Abstract
Purpose
Hypothalamic kisspeptin neurons are considered to play a critical role in regulating mammalian reproduction and integrating humoral and neuronal inputs that control gonadotropin‐releasing hormone (GnRH)/gonadotropin release. The present study aimed to investigate the upstream regulator candidates for kisspeptin neurons.
Methods
Visualized kisspeptin neurons that were taken from the arcuate nucleus (ARC) of Kiss1‐tdTomato rats were subjected to next‐generation sequencing (NGS) analysis. In situ hybridization (ISH) for the calcitonin receptor gene (Calcr) was performed throughout the whole forebrain of ovariectomized wild‐type female rats that had been implanted with a negative feedback level of estrogen, because the Calcr expression was evident in the ARC kisspeptin neurons from the NGS analysis. Then, a double ISH was performed for the Calcr and kisspeptin gene (Kiss1) in the brain regions, containing either the anteroventral periventricular nucleus (AVPV) or ARC of the female rats.
Results
The NGS analysis revealed that the Calcr was highly expressed in the ARC kisspeptin neurons. It was found that the Calcr was co‐expressed in 12% and 22% of the Kiss1‐expressing cells in the ARC and AVPV, respectively.
Conclusion
The present study suggests that calcitonin receptor signaling could be involved in the regulation of reproductive function through the direct control of the ARC and/or AVPV kisspeptin neurons, and then GnRH/gonadotropin release.
Keywords: anteroventral periventricular nucleus, arcuate nucleus, calcitonin receptors, in situ hybridization, kisspeptin‐1
1. INTRODUCTION
Increasing evidence suggests that kisspeptin–GPR54 (kisspeptin receptor) signaling plays a critical role in the regulation of reproductive functions via the direct stimulation of gonadotropin‐releasing hormone (GnRH) and then gonadotropin release in mammals, including rodents, ruminants, primates, pigs, and musk shrews.1, 2, 3, 4, 5, 6 Mutations of Kiss1, a gene encoding kisspeptin, or Gpr54 cause hypogonadotropic hypogonadism in female rodents and humans,1, 2, 7, 8, 9 suggesting that kisspeptin–GPR54 signaling is indispensable for puberty and normal reproductive function at adulthood in mammals. The expression of Kiss1 and kisspeptin has been found in two hypothalamic regions: the arcuate nucleus (ARC) and anteroventral periventricular nucleus (AVPV) in rodents.10, 11, 12 Both the ARC and AVPV kisspeptin neurons express estrogen receptor alpha in female rodents10, 12, 13 and estrogen negatively and positively regulates Kiss1 expression in the ARC and AVPV kisspeptin neurons, respectively.10, 12, 13 Thus, the ARC and AVPV kisspeptin neurons are considered to mediate estrogen negative and positive feedback effects on GnRH/gonadotropin release and therefore they are suggested to govern GnRH/gonadotropin pulses and surges, respectively10, 11, 13 Hypothalamic kisspeptin neurons are suggested to be hub neurons that receive inputs from other neurons to mediate the influence of the exogenous or endogenous environment on reproduction through controlling GnRH release. Indeed, leptin receptors are expressed in the ARC kisspeptin neurons and these receptors are suggested to be involved in the facilitation of the ARC kisspeptin neurons under a high available nutritional status14 because the secretion of leptin, an anorexigenic hormone from the adipose tissue, is stimulated by food intake to exert several physiological roles, such as the inhibition of feeding behavior and the facilitation of gonadotropin release.14, 15 Furthermore, the authors’ previous study showed that glutamatergic agonists, which are potent stimulators for gonadotropin release, failed to stimulate luteinizing hormone (LH) release in Kiss1 KO rats,1 suggesting that kisspeptin neurons mediate the glutamatergic stimulation of gonadotropin release.
The present study aimed to investigate the upstream regulator candidates for the kisspeptin neurons located in the ARC. To this end, the receptors that are exclusively expressed in kisspeptin neurons were analyzed by using next‐generation sequencing (NGS) on isolated visualized kisspeptin neurons that had been taken from the ARC of Kiss1‐tdTomato heterozygous rats. In order to investigate the ligand‐receptor signaling candidates that affect kisspeptin neurons, and consequently tonic GnRH/gonadotropin release, the focus was on the G‐protein‐coupled receptors (GPCRs) that were highly expressed in the ARC kisspeptin neurons. The NGS analysis revealed that considerable levels of calcitonin receptor (CALCR) messenger (m)RNA were expressed in the ARC kisspeptin neurons in the Kiss1‐tdTomato female rats. Thus, the localization of Calcr in rat forebrains was investigated and it was examined if Calcr is co‐expressed in the ARC, as well as in the AVPV kisspeptin neurons, in female rats by using double in situ hybridization (ISH).
2. MATERIALS AND METHODS
2.1. Animals
Adult female Wistar–Imamichi rats at 8‐9 weeks of age (206‐225 g body weight [BW]) and genetically modified rats carrying the tandem dimer Tomato (tdTomato; Clontech, Mountain View, CA, USA) in the Kiss1 locus (Kiss1–tdTomato) at 12 weeks of age (244‐288 g BW)1 were maintained under a controlled environment (14 hours light and 10 hours darkness; lights on at 05:00 hours; 23 ± 3°C) with free access to food (CE2; Clea, Tokyo, Japan) and water. The vaginal smears were checked daily to determine their estrous cyclicity and females with at least two consecutive estrous cycles were used. The Kiss1‐tdTomato heterozygous rats that had been ovariectomized (OVX) 2 weeks prior were used for the NGS analysis of the ARC kisspeptin neurons. The OVX Kiss1‐tdTomato rats without estradiol‐17β (E2) treatment were chosen because E2 treatment suppresses Kiss1 and kisspeptin, as well as tdTomato expression in the ARC. The Wistar–Imamichi rats were OVX and then immediately received subcutaneous Silastic implants (internal diameter: 1.57 mm; outer diameter: 3.18 mm; 25 mm in length; Dow Corning, Midland, MI, USA) that were filled with E2 (20 μg/mL in peanut oil) for 1 week to serve as the OVX + low E2 rats. The low E2 treatment was previously confirmed to produce a plasma E2 level of 35.8 pg/mL and to produce a negative feedback effect on LH pulses, but not to induce LH surges.16 The OVX + low E2 rats were chosen so as to analyze the co‐expression of the Calcr and Kiss1 in both the AVPV and ARC because the OVX + low E2 rat model was suitable to detect Kiss1 in these nuclei. All the operations were performed under ketamine (27 mg/kg) and xylazine (5.3 mg/kg) anesthesia and aseptic conditions. All the rats were injected with antibiotics (Mycillin Sol; Meiji Seika, Tokyo, Japan) after surgery. One week after the OVX and E2 treatment, the animals were deeply anesthetized with sodium pentobarbital (40 mg/kg, Kyoritsu Seiyaku Co., Tokyo, Japan) and perfused with 0.05 mol L–1 phosphate‐buffered saline (PBS), followed by 4% paraformaldehyde in 0.05 mol L–1 PB. The brain was immediately removed from the skull, postfixed with the same solution for 6‐7 hours at 4°C, and then kept in 30% sucrose in 0.05 mol L‐1 PB for 3‐4 days at 4°C under ribonuclease (RNase)‐free conditions. Serial coronal sections (50 μm in thickness) of the forebrain that contained the hypothalamus and the mid‐brain (from 0.0 mm to 4.2 mm posterior to the bregma) were obtained by using a cryostat on the day before ISH and then were stored at 4°C in PBS.
2.2. Expression analysis of the G‐protein‐coupled receptors in isolated tdTomato‐positive cells taken from Kiss1‐tdTomato rats
The ARC tissues were dissected out from the OVX Kiss1‐tdTomato heterozygous rats. Acutely dissociated cells were prepared, as described previously,17 with slight modifications. Briefly, the tissues were treated with 10 U/mL papain (Worthington, Lakewood, NJ, USA) for 50 minutes at 30°C in Dulbecco's modified Eagle medium (Life Technologies, Carlsbad, CA, USA), gently dispersed by repetitive pipetting, then filtered through a 70‐μm pore size nylon mesh (BD Biosciences, San Jose, CA, USA). Cell suspensions were applied to a discontinuous Percoll (Sigma‐Aldrich, St. Louis, MO, USA) density gradient that was composed of 0.28 and 0.75 g/mL layers, and then centrifuged. The cells were obtained from the middle layer. The cell suspension was placed onto plastic dishes. The tdTomato‐positive cells were collected under a fluorescent microscope by pipettes (inner diameter: 20‐30 μm) that were filled with 150 mmol L–1 KCl, 3 mmol L–1 MgCl2, 5 mmol L–1 EGTA, and 10 mmol L–1 HEPES solution. Ten positive cells were pooled into a polymerase chain reaction (PCR) tube containing a RNase inhibitor (RNasin Plus; Promega, Madison, WI, USA).
The complementary (c)DNAs were synthesized with a SMARTer Pico PCR cDNA Synthesis kit (Clontech) and amplified by universal primers in accordance with the manufacturer's instructions. The first‐round PCR was performed at 94°C for one minute, then 21 cycles of 94°C for 30 seconds, 65°C for 30 seconds, 72°C for six minutes, and finally 72°C for 10 minutes. A 100‐μL aliquot of PCR mixture contained 61 μL of the template cDNA, 2 μL of 12 μmol L–1 cDNA synthesis primer (Takara Bio, Kusatsu, Japan), 10 μL of 25 mmol L–1 MgCl2, 16 μL of 2.5 mmol L–1 dNTP mix, and 1 μL of Takara LA‐taq DNA polymerase (Takara Bio). The products of the first‐round PCR were fragmented and used to generate a library for the RNA‐seq. Barcoded SOLiD 3’ primers from the SOLiD RNA Barcoding Kit (Life Technologies) were used to distinguish between the samples. The library was sequenced by multiplex paired‐end sequencing on a flow cell using the 5500xl SOLiD system (Life Technologies). Mapping of the paired‐end reads to the rat reference genome (RGSC 3.4 assembly) was performed. The mRNA expression levels were normalized by calculating the reads per kilo base per million mapped reads (RPKM) for each mRNA. The mRNAs coding for GPCR were extracted from the RNA‐seq data by using the rat GPCR dataset that had been obtained from the SEVENS GPCR database (http://sevens.cbrc.jp).
2.3. In situ hybridization for Calcr
In order to detect Calcr mRNA, a Calcr‐specific digoxigenin (DIG)‐labeled probe (position 293‐1303; GenBank accession no. NM_053816) was synthesized by using a DIG‐labeling kit (Roche Diagnostics Corporation, Indianapolis, IN, USA). The sections were washed with PBS and treated with 1 μg/mL protease K for 15 minutes at 37°C. Then, they were incubated with 0.25% acetic anhydride in 0.1 mol L–1 triethanolamine for 10 minutes. Finally, the sections were hybridized overnight at 60°C with 1 μg/μL DIG‐labeled anti‐sense cRNA probes. The specificity of the cRNA probe was confirmed by performing ISH by using anti‐sense and sense probes. No signal was detected in the brain sections that had been treated with a sense probe. After hybridization, the sections were washed with 2 × saline‐sodium citrate (SSC) containing 50% formamide for 15 minutes at 60°C twice. The sections then were treated with 20 μg/mL RNase A for 30 minutes at 37°C and then immersed sequentially in 2 × SSC, 0.5 × SSC, and DIG‐1 buffer (100 mmol L–1 Tris‐HCl [pH = 7.5], 150 mmol L–1 NaCl, and 0.01% Tween20) for 15 minutes, twice each. Following this, the sections were immersed in 1.5% blocking reagent (Roche Diagnostics Corporation) in DIG‐1 buffer for one hour at room temperature and incubated with an alkaline phosphatase‐conjugated anti‐DIG antibody (1:1000; Roche Diagnostics Corporation) overnight at 4°C. Then, the sections were washed with DIG‐1 buffer and treated with DIG‐3 buffer (100 mmol L–1 Tris‐HCl [pH = 9.5], 100 mmol L–1 NaCl, and 50 mmol L–1 MgCl2). Following this, the sections were treated with a chromagen solution (337 μg/mL 4‐nitroblue tetrazolium chloride and 175 μg/mL 5‐bromo‐4‐cloro‐3‐indolyl‐phosphate in DIG‐3 buffer) at 37°C until a visible signal was detected. The reaction was stopped by adding a reaction stop solution (10 mmol L–1 Tris‐HCl [pH = 7.6] and 1 mmol L–1 EDTA [pH = 8.0]), then washed two times with 0.05 mol L‐1 PBS. After processing, the sections were mounted and examined by light microscopy (BX53; Olympus, Tokyo, Japan).
2.4. Double in situ hybridization for Calcr and Kiss1
In order to detect the Calcr and Kiss1 mRNA, the Calcr‐specific DIG‐labeled cRNA probe and the Kiss1‐specific fluorescein isothiocyanate (FITC)‐labeled cRNA probe (position 33‐348; GenBank accession no. AY196983) that had been synthesized from the rat hypothalamic cDNA by using a FITC‐labeling kit (Roche Diagnostics) were used. Briefly, the sections were washed with PBS, treated with 1 μg/mL protease K for 15 minutes at 37°C, and then incubated with 0.25% acetic anhydride in 0.1 mol L–1 triethanolamine for 10 minutes. Finally, the sections were hybridized overnight at 60°C with 1 μg/μL DIG‐labeled Calcr anti‐sense cRNA probes and 1 μg/μL FITC‐labeled Kiss1 anti‐sense cRNA probes. After hybridization, the sections were washed twice with 2 × SSC containing 50% formamide for 15 minutes at 60°C. The sections then were treated with 20 μg/mL RNase A for 30 minutes at 37°C and immersed sequentially with 2 × SSC, 0.5 × SSC, and 1% H2O2 in 0.05 mol/L PBS for 30 minutes at 30°C. Next, the sections were washed once with 0.05 mol L–1 PBS and DIG‐1 buffer (100 mmol L–1 Tris‐HCl [pH = 7.5], 150 mmol L–1 NaCl, and 0.01% Tween‐20) for five minutes. Subsequently, the sections were immersed with 1.5% blocking reagent (Roche Diagnostics) in the DIG‐1 buffer for one hour at room temperature and incubated with a peroxidase (POD)‐conjugated anti‐FITC antibody in 1.5% blocking reagent (1:1000) for two hours at room temperature. Then, the sections were washed with DIG‐1 buffer for five minutes two times and treated with tyramide signal amplification (TSA) plus FITC (1:100; PerkinElmer, Inc., Shelton, CT, USA) for 30 minutes at room temperature. The sections were observed under a fluorescence microscope in a dark room for the Kiss1 mRNA visible signal to be detected. The reaction was stopped by washing two times with DIG‐1 buffer for five minutes at room temperature, then treated with 0.1 N HCl for 15 minutes at room temperature. The sections were washed two times with DIG‐1 buffer for five minutes at room temperature and incubated with a POD‐conjugated anti‐DIG antibody in 1.5% blocking reagent (1:1000) overnight at 4°C, then washed two times with DIG‐1 buffer at room temperature and treated with TSA plus Biotin (1:100; PerkinElmer) for 30 minutes at room temperature, followed by being washed two times with DIG‐1 buffer at room temperature. Subsequently, the sections were treated with Dylight 594‐conjugated streptavidin (Thermo Fisher Scientific, Waltham, MA, USA) in DIG‐1 buffer (1:500) for 20 minutes at room temperature. All the sections were mounted to gelatin‐coated slides and the photomicrographs were taken under a fluorescence microscope by using ApoTome (Carl Zeiss, Oberkochen, Germany), where the number of Kiss1‐expressing cells and Calcr‐expressing cells were counted. The visualized cells throughout the AVPV (five‐to‐seven sections divided into two parts, from the 0.0‐0.6 mm posterior to the bregma according to a rat brain atlas18) and ARC (10‐12 sections divided into three parts, from the 1.8‐4.2 mm posterior) were counted bilaterally twice by one of the authors under a microscope and the average was calculated. No positive signal for Calcr or Kiss1 mRNA was detected in the brain sections that had been hybridized with the corresponding sense probes.
3. RESULTS
3.1. Discovery of Calcr expression in the arcuate kisspeptin neurons by a next‐generation sequencing analysis
The RNA‐seq of kisspeptin neurons that had been derived from the ARC of the OVX Kiss1‐tdTomato heterozygous female rats (total reads: 66,840,006; mean read length: 53 bp; aligned base rate: 88%) showed that about 8200 genes possibly were expressed in the ARC kisspeptin neurons, in terms of the RPKM values. The list included well‐known genes, such as Kiss1 (2252 RPKM), Tac3 (encoding neurokinin B, 4384 RPKM), and Pdyn (encoding dynorphin, 443 RPKM). In addition, several mRNAs that encode GPCR, including Tacr3 ( encoding neurokinin 3 receptor, 199 RPKM) and Calcr (83 RPKM), were detected. The RPKM value of Calcr ranked second among the GPCR mRNAs that were expressed in the ARC Kiss1‐tdTomato cells. No Gpr54, Oprk1 (encoding dynorphin receptor), and Gnrhr (encoding GnRH receptor) expression was detected in the ARC kisspeptin neurons in the rats.
3.2. Distribution of the Calcr‐expressing cells in the forebrain of the female rat
Figure 1 indicates a schematic illustration that shows the sites of Calcr mRNA expression throughout the forebrain of the OVX + low E2 rats. Calcr‐expressing cells were found in the AVPV, ARC, medial preoptic area (mPOA), and dorsomedial hypothalamus (DMH). More specifically, a large number of Calcr‐expressing cells were found in the whole ARC and Calcr‐expressing cells were located abundantly in the central‐to‐caudal parts of the ARC. In addition, a number of Calcr‐expressing cells were located in the region close to the cerebral ventricles, such as the lateral and third cerebroventricles.
Figure 1.

Schematic illustration showing the distribution of Calcr messenger RNA expression in the forebrain of ovariectomized + low estradiol rats, according to the rat brain map.18 The Calcr‐expressing cell bodies are indicated by red dots (one dot corresponding to 25 cells). 3V, third ventricle; aca, anterior commissure; ARC, arcuate nucleus; AVPV, anteroventral periventricular nucleus; DMH, dorsomedial hypothalamus; LS, lateral septum; MM, mammillary nucleus; mPOA, medial preoptic area; och, optic chiasm; PVN, paraventricular nucleus; SCh, suprachiasmatic nucleus; SFO, subfornical organ; VMH, ventromedial hypothalamic nucleus
Figure 2 shows the Calcr mRNA expression that was detected by ISH in the forebrain of the representative OVX + low E2 rats. A number of Calcr‐expressing cells was found in the AVPV (Figure 2A), mPOA (Figure 2B), DMH (Figure 2C) and anterior (Figure 2D), middle (Figure 2E), and posterior of the ARC (Figure 2F).
Figure 2.
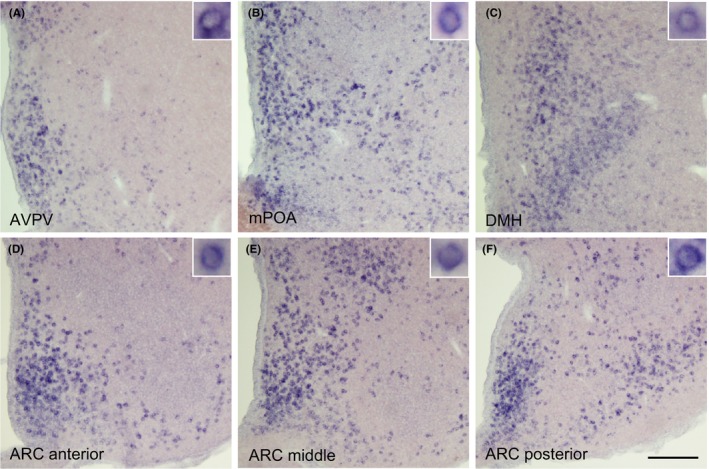
Calcr‐expressing cells that were detected by in situ hybridization in the hypothalamus of the representative ovariectomized + low estradiol rats. The Calcr‐expressing cells were significantly found in the (A) anteroventral periventricular nucleus (AVPV), (B) medial preoptic area (mPOA), (C) dorsomedial hypothalamus (DMH), and (D) anterior, (E) middle, and (F) posterior parts of the arcuate nucleus (ARC). The insets indicate Calcr‐expressing cells at a higher magnification. Scale bar: 200 μm
3.3. Colocalization of Calcr and Kiss1 expression in the arcuate nucleus and anteroventral periventricular nucleus in the female rat
Figure 3 shows the colocalization of Calcr and Kiss1 mRNA expression in the ARC and AVPV of the representative OVX + low E2 rats. Calcr expression was found in some Kiss1‐expressing cells in the ARC (Figure 3A‐C). Calcr expression also was detected in a part of the Kiss1‐expressing cells in the AVPV (Figure 3D‐F). Figure 4 shows the quantitative analysis of the Kiss1‐ and Calcr‐expressing cells in the ARC and AVPV in the OVX + low E2 rats (n = 4; note that the y‐axis is expressed as a logarithmic scale). It was found that some Kiss1‐expressing cells that were located in the rostral‐to‐caudal parts of the ARC and AVPV co‐expressed Calcr; similarly, a part of the Calcr‐expressing cells co‐expressed Kiss1. In total, 12% of the ARC Kiss1‐expressing cells showed Calcr co‐expression, while 35% of the Calcr‐expressing cells in the ARC showed co‐expression of Kiss1 (Figure 4A,B); 22% of the AVPV Kiss1‐expressing cells showed Calcr mRNA expression, while 12% of the AVPV Calcr‐expressing cells showed co‐expression of Kiss1.
Figure 3.
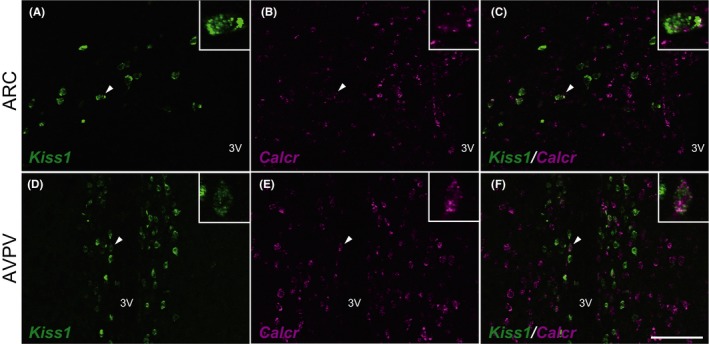
Kiss1 (green)‐ and Calcr (magenta)‐expressing cells in the arcuate nucleus (ARC) (A‐C) or anteroventral periventricular nucleus (AVPV) (D‐F) in the representative ovariectomized + low estradiol rats. The insets indicate the Kiss1‐ and Calcr‐co‐expressing cells (arrowheads) at a higher magnification. Scale bar: 100 μm. 3V, third ventricle
Figure 4.
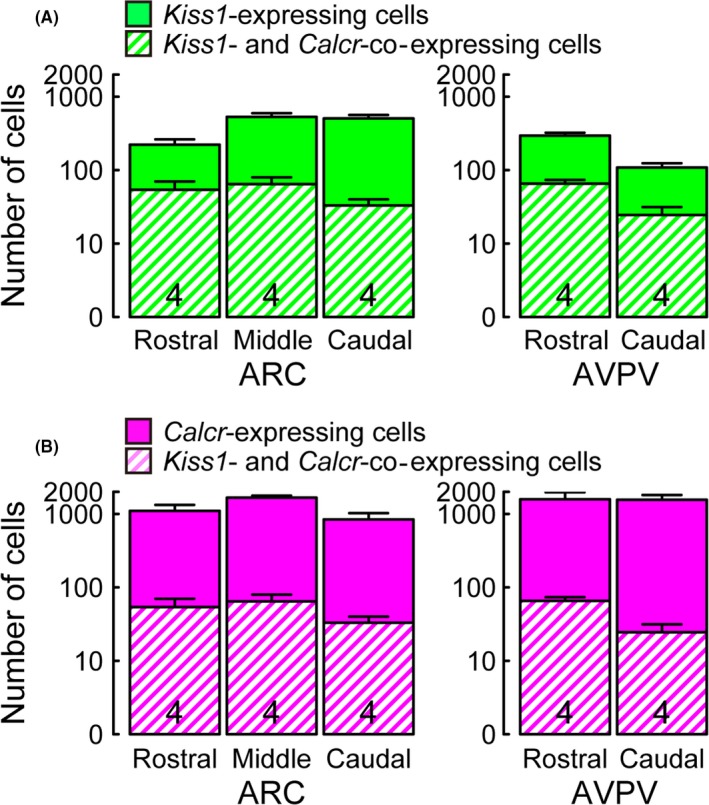
Numbers of Kiss1‐ and Calcr‐expressing cells in the ARC or AVPV. The number of Kiss1‐expressing cells with or without Calcr expression in the ARC or AVPV in OVX+low E2 rats (A). The number of Calcr‐expressing cells with or without Kiss1 expression in the ARC or AVPV in OVX + low E2 rats (B). Values are mean ± SEM and are represented in a logarithmic scale on the Y‐axis. Numbers in each column indicate the number of animals used. The ARC was divided into three parts (from 1.8 to 2.6 mm, from 2.6 mm to 3.4 mm and from 3.4 to 4.2 mm posterior to the bregma) while the AVPV was divided into two parts (from 0.0 to 0.3 mm and from 0.3 to 0.6 mm posterior to the bregma)
4. DISCUSSION
In the present study, the NGS analysis revealed that considerable levels of Calcr are expressed in the ARC kisspeptin neurons. The further histological analysis with double ISH for Calcr and Kiss1 revealed that Calcr was co‐expressed in a part of the ARC Kiss1‐expressing cells, as well as the AVPV Kiss1‐expressing cells. Kisspeptin neurons have been well accepted to be a master regulator for reproduction via stimulation of GnRH and the consequent gonadotropin release in mammals.1, 2, 3, 4, 5, 6 The ARC kisspeptin neurons are considered to be involved in GnRH/gonadotropin pulse generation, which is involved in folliculogenesis, spermatogenesis, and steroidogenesis, while AVPV kisspeptin neurons are considered to be responsible for GnRH/LH surge generation, which in turn regulates ovulation. Thus, CALCR signaling could have a role in affecting kisspeptin neurons in the ARC and/or AVPV to regulate GnRH/gonadotropin pulses and/or surges, respectively. In addition to the AVPV and ARC, Calcr was observed in several regions of the hypothalamus, such as the mPOA, DMH, and regions close to the cerebral ventricles, such as the lateral and third cerobroventricles. The results are largely consistent with previous reports in rats using ISH and immunohistochemistry to detect Calcr and CALCR.19, 20
The CALCR is a member of a subfamily of the seven‐transmembrane domain GPCR superfamily. Calcitonin, an endogenous ligand for CALCR21 that is secreted from parafollicular cells of the thyroid gland, is known to be involved in regulating peripheral calcium metabolism during pregnancy and lactation in mammals22, 23, 24 to protect the maternal skeleton by direct action on osteocytes to inhibit osteolysis.24, 25, 26 Interestingly, previous studies demonstrated that a peripheral administration of calcitonin decreased the plasma LH and testosterone levels in rats.27, 28 Furthermore, a central injection of a calcitonin family peptide hormone, such as calcitonin gene‐related peptide, suppressed LH pulses in female rats.29 It is reported that LH release is suppressed during lactation in rats and cows.30, 31, 32, 33 These results suggest that calcitonin–CALCR signaling in hypothalamic kisspeptin neurons could be involved in the suppression of LH release in lactating animals, in which calcium availability for milk synthesis is essential,24 and plasma calcitonin levels increase in humans and rats.22, 26, 34 Thus, it is speculated that calcium availability could affect the reproductive function in mammals via the effect of calcitonin–CALCR signaling on the ARC and/or AVPV kisspeptin neurons.
Amylin has been demonstrated as another endogenous ligand for CALCR and it was originally found as a product in the pancreatic B cells. It is coreleased with insulin under hyperglycemic conditions or after feeding.35 Amylin expression is also evident in the brain in rats.36 Amylin is known to be capable of binding to CALCR, when it forms a complex with receptor activity‐modifying proteins.37, 38 Thus, amylin that was derived in the peripheral organs and/or brain could be involved in regulating GnRH/LH release via CALCR that is expressed in the hypothalamic kisspeptin neurons. It is demonstrated that a central injection of amylin inhibits sexual behavior in male rats.39 Interestingly, amylin mRNA levels increased in the mPOA in rats at lactation,40 during which LH release was profoundly suppressed. Taken together with the present study, these results suggest that calcitonin or amylin could exert an inhibitory role in GnRH/gonadotropin release via the activation of CALCR in the ARC and AVPV kisspeptin neurons.
The present study showed that a large population of the ARC Kiss1‐expressing cells did not express Calcr. The ARC Kiss1‐expressing cells without Calcr could be affected indirectly via the neighboring Kiss1‐expressing cells because the authors’ previous study showed that kisspeptin neurons in the ARC interact with each other via gap junctional connections in mice.41 A number of Calcr‐expressing cells without Kiss1 expression in the ARC also was found. Those cells might be dopaminergic cells because the central and peripheral administration of calcitonin decreased the plasma prolactin levels in the lactating rats.42 Thus, it is speculated that calcitonin and/or amylin could affect the tubuloinfundibular dopaminergic (TIDA) neurons, which in turn, might contribute to controlling prolactin release from the anterior pituitary during lactation. Interestingly, GPR54 expression was evident in the TIDA neurons in rats,43 suggesting that ARC kisspeptin and TIDA neurons could interact with each other through kisspeptin and CALCR signaling. Furthermore, the Calcr‐expressing cells in the brain regions close to the cerebral ventricles might have a role as an interface of peripheral and central interaction. Indeed, the circulating calcitonin is capable of binding to the CALCR around the ventricles.44 Further studies are required to clarify the role of the CALCR around the ventricles.
In conclusion, the present study suggests that the ARC and AVPV kisspeptin neurons could be direct action sites of calcitonin and/or amylin and that the CALCR signaling might have a role in regulating GnRH/gonadotropin release because Calcr expression was evident in some of the kisspeptin neurons in these nuclei. Further studies are required to clarify if the CALCR signaling exerts inhibitory regulation of GnRH/gonadotropin release.
DISCLOSURES
Conflict of interest: The authors declare no conflict of interest. Human rights statement and informed consent: This article does not contain any study with human participants that was performed by any of the authors. Animal studies: All the institutional and national guidelines for the care and use of laboratory animals were followed. All the experiments were approved and conducted in accordance with the guidelines of the Committee of Animal Experiments of the Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan.
ACKNOWLEDGEMENTS
We are grateful to Drs. Chengzhu Yin, Masakatsu Kato, and Yasuo Sakuma (Nippon Medical School) for their technical support with the fluorescent‐cell collection. We thank Makiko Suwa for her technical advice on the usage of the SEVENS GPCR database that was produced in the Computational Biology Research Center at the National Institute of Advanced Industrial Science and Technology, Tokyo, Japan. We thank Dr. Nicola Skoulding for her English edition.
Assadullah, Ieda N, Kawai N, et al. Co‐expression of the calcitonin receptor gene in the hypothalamic kisspeptin neurons in female rats. Reprod Med Biol. 2018;17:164–172. https://doi.org/10.1002/rmb2.12085
Funding information
The present study was supported in part by a grant of the Project for the Promotion and Enhancement of the Afghan Capacity for Effective Development (PEACE) of the Japan International Cooperation Agency (JICA) and Japan Society for the Promotion of Science (JSPS) KAKENHI Grant nos 16K18783 to NIe, 16K07987 to NIn, and 26252046 to HT.
REFERENCES
- 1. Uenoyama Y, Nakamura S, Hayakawa Y, et al. Lack of pulse and surge modes and glutamatergic stimulation of luteinising hormone release in kiss1 knockout rats. J Neuroendocrinol. 2015;27:187‐197. [DOI] [PubMed] [Google Scholar]
- 2. d'Anglemont de Tassigny X, Fagg LA, Dixon JP, et al. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci USA. 2007;104:10714‐10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Okamura H, Tsukamura H, Ohkura S, et al. Kisspeptin and GnRH pulse generation. Adv Exp Med Biol. 2013;784:297‐323. [DOI] [PubMed] [Google Scholar]
- 4. Topaloglu AK, Tello JA, Kotan LD, et al. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med. 2012;366:629‐635. [DOI] [PubMed] [Google Scholar]
- 5. Tomikawa J, Homma T, Tajima S, et al. Molecular characterization and estrogen regulation of hypothalamic KISS1 gene in the pig. Biol Reprod. 2010;82:313‐319. [DOI] [PubMed] [Google Scholar]
- 6. Inoue N, Sasagawa K, Ikai K, et al. Kisspeptin neurons mediate reflex ovulation in the musk shrew (Suncus murinus). Proc Natl Acad Sci USA. 2011;108:17527‐17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seminara SB, Messager S, Chatzidaki EE, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614‐1627. [DOI] [PubMed] [Google Scholar]
- 8. de Roux N, Genin E, Carel JC, et al. Hypogonadotropic hypogonadism due to loss of function of the KiSS1‐derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972‐10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lapatto R, Pallais JC, Zhang D, et al. Kiss1‐/‐ mice exhibit more variable hypogonadism than Gpr54‐/‐ mice. Endocrinology. 2007;148:4927‐4936. [DOI] [PubMed] [Google Scholar]
- 10. Adachi S, Yamada S, Takatsu Y, et al. Involvement of anteroventral periventricular metastin/kisspeptin neurons in estrogen positive feedback action on luteinizing hormone release in female rats. J Reprod Dev. 2007;53:367‐378. [DOI] [PubMed] [Google Scholar]
- 11. Gottsch ML, Cunningham MJ, Smith JT, et al. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073‐4077. [DOI] [PubMed] [Google Scholar]
- 12. Smith JT, Cunningham MJ, Rissman EF, et al. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005;146:3686‐3692. [DOI] [PubMed] [Google Scholar]
- 13. Kinoshita M, Tsukamura H, Adachi S, et al. Involvement of central metastin in the regulation of preovulatory luteinizing hormone surge and estrous cyclicity in female rats. Endocrinology. 2005;146:4431‐4436. [DOI] [PubMed] [Google Scholar]
- 14. Quennell JH, Howell CS, Roa J, et al. Leptin deficiency and diet‐induced obesity reduce hypothalamic kisspeptin expression in mice. Endocrinology. 2011;152:1541‐1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagatani S, Guthikonda P, Thompson RC, et al. Evidence for GnRH regulation by leptin: leptin administration prevents reduced pulsatile LH secretion during fasting. Neuroendocrinology. 1998;67:370‐376. [DOI] [PubMed] [Google Scholar]
- 16. Cagampang FR, Maeda KI, Tsukamura H, et al. Involvement of ovarian steroids and endogenous opioids in the fasting‐induced suppression of pulsatile LH release in ovariectomized rats. J Endocrinol. 1991;129:321‐328. [DOI] [PubMed] [Google Scholar]
- 17. Tanaka N, Ishii H, Yin C, et al. Voltage‐gated Ca2 + channel mRNAs and T‐type Ca2 + currents in rat gonadotropin‐releasing hormone neurons. J Physiol Sci. 2010;60:195‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates, 6th edn San Diego, CA: Academic Press; 2007. [Google Scholar]
- 19. Sheward WJ, Lutz EM, Harmar AJ. The expression of the calcitonin receptor gene in the brain and pituitary gland of the rat. Neurosci Lett. 1994;181:31‐34. [DOI] [PubMed] [Google Scholar]
- 20. Becskei C, Riediger T, Zund D, et al. Immunohistochemical mapping of calcitonin receptors in the adult rat brain. Brain Res. 2004;1030:221‐233. [DOI] [PubMed] [Google Scholar]
- 21. Sexton PM, Houssami S, Brady CL, et al. Amylin is an agonist of the renal porcine calcitonin receptor. Endocrinology. 1994;134:2103‐2107. [DOI] [PubMed] [Google Scholar]
- 22. Toverud SU, Cooper CW, Munson PL. Calcium metabolism during lactation: elevated blood levels of calcitonin. Endocrinology. 1978;103:472‐479. [DOI] [PubMed] [Google Scholar]
- 23. Woodrow JP, Sharpe CJ, Fudge NJ, et al. Calcitonin plays a critical role in regulating skeletal mineral metabolism during lactation. Endocrinology. 2006;147:4010‐4021. [DOI] [PubMed] [Google Scholar]
- 24. Kovacs CS, Kronenberg HM. Maternal–fetal calcium and bone metabolism during pregnancy, puerperium, and lactation. Endocr Rev. 1997;18:832‐872. [DOI] [PubMed] [Google Scholar]
- 25. Clarke MV, Russell PK, Findlay DM, et al. A role for the calcitonin receptor to limit bone loss during lactation in female mice by inhibiting osteocytic osteolysis. Endocrinology. 2015;156:3203‐3214. [DOI] [PubMed] [Google Scholar]
- 26. Silva OL, Titus‐Dillon P, Becker KL, et al. Increased serum calcitonin in pregnancy. J Natl Med Assoc. 1981;73:649‐652. [PMC free article] [PubMed] [Google Scholar]
- 27. Tsai SC, Lu CC, Chen JJ, et al. Inhibition of salmon calcitonin on secretion of progesterone and GnRH‐stimulated pituitary luteinizing hormone. Am J Physiol. 1999;277:E49‐E55. [DOI] [PubMed] [Google Scholar]
- 28. Wang PS, Tsai SC, Hwang GS, et al. Calcitonin inhibits testosterone and luteinizing hormone secretion through a mechanism involving an increase in cAMP production in rats. J Bone Miner Res. 1994;9:1583‐1590. [DOI] [PubMed] [Google Scholar]
- 29. Bowe JE, Li XF, Kinsey‐Jones JS, et al. Calcitonin gene‐related peptide‐induced suppression of luteinizing hormone pulses in the rat: the role of endogenous opioid peptides. J Physiol. 2005;566:921‐928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsukamura H, Maeda K. Non‐metabolic and metabolic factors causing lactational anestrus: rat models uncovering the neuroendocrine mechanism underlying the suckling‐induced changes in the mother. Prog Brain Res. 2001;133:187‐205. [DOI] [PubMed] [Google Scholar]
- 31. Yamada S, Uenoyama Y, Deura C, et al. Oestrogen‐dependent suppression of pulsatile luteinising hormone secretion and kiss1 mRNA expression in the arcuate nucleus during late lactation in rats. J Neuroendocrinol. 2012;24:1234‐1242. [DOI] [PubMed] [Google Scholar]
- 32. Ohkura S, Uenoyama Y, Yamada S, et al. Physiological role of metastin/kisspeptin in regulating gonadotropin‐releasing hormone (GnRH) secretion in female rats. Peptides. 2009;30:49‐56. [DOI] [PubMed] [Google Scholar]
- 33. Carruthers TD, Hafs HD. Suckling and four‐times daily milking: influence on ovulation, estrus and serum luteinizing hormone, glucocorticoids and prolactin in postpartum holsteins. J Anim Sci. 1980;50:919‐925. [DOI] [PubMed] [Google Scholar]
- 34. Stevenson JC, Hillyard CJ, MacIntyre I, et al. A physiological role for calcitonin: protection of the maternal skeleton. Lancet. 1979;2:769‐770. [DOI] [PubMed] [Google Scholar]
- 35. Moore CX, Cooper GJ. Co‐secretion of amylin and insulin from cultured islet beta‐cells: modulation by nutrient secretagogues, islet hormones and hypoglycemic agents. Biochem Biophys Res Commun. 1991;179:1‐9. [DOI] [PubMed] [Google Scholar]
- 36. Sexton PM, Paxinos G, Kenney MA, et al. In vitro autoradiographic localization of amylin binding sites in rat brain. Neuroscience. 1994;62:553‐567. [DOI] [PubMed] [Google Scholar]
- 37. Christopoulos G, Perry KJ, Morfis M, et al. Multiple amylin receptors arise from receptor activity‐modifying protein interaction with the calcitonin receptor gene product. Mol Pharmacol. 1999;56:235‐242. [DOI] [PubMed] [Google Scholar]
- 38. Lutz TA, Tschudy S, Rushing PA, et al. Amylin receptors mediate the anorectic action of salmon calcitonin (sCT). Peptides. 2000;21:233‐238. [DOI] [PubMed] [Google Scholar]
- 39. Clementi G, Busa L, de Bernardis E, et al. Effects of centrally injected amylin on sexual behavior of male rats. Peptides. 1999;20:379‐382. [DOI] [PubMed] [Google Scholar]
- 40. Dobolyi A. Central amylin expression and its induction in rat dams. J Neurochem. 2009;111:1490‐1500. [DOI] [PubMed] [Google Scholar]
- 41. Ikegami K, Minabe S, Ieda N, et al. Evidence of involvement of neurone‐glia/neurone–neurone communications via gap junctions in synchronised activity of KNDy neurones. J Neuroendocrinol. 2017;29. doi: /10.1111/jne.12480. [DOI] [PubMed] [Google Scholar]
- 42. Tohei A, VandeGarde B, Arbogast LA, et al. Calcitonin inhibition of prolactin secretion in lactating rats: mechanism of action. Neuroendocrinology. 2000;71:327‐332. [DOI] [PubMed] [Google Scholar]
- 43. Higo S, Iijima N, Ozawa H. Characterisation of Kiss1r (Gpr54)‐expressing neurones in the arcuate nucleus of the female rat hypothalamus. J Neuroendocrinol. 2017;29 https://doi.org/10.1111/jne.12452. [DOI] [PubMed] [Google Scholar]
- 44. van Houten M, Rizzo AJ, Goltzman D, et al. Brain receptors for blood‐borne calcitonin in rats: circumventricular localization and vasopressin‐resistant deficiency in hereditary diabetes insipidus. Endocrinology. 1982;111:1704‐1710. [DOI] [PubMed] [Google Scholar]


