathogenic bacteria.
Antibacterial effectors translocated through this system include phospholipases that destroy cell membranes, glycohydrolase toxin that degrades NAD(P)+, and toxins that form pores in the target cell membrane,79 as well as nucleases that degrade plasmid and chromosomal DNA, and amidases that degrade the peptidoglycan cell wall.76, 78, 79 Following phagocytosis or endocytosis, pathogenic bacteria translocate an effector protein that results in actin cross-linking, which protects the remaining pathogenic bacteria from being engulfed.78 These effectors and toxins do not appear to have an amino-terminal signal sequence, and it is not clear how they are directed into the secretion mechanism.77, 87
Among Gram-negative bacteria, this system appears to function as an interbacterial defense system or to confer competitive advantage during bacterial invasion.76, 78, 79 Without exception, the genes for every cytotoxic effector protein are found adjacent to genes for an immunity protein that binds and inactivates the effector, thus preventing self-intoxication.76 These immunity proteins are also found in bacteria from other species that do not produce the corresponding effector proteins, indicating that they are needed to defend against attack.76, 78, 79 It is possible that the T6SS is used to distinguish between friend and foe in areas with high bacterial density, or to remove defective or infected bacteria to protect the colony.76, 79 Recent research has revealed that some probiotic bacteria also use the T6SS to create or protect their environmental niche; in these bacteria, the Type 6 loci are highly conserved with three distinct genetic signatures; two are found in most Bacteroidales species, but a third structure is only found in B. fragilis.79
The T7SS found in Gram-positive bacteria was first identified in Mycobacterium tuberculosis; mycobacteria have a thick hydrophobic layer of mycolic acids, called a mycomembrane, made of hydroxylated branched-chain fatty acids covalently bound to the peptidoglycan layer.77 The T7SS translocates proteins with a C-terminal signal sequence, and appears to be similar in structure to the Type 1 through Type 4 secretion systems, with a multi-subunit mechanism that is thought to span the inner cell membrane, the peptidoglycan layer (and in mycobacteria, the mycomembrane), as well as the capsule or outer layer of polysaccharides, although a two-step mechanism has also been proposed.77, 82
Mycobacteria use five homologous so-called ESX secretion systems that appear to function independently of each other; in addition to mycobacteria, which belong to the Actinobacteria phylum, the T7SS has been identified in other high G+C Gram-positive species that do not have a mycomembrane, such as Streptomyces, as well as in members of the Firmicutes phylum, including Listeria monocytogenes, Streptococcus agalactiae, Staphylococcus aureus, and other Bacillus and Clostridium species.77, 82 Based on genomic analysis, some researchers have proposed that bacteria using the T7SS should be segregated into subtypes, with those in the Actinobacteria phylum designated as Type 7A and those in Firmicutes designated as Type 7B,77 whereas other researchers think that the T7SS is unique to the high G+C species with a mycomembrane and that the similar secretion system used by those without a mycomembrane should be classified as a separate WXG100 system.82
Adhesins and Receptors
Adhesins and receptors are the mechanisms by which bacteria and their hosts interact; as with immunoglobins and enzymes, adhesins bind with their specific “substrate,” which is their receptor molecule.51, 52, 92 Fimbrial adhesins that bind with carbohydrate receptor molecules are referred to as lectins.51 Soluble molecules in serum, interacting with proteins on the surface of bacteria, and bacterial adhesins interacting with cell surface molecules can initiate signaling events that result in activation of the innate immune system, generating either proinflammatory or anti-inflammatory responses.52 They can also mediate invasion of or uptake by the host cell, production of antibacterial peptides, and coagulation events.52 Cell surface structures on the intestinal mucosal layer, including mucins and glycans, act as receptors for bacterial ligands and provide adhesin binding sites.36 C-type lectin receptors, found on dendritic cells and macrophages, and TLRs are among the known intestinal receptors that recognize glycolipids, lipoproteins, glycoproteins, peptidoglycans, and other extracellular structures that may be present, such as flagella, pili, and fimbria.36, 46, 50
The bacterial surface structures produced by many of these secretion systems are organelles that have traditionally been known as pili; while the terms pili and fimbria are often used interchangeably, they actually designate functions.92 To distinguish those that transfer bacterial DNA during conjugation, still known as pili, those used for adhesion to surfaces for any other purpose are now more commonly called fimbria;92 despite this, the traditional terms Type IV pili, Type I pili, and P pili have been retained by researchers.
While adhesins can be categorized by the secretion system that transports them across the inner and outer bacterial cell membranes, they are perhaps better classified into three broad categories based on their structure and adhesion system. Polymeric fimbria have different subunits in the shaft and the adhesion protein at its tip, and are generally assembled through the chaperone/usher system; in contrast, monomeric/oligomeric filaments, and secreted soluble molecules that enable adhesion are primarily generated through T1SS and T5SS.52, 75, 92 Fimbria can be further classified by their structural subunits, which distinguish their method of attachment to the mucosal or epithelial cell surface and their binding domains.46, 48, 75 While it is impossible to include every type of molecule secreted by bacteria, examples of some adhesins and their receptors are shown in Table 2.51, 52, 54, 75, 85, 92-95
Table 2. Examples of known classes of adhesion molecules of Gram negative and Gram-positive bacteria.
| Adhesion System | Adhesin Produced | Secretion System | Host Tissue | Receptor | Type of receptor | Ref |
|---|---|---|---|---|---|---|
| Fimbrial | Type P pili | Chaperone/ Usher | Upper urinary tract | α-D-galactopyranosyl-(1-4)-β-D-galactopyranoside; Toll-like receptor 4 | Glycolipids; proteins | 52, 92 |
| Type I pili | Chaperone/ Usher | Urinary tract, intestinal tract | monomannose; trimannose; fibronectin; plasminogen; laminin | Glycoproteins; ECM proteins | 51, 52, 54, 92 | |
| Coli surface antigen I | Alternate Chaperone/ Usher | Epithelial cells | Unknown | Unknown | 75 | |
| Afa/Dr adhesins | Chaperone/ Usher | Various | Dra-blood group antigen of CD55 (DAF); CEACAMs; Type IV collagen; α5β1 integrins | Proteins | 75, 92 | |
| Type IV pili | T2SS | Mucosal epithelial cells | Possibly integrins; CD46 | Glycolipids | 52, 54, 75 | |
| Flagella | T3SS | Intestinal tract | Toll-like receptor 5 | Proteins | 52 | |
| Curli | Curli Biogenesis System | Intestinal tract | Human contact phase proteins; fibronectin; plasminogen | ECM proteins | 51, 52, 89 | |
| SRRPs | Sec2 | Epithelial cells | Sialylated glycoconjugates | Glycoconjugates | 52 | |
| Sortase-assembled pili | Sec | Epithelial cells; brain endothelial cells | Proline-rich proteins; Gal-β(1-3)-GalNAc; Gal-β(1-3)-Gal; fibronectin; collagen | Proteins; saccharides; ECM proteins | 52, 92 | |
| Filamentous | OMPs | |||||
| OmpA | T5SS | Brain endothelial cells | Ecgp | Glycoprotein | 75 | |
| OpcA | T5SS | Endothelial and epithelial cells | Heparan sulfates; pyranose saccharides; sialic acids | Glycoproteins; proteoglycans; saccharides | 75 | |
| Invasin | T5SS | M-cells | β1 integrins | Integrins | 75 | |
| Intimin/Tir | T5SS/T3SS | Intestinal tract | Tir; integrin; nucleolin | Proteins | 75, 95 | |
| TAA | T5SS | Epithelial cells | Collagen; laminin; fibronectin | ECM proteins | 52, 75 | |
| Classical AT | T5SS | Epithelial cells | Fibronectin; Lewis blood group antigens | ECM protein; saccharides | 75, 92 | |
| TPS | T5SS | Mucosal epithelial cells | Various | Various | 75 | |
| Soluble | TPS | T5SS | Epithelial cells, dendritic cells | Various | Various | 75 |
| BAP family | T1SS | Epithelial cells | Calcium ions | Calcium ions | 75 | |
| α-hemolysin | T1SS | RBCs; WBCs; fibroblasts; endothelial and epithelial cells | Glycophorin; β-2 integrins | Glycoprotein; integrins | 75, 94 | |
| SiiE | T1SS | Epithelial cells | Various | Various | 75 | |
| GbpA | T2SS | Epithelial cells | N-acetylglucosamine residues | Saccharides | 75 |
Adhesion molecules are secreted by both pathogenic and commensal bacteria. Their polymeric fimbrial structures are primarily assembled via the chaperone/usher secretion system, while monomeric or oligomeric filamentous structures are associated with the Type 5 secretion system and many soluble adhesion molecules are secreted via the Type 1 secretion system. Most receptors, whether found on host tissues or other bacterial species, are surface-bound molecules that have a conformation which is compatible with a given adhesion molecule.
AT – Autotransporter; BAP – Biofilm-associated protein; CD55 (DAF) – complement delay-accelerating factor; CEACAMS – carcinoembryonic antigen-related cell adhesion molecule; ECM – extracellular matrix; OMPs – outer membrane proteins; RBCs – red blood cells; SRRPs – serine-rich repeat proteins; TAA – trimeric autotransporter adhesins; TPS – two-partner-secreted filamentous adhesins; WBCs – white blood cells
Adhesins determine survival as well as virulence, and many bacteria express more than one type of adhesin in response to the tissue or environment in which they find themselves, however each adhesin is specific to its ligand on a given receptor, known as tissue tropism, and bacteria selectively and sequentially express a series of adhesion molecules during infection.51, 75, 85 Microbial adhesion mechanisms include non-specific factors such as hydrophobic and/or electrostatic forces, steric hindrance, and secretion of lipoteichoic acids, as well as specific structures found in cell wall properties, and secretion of adhesins or lectin-like proteins;44, 48 flagellar-mediated adhesion has also recently been recognized.52 The most-studied adhesins are proteins found at the distal end of polymeric fimbria anchored in the outer membrane of Gram-negative bacteria; polymeric fimbria are now known to also exist in the cell wall of some Gram-positive bacteria.52, 92 These hair-like extensions are composed of hundreds or even thousands of secreted subunits that are assembled into polymeric structures as they are translocated into the extracellular environment.52, 75
The β, γ, κ, and π fimbriae that are classified as Type I and P pili are generated through the chaperon/usher system, and are anchored in the outer membrane of the bacteria; these fimbriae are particularly abundant in E. coli, but although they have different adhesin proteins and bind to different receptors, they result in agglutination of human RBCs.52, 75 Class III adhesins of P pili bind specifically to the terminal gal-α(1-4)-βgal unit, found on the globo-A antigen of the P-blood group or the globopentosylceramide antigen of the Forssman blood group,51, 52, 75 whereas class I and class II adhesins of P pili bind to globotriaosylceramides or globosides, respectively.51
Adhesins on Type I pili of pathogenic E. coli strains recognize mannose monomers, which are abundant glycoproteins found on the cells of the urinary tract, however, those on non-pathogenic strains bind with receptors containing trimannose structures.75 In addition, adaptations have occurred among bacterial species; while Type I pili are found in Salmonella enterica, genome sequencing of just serovar Typhimurium alone revealed 13 homologous fimbrial gene sequences and at least 11 serovar-specific fimbria with phase variations of some operons.75
The α-fimbriae classified as Coli surface antigen pili are generated through the alternate chaperon/usher system, and like those of the Type I and P pili, are anchored in the outer membrane of the bacteria, however, although these pili have very similar functions, the Coli surface antigen pili are genetically discrete.75 Further, these α-fimbriae are only found in enterotoxigenic E. coli and other bacteria which bind to intestinal epithelial cells, but to date the host cell receptor molecule has not been determined.75
The family of diffuse adherence fibrillar adhesin/Dr blood group antigen adhesins (Afa/Dr adhesins) are generated through the chaperon/usher system and form a variety of outer-membrane structures, which differ depending on the genetic sequence of the adhesin itself, resulting in non-fimbrial, afimbrial, as well as fimbrial structures.75, 92 Afa/Dr adhesins are thought to interact with α5β1 integrins and with various carcinoembryonic antigen-related cell adhesion molecule (CEACAM) surface molecules.75, 92 Dr adhesins primarily form fimbrial adhesins which bind with the Dra-blood group antigen of complement delay-accelerating factor CD55 (DAF), a complement regulatory molecule, and with collagen IV, while Afa adhesins form afimbrial sheaths.75, 92 The host signaling cascade that follows Afa/Dr adhesin interaction with surface receptors on epithelial cells may have a zipper-like effect that enables cell invasion.75
The Type IV pili are generated by a multi-subunit mechanism that is similar to yet distinct from the T2SS; unlike that system's pseudopilus, which is anchored in the inner membrane but only extends across the periplasm, the Type IV pilus extends across the outer membrane.75, 87, 96 The Type IV pili enable cell motility, adhesion, and signaling,88 aggregation and biofilm formation,90 and DNA uptake from the external environment (termed “competence”).90, 96
Bundle-forming pili occur when Type IV pili aggregate laterally in some bacterial species like Vibrio cholera and enteropathogenic E. coli.92 The Type IV pilus should not be confused with the T4SS; the Type IV pilus does not secrete anything and it does not penetrate the host or target cell wall, unlike the pili of the T4SS, which are used for DNA conjugation and release, and transmembrane protein translocation.75, 82, 87 Adherence by Type IV pili induces changes in the host cell cytoskeleton that result in formation of filopodium that protect the bacteria and allow formation and survival of microcolonies.52
Two subclasses of Type IV pili are distinguished by their functions, their assembly systems, and their pilin subunit sequences and lengths: in addition to competence, Type IVa pili provide both adhesion and motility functions, while Type IVb pili are more associated with adherence and aggregation of bacterial cells96 and have only been found in bacteria that colonize the intestinal tract in humans.75 Once the Type IVa pilus is attached to a receptor or cell surface, it can be forcefully retracted, released, and then extended and reattached, which results in a distinctive type of movement termed “twitching motility.”75, 96 It is not clear whether bacteria with the Type IVb pilus have this capacity.96 The cystic fibrosis transmembrane conductance regulator (CFTCR) is a Type IVb epithelial apical membrane receptor whose concentrations are thought to be increased when activated by binding with S. Typhi, which results in greater adhesion and increased uptake of the bacteria.75
Curli functional amyloids are non-branching fibers secreted by many enteric bacteria through extracellular nucleation, which are important for surface adhesion, surface colonization of host tissues,89 and host immune system interactions.87 Curli amyloids are extremely sticky aggregative adhesins that do not require a receptor and do not bind to specific ligands; they play a major role in stability of the bacterial biofilm, and establishment of enteric populations.52, 89 Curli amyloid production within a biofilm is regulated by temperature, oxygen, osmolarity, oxidative stress, and other environmental signals.89 In humans, curli are known to interact with plasminogen, laminin, fibronectin, MHC class I molecules, and contact phase proteins,75 and may interact with cellulose.52 Curli induce cytokine production leading to an inflammatory response and, in addition to releasing bradykinin, curliated E. coli and Salmonella species can also induce expression of nitric oxide synthase 2, resulting in serious hypotension.52
After translocation through the Sec system, some periplasmic proteins form integral outer membrane proteins (Omp) that are anchored with a β-barrel, and some of these Omps are important pathogenic adhesins.75 Meningitis in newborns is a result of the adhesin OmpA recognizing the receptor Ecgp on microvascular endothelial cells in the human brain, allowing E. coli to cross the blood-brain barrier, whereas the adhesin OpcA, the Omp found in Neisseria meningitidis, recognizes integrins, proteoglycans, and saccharides, which enables invasion of both endothelial and epithelial cells.75 Invasin is an Omp found in Yersinia pseudotuberculosis, which has a lectin adhesin that recognizes β1-integrin receptors on the host cell; adhesion to the host cell results in a signaling cascade that produces pseudopods which enable the bacteria to invade M cells.75
Although the intimin Omp is genetically and structurally similar to the invasin Omp, the Intimin/Tir system is unusual in that it consists of an Omp adhesin (intimin) and a cognate receptor.75 The translocated intimin receptor (Tir) is actually provided to the host cell by the bacteria using its T3SS; after the effector molecule is injected into the host cell, it is phosphorylated and inserted in the cell membrane, whereupon the intimin lectin adhesin binds to it.75 This type of system is used by enteropathogenic and enterohemorrhagic E. coli, and by Citrobacter species.75
The trimeric autotransporter adhesins (TAAs) have varying lengths, but all share the same general structure: a β-barrel in the outer membrane anchors a complex of proteins that are secreted through this pore, consisting of a filamentous trimer made up of repeating coiled coils, with an adhesin molecule at the distal end, similar to a balloon on a string.75 TAAs bind to complement proteins, enable binding to extracellular membrane proteins and host cells, and can also result in target cell invasion.75 TAAs are highly-stable and have been found in bacteria ranging from Bartonella henselae (BadA) to N. meninigitidis, enteropathogenic Yersinia species (YadA), and Haemophilus influenza.52, 75 YadA is the primary adhesin in Yersina species and the most studied, however, it is known that there are significant differences in the length of Yad filaments among various Yersinia serotypes; further, while YadA filaments extend about 28 nm from the outer membrane, BadA filaments extend 100-300 nm from the bacterial surface.75
Classical autotransporters (ATs) consist of a translocation component and a passenger component translated in the same protein, and have an adhesin at the distal end of a monomeric filament that remains attached to the β-barrel in the outer membrane.75 The adhesin MisL, found in Salmonella enterica serovar Typhimurium, and ShdA, an AT also found in Salmonella species, appear to be encoded in two different pathogenicity islands, namely, SPI3 and SC54; ShdA was found to bind with fibronectin and to play a role in both adhesion and virulence.75
Two-partner system autotransporters (TPS) differ from ATs in two significant ways. First, the translocation component (β-barrel) and the passenger component (monomeric filament) are translated as two separate proteins which recognize each other in the periplasmic space.75 Second, once translocated, the monomeric filament can either remain attached or be cleaved and released to function as a secreted protein molecule.75 One example of a TPS is FHA, which in B. pertussis is found in both a membrane bound and a secreted form.75 A significant portion of the translocated monomeric filaments are cleaved and released into the extracellular space and only a small portion remains bound to the bacterial outer membrane; while surface-bound FHA may enable binding to several ligands and play more than one role in adhesion, it is thought that secreted FHA may act to influence immune response by dendritic cells and modulate cytokine release.75
Some soluble proteins secreted by the T1SS are members of the biofilm-associated proteins (BAP) family, involved primarily in biofilm formation but also in adhesion.75 The common factors in these large proteins are that they contain a signal sequence that results in extracellular secretion, they have a glycine-rich motif that is usually a repeated sequence at the C-terminal, and they bind calcium ions.75, 87 BAP family proteins are found in Gram-positive as well as Gram-negative bacteria; those found in S. enterica, S. aureus, Staphylococcus epidermis, Burkholderia cepacia, Enterococcus faecalis, and Vibrio parahaemolyticus enable biofilm formation, whereas those found in E. coli, Pseudomonas putida, S. pyogenes, Lactobacillus reuteri, and Enterococcus faecium enable adhesion.75
The soluble adhesin molecule α-hemolysin, which is secreted through the T1SS of certain uropathogenic E. coli bacteria, acts as a toxin by forming a pore in the membrane of the host cell that causes cell death as a result of Ca2+ influx.94 Because α-hemolysin undergoes acylation at two lysine residues before it is secreted, it is thought that these lipid chains allow it to penetrate the lipid membrane of the host cell, however, some research has shown that β2-integrin receptors or endocytosis of outer membrane vesicles containing α-hemolysin also play a role.94 Also secreted by the T1SS, SiiE is an adhesin produced by S. enterica that is not used for biofilm formation but is needed for adhesion to epithelial cells during bacterial invasion; secretion of SiiE is co-regulated with expression of the T3SS of S. enterica.75 Although most soluble adhesion molecules are secreted through the T1SS, GbpA is an adhesion molecule secreted by the T2SS of V. cholera that binds to epithelial cells through surface associated N-acetylglucosamine residues.75
Bacteria do not use the translocation structures of T3SS, T4SS, and T6SS to secrete adhesins, because the translocated effector molecules are delivered directly into the host cell, but they do use other secretion systems in their outer membrane to facilitate adhesion to the target cell, via adhesins such as intimin, curli, and TAAs.52, 75 For example, the T4SS used by Helicobacter pylori appears to be capable of coordinated adhesion as well as secretion of effector molecules: its CagL protein binds with α5β1 integrins on the surface of gastric epithelial cells to ensure delivery of its CagA effector molecule, which upregulates glycosyltransferases that synthesize Lewis antigens, whereas its autotransporter BabA and SabA adhesins recognize and bind with nonsialylated and sialylated Lewis blood group antigens, respectively.52, 92
Type IV pili can also be encoded on plasmids, which can then be transferred via the T4SS; plasmid-encoded fimbria adhere specifically to the terminal Gal-β(1-4)-Fuc-α(1-3)-GlcNAc moiety of the Lewisx histo-blood group antigen, which is found primarily on the crypt cells of the GI tract.54 These plasmid-encoded fimbria are known to be expressed by S. enterica serovar Typhimurium, which causes numerous crypt abscesses in those who are infected by this pathogen.54 There are additional adhesive pili which appear to provide additional ways for both pathogenic and commensal bacterial to establish microcolonies and biofilms, such as the bundle-forming pili of the T2SS, and the E. coli common pili, which have been described but not classified into one of the secretion systems.52, 95
Serine-rich repeat proteins (SRRPs) are a family of large glycosylated proteins exported by the Sec2 pathway in pathogenic Gram-positive bacteria which remain anchored to the cell wall; although not technically containing an adhesin molecule, SRRPs act as adhesins because the N-terminal is quite basic and binds to sialylated glycoconjugates on the host cell surface.52 SRRPs contribute to the invasiveness and persistence of Streptococcus pneumoniae in the lungs, and are one of the reasons for the virulence and comorbidities of other Streptococcus and Staphylococcus species.52
Sortase-assembled pili are also anchored to the peptidoglycan cell wall of pathogenic Gram-positive bacteria, and consist of covalently-linked pilin subunits forming the main pilus, with minor pilin subunits that function as adhesin molecules.52 Sortase-assembled pili are important for adhesion, aggregation, biofilm formation, and invasion of host cells by pathogens as varied as Actinomyces naeslundii (dental caries and gingivitis); E. faecalis (urinary tract infections, endocarditis); Corynebacterium diphtheriae and S. pneumoniae (severe respiratory infections); S. agalactiae (meningitis, sepsis, and pneumonia in newborns);52 S. pyogenes (necrotizing fasciitis); and S. aureus (toxic shock syndrome, endocarditis, mastitis, septic arthritis, and hospital-acquired pneumonia).92, 97
Fibronectin-binding proteins (FnBPs) are adhesins secreted by bacteria, which bind to fibronectin glycoproteins found on epithelial cells, in the ECM, and in body fluids of the host; in Gram-negative bacteria, many FnBPs are autotransporters, whereas in Gram-positive bacteria, FnBPs are sortase-assembled structures.97 Plasma fibronectin, which is produced by hepatocytes, is found in blood, saliva, and other body fluids and is an important component of blood clotting and wound healing.97 Cellular fibronectin, a dimer secreted by most cells, acts as a ligand that binds with β-1 integrins to facilitate adhesion to the ECM and also is known to bind with DNA, tissue transglutaminase, fibrin, glycosaminoglycans such as heparin, and various collagens.97 Fibronectin also plays a role in inflammatory responses and macrophage activation, and mediates leucocyte function.97 FnBPs enable pathogen-specific binding to the ECM, laminin, collagen, fibroblasts, osteoblasts, RBCs, endothelial cells, or epithelial cells of the host, followed by invasion of the host cell.97 Although FnBPs contribute to the virulence of the pathogen, if the bacteria happens to bind to the fibronectin receptor on macrophages, it is ingested.97
Moonlighting proteins, which are not adhesins, are also found on the surface of bacteria, and although their normal function may be as housekeeping, ribosomal, or glycolytic proteins, once they are exposed to the external environment, they can assume other functions, similar to adhesins.46, 48 Some are involved in attachment to mucus or mucosal components, or have an affinity for plasminogen, fibronectin, or other extracellular matrices,48 however, not all strains have the same adhesive capacity. For example, Lactobacilli have evolved mucus binding adhesin proteins consisting of long fibrils that have Ig and mucin-binding domains repeated in tandem along the length, which hold them securely in the mucosal layer and, like most commensal bacteria, do not penetrate to the epithelium.36 In contrast, some pathogenic Gram-positive bacteria appear to have only one or two mucus binding repeats at the end of their adhesin proteins, but have evolved the ability to penetrate the mucosal layer and adhere to the epithelial surface with lectins or lectin domains.36
Enterotypes
Secretors have an abundance of bacteria with glycan-degrading enzymes such as α-L-fucosidase and β-galactosidase that enable them to degrade the HBGAs found in the intestinal mucosa, thus taking advantage of a readily-available food source as well as making the resulting monosaccharides available to co-occurring species that do not have these enzymes.98 In contrast, bacteria that are known to be abundant in non-secretors have also been associated with irritable bowel syndrome (IBS) and autoimmune conditions such as IBD, celiac disease, and T1DM,98 as well as chronic inflammatory diseases like primary sclerosing cholangitis and Crohn's disease.74 These associations might be due to the fact that non-secretors also have fewer commensal bacteria such as Bifidobacteria, which generally play a protective role in the GI tract by preventing pathogens from achieving colonization.99
A recent study used reference genomes and metagenomic techniques to map known species by their phylogenetic profiles; the data were sorted and analyzed, and the resulting three clusters were designated as “enterotypes” composed of groups of species that were found to contribute specific functions to what could be considered a microbial community.100 The main contributors to each enterotype were the Bacteroides, Prevotella, and Ruminococcus genera, with each community composed of a network of co-occurring groups of species.100
Although HBGAs were not identified as host properties in this study, degradation of mucosal glycoproteins played a fundamental role.100 Further analysis showed that each enterotype derived energy from specific fermentable substrates; one degraded carbohydrates and proteins, while the other two degraded glycoproteins found in the mucosal lining of the intestine.100 It is possible that these enterotypes reflect the method of hydrogen disposal utilized during fermentation; enterotype 1 (Prevotella) is enriched with a sulfate reducer and enterotype 3 (Bacteroides) is enriched with a methanogen, and these just happen to be two of the ways by which hydrogen is removed.100
Also of interest are the findings that secretors were more likely to have enterotype 3, and have a higher risk of C. difficile or Salmonella infection following antibiotic treatment, whereas non-secretors were more likely to have enterotype 1, and have a higher risk of urinary tract infections, vaginal candidiasis, T1DM, or necrotizing enterocolitis.98 Secretor status also determines susceptibility to a variety of other pathogens: non-secretors have a lower risk of Norovirus infection and a greater risk of H. pylori, V. cholera, and C. jejuni infection.74 These differences in risk are thought to be due to differences in the gut microbial composition as a result of availability of the mucosal carbohydrate antigens in the intestinal lumen of secretors and non-secretors.60, 74, 98
Carbohydrates are the primary energy source for the microbiota, and aside from diet, one of the main sources of glycosides are the oligosaccharides which make up 50-80% of mucins by weight; in secretors, fucose residues are a major component of the mucosal glycans but these residues are not present in non-secretors.74 B. fragilis is known to incorporate fucose residues into its polysaccharide layer, and Bacteroides thetaiotaomicron, a prominent Gram-negative commensal bacteria, not only metabolizes fucose residues, it actually induces fucosylation of intestinal mucins.74 Although the greatest influence appears to be secretor/non-secretor status as a result of FUT2 allelic expression, in addition to HBGAs, long-term diet is known to influence the microbial profile for each enterotype.98, 99
Dietary and genetic factors
In a human study that assessed the effect of long-term diet on microbiota composition,18 enterotype 1 (Prevotella) was strongly associated with a high-carbohydrate and low animal protein diet, such as that found in agrarian populations. However, enterotype 3 (Bacteroides) was strongly associated with a diet high in animal fat and animal proteins, typical of the Western diet. While short-term dietary changes had a significant effect within 24 hours on microbiome composition of the subjects, the effect was not strong enough to change their enterotype classification.18
Another study found that variation in bacterial diversity was distributed along a Prevotella/Bacteroides gradient that was strongly affected by the presence of Bifidobacteria.24 Studies with humanized gnotobiotic mice have shown that non-secretors have greater abundance of Bacteroidetes and decreased diversity within their bacterial microbiota, simply because of the lack of fucosylation of intestinal mucosa.74 However, substituting a diet deficient in plant polysaccharides but rich in glucose resulted in microbial diversity similar to secretors; these findings indicate that although the microbiota adapt to the environment dictated by the host genotype, diet has a greater impact than genotype.74
Microbial ability to rapidly adjust to dietary changes gave selective advantage to ancestral populations whose diet depended on success in foraging and hunting, and was further affected by season, climate, and temperature.101 A short-term dietary intervention in humans found that significant changes in the structure of the gut microbiota were observed within 24 hours of introducing an animal-based diet, which reverted to the baseline profile within 2 days of the end of the intervention.101 In this study, the high-fat diet resulted in increased microbial production of the secondary bile acid, deoxycholic acid (DCA), which is known to inhibit Bacteroidetes and Firmicutes growth and may explain the change in the microbiota profile.101
A high-fat diet is also associated with Bilophila wadsworthia, a pathogen whose growth is stimulated if secretion of bile acids occurs while consuming a diet high in saturated milk fats.101 B. wadsworthia is in the Desulfovibrionaceae family that reduces sulfite to hydrogen sulfide, a genotoxic and cytotoxic gas which can cause impaired barrier function, endotoxemia, and inflammatory bowel disease, and contribute to the development of cancer.102 Hydrogen sulfide reduces beneficial butyrate oxidation, causing altered redox status and increased MAP kinase activation, and is extremely toxic to colonocytes, resulting in increased colonocyte turnover, thereby increasing intestinal permeability and reducing integrity of the epithelial barrier.102 Proinflammatory microbial products from a high-fat diet can also result in metabolic changes in the host leading to increased energy harvest, obesity, and insulin resistance.102
Diets high in plant polyphenols and polysaccharides such as dietary fiber, which are naturally resistant to digestion and absorption in the small intestine, provide important substrates for colonic bacteria, which produce small phenolic compounds and short chain fatty acids (SCFAs).103 Once absorbed, SCFAs provide energy for colonocytes as well as heart, brain, and muscle cells; regulate immune function; and play important roles in lipid metabolism, thermogenesis, cell differentiation, proliferation, and apoptosis.103 Plant polyphenols are antioxidants and free radical scavengers, while small phenolic acids are known to participate in biological processes that include antibacterial, anti-inflammatory, and phytoestrogenic activities; anti-AGE formation; and stimulation of enzymes needed for detoxification and degrading xenobiotics.103
Prebiotic dietary fiber is the primary energy source that supports the composition and metabolic activity of the colonic microbiota and maintains human health by protecting against obesity, T2DM, metabolic syndrome, IBD, and colon cancer.103 Although consumption of dietary fiber ranges between 70 g and 120 g per day in populations with a more traditional plant-based diet, in populations with a Western diet, intake averages just 20 g per day.103 When the availability of fermentable polysaccharide substrate is inadequate, colonic bacteria substitute amino acid fermentation, which generates potentially harmful metabolites that can be cytotoxic, genotoxic, or carcinogenic.103 Bacteroidetes have developed an ecological network that enables them to maximize polysaccharide substrate availability: strains and species that can utilize one type of polysaccharide structure, such as inulin, provide breakdown products that support others that need a different type of polysaccharide structure, such as fructose.104
Tissues that are not in direct contact with intestinal microorganisms can be profoundly affected by changes in the composition and density of bacterial species in the gut which alter immune responses; this should be considered when inflammatory processes are being investigated, including chronic inflammatory responses and metabolic syndrome.13 Commensal bacteria prevent liver inflammation and fibrosis as a result of translocation of pathogenic bacteria and their products across the intestinal barrier.105 There is evidence that both beneficial and pathogenic bacteria may respond directly to stress-related catecholamines, which can affect their growth, motility, and virulence.106 Exposure to stress not only modifies the gut microbiota, it also enhances bacterial mucosal adherence, reduces sIgA secretions, and increases paracellular permeability of the epithelium, which may seriously influence the outcome of infections.15, 106, 107
In addition, changes in the intestinal microbiota directly correlate with the appearance of stress-related disorders,106 including such affective states as anxiety and depression.107 Physiological responses to inflammation and stress also affect a variety of gastrointestinal functions as diverse as visceral sensitivity, mucosal blood flow, gut motility, acid secretion, and release of various hormones and neuropeptides, which can result in dyspepsia, abdominal pain, diarrhea, and other unpleasant symptoms.15, 106 Thus, effective treatment of inflammatory, stress-related, and functional disorders of the GI tract requires a thorough understanding of how the physiological responses of the gut are regulated.
The Enteric Nervous System
The uniqueness of the enteric nervous system (ENS) and the central role it plays in the proper physiological functioning of the GI tract cannot be fully described without a brief review of the distinctions between the CNS, the peripheral nervous system (PNS), and the ENS. The CNS consists of the brain and the nerves of the spinal cord, while the PNS consists of all other nerves in the body that communicate with the CNS.108 Thus the efferent nerves of the PNS carry all nerve signals from the CNS to muscles, tissues, and glands, which evokes their response, and information gathered by sensory receptors in the body is carried by the afferent nerves of the PNS back to the CNS, where it is processed.108
The nerves of the skeletal motor system carry signals from the CNS directly to the skeletal muscles, while the autonomic nervous system (ANS) carries the nerve signals that control the behavior of the heart, blood vessels, glands, and visceral muscles.108 However, unlike the skeletal motor nerves, the nerves of the ANS are characterized by ganglionic synapses, thus the signals they carry can be amplified, weakened, or otherwise modified by processes that occur at those synapses, which allows for instant adaptation to changing circumstances.108
The efferent nerves of the ANS are further classified as either sympathetic or parasympathetic, based on anatomical characteristics of the nerve fibers and the proximity of their ganglia to their target organ or tissue.108 The parasympathetic division consists of the cranial and sacral nerves, with their ganglia located just outside of or within the organ or tissue they innervate, whereas the sympathetic division is made up of the thoracic and lumbar nerves, with their ganglia located immediately adjacent to the spinal column.108 Parasympathetic responses are usually faster and more precisely targeted, and sympathetic responses are usually slower and more diffuse, but these are tendencies and not absolute distinctions.108
The GI tract is innervated with efferent nerves from both the sympathetic and parasympathetic divisions of the PNS: it is supplied with a set of nerves from the sympathetic ganglia associated with the thoracic and abdominal nerves, while the vagus nerves (the large cranial nerves that connect the brain to the gut) innervate it from the esophagus to the middle of the colon and the sacral nerves innervate the distal colon.108 As with other organs, the efferent nerves of the PNS deliver catecholamines to the GI tract, and the afferent nerves of the PNS carry sensory information, such as pain and pressure, from the GI tract back to the CNS.108 However, the minute-to-minute control, coordination, and regulation of the myriad physiological functions of the GI tract falls to the ENS.
Auerbach discovered the existence of the ENS in the 1860s, which is now known to include not only Auerbach's (myenteric) nerve plexus but also Meissner's (submucosal) nerve plexus; these are complex networks of nerve cells and intrinsic nerve fibers108 which contain sensory neurons, interneurons, and motor neurons.15 The interneurons are serotonin-secreting nerve cells that enable communication between other nerve cells; interneurons are only found in the CNS and ENS, and add layers of complexity and sophistication to neural communication that allow the brain to modulate and process the information it receives.108
However, the number of motor nerve fibers connecting the CNS to the GI tract is incredibly small in comparison to the number of ENS nerve cells in the gut; for example, there are only about 2000 nerve fibers in the vagus nerves at the point where they enter the abdomen but there are over 100 million ENS nerve cells in the human small intestine alone.108 Given this overwhelming disparity, it seems highly unlikely that such a small number of motor nerve fibers can innervate so many ENS nerve cells, along with the many hundreds of millions of intrinsic nerve fibers that these cells put out to communicate with each other—and indeed, this is true.108
In a simple yet elegant experiment,108 an isolated section of intestine suspended in an organ bath detected and responded to pressure-inducing stimuli with a coordinated descending oral-to-anal wave of contraction and relaxation. This reproducible result was especially startling because the brain, spinal cord, and sensory ganglia had been discarded, leaving only the segment of gut. Such groundbreaking research by Trendelenburg in the early 1900s, confirmed by Langley and others in subsequent investigations, established that—unlike any other organ—the gut could manifest reflex activity independently of input from the CNS, and that the ENS did not require commands from the CNS in order to function. Trendelenburg introduced the term “peristaltic reflex” to describe the pressure-induced, unidirectional propulsive activity of the gut.108
Thus while the nerves of the PNS do communicate between the CNS and the GI tract itself, the nerves of the PNS are virtually irrelevant to many of the behaviors of the gut because the ENS is not affected when these motor nerves are severed; the smooth muscle and glands of the gut are not supplied by them but by complex intrinsic enteric neural circuits that may involve many nerve cells.108 The nerve cells of the ENS are not just post-ganglionic links in the parasympathetic nerve pathway; the ENS is actually an integrated system of neural communication and processing that is anatomically and functionally independent from the CNS and therefore can be considered neither sympathetic nor parasympathetic.108
The sensory receptor nerves of the ENS independently detect, process, and act on data from the gut to activate a set of effector nerves that it alone controls, which means that all of the necessary elements of this neural apparatus are intrinsic components of the wall of the gut, and that the intrinsic nervous system of the gut has properties that are like those of the brain.108 In fact, over 95% of the body's serotonin is made in the nerve cells of the ENS, and more importantly, every one of the classes of neurotransmitters found in the brain is represented in the ENS.108 The proper understanding of the ENS as a separate but equal division of the ANS, and as a “second brain” that is independent of the CNS, holds great potential for clinical medicine and may hold the key to understanding some of our most challenging conditions.
The Brain-Gut-Microbiota Axis
The term brain-gut microbiota axis refers to the multiple parallel pathways by which the brain and the gut communicate, which include the splanchnic and vagus pathways of the ANS, the hormonal pathways of the hypothalamic-pituitary-adrenal (HPA) axis, and the various components of the immune system.106 Because this is a bi-directional axis, the term also refers to the various mechanisms by which the gut microbiota communicates with the mucosal cells, immune cells, and neural endings of the intestines, which in turn modulate permeability, visceral sensitivity, and motility of the GI tract.106 Through these mechanisms, changes in the bacterial status of the intestinal lumen can also activate signaling pathways between the ANS (either the ENS or the vagus nerves, or both) and the brain, thus this axis plays an important role in modulating the stress response of the gut.107 It is also possible that psychosocial or life stressors which induce changes in the CNS and alter brain chemistry may in turn result in disruptions to the gut microbiota, and that these changes could then modulate host responses and behaviors.109
The enteroendocrine cells of the epithelium are also an important part of the brain-gut-microbiota axis, and they too communicate through multiple parallel pathways: their primary influence on the axis is through secretion of serotonin, which activates neural pathways in the ENS, PNS, and CNS.15 Serotonin plays a role in regulating cognition and mood in the brain, but peripherally it is involved in perception of pain, gut motility (contraction and relaxation of smooth muscles), and regulating secretions in the GI tract.110 Of note, certain bacteria can promote intestinal synthesis of serotonin.109, 110 The enteroendocrine cells of the epithelium also act as highly specialized chemoreceptors, which sense changes in the pH, osmolarity, and nutrient composition of the lumen.15 As the largest endocrine organ of the human body, they secrete hormones such as ghrelin, somatostatin, cholecystokinin, glucagon-like peptide, gastric inhibitory peptide, and peptide YY, which affect both the brain and the gut.15
The role of the intestinal microbiota in the development of metabolic diseases (discussed later) and neuropsychiatric disorders has become an area of intense research interest since 2010.111 CNS receptors for cytokines and other immune system molecules suggest the importance of the immune system in normal brain function; the discovery of anti-brain antibodies in conditions as diverse as autism spectrum disorder (ASD), obsessive-compulsive disorder, and schizophrenia indicates that autoimmunity may be a factor in the pathogenesis of some neuropsychiatric conditions.112 In addition to possibly contributing to neurodevelopmental disorders such as ASD, intestinal microbiota may be involved in the pathophysiology of depression, anxiety, and other mood disorders.5, 15, 113-115
The brain-gut-microbiota axis is also thought to be involved in functional GI disorders, which have been associated with psychiatric comorbidities.15 Major depressive disorder (MDD) may be associated with epithelial barrier dysfunction and greater permeability, known as “leaky gut,” which increases the probability of intestinal inflammation, followed by activation of the immune system and upregulation of IL-6 and IFN-γ production.110 MDD patients have significantly higher serum concentrations of IgA and IgM antibodies against LPS from enterobacteria, suggestive of leaky gut as a possible contributor to MDD development.110
Fecal Transfers
Historic records document that fecal transfers have occasionally been done but there have been no fecal transfer studies in humans that specifically focused on neuropsychiatric diseases.116 Fecal transfer studies with germ-free mice are of limited use, due to known alterations in physiology and immune development.109 A few fecal transplant studies with wild-type mice have investigated the associations, if any, between diet, intestinal permeability, inflammatory markers, and behavior.110 In one study, chow-fed mice, treated with antibiotics to deplete their intestinal bacteria, received microbiota from donor mice fed a high-fat diet; the recipient mice were subsequently found to have increased gut permeability and circulating inflammatory markers, and exhibited selective yet significant disturbances in cognitive, exploratory, and stereotypical behaviors.109, 110 Experimental fecal transfers in humans to treat IBS, chronic fatigue syndrome, MS, ASD, and Parkinson's disease have had limited success in some cases, however, much more research is needed to obtain reliable evidence of the efficacy of fecal transplants for treatment of neuropsychiatric disorders.116
Probiotic Effects
There is significant evidence that probiotics can prevent intestinal barrier damage from inflammatory conditions such as colitis or IBD, and can lessen stress-induced intestinal damage, by minimizing gut hypersensitivity and mucosal barrier disruption, thereby preventing the cascade of hyperpermeability and endotoxemia, HPA axis response, and neuroinflammation.15 In addition, several studies have found that direct manipulation of the intestinal microbiota, including with probiotic supplementation, can modulate behavior.3, 5, 8, 107 Thus, addressing functional gastrointestinal disorders with probiotics may better regulate intestinal and brain barrier function, which may in turn alleviate psychiatric comorbidities.15
Depressive behavior, social avoidance, inactivity, and impaired appetite are frequently associated with systemic viral infections, and conversely, treatment of hepatitis C, MS, and metastatic cancer with INF-α and IL-2 often results in development of MDD.113, 114 It is thought that pro-inflammatory cytokines induce secretion of CXCL10 by the epithelial and endothelial cells of the meninges in the brain, which results in activation of CXCR3 signaling followed by a subsequent decrease in synaptic activity.113, 114 Recently, research has established that lymphatic vessels exist in the meninges of the brain, which appear to play a role in drainage of cerebral spinal fluid and may be responsible for immune surveillance of the CNS as well as removal of waste products from the brain parenchyma.117, 118 Impaired function of these lymph vessels may be associated with neurological disorders such as cerebral amyloid angiopathy, neuromyelitis optica, Alzheimer's disease, or MS.117, 119
In a mouse model of MS, concurrent treatment with several Lactobacillus strains decreased proinflammatory responses and reduced clinical symptoms, and treatment with B. fragilis induced protection against demyelination.120 In a mouse model of ASD, treatment with B. fragilis has been found to change microbial composition; restore gut permeability; decrease repetitive, anxiety-like, sensorimotor behaviors; and improve social behaviors.15 These findings suggest that ASD behaviors may be a result of viral infection during pregnancy, at least some of which may be reversible with B. fragilis administration.109 Recall that B. fragilis colonization was seriously impaired and remained depressed in babies following Cesarean birth, when compared to babies delivered vaginally.25 Stool samples from children with ASD have a significantly greater prevalence of Clostridium species than samples from children without ASD.120 Further studies are clearly warranted to assess whether there is any correlation between intestinal dysbiosis and ASD or MS, but targeted probiotics appear to have some prophylactic value.
Most Gram-positive commensal or probiotic bacteria have the same secretion systems and adhesins as Gram-positive pathogenic bacteria, and recognize the same receptor molecules.46, 48 Over-competition and competitive exclusion by such probiotic bacteria can thus prevent infection by both Gram-negative and Gram-positive pathogenic bacteria by ensuring that adhesion and colonization cannot occur.46, 48 Extracellular proteins and peptides produced by probiotic bacteria, such as Lactobacilli and Bifidobacteria, are important for maintaining intestinal health and homeostasis.46, 48 Bifidobacteria are known to secrete a serine protease inhibitor (serpin) that prevents inflammation by inhibiting secretion of inflammatory cytokines and pancreatic and neutrophil elastases.46 Various species of Bifidobacteria and Lactobacilli secrete other extracellular proteins that regulate cell proliferation and apoptosis; modulate immune function and activity; promote dendritic cell maturation and survival; induce production of defensins; strengthen the mucosal barrier; increase production of tight-junction proteins; reduce intestinal permeability; and regulate transcription of proto-oncogenes.46
Of note, the Gram-positive Lactobacilli strain, L. rhamnosus GG (LGG), has pili which form strong attachments to the intestinal mucus layer and the epithelial cells beneath, as well as between bacteria within the colony, enabling it to form biofilms and resist intestinal shear from peristalsis.121 The pili can form a zipper-like mechanism that brings the bacteria into closer interaction with host cells, enabling attachment of surface adhesins and immune modulation by effector molecules.121, 122 Although lipoteichoic acid molecules from the Gram-positive bacterial surface can stimulate pro-inflammatory IL-8 cytokines and induce NF-KB activation, the LGG pili directly induce anti-inflammatory responses and suppress NF-KB activity, and indirectly bring about the secretion of anti-inflammatory proteins p40 and p75, which are known to have an anti-apoptotic effect and reduce cell damage.121, 122 As a probiotic, LGG has been shown to be safe, was effective in clearing vancomycin-resistant Enterococcus infections, and showed promising results in treating C. difficile infections, preventing Candida colonization, and reducing intensity and frequency of pain in patients with IBS.121
Perhaps not surprisingly, there is growing interest in the targeted use of prebiotics and probiotics as therapeutic approaches to the prevention or treatment of a variety of diseases,123, 124 and to offset the deleterious effects of stress on the intestinal microbiota which contribute to dysregulation of the brain-gut-microbiota axis.106 This approach may be of particular interest to those exploring a possible treatment for the high prevalence of Clostridium bolteae found in some children with ASD.8 Although extensive research with mice has shown that probiotics have a consistent anti-obesity effect,125 further trials with humans are now needed. When mouse and human bacterial diversity were compared using distal gut rRNA sequences, a high degree of taxonomic similarity was seen at the division level but a full 85% of the mouse sequences belonged to genera that have not been found in humans.126 Despite the fact that there are large differences in the composition of their gut microbiota at lower levels, because mice and humans are genetically similar host organisms with highly conserved metagenomic core functions and similar microbiotic composition at higher taxonomic levels,4 the use of germ-free or humanized gnotobiotic mice is an important research tool.
It should also be noted that, although the dynamics of the microbiota and the immune system have been studied, the inter-kingdom signaling pathways between intestinal bacteria, fungi, parasites, and viruses are not well understood.8 One virome study with monozygotic twins and their mothers did not find the expected viral-microbial dynamic typical of other ecosystems.8 The gut virome of the participants was not influenced by their genetic relatedness, but was found to be unique to each individual, and the diversity of each person's virome remained virtually unchanged over the research period (95% of virotypes were retained).8
Disease Mechanisms
Although there are still significant gaps in our knowledge, completion of the HMP has resulted in much greater understanding of the mechanisms by which the microbiota influence health and disease, some of which are reviewed here. Certain diseases associated with microbiome characteristics are also shown in Table 3. The human microbiota influences our mood, alters our behavior, and affects our physiology in a variety of ways, including modulating predisposition to obesity, diabetes, colitis, cancer, skin and mucosal disorders, and infection.3, 8, 20, 107 Tight junction dysfunction has been associated with conditions as diverse as autoimmune thyroiditis, IgA nephropathy, chronic kidney disease, T1DM, primary biliary cirrhosis, primary sclerosing cholangitis, alcoholic liver disease, and cirrhosis of the liver.15
Table 3. Association of diseases with characteristics of the microbiome.
| Microbiome | Disease or Condition | Relevant Characteristics |
|---|---|---|
| Cutaneous | Psoriasis | Significantly increased ratio of Firmicutes to Actinobacteria in lesions4 |
| Acne | Propionibacterium acnes infection of pilosebaceous units4 | |
| Chronic skin ulcers | Increased Pseudomonadaceae following antibiotic treatment4 | |
| Diabetic skin ulcers | Increased Streptococcaceae4 | |
| Gastric |
Increased risk of peptic ulcer disease, gastric mucosa-associated lymphoid tissue tumors, and non-cardia gastric adenocarcinomas Decreased risk of reflux esophagitis and childhood-onset asthma |
Presence of H. pylori in gastric microbiota4 |
| Increased risk for childhood-onset asthma, reflux esophagitis, gastroesophageal reflux disease, Barrett's esophagus, and esophageal and gastric cardia adenocarcinomas | Absence of H. pylori in gastric microbiota2 | |
| Reflux esophagitis | Esophageal microbiota dominated by Gram negative anaerobes; gastric microbiota has low or absent H. pylori4 | |
| Childhood-onset asthma | Absence of H. pylori in gastric microbiota4 | |
| Age-related gastric atrophy | Enhanced by presence of H. pylori in gastric microbiota3 | |
| Upper GI | Obesity | Reduced ratio of Bacteroidetes to Firmicutes; enrichment of genes related to lipid and carbohydrate metabolism; risk significantly increased with antibiotic use prior to 6 months of age4 Successful weight loss associated with higher levels of Bacteroides fragilis, Lactobacilli, and Bifidobacteria5 |
| Cardiovascular disease | Gut-microbiota-dependent metabolism of phosphatidylcholine4 | |
| Diseases of the liver: non-alcoholic fatty liver disease, alcoholic steatosis, hepatocellular carcinoma | Exposure to metabolic products of microbiome: acetaldehyde; phenols; ammonia4 | |
| Cirrhosis | Substantially altered microbiome; with enrichment of Proteobacteria and Fusobacteria phyla, and Enterobacteriaceae, Veillonellaceae, and Streptococcaceae families4 | |
| ASD | Significantly increased presence of Clostridium bolteae8 | |
| Small intestine bacterial overgrowth | Associated with recurrent antibiotics, gastric acid inhibitors, Crohn's disease, cirrhosis, chronic pancreatitis, end stage renal disease12 | |
| Colonic | Inflammatory bowel disease | Host polymorphisms in bacterial sensor genes (NOD2; CARD15; TLR4); symptoms may improve with antibiotic treatment4 Bacterial sensor genes (NOD2, TLR4, CARD8, CARD9, NLRP3) and autophagy genes (ATG16L1, IRGM, LRRK2)5 Reduced butyrate, acetate, methylamine, trimethylamine; elevated amino acids; metabolites from arachidonic acids and bile acids were most affected3 Impaired production of α-defensin40 |
| Functional bowel diseases | Larger populations of Veillonella and Lactobacillus4 | |
| Ulcerative colitis | Large populations of Enterobacteriaceae, increased proportions of Actinobacteria and Proteobacteria4 Increased taurine and cadaverine3 | |
| Crohn's disease | Increased risk with early childhood exposure to antibiotics; Significantly diminished microbial diversity; Large populations of Enterococcus faecium and several Proteobacteria4 Genes involved in epithelial barrier integrity (IBD5, DLG5, PDGER4, DMBT1, XBPI); decreased Faecalibacterium prausnitzii and Roseburia hominis5 Linked to disruption of duodenal microbiota12 Higher amounts of bacteriophages3 |
|
| Colorectal cancer | Larger populations of Fusobacterium spp.; significantly lower Desulfovibrio spp.4, 5, 8 Invasive CRC associated with Escherichia coli NC1015 Lower butyrate and acetate, higher proline and cysteine3 |
|
| Irritable bowel syndrome | Distinctive alterations in Coprococcus, Collinsella, and Coprobacillus genera5 Higher levels of Pseudomonas aeruginosa and lower levels of Bifidobacterium catenulatum in upper GI12 Higher bile acid concentration, lower levels of branched chain fatty acids3 May follow infection by Campylobacter enteritis, Shigella, or Salmonella40 |
The presence or absence of certain human diseases is associated with and influenced by genetic and bacterial conditions of the relevant microbiome.
Life-threatening conditions such as multiple severe injuries, hemorrhagic shock, splanchnic ischemia-reperfusion injury, and severe burns can result in increased intestinal permeability, gut inflammation, loss of intestinal barrier function, and translocation of endotoxins and/or bacteria from the GI tract.15 Inflammatory cytokines such as IFN-γ and TNF-α can up-regulate expression of channel-forming proteins found in tight junctions, whereas butyrate, glutamine, and estrogens protect the intestinal barrier by increasing mucus production and enhancing expression of tight junction proteins that act to reduce permeability.15
Autoimmune diseases
Dysregulation of the intestinal immune system and activation of TH1 cells have been associated with T1DM, Crohn's disease, MS, and other autoimmune diseases, whereas activated TH2 cells are associated with allergic disorders, asthma, and ulcerative colitis.30, 120 TH17 cells may contribute to autoimmune disorders such as psoriasis and rheumatoid arthritis.30 In autoimmune diseases such as celiac disease and T1DM, the protein zonulin plays a critical role in tight junction regulation, but wheat gluten is the dietary trigger that induces increased intestinal permeability and inflammation in those who are genetically predisposed to celiac disease.15 Recall that the hygiene hypothesis suggests that some allergies and autoimmune disorders could be avoided with more exposure to bacteria in early life, which is less common in our modern Western society than in more rural, bacteria-filled lifestyles.5 Lack of exposure during development of the immune system may result in alterations that lead to loss of self-tolerance, which may be irreversible.111
It should be noted that the pathophysiology of food allergies can involve enhanced intestinal permeability even in the absence of food allergens,15 and antibodies are known to exist against some proteins secreted by commensal bacterial in the gut.46 Drugs and toxins can also induce intestinal barrier dysfunction: for example, tacrolimus treatment can result in new-onset food allergies by increasing intestinal permeability; NSAIDs can increase intestinal permeability, inhibit epithelial repair, and increase intestinal nitric oxide synthesis; and ethanol is oxidized by intestinal bacteria to acetaldehyde, which is a toxic product that increases intestinal permeability.15
In individuals who are genetically susceptible to celiac disease, the transferrin receptor CD71 is expressed at high levels on the apical surface of intestinal epithelial cells.34 When antigen-specific sIgA antibodies bind with gluten-derived deamidated gliadin peptides in the lumen, this receptor recognizes the complex, which is then transcytosed and delivered intact to the basolateral side, where these highly reactive peptides activate CD4 T cells and stimulate inflammatory processes.34 Some bacteria secrete proteases that degrade the gliadin protein in the duodenum, and may therefore contribute to the severity of symptoms.45 However, there is a subset of celiac disease patients whose symptoms do not resolve with a gluten-free diet, indicating that microbial dysbiosis may be an independent risk factor.127 If commensal bacteria from the lumen penetrate the mucosal barrier, LPS from the Gram-negative bacterial cell wall would be recognized by the CD14-TLR-4 complex and would activate the innate immune system, resulting in release of pro-inflammatory cytokines.127
T1DM is not classified as a metabolic disorder, but instead is considered an autoimmune disease; it appears to be a consequence of disruption of the mucosal microbiota and the intestinal epithelial cells, which results in intestinal immune activation and eventually leads to destruction of the pancreatic beta cells by the autoimmune system.128 A large genetic study has established that non-secretors are more susceptible to T1DM (OR=1.29, 95% CI 1.20-1.37; p=7.3 × 10-14).129, 130 It is thought that the associated decrease in diversity and stability of intestinal microbiota results in decreased mucus production and increased gut permeability followed by translocation of bacteria, with corresponding extra-intestinal inflammation and up-regulation of T cell response.128, 131
Chronic pancreatitis
Unlike acute pancreatitis, no serum tests exist for diagnosis of chronic pancreatitis, because elevated serum lipase activity, which also occurs with a number of critical illnesses, typically does not meet the threshold of three times the upper normal limit and may even be within normal limits.132 A recent GWAS study conducted on chronic pancreatitis patients and volunteer blood donors132 reported that chronic pancreatitis was significantly associated with non-secretor status (OR=1.53; p=8.56 × 10-4) and ABO blood type B (OR=1.69; p=1.0 × 10-4). Surprisingly, asymptomatic elevated serum lipase in volunteer blood donors was also significantly associated with non-secretor status (OR=1.49; p=0.012) and ABO blood type B (OR=2.48; p=7.29 × 10-8). Those with ABO blood type O had a significantly reduced risk of chronic pancreatitis (OR=0.62; p=1.22 × 10-05) and elevated serum lipase activity (OR=0.59; p=8.14 × 10-05).132
Non-secretors with ABO blood type B have a genetic predisposition to chronic pancreatitis; asymptomatic patients with elevated serum lipase who have this phenotype may have subclinical pancreatic injury, even when serum lipase levels are within the normal range, and also have an increased risk of developing chronic pancreatitis.132 Although non-secretors make up about 20% of most ethnic populations, because ABO blood type frequencies vary by population (ABO blood type B ranges from 8% in Norwegians to 12-15% in Western populations and 32% in Kashmiris) and genetic background varies with ethnicity, future genetic and environmental studies will need to adjust for these confounding factors in each particular cohort.132
Irritable bowel syndrome
Anxiety, depression, post-traumatic stress disorder, sexual abuse, and other life stressors are psychosocial risk factors for the development, worsening, and persistence of functional GI disorders such as IBS and functional dyspepsia.15 However, it is now known that these disorders involve both intestinal barrier dysfunction and dysregulation of the brain-gut-microbiota axis.15 Psychological and physical stress are associated with increased small intestine motility (in IBS) and enhanced visceral sensitivity, but clinical manifestations of these functional disorders also include mucosal inflammation and increased intestinal permeability.15
Further, it should be noted that low-grade mucosal inflammation causes increased infiltration of mast cells, which affect intestinal permeability and activate visceral afferent nerves that signal abdominal pain.15 Indeed, new imaging techniques have shown significant overlap between regions of the brain which modulate gut motility and visceral pain, and regions involved in processing emotion, with greater activation of the latter in response to GI distress than in healthy subjects.15 Enteric mast cells are able to communicate bidirectionally with the ENS, ANS, and CNS, and the presence of inflammation increases contacts between mast cells and enteric nerve fibers, which has been positively associated with frequency and severity of abdominal pain.15
Inflammatory bowel disease
Ulcerative colitis and Crohn's disease are the two most common forms of IBD, and are distinguished by being both chronic and relapsing.5 Such inflammatory processes may be due to functional interactions of an entire subset of the microbial community, rather than a single microorganism; known as ‘community as a pathogen,’ this is an emerging concept in the study of microbial pathogenesis.2, 42 Alternatively, a host-specific genetic effect on the composition of the microbiota could explain some of the dysbiosis that is associated with these conditions.4 Autophagy, an important component of physiological processes involved in cell growth and homeostasis, is required for bacterial degradation; mutations in several genes in the autophagy pathway have been linked with susceptibility to Crohn's disease.42
At least 160 single nucleotide polymorphisms (SNPs) are associated with increased risk of developing one or both forms of IBD, however the genes involved are not disease specific but are instead genes associated with pattern recognition receptors, defensins, and maintenance of epithelial barrier integrity.5 Increased antigen permeability and uptake of hepatotoxins, whether due to IgA deficiency or altered intestinal mucosa, can result in damage to hepatocytes and the chronic active (non-viral) hepatitis associated with IBD.39 Removal of inflamed bowel tissue, which concordantly reduces uptake of toxins, has resulted in improvement in the liver condition.39
Receptor polymorphisms are associated with deregulation of resident microbial tolerance by the adaptive immune system, reduced expression of antimicrobial peptides, impaired innate immunity, and severe intestinal inflammation.21, 27 Not surprisingly, such polymorphisms are associated with increased susceptibility to IBD, and symptoms sometimes improve following antibiotic treatment, however, it should be noted that exposure to antibiotics in early childhood has been significantly associated with increased risk for Crohn's disease.4 Non-secretor status has been associated with Crohn's disease but not ulcerative colitis,40, 132 and would account for the reduced microbial diversity found in Crohn's disease patients, 5 which has been associated with increased intestinal permeability.
Sidebar One: Lesion Topology and Secretor Status in Crohn's Disease
When Japanese researchers133 stratified inflammatory lesions by intestinal region, secretor status was associated with colonic Crohn's disease (CD) lesions, but not with ileal or ileocolonic lesions or with ulcerative colitis (UC). The nonsense mutation (G428A) in the FUT2 gene of non-secretors, found in about 20% of those with Northern European heritage, is not found in the Japanese population, which instead has a different nonsense mutation (A385T), two missense mutations (C571T and C628T), and a fusion gene, with a non-secretor prevalence of about 25%. All study subjects had the A385T mutation and ABO blood type A; whereas 80% of healthy controls and patients with UC and ileocolonic CD were secretors, only 67% of ileal CD patients were secretors, but all colonic CD patients were secretors (p=0.036). There is normally almost no expression of ABO blood group antigens in the rectum, yet staining of rectal tissue specimens found abnormally high expression of blood group A antigen in some colonic CD patients. Colonic tissue from experimental IBD mice used in the study was stained for blood group H1 antigens and was strongly positive in symptomatic 10-week old IL-10-/- mice. Of note, similar staining in asymptomatic 8-week old IL-10-/- mice was also strong for H1 antibodies, although inflammation was not present. Although colonic CD is less prevalent, abnormal colonic expression of HBGAs in secretors may be a diagnostic indicator. Further studies on populations with different FUT2 mutations and ABO blood group antigens, stratified by topology, would be highly instructive.
Metabolic diseases
A metagenomic study found a strong correlation between gut microbial communities, certain associated enzyme profiles, and obesity and IBD phenotypes in the hosts.134 A large proportion of these host-state associated enzymes are involved in either the phosphotransferase system, which regulates carbohydrate uptake and has been noted as a biomarker for IBD, or the nitrate reductase pathway, which converts nitrate to nitrite, nitric oxide, and other nitrogenous end-products.134 Increased nitric oxide levels are carcinogenic, have inflammatory effects, and have been linked with both insulin resistance in the obese and IBD.134 A study with monozygotic twins found that although the proportion of microbes in fecal samples were not significantly affected by probiotics and prebiotics, RNA sequencing showed altered expression of microbial genes that control carbohydrate metabolism.135 Metabolomic analysis of the less diverse microbiota has confirmed lower levels of butyrate, increased potential for production of hydrogen sulfide, and less capability to manage oxidative stress.7
Other studies with lean and obese twins have found an association between obesity and decreased numbers and diversity of bacterial species, and have identified increased energy-harvesting abilities of the bacteria that function within the obese person's microbiome.8, 32, 134, 136, 137 There appears to be a direct correlation between the amount of weight loss and the initial bacterial makeup of the gut microbiota before dieting,138 and people with plentiful B. fragilis, Lactobacilli, and Bifidobacteria were more successful in losing weight.5 Weight gain after a smoking cessation intervention was associated with a decrease in Proteobacteria and Bacteroidetes and a significant increase in Firmicutes and Actinobacteria, even though caloric intake was stable or even lower than when subjects smoked.5
In a small study comparing bacterial diversity in normal-weight, obese, and post-gastric bypass patients,8 phylogenetic analysis found that obese patients had enhanced levels of Prevotellaceae, gastric bypass patients had higher levels of Gammaproteobacteria, whereas Firmicutes were significantly decreased in the gastric bypass patients but were dominant in obese and normal-weight patients. In addition, obese patients had significantly higher levels of hydrogen-oxidizing methanogens, mainly Methanobacteriales, resulting in increased energy uptake in the colon.8
Gastric bypass surgery permanently alters the gut microbiota; this change may be the driver behind complete resolution of T2DM reported in these patients before surgery-associated weight loss has occurred.8 Recent studies with mice found that alterations in the innate immune system of the gut mucosa correlate with hyperphagia and development of T2DM and metabolic syndrome, and these metabolic changes are associated with modifications in gut microbial composition.8 If such dysregulation of the intestinal microbiota is a central mechanism in the development of obesity and T2DM, it may be possible to reverse these condition by changing the composition of the gut microbiota without resorting to surgery.8
Researchers are using fecal transplant studies with mice to investigate possible factors involved in metabolic dysfunction, ranging from insulin resistance, glucose intolerance, and cholesterol metabolism, to obesity, T2DM, and metabolic syndrome.8 These last three conditions are all associated with low-grade inflammation, indicating a possible activation of the innate immune system in response to the intestinal microbiota.32, 139 Chronic low-grade inflammation, which is associated with both obesity and metabolic syndrome, increases expression of TNF-α, which inactivates insulin receptor substrate-1, leading to insulin resistance, hyperinsulinemia, T2DM, fatty liver, and excess adiposity.140 Transplantation of the microbiota has resulted in transfer of the donor's altered phenotypic expression for cholesterol metabolism, colitis, and obesity.8, 136
Two small but intriguing studies looked at the difference in microbial gene richness among obese and overweight people. They found that people with low gene count (LGC) were more likely to have more pronounced low-grade inflammation, more disturbed metabolism, and more overall body fat (adiposity), and were more likely to gain more body weight than people with high gene count (HGC).141, 142 Gene richness represents the abundance and diversity of bacterial species, or the complexity of the microbiota.141-143 At baseline in one study, 40% of participants were LGC, having only two-thirds of the gene richness of HGC subjects; after a 6-week calorie-restriction intervention, LGC subjects had a significant increase in gene richness that was not seen in HGC participants, accompanied by a significant decrease in adiposity and serum cholesterol and a less-robust decrease in inflammation.141, 143 In the other study, although only 23% of the participants were LGC, this group had a significantly larger proportion of obese people who had gained significantly more weight during the previous 9 years than participants in the HGC group, and their most abundant microbial species were associated with inflammation.142 Stratification of obese and overweight people by LGC and HGC may be a way to define subsets with different metabolic risk profiles and severity of inflammation-associated comorbidities.142
Human fecal transplants have also shown promise as a possible treatment for people with conditions such as antibiotic-induced diarrhea or C. difficile infection,8, 116 and fecal transplants from lean donors to recipients with metabolic syndrome resulted in statistically significant increases in insulin sensitivity after just six weeks.124 Building on these studies, fecal transplants could become a mainstream treatment following intensive or long-term antibiotic therapy, to restore the microbiota to healthy proportions and to prevent or correct dysregulation of the epithelial membrane. It is also possible that the structure and metabolic function of the “obese” microbiota could be influenced through targeted antibiotic treatment.4
Cardiovascular disease is also associated with the intestinal microbiome. Gut microbes play an obligate role in the process of converting dietary phospholipids to trimethlyamine N-oxide (TMAO), which leads to formation of atherosclerotic plaques.144 The current medical approach is to take drugs and change the diet, but it may be possible to prevent or treat cardiovascular disease by changing the gut microbiome instead. One study reported statistically significant reductions of plasma lipoproteins and liver concentrations of di- and trimethylamine (TMOA precursors) following dietary supplementation with Lactobacillus paracasei when compared to controls,9 which suggests that targeted probiotic consumption can induce modulation of bacterial metabolism with consequent changes in host metabolic phenotypes and health status.
Effect of vaccines and antibiotics
There is no doubt that vaccinations and antibiotics have saved countless lives, and there is also no doubt that they have had unintended consequences. Loss of one pathogenic species may give opportunity to an ecological competitor, which may be more virulent and more easily transmissible.2 For example, following successful vaccination programs targeting S. pneumoniae, replacement with S. aureus has occurred, concurrent with an unprecedented surge in community-acquired methicillin-resistant S. aureus infections.2 And while it is not yet known whether low bacterial diversity associated with some diseases is a side effect of the specific disorder or a causative factor,7 it is known that antibiotic usage affects the diversity of the microbiota.
Ciprofloxacin is a commonly used, broad-spectrum antibiotic that does not have some of the more unpleasant side effects of clindamycin or amoxicillin-clavulanic acid and was thought to be less disturbing to gut microbial diversity.145 However, pyrosequencing of the distal gut bacteria prior to and following a short course of treatment revealed that it rapidly and profoundly affected the abundance of 30% of the bacterial taxa, with additional individualized effects in each participant, and was pervasive in its effects, decreasing richness, diversity, and evenness.145 Although no functional disturbances were reported, and the composition at 4 weeks post-treatment was similar to its pre-treatment state, some taxa still had not recovered 6 months post-treatment.145 Despite functional redundancy, continuity of metabolic activity does not inform about more specialized functions that may have been affected, such as immune modulation, pathogen resistance, or bile transformation.145
Two courses of ciprofloxacin separated by 6-months were each followed by a return to a state approximating the pre-treatment state, however, recovery was incomplete and although the composition was stable, it was different from the original pre-treatment composition.146 The long-term effects of a persistent altered state are unknown,146 but it is known that the effects of one course of antibiotics can persist for years, that some effects may not be apparent right away, 145 and that antibiotics are associated with both acute and chronic health conditions.146 One of the consequences that may have the most long-term ramifications is acquired antibiotic resistance in the intestinal microbiota; another is replacement of a mutualist that provides a specialized function with a commensal species or strain that does not.146
Since the 1950s, one of the primary sources of antibiotic intake in humans has been the U.S. food supply; sub-therapeutic antibiotic treatment (STAT) of mammals and birds used for food production has reduced infection and disease rates and increased weight gain in farm animals, but there may have been unintended long-term effects on human consumers and their microbiota.147 In a murine model, significant alterations in the gut microbiome due to early-life STAT exposure resulted in increased production of hormones that regulate metabolism; changes in regulation of lipids and cholesterol in the liver; changes in genes involved in metabolism of carbohydrates to SCFAs and increased SCFAs in the colon; and not surprisingly, increased adiposity.147 It is not unreasonable to think that repeated daily STAT intake through our food supply could have similar effects in humans.
Sidebar Two: Antibiotics and Type I Diabetes Mellitus
In non-obese diabetic (NOD) mice, autoimmune diabetes spontaneously develops as a result of immune-mediated destruction of pancreatic β-cells, similar to type 1 diabetes mellitus (T1DM) in humans.148 In post-pubertal, 6-week old NOD mice, the incidence of T1DM is 2:1 female-to-male, which is thought to be due to protective effects of testosterone.148 Male 6-week old NOD mice exposed to pulsed therapeutic-dose antibiotics (PAT) in early life 149 had higher T1DM incidence compared to male controls (53% vs. 26% respectively, p<0.05). They also had reduced TREG and TH17 populations in the small intestinal lamina propria; upregulated gene expression for cholesterol biosynthesis and for steroid, sterol, lipid, and cholesterol metabolism; and downregulated serum amyloid A (SAA) gene expression.149 In addition, they had reduced intestinal microbiota diversity with a distinct community structure, nearly complete loss of Bacteroidetes and Actinobacteria but increased Proteobacteria and Akkermansia mucinophila; and accelerated development of T1DM and insulitis.149 Reduced SAA gene expression may be associated with impaired intestinal barrier function, translocation of gut bacteria, and inflammation.149 In humans, increased risk for T1DM is associated with barrier function loss and prior antibiotic exposure, and the PAT model was chosen because its dosing and pharmacokinetics are representative of the type and frequency of antibiotic treatments received annually by U.S. children.149 Of note, although incidence of T1DM in humans is not sex-biased, women have a higher incidence of autoimmune diseases; the female-to-male ratio is inversely associated with age of onset for rheumatoid arthritis and MS, which is consistent with declining testosterone levels in men.148
A new vaccine for N. meningitidis serogroup B is a result of completion of the HMP, which has given drug developers the ability to reverse-engineer vaccines by analyzing whole genome sequences, selecting one or more bacterial surface-exposed proteins, and developing a vaccine that includes these bacterial antigens.150 Use of such a targeted vaccine will result in production of bactericidal antibodies that recognize those proteins and generate an immune response.150 However, due to antibody cross-reactivity, any microorganisms with antigen sequences that are highly similar to the selected proteins will also be targeted by the immune response, even if they are beneficial and protect against pathogens.150
Indeed, about 5% of the total human microbiota are non-pathogenic Neisseria species that share some of these very same sequences; found in the mouth and airway, some protect against dental caries, and some are thought to promote natural immunity to N. meningitidis.150 Given existing concern about the effects of vaccines, vaccine developers will need to be careful that new reverse-engineered vaccines do not have unintended consequences for our beneficial, health-promoting bacteria.
Although no credible evidence has been found that establishes an association between childhood vaccinations and development of ASD,151 other vaccines have occasionally been associated with serious adverse events. The live virus smallpox vaccine given to active duty U.S. military service members sometimes results in myopericarditis, which is inflammation of the pericardium and/or the myocardium.152 Among the millions of U.S. military service members who received the vaccine since 2002, occurrence of myopericarditis and subclinical myopericarditis is estimated at 0.46% and 2.87%, respectively.152 Preliminary metabolomic analysis distinguished unique pre- and post-vaccine profiles in those who experienced adverse effects.152 With the refinement of case definitions for this and other vaccines, metabolomics may be able to identify a metabotype or metabolic signature that could predict who might experience adverse events from their pre-vaccine profile alone.152
Metabolomics and the Microbiome
Metabolomics is used to assess the presence and quantity of thousands of metabolites simultaneously, identify biomarkers of disease, and explore genomic variations that affect function.7 Unlike genomic analysis, which delineates potential for metabolism, metabolomic analysis provides information about actual metabolism and elucidates possible mechanisms; metabolic outcomes in the host can be directly compared with the metabolism of the intestinal microbiota.7 The emerging field of metabolomics provides an invaluable tool to explore how disease may be associated with perturbations in either the metabotype or gut microbial populations; how bacterial metabolites can contribute to inflammation or disease processes if the human host is unable to adequately metabolize them; and how nutritional and metabolic imbalances influence health and disease in previously unknown ways.
In a metabolomics study comparing germ-free and conventional mice,153 metabolism of certain dietary compounds by enteric bacteria resulted in higher serum concentrations in the conventional mice. When provided with dietary tryptophan, mice with enteric bacteria that expressed the enzyme tryptophanase had decreased serum tryptophan concentrations and increased concentrations of indole, ammonia, and pyruvate. In metabolomics studies of human patients with chronic kidney failure, elevated indole was transformed by the liver into indoxyl sulfate, which is a nephrotoxin.153
Conversely, in mice colonized by different subsets of enteric bacteria, indole was transformed into indole-3-propionic acid, which is a powerful antioxidant. The tyrosine metabolites phenyl and p-cresol sulfate were found to be 1.4-fold higher, which was thought to be due to activation of the sulfation mechanism used by the body during phase II drug metabolism.153 However, glycine conjugation, another phase II drug metabolism mechanism, resulted in elevated serum concentrations of hippuric acid without reducing serum phenylalanine.153 These findings demonstrate a significant effect on the host's health from microbial metabolic products, and indicate the complexity of the interaction between the host and the microbiota.
A metabolomics study of progressive knee osteoarthritis in overweight and obese people154 identified distinct metabolomics profiles at baseline and at 18 months. Different metabolite patterns distinguished between progressors and non-progressors at baseline for glycolate, Hippurate, and trigonelline, and at 18 months for alanine, N,N-dimethylglycine, glycolate, Hippurate, histidine, and trigonelline, despite BMI being similar in subjects at baseline, at 18 months, and when comparing baseline and 18 months.154 In addition, while the mean plasma level of IL-6 was similar at baseline, it decreased by 18 months in non-progressors but increased in progressors.154 Hippurate and trigonelline are believed to be metabolites produced by enteric bacteria,154 indicating that the makeup of the microbiome plays an important role in metabolic differences associated with progression of inflammation and degenerative diseases.
Future Research
Completion of the HMP has resulted in a comprehensive catalog of reference data, ranging from taxonomically characterized discrete microbial communities to the overall function and metabolism of the healthy human microbiome.6 Not only are these data an invaluable resource that can be used to correlate and compare with data from future studies, but the protocols and methods developed and refined during this project can serve as a model for future research efforts.6 A factor that appears to have relevance for future studies is the effect of racial and ethnic heritage on the microbial communities within the human microbiota. In reporting on the progress of the HMP, areas that required further exploration included host ancestry and genetics, non-European heritage, effects of environment, and microbiome interactions with the host.1 In a follow-up HMP analysis, microbial metabolic and functional pathways were found to correlate with clinical metadata and host phenotypes, and were strongly associated with racial and ethnic heritage.10
There are still significant challenges and limitations associated with identifying the intestinal microbiota that play a significant role in metabolism of nutrients in the small intestine and research in this area should provide much-needed understanding of some of our most difficult and hard-to-treat diseases. Although the microbiota found in stool samples is generally considered to be representative of the lower intestinal microbiome, it does not accurately represent the flora found in the upper GI tract, and further, in the distal GI, the composition of the fecal microbiota differs substantially from mucosa-associated microbiota.14 Despite recent advances in sequencing technology and metagenomic analysis of the microbiota of the large intestine, research into the small intestinal microbiota continues to be hampered.12
Owing to the discrete functions and pH levels of the esophagus, stomach, and small intestine, different microorganisms are associated with each of these environments, however, relatively little research has been done into upper GI conditions, because each region must be sampled separately and there is no easy method to obtain uncontaminated specimens of these mucosal microbes.12 Studies have reported that the most common genera in the normal esophagus are Streptococcus, Prevotella, and Veillonella, with Streptococcus viridans being the most common microorganism.12 However, Gram-negative anaerobes dominated in Barrett's esophagus and esophagitis, and aerobic Gram-positive and anaerobic bacteria were most common with megaesophageal disease.12 Unless H. pylori is present, many of these esophageal bacteria are also found in the stomach.4
Conventional testing has included cultures or biochemical assays, tissue histology, and host serological responses, which may or may not reveal the presence of a particular organism, and polymerase chain reaction-based techniques, which can detect low levels of microbes belonging to minor populations.2 It must then be determined whether the organism is part of the resident population of that region, or transient from another proximal region of the GI tract.2 For example, one study of 17 patients receiving small intestine transplants found that the normal ileal population of strict anaerobic Bacteroides and Clostridium species was temporarily replaced by facultative anaerobic species of Lactobacillus and Enterobacteriacae until their ileostomies were closed, whereupon the normal community structure was restored.8
The influence of diet on the microbiome is another emerging area of research. A well-designed study correlated changes in the diet with changes in the intestinal microbiota;18 although there were immediate effects on the microbiota (within 24 hours of dietary changes), short-term dietary changes did not correlate with reclassifications from one enterotype to another. However, this study18 found a very strong association between long-term dietary patterns and proposed enterotype classifications.100 If future research shows similar strong associations between these enterotypes and certain disease states, then long-term dietary changes may become the intervention of choice for both preventing and treating these diseases.
In addition to dietary changes or enteric infections, antibiotics induce functional changes in the microbiota, which are mediated by changes in population structure, or by introduction or extinction of specific microbial groups.4 Thus, the effect of long-term sub-therapeutic antibiotic levels in the diet and the evolution of antibiotic resistant bacteria are both of increasing concern. As we have seen, low-dose antibiotic administration modulates the gut microbiota as well as the host metabolism and phenotype.147 Recent research has also documented the destabilizing effect on the microbiota of repeated or extensive antibiotic use145, 146 as well as the increase in adiposity associated with Fermicutes dominance.4, 146 In Italy, probiotics such as Bifidobacterium bifidum and Lactobacillus acidophilus are routinely prescribed following antibiotic treatment.16
With further delineation of the various bacterial secretion systems and greater understanding of the role of adhesins in colonization and survival of pathogenic and beneficial bacteria, anti-virulence compounds have been developed that target key proteins needed for pilus biosynthesis and curli biogenesis, or impair dimerization needed for T3SS needle production or assembly of the T4SS.87 The use of anti-adhesion vaccines that are carefully targeted to specific adhesion molecules used by specific pathogens, and which do not impair the protective functioning of probiotic or commensal bacteria, have been shown to be effective in both cattle and pigs; development of similar vaccines for human use is a research area that would have potentially life-saving benefits.51 Future research could also focus on the development of targeted drugs that block signal peptidases or impair translocase function along with the selective use of probiotics to compete with pathogens and rebalance the intestinal population.82
The growing number of children with ASD is another area of intense research. Various metabolomics studies of people with ASD have found changes in the enteric microbiota; decreased glutamate and increased taurine levels in urine; decreased plasma levels of polyunsaturated fatty acids and increased saturated fatty acids; abnormal amino acid metabolism; greater oxidative stress; and changes in plasma metabolites associated with mitochondrial dysfunction.155 A recent metabolomics study using independent discovery and validation case-control cohorts found 17 serum metabolites associated with ASD, validated 11 metabolites with univariate analysis, and in a logistic regression model, identified 2 metabolites, docosahexaenoic acid (DHA) and sphingosine 1-phosphate, that were significant predictors of ASD.155 Polyunsaturated fatty acids (PUFAs) such as DHA are necessary for the structural and functional integrity of the lipid membranes of the CNS, while sphingomyelin metabolism is associated with cerebral white matter, and abnormal white matter has been associated with ASD.155
Among the metabolites associated with ASD, this study155 found significant differences between cases and controls in serum L-acetylcarnitine and pregnanetriol levels, and abnormal lysophosphatide metabolism in cases. It was speculated that the abnormal fatty acid metabolism associated with ASD may be related to disturbed metabolism of estrogen; reduced L-acetylcarnitine and decanoylcarnitine metabolism may be associated with mitochondrial dysfunction; and abnormal lysophosphatide metabolism may be associated with abnormalities in metabolism of sphingomyelin and PUFAs.155 Future metabolomics research investigating the role of the enteric microbiota and their metabolites may reveal additional associations with ASD, and could also provide a mechanism for evaluating the effectiveness of dietary, probiotic, or other therapeutic treatment of ASD.
Perhaps the most pressing concern that could be addressed by future research is the obesity epidemic, because of its associated health implications. The common wisdom has been that obesity is no more than the result of not exercising enough to burn off the calories that are consumed, and that the solution is simply to eat less and exercise more. However, millions of frustrated dieters can attest that it isn’t that simple. And they are correct. As this review has made clear, many factors contribute to obesity, and the unique microbiome and genetic makeup of each person must be taken into account. Here again, metabolomics analysis, as a data-driven diagnostic and predictive tool that can identify metabolic and pathway biomarkers as well as potential pathophysiologic mechanisms and phenotypes, would add a dimension to obesity research that complements, clarifies, and strengthens our ability to understand the still-emerging dynamics of host-microbiome symbiosis and may aid in changing the common belief that weight loss is simply a matter of lack of willpower.
Metabolomics research will also help us better understand functional GI disorders, unravel the mysteries of the brain-gut-microbiome axis, and explore the metabolites associated with depression, anxiety, and stress-related disorders. A small study used metabolomics to evaluate an immersion treatment program for weight loss in obese adolescents that included psychosocial inventories of depression and self-esteem.156 The urinary metabolites Hippurate, leucine, and 2-oxoisocaproate differentiated responders and non-responders to weight loss efforts, as measured by change in BMI.156 Participants with healthy and impaired depression and self-esteem classifications on the psychosocial inventories could also be distinguished by the score plots of certain metabolites at baseline and conclusion of the study.156 The ability to integrate metabolomic signatures, psychosocial inventory classifications, and metabolic responses to dietary changes and exercise is an invaluable contribution to developing successful weight loss interventions and personalizing weight management.156 The identification of obesity-associated metabotypes consisting of specific metabolites may also help researchers understand why certain people are non-responders and then develop weight-loss strategies that overcome those obstacles to weight loss.
Conclusion
The human intestinal microbiota plays such an important role in the healthy functioning of our bodies that it is hard to imagine how much our approach to illness and disease will change as the results of current and future research are translated into mainstream medicine and education. The current medical model and standard treatments use a “one size fits all” approach for most conditions, but as genomic and metagenomic research unravels the mystery of each of us, personalized medicine becomes more and more likely, necessary, and realistic. This review has explored how the enteric microbiota modulates mucosal barrier function and immune response, influences the host metabolic functions, and stimulates host gene expression. We have reviewed the mechanisms used by bacteria for communication, colonization, and pathogenesis, as well as the ways the human intestine establishes and maintains homeostasis, and the role the ENS plays. We have seen how delicate the balance is between health and disease, and discussed how complex and challenging it can be to regain homeostasis and health once it is lost. The HMP project and the advances in genetic and genomic sequencing technologies have created exciting opportunities for research using newer technologies like metabolomics.
One focus for future research that may not be as obvious yet could be groundbreaking is the influence of the host genotype on the composition, function, and metabolic activity of the mucosal microbiome. Although the presence or absence of HBGAs have not been determined in most microbiome or metabolome research, they play such an important role in human health and disease that they should be controlled for as confounders or modifiers. Further, analysis of results by including HBGAs as a variable may reveal associations with statistical significance that have been missed. There is a perception that the intestinal microbiota is a controlling factor in human health, however, the HBGAs predate the microbiome. These antigens are present at birth, before the microbiome has been established and before any nutrients have passed the lips of the newborn. The glycans that form these antigens are receptors for the adhesion molecules of the assembling bacterial community, and ultimately determine which bacterial species find a supporting environment, and which do not. You are what you eat, but only because you are who you are!
Acknowledgments
The preparation of this article was funded, in part, by NIH Common Fund Grant (U24 DK097193; Sumner, PI)
Footnotes
Conflict of Interest: None.
References
- 1.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10:287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Micro. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lepage P, Leclerc MC, Joossens M, Mondot S, Blottière HM, Raes J, Ehrlich D, Doré J. A metagenomic insight into our gut's microbiome. Gut. 2013;62:146–158. doi: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 4.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biedermann L, Rogler G. The intestinal microbiota: its role in health and disease. Eur J Pediatr. 2015;174:151–167. doi: 10.1007/s00431-014-2476-2. [DOI] [PubMed] [Google Scholar]
- 6.Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, Gevers D, Petrosino JF, Abubucker S, Badger JH, et al. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ursell LK, Haiser HJ, Van Treuren W, Garg N, Reddivari L, Vanamala J, Dorrestein PC, Turnbaugh PJ, Knight R. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014;146:1470–1476. doi: 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin FPJ, Wang Y, Sprenger N, Yap IKS, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB, et al. Probiotic modulation of symbiotic gut microbialhost metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J, Li R, Raes J, Arumugam M, Burgdorf K, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang ZK, Yang YS. Upper gastrointestinal microbiota and digestive diseases. World J Gastroenterol. 2013;19:1541–1550. doi: 10.3748/wjg.v19.i10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunil. 2011;23:353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouwerkerk JP, de Vos WM, Belzer C. The Gut M. Glycobiome: Bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol. 2013;27:25–38. doi: 10.1016/j.bpg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Alonso C, Vicario M, Pigrau M, Lobo B, Santos J. Intestinal barrier function and the brain-gut axis. In: Lyte M, Cryan JF, editors. Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease. New York: Springer; 2014. pp. 73–113. [DOI] [PubMed] [Google Scholar]
- 16.Bocci V. The neglected organ: bacterial flora has a crucial immunostimulatory role. Perspect Biol Med. 1992;35:251–260. doi: 10.1353/pbm.1992.0004. [DOI] [PubMed] [Google Scholar]
- 17.Guzman JR, Conlin VS, Jobin C. Diet, microbiome, and the intestinal epithelium: an essential triumvirate? Biomed Res Int. 2013;2013:1–12. doi: 10.1155/2013/425146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu GD, Chen J, Hoffmann C, Bittinger K, Ying-Yu C, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly D, Mulder IE. Microbiome and immunological interactions. Nutr Rev. 2012;70:18–30. doi: 10.1111/j.1753-4887.2012.00498.x. [DOI] [PubMed] [Google Scholar]
- 20.Ohnmacht C, Marques R, Presley L, Sawa S, Lochner M, Eberl G. Intestinal microbiota, evolution of the immune system and the bad reputation of pro-inflammatory immunity. Cell Microbiol. 2011;13:653–659. doi: 10.1111/j.1462-5822.2011.01577.x. [DOI] [PubMed] [Google Scholar]
- 21.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 22.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota Across Multiple Body Habitats in Newborns. Proc Natl Acad Sci U S A. 2010;107:11971. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houghteling PD, Walker WA. Why is initial bacterial colonization of the intestine important to infants' and children's health? J Pediatr Gastroenterol Nutr. 2015;60:294–307. doi: 10.1097/MPG.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grönlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28:19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Rescigno M. Mucosal immunology and bacterial handling in the intestine. Best Pract Res Clin Gastroenterol. 2013;27:17–24. doi: 10.1016/j.bpg2013.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2011;108:4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delves PJ. Overview of the immune system. Merck Manual for Healthcare Professionals [On-line information] 2017 Available at: http://www.merckmanuals.com/professional/immunology-allergic-disorders/biology-of-the-immune-system/overview-of-the-immune-system.
- 29.Delves PJ. Molecular components of the Immune system. Merck Manual for Healthcare Professionals [On-line information] 2017 Available at: http://www.merckmanuals.com/professional/immunology-allergic-disorders/biology-of-the-immune-system/molecular-components-of-the-immune-system.
- 30.Delves PJ. Cellular components of the immune system. Merck Manual for Healthcare Professionals [On-line information] 2017 Available at: http://www.merckmanuals.com/professional/immunology-allergic-disorders/biology-of-the-immune-system/cellular-components-of-the-immune-system.
- 31.Rescigno M. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol. 2011;32:256–264. doi: 10.1016/j.it.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benckert J, Schmolka N, Kreschel C, Zoller MJ, Sturm A, Wiedenmann B, Wardemann H. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J Clin Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantis NJ, Rol N, Corthesy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagnini C, Saeed R, Bamias G, Arseneau KO, Pizarro TT, Cominelli F. Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci U S A. 2010;107:454–459. doi: 10.1073/pnas.0910307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etzold S, Kober OI, MacKenzie DA, Tailford LE, Gunning AP, Walshaw J, Hemmings AM, Juge N. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ Microbiol. 2014;16:888–903. doi: 10.1111/1462-2920.12377. [DOI] [PubMed] [Google Scholar]
- 37.Varum FJO, Basit AW. Gastrointestinal Mucosa and Mucus. Mucoadhesive Materials and Drug Delivery Systems. 2014:83–98. Available at: http://public.eblib.com/choice/publicfullrecord.aspx?p=1712885.
- 38.Moncada D, Chadee K. Production, structure, and function of gastrointestinal mucins. In: Blaser MJ, Smith PD, Ravdin JI, Greenberg HB, Guarrant Rl, editors. Infections of the gastrointestinal tract. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 57–79. [Google Scholar]
- 39.Walker WA. Antigen handling by the gut. Arch Dis Child. 1978;53:527–531. doi: 10.1136/adc.53.7.527. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1545000/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol Rev. 2012;245:147–163. doi: 10.1111/j.1600-065X.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- 41.Atuma A, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. Available at: http://ajpgi.physiology.org/content/280/5/G922.full. [DOI] [PubMed] [Google Scholar]
- 42.Swiatczak B, Rescigno M. How the interplay between antigen presenting cells and microbiota tunes host immune responses in the gut. Sem Immunol. 2012;24:43–49. doi: 10.1016/j.mim.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Toivanen P, Vaahtovuo J, Eerola E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect Immun. 2001;69:2372–2377. doi: 10.1128/IAI.69.4.2372-2377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz L, Hevia A, Bernardo D, Margolles A, Sánchez B. Extracellular molecular effectors mediating probiotic attributes. FEMS Microbiol Lett. 2014;359:1–11. doi: 10.1111/1574-6968.12576. [DOI] [PubMed] [Google Scholar]
- 46.Sánchez B, Urdaci MC, Margolles A. Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiology. 2010;156:3232–3242. doi: 10.1099/mic.0.044057-0. [DOI] [PubMed] [Google Scholar]
- 47.Round JL, Mazmanian SK. The gut microbiome shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.González-Rodríguez I, Ruiz L, Gueimonde M, Margolles A, Sánchez B. Factors involved in the colonization and survival of bifidobacteria in the gastrointestinal tract. FEMS Microbiol Lett. 2013;340:1–10. doi: 10.1111/1574-6968.12056. [DOI] [PubMed] [Google Scholar]
- 49.Nwodo UU, Green E, Okoh AI. Bacterial Exopolysaccharides: Functionality and Prospects. Int J Mol Sci. 2012;13:14002–14015. doi: 10.3390/ijms131114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Didierlaurent A, Sirard JC, Kraehenbuhl JP, Neutra MR. How the gut senses its content. Cell Microbiol. 2002;4:61–72. doi: 10.1046/j.1462-5822.2002.00177.x. [DOI] [PubMed] [Google Scholar]
- 51.Klemm P, Schembri MA. Bacterial adhesins: function and structure. Int J Med Microbiol. 2000;290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- 52.Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 53.Kazeeva T, Shevelev A. IgA-specific proteins of pathogenic bacteria. Biochem J. 2009;74:12–21. doi: 10.1134/S0006297909010027. [DOI] [PubMed] [Google Scholar]
- 54.Kato K, Ishiwa A. The role of carbohydrates in infection strategies of enteric pathogens. Trop Med Health. 2015;43:41–52. doi: 10.2149/tmh.2014-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cress BF, Englaender JA, He W, Kasper D, Linhardt RJ, Koffas MAG. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev. 2014;38:660–697. doi: 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Proft T, Fraser JD. Bacterial superantigens. Clin Exp Immunol. 2003;133:299–306. doi: 10.1046/j.1365-2249.2003.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daniels G. Human blood groups. 3rd. Somerset, NJ: John Wiley and Sons; 2013. [Google Scholar]
- 58.Lloyd KO. Blood Group Antigens as Markers for Normal Differentiation and Malignant Change in Human Tissues. Am J Clin Pathol. 1987;87:129–139. doi: 10.1093/ajcp/87.1.129. [DOI] [PubMed] [Google Scholar]
- 59.Ravn V, Dabelsteen E. Tissue Distribution of Histo-Blood Group Antigens. APMIS. 2000;108:1–28. doi: 10.1034/j.1600-0463.2000.d01-1.x. [DOI] [PubMed] [Google Scholar]
- 60.Ewald DR, Sumner SCJ. Blood type biochemistry and human disease. WIREs Syst Biol Med. 2016;8:517–535. doi: 10.1002/wsbm.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stowell SR, Arthur CM, Dias-baruffi M, Rodrigues LC, Gourdine Jp, Heimburg-molinaro J, Ju T, Molinaro RJ, Rivera-marrero C, Xia B, et al. Innate Immune Lectins Kill Bacteria Expressing Blood Group Antigen. Nat Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weng M, Walker WA. The role of gut microbiota in programming the immune phenotype. J Dev Orig Hlth Dis. 2013;4:203–214. doi: 10.1017/S2040174412000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makivuokko H, Lahtinen SJ, Wacklin P, Tuovinen E, Tenkanen H, Nikkila J, Bjorklund M, Aranko K, Ouwehand AC, Matto J. Association between the ABO blood group and the human intestinal microbiota composition. BMC Microbiol. 2012;12:94–105. doi: 10.1186/1471-2180-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watkins WM. The ABO blood group system: historical background. Transfusion Med. 2001;11:243–265. doi: 10.1046/j.1365-3148.2001.00321.x. [DOI] [PubMed] [Google Scholar]
- 65.Franchini M, Bonfanti C. Evolutionary aspects of ABO blood group in humans. Clin Chim Acta. 2015;444:66–71. doi: 10.1016/j.cca.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 66.Henry S, Oriol R, Samuelsson B. Lewis histo-blood group system and associated secretory phenotypes. Vox Sang. 1995;69:166–182. doi: 10.1111/j.1423-0410.1995.tb02591.x. [DOI] [PubMed] [Google Scholar]
- 67.Henry SM. Review: phenotyping for Lewis and secretor histo-blood group antigens. Immunohematology. 1996;12:51–61. [PubMed] [Google Scholar]
- 68.Lee JW, Epardaud M, Sun J, Becker JE, Cheng AC, Heath JK, Turley SJ. Peripheral Antigen Display by Lymph Node Stroma Promotes T Cell Tolerance to Intestinal Self. Nat Immunol. 2007;8:181–190. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- 69.Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, et al. Group 3 Innate Lymphoid Cells Mediate Intestinal Selection of Commensal Bacteria-Specific CD4+ T Cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson RC, Dalziel JE, Gopal PK, Bassett S, Ellis A, Roy NC. The Role of Intestinal Barrier Function in Early Life in the Development of Colitis. In: Fukata D, editor. Colitis. Rijeka, Croatia: InTech; 2012. pp. 1–30. [Google Scholar]
- 71.Gritz EC, Bhandari V. The human neonatal gut microbiome: a brief review. Front Pediatr. 2015;3 doi: 10.3389/fped.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc Biol Sci. 2015;282:20143085. doi: 10.1098/rspb.2014.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Battersby AJ, Gibbons DL. The gut mucosal immune system in the neonatal period. Pediatr Allergy Immunol. 2013;24:414–421. doi: 10.1111/pai.12079. [DOI] [PubMed] [Google Scholar]
- 74.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013;110:17059–17064. doi: 10.1073/pnas.1306070110/-/DCSupplemental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerlach RG, Hensel M. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297:401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 76.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Micro. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abdallah AM, Gey van Pittius NC, DiGiuseppe Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CMJE, Appelmelk BJ, Bitter W. Type VII secretion - mycobacteria show the way. Nat Rev Micro. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 78.Coulthurst SJ. The Type VI secretion system - a widespread and versatile cell targeting system. Res Microbiol. 2013;164:640–654. doi: 10.1016/j.resmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 79.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. PNAS USA. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amor JC, Swails J, Zhu X, Roy CR, Nagai H, Ingmundson A, Cheng X, Kahn RA. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem. 2005;280:1392–1400. doi: 10.1074/jbc.M410820200. [DOI] [PubMed] [Google Scholar]
- 81.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freudl R. Leaving home ain't easy: Protein export systems in Gram-positive bacteria. Res Microbiol. 2013;164:664–674. doi: 10.1016/j.resmic.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 83.Natale P, Bruser T, Driessen AJM. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane--distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 84.Rennoll-Bankert KE, Rahman MS, Gillespie JJ, Guillotte ML, Kaur SJ, Lehman SS, Beier-Sexton M, Azad AF. Which Way In? The RalF Arf-GEF Orchestrates Rickettsia Host Cell Invasion. PLoS Pathog. 2015;11:e1005115. doi: 10.1371/journal.ppat.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Korea CG, Ghigo JM, Beloin C. The sweet connection: Solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli. BioEssays. 2011;33:300–311. doi: 10.1002/bies.201000121. [DOI] [PubMed] [Google Scholar]
- 86.Nuccio SP, Bdumler AJ. Evolution of the Chaperone/Usher Assembly Pathway: Fimbrial Classification Goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Micro. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 88.Nivaskumar M, Francetic O. Type II secretion system: A magic beanstalk or a protein escalator. Biochim Biophys Acta. 2014;1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 89.Evans ML, Chapman MR. Curli biogenesis: Order out of disorder. Biochim Biophys Acta. 2014;1843:1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Campos M, Cisneros DA, Nivaskumar M, Francetic O. The type II secretion system – a dynamic fiber assembly nanomachine. Res Microbiol. 2013;164:545–555. doi: 10.1016/j.resmic.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 91.Schneewind O, Missiakas D. Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim Biophys Acta. 2014;1843:1687–1697. doi: 10.1016/j.bbamcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pizarro-Cerdá J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 93.van Wely KHM, Swaving J, Freudl R, Driessen AJM. Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol Rev. 2001;25:437–454. doi: 10.1016/S0168-6445(01)00062-6. [DOI] [PubMed] [Google Scholar]
- 94.Thomas S, Holland IB, Schmitt L. The Type 1 secretion pathway - The hemolysin system and beyond. Biochim Biophys Acta. 2014;1843:1629–1641. doi: 10.1016/j.bbamcr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 95.Rendón MA, Saldaña Z, Erdem AL, Monteiro-Neto V, Vázquez A, Kaper JB, Puente JL, Girón JA. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc Natl Acad Sci U S A. 2007;104:10637–10642. doi: 10.1073/pnas.0704104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ayers M, Howell PL, Burrows LL. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 2010;5:1203–1218. doi: 10.2217/fmb.10.76. [DOI] [PubMed] [Google Scholar]
- 97.Henderson B, Nair S, Pallas J, Williams MA. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol Rev. 2011;35:147–200. doi: 10.1111/j.1574-6976.2010.00243.x. [DOI] [PubMed] [Google Scholar]
- 98.Wacklin P, Tuimala J, Nikkiä J, Sebastian T, Mäkivuokko H, Alakulppi N, Laine P, Rajilic-Stojanovic M, Paulin L, de Vos WM, et al. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS One. 2014;9:e94863. doi: 10.1371/journal.pone.0094863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wacklin P, Mäkivuokko H, Alakulppi N, Nikkilä J, Tenkanen H, Räbinä J, Partanen J, Aranko K, Mättö J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen W, Gaskins HR, McIntosh MK. Influence of dietary fat on intestinal microbes, inflammation, barrier function and metabolic outcomes. J Nutr Biochem. 2014;25:270–280. doi: 10.1016/j.jnutbio.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 103.Tuohy KM, Conterno L, Gasperotti M, Viola R. Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. J Agric Food Chem. 2012;60:8776–8782. doi: 10.1021/jf2053959. [DOI] [PubMed] [Google Scholar]
- 104.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human Intestinal symbionts. Curr Biol. 2014;24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mazagova M, Wang LR, Anfora AT, Wissmueller M, Lesley SA, Miyamoto Y, Eckmann L, Dhungana S, Pathmasiri W, Sumner S, et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB J. 2015;29:1043–1055. doi: 10.1096/fj.14-259515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Konturek PC, Brzozowski T, Konturek SJ. Stress and the gut: pathophysiology, clinical consequences, diagnostic approach and treatment options. J Physiol Pharmacol. 2011;62:591–599. Available at: www.jpp.krakow.pl/journal/archive/12_11/pdf/591_12_11_article.pdf. [PubMed] [Google Scholar]
- 107.Foster JA, McVey Neufeld KA. Gutbrain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36:305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 108.Gershon MD. The Second Brain: A Groundbreaking New Understanding of Nervous Disorders of the Stomach and Intestine. New York, NY: HarperCollins; 1998. [Google Scholar]
- 109.Forsythe P, Kunze W, Bienenstock J. Moody microbes or fecal phrenology: what do we know about the microbiota-gut-brain axis? BMC Medicine. 2016;14 doi: 10.1186/s12916-016-0604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Foster JA, Rinaman L, Cryan JF. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiol Stress. 2017;7:124–136. doi: 10.1016/j.ynstr.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khanna S, Tosh PK. A Clinician's Primer on the role of the Microbiome in Human Health and Disease. Mayo Clinic Proceedings. 2014;89:107–114. doi: 10.1016/j.mayocp.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 112.Hornig M. The role of microbes and autoimmunity in the pathogenesis of neuropsychiatric illness. Current Opinion in Rheumatology. 2013;25:488–495. doi: 10.1097/BOR.0b013e32836208de. [DOI] [PubMed] [Google Scholar]
- 113.Herz J, Kipnis J. Bugs and Brain: How Infection Makes You Feel Blue. Immunity. 2016;44:718–720. doi: 10.1016/j.immuni.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 114.Blank T, Detje Claudia N, Spieß A, Hagemeyer N, Brendecke Stefanie M, Wolfart J, Staszewski O, Zِöller T, Papageorgiou I, Schneider J, et al. Brain Endothelial- and Epithelial-Specific Interferon Receptor Chain 1 Drives Virus-Induced Sickness Behavior and Cognitive Impairment. Immunity. 2016;44:901–912. doi: 10.1016/j.immuni.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 115.Bradstreet JJ, Ruggiero M, Pacini S. Commentary: Structural and functional features of central nervous system lymphatic vessels. Front Neurosci. 2015;9:485–487. doi: 10.3389/fnins.2015.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Evrensel A, Ceylan ME. Fecal Microbiota Transplantation and Its Usage in Neuropsychiatric Disorders. Clin Psychopharmacol Neurosci. 2016;14:231–237. doi: 10.9758/cpn.2016.14.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, Derecki NC, Castle D, Mandell JW, Lee KS, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523:337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raper D, Louveau A, Kipnis J. How Do Meningeal Lymphatic Vessels Drain the CNS? Trends Neurosci. 2016;39:581–586. doi: 10.1016/j.tins.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ueno M, Chiba Y, Murakami R, Matsumoto K, Kawauchi M, Fujihara R. Blood–brain barrier and blood–cerebrospinal fluid barrier in normal and pathological conditions. Brain Tumor Pathol. 2016;33:89–96. doi: 10.1007/s10014-016-0255-7. [DOI] [PubMed] [Google Scholar]
- 120.Ochoa-Reparaz J, Mielcarz DW, Begum- Haque S, Kasper LH. Gut, bugs, and brain: role of commensal bacteria in the control of central nervous system disease. Ann Neurol. 2011;69:240–247. doi: 10.1002/ana.22344. [DOI] [PubMed] [Google Scholar]
- 121.Segers ME, Lebeer S. Towards a better understanding of Lactobacillus rhamnosus GG - host interactions. Microb Cell Fact. 2014;13:S7–S7. doi: 10.1186/1475-2859-13-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lebeer S, Claes I, Tytgat HLP, Verhoeven TLA, Marien E, von Ossowski I, Reunanen J, Palva A, de Vos WM, De Keersmaecker SCJ, et al. Functional Analysis of Lactobacillus rhamnosus GG Pili in Relation to Adhesion and Immunomodulatory Interactions with Intestinal Epithelial Cells. Appl Environ Microbiol. 2012;78:185. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dominguez-Bello MG, Blaser MJ. Do you have a probiotic in your future? Microb Infect. 2008;10:1072–1076. doi: 10.1016/j.micinf.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kootte RS, Vrieze A, Holleman F, Dallinga-Thie GM, Zoetendal EG, de Vos WM, Groen AK, Hoekstra JB, Stroes ES, Nieuwdorp M. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes Metab. 2012;14:112–120. doi: 10.1111/j.1463-1326.2011.01483.x. [DOI] [PubMed] [Google Scholar]
- 125.Arora T, Singh S, Sharma RK. Probiotics: Interaction with gut microbiome and antiobesity potential. Nutrition. 2013;29:591–596. doi: 10.1016/j.nut.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 126.Ley RE, Backhed F, Turnbaugh P. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cenit MC, Olivares M, Codoñer-Franch P, Sanz Y. Intestinal microbiota and celiac disease: cause, consequence or co-evolution? Nutrients. 2015;7:6900–6923. doi: 10.3390/nu7085314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vaarala O. Human intestinal microbiota and type 1 diabetes. Curr Diab Rep. 2013;13:601–607. doi: 10.1007/s11892-013-0409-5. [DOI] [PubMed] [Google Scholar]
- 129.Smyth DJ, Cooper JD, Howson JM, Clarke P, Downes K, Mistry T, Stevens H, Walker NM, Todd JA. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011;60:3081–3084. doi: 10.2337/db11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang P, Li HL, Wang CY. FUT2 nonfunctional variant: a “missing link” between genes and environment in type 1 diabetes? Diabetes. 2011;60:2685–2687. doi: 10.2337/db11-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyِöty H, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weiss FU, Schurmann C, Guenther A, Ernst F, Teumer A, Mayerle J, Simon P, Vöِlzke H, Radke D, Greinacher A, et al. Fucosyltransferase 2 (FUT2) non-secretor status and blood group B are associated with elevated serum lipase activity in asymptomatic subjects and an increased risk for chronic pancreatitis a genetic association study. Pancreatology. 2014;14:S14. doi: 10.1136/gutjnl-2014-306930. [DOI] [PubMed] [Google Scholar]
- 133.Miyoshi J, Yajima T, Okamoto S, Matsuoka K, Inoue N, Hisamatsu T, Shimamura K, Nakazawa A, Kanai T, Ogata H, et al. Ectopic expression of blood type antigens in inflamed mucosa with higher incidence of FUT2 secretor status in colonic Crohn's disease. J Gastroenterol. 2011;46:1056–1063. doi: 10.1007/s00535-011-0425-7. [DOI] [PubMed] [Google Scholar]
- 134.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Gordon JI, Henrissat B, Oozeer R, Cools-Portier S, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 137.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Santacruz A, Marcos A, Wärnberg J, Martí A, Martin-matillas M, Campoy C, Moreno LA, Veiga O, Redondo-figuero C, Garagorri JM, et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity. 2009;17:1906–1915. doi: 10.1038/oby.2009.112. [DOI] [PubMed] [Google Scholar]
- 139.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kallus SJ, Brandt LJ. The intestinal microbiota and obesity. J Clin Gastroenterol. 2012;46:16–24. doi: 10.1097/MCG.0b013e31823711fd. [DOI] [PubMed] [Google Scholar]
- 141.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 142.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 143.Fang S, Evans RM. Microbiology: wealth management in the gut. Nature. 2013;500:538–539. doi: 10.1038/500538a. [DOI] [PubMed] [Google Scholar]
- 144.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 149.Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, McRitchie S, Li H, Chung J, Sohn J, Kim S, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nature microbiology. 2016;1:16140. doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gebhardt M, Hutchins E, Comella P, Aho E. Genes encoding meningococcal vaccine antigens are present in nonpathogenic bacteria found in the human microbiome. Bios. 2014;85:142–150. Available at: http://www.jstor.org/stable/24367863. [Google Scholar]
- 151.Mnookin S. The Panic Virus: A True Story of Medicine, Science, and Fear. New York: Simon & Schuster; 2011. [Google Scholar]
- 152.McClenathan BM, Stewart DA, Spooner CE, Pathmasiri WW, Burgess JP, McRitchie SL, Choi YS, Sumner SCJ. Metabolites as biomarkers of adverse reactions following vaccination: A pilot study using nuclear magnetic resonance metabolomics. Vaccine. 2017;35:1238–1245. doi: 10.1016/j.vaccine.2017.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Loeser RF, Pathmasiri W, Sumner SJ, McRitchie S, Beavers D, Saxena P, Nicklas BJ, Jordan J, Guermazi A, Hunter DJ, et al. Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: an exploratory study. Osteoarthritis Cartilage. 2016;24:1479–1486. doi: 10.1016/j.joca.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang H, Liang S, Wang MQ, Gao JQ, Sun CH, Wang J, Xia W, Wu SY, Sumner SJ, Zhang FY, et al. Potential serum biomarkers from a metabolomics study of autism. J Psychiatry Neurosci. 2016;41:27–37. doi: 10.1503/jpn.140009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pathmasiri W, Pratt KJ, Collier DN, Lutes LD, McRitchie S, Sumner SCJ. Integrating metabolomic signatures and psychosocial parameters in responsivity to an immersion treatment model for adolescent obesity. Metabolomics. 2012;8:1037–1051. doi: 10.1007/s11306-012-0404-x. [DOI] [Google Scholar]