ABSTRACT
The molecular mechanisms underlying resistance to radiotherapy in breast cancer cells remain elusive. Previously, we reported that elevated β1-integrin is associated with enhanced breast cancer cell survival postirradiation, but how β1-integrin conferred radioresistance was unclear. Ionizing radiation (IR) induced cell killing correlates with the efficiency of DNA double-strand break (DSB) repair, and we found that nonmalignant breast epithelial (S1) cells with low β1-integrin expression have a higher frequency of S-phase-specific IR-induced chromosomal aberrations than the derivative malignant breast (T4-2) cells with high β1-integrin expression. In addition, there was an increased frequency of IR-induced homologous recombination (HR) repairosome focus formation in T4-2 cells compared with that of S1 cells. Cellular levels of Rad51 in T4-2 cells, a critical factor in HR-mediated DSB repair, were significantly higher. Blocking or depleting β1-integrin activity in T4-2 cells reduced Rad51 levels, while ectopic expression of β1-integrin in S1 cells correspondingly increased Rad51 levels, suggesting that Rad51 is regulated by β1-integrin. The low level of Rad51 protein in S1 cells was found to be due to rapid degradation by the ubiquitin proteasome pathway (UPP). Furthermore, the E3 ubiquitin ligase RING1 was highly upregulated in S1 cells compared to T4-2 cells. Ectopic β1-integrin expression in S1 cells reduced RING1 levels and increased Rad51 accumulation. In contrast, β1-integrin depletion in T4-2 cells significantly increased RING1 protein levels and potentiated Rad51 ubiquitination. These data suggest for the first time that elevated levels of the extracellular matrix receptor β1-integrin can increase tumor cell radioresistance by decreasing Rad51 degradation through a RING1-mediated proteasomal pathway.
KEYWORDS: β1-integrin, homologous recombination, HR, Rad51, breast cancer, radioresistance
INTRODUCTION
Cell attachment to extracellular matrix (ECM) components triggers biochemically and mechanically induced signaling for the stimulation of growth factor receptors, ion channels, and cytoplasmic protein kinases, etc. Cell-matrix binding is mediated primarily by integrins, a family of 18 α and 8 β transmembrane glycoproteins that form 24 distinct heterodimeric receptors (1). Most integrins recognize several ECM proteins, and individual matrix proteins bind to several integrins. Activation of integrin-mediated intracellular pathways regulates signaling elements in proliferation, survival, apoptosis, invasion, metastasis, and tissue organization (2). Similar to other integrin receptors, β1-integrins are overexpressed in various cancers, including breast cancers, where they indicate a poor prognosis (3), and have been shown to affect mammary tumor induction and proliferation of metastatic mammary carcinoma cells disseminated in the lungs (4). Overexpression of β1-integrin has also been shown to mediate tumor cell resistance to chemo- and radiotherapy (5, 6). Since about 50% of all cancer patients are treated with radiotherapy, understanding the role of β1-integrin in DNA damage repair is crucial for the optimization and development of innovative drugs for improving radiotherapy response rates.
β1-Integrin inhibitors have been reported to sensitize cancer cells to conventional radio- and chemotherapies (7). Several investigators have reported that targeting of β1-integrin disrupts resistance to anti-epidermal growth factor receptor (anti-EGFR) (8) and antiangiogenic therapies (9). In addition to chemo- and radioresistance, β1-integrin targeting has demonstrated an inhibitory effect on metastasis of colon, breast, and refractory tumors (10). However, the underlying molecular mechanisms of β1-integrin-mediated resistance to radiotherapy remain largely unclear.
One key determinant of cellular radiosensitivity is DNA double-strand break (DSB) repair (11). Cells have evolved two main repair pathways, nonhomologous end joining (NHEJ) (12) and homologous recombination (HR) (13), to cope with DSB genotoxicity. Nonhomologous end joining, an intrinsically error-prone repair system that operates throughout the cell cycle, involves direct sealing of DSB ends. Homologous recombination, an error-free repair system, requires sister chromatids and therefore is restricted to S and G2 phases of the cell cycle. Homologous recombination starts with 5′-to-3′ resection of DNA ends that generates single-stranded DNA (ssDNA) ends. The ssDNA is rapidly coated by replication protein A (RPA), which is then replaced by RAD51 (14). RAD51 filaments are critical to promote DNA strand invasion and ensure HR. Although the HR process for DNA DSB repair has been examined in various contexts, regulation of Rad51 is not well understood.
Rad51, a key component of the HR pathway, is overexpressed in multiple tumor types, including breast cancer (15). Cells overexpressing Rad51 exhibit cell cycle disruption, resistance to apoptotic signals, and associated resistance to DNA-damaging agents (16, 17). It is thought that Rad51 overexpression may provide a mechanism to compensate for any deficiency in alternative DNA repair pathways and thereby help cancer cells to survive (18), and deletion of Rad51 results in mouse embryo and chicken cell lethality (19). Cell lines lacking Rad51 are defective in DNA repair, which promotes genomic instability and increases cancer risk (20). Depletion of nuclear Rad51 expression in breast cancers was significantly associated with an aggressive phenotype (21). In addition, a relationship between Rad51, activation of the HR repair pathway, and radio- and chemoresistance has been reported in breast and other carcinoma cell lines (20, 22). Thus, the elevated Rad51 levels in irradiated human tumor cells may lead not only to HR activation but also to radiation therapy resistance. However, the cooperative function of Rad51 with other key stress elements in the activation of HR and conferral of radioresistance remains to be elucidated.
A connection between β1-integrin and impaired DNA repair in head and neck cancer cells has been reported (23), but whether integrins participate directly in DNA repair regulation is currently unclear. Two groups have reported that β1-integrins activate PARP-1/ligase III α/XRCC1-dependent base excision repair of bleomycin-induced DNA damage (24, 25). However, there are no reported studies yet on the role of β1-integrin in HR-mediated repair of DNA DSBs. Using an isogenic cell line pair, nonmalignant S1 cells and the derivative malignant breast T4-2 cells, we show here that HR repair and cell survival are increased in β1-integrin-overexpressing cells as β1-integrin stabilizes Rad51 levels via inhibition of ubiquitination by RING1, resulting in protection from proteasome-mediated Rad51 degradation.
RESULTS
IR-induced genomic instability and 53BP1/RIF1 foci are increased in nonmalignant S1 cells compared to their derivative radioresistant malignant breast T4-2 cells.
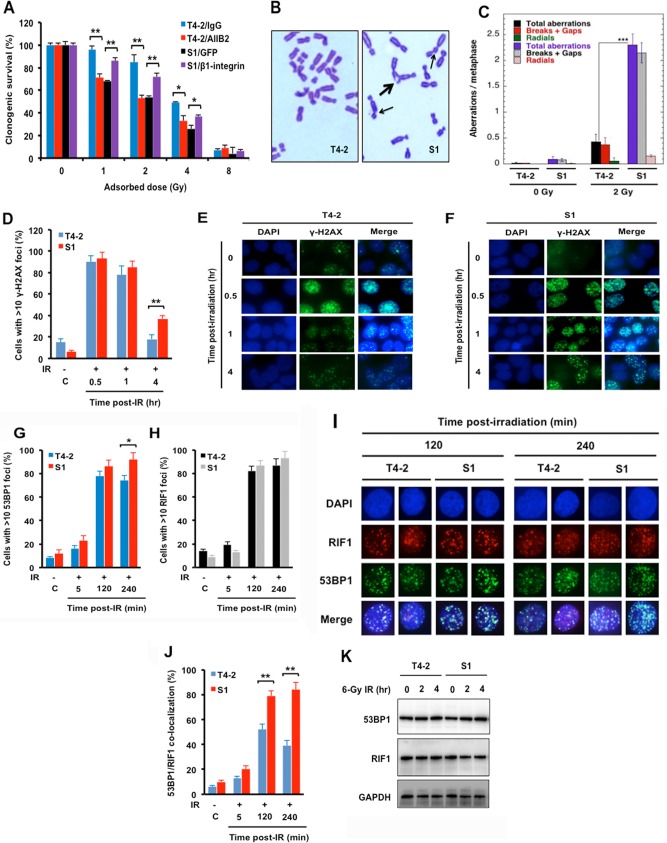
We have previously shown that malignant T4-2 cells had increased β1-integrin levels and were resistant to ionizing radiation (IR)-induced death in comparison to their parental counterpart, nonmalignant breast epithelial S1 cells (26). To verify that T4-2 cells indeed had increased radioresistance to ionizing radiation, compared to S1 cells, we preformed clonogenic survival assays (Fig. 1A). As expected, the T4-2 cells displayed greater radioresistance than did the S1 cells. In addition, inhibition of β1-integrin with AIIB2 monoclonal antibody increased cell killing in T4-2 cells, while overexpression of β1-integrin in S1 cells significantly decreased IR-induced cell killing (Fig. 1A), providing further evidence for β1-integrin as an important determinant of cellular radiosensitivity. To determine whether the increased radiosensitivity of S1 cells was due to defective DNA damage repair, we measured IR-induced chromosome aberrations in S1 and T4-2 cells. Cell cycle phase-specific chromosome aberrations after IR exposure revealed no significant differences in G1- or G2-phase-specific aberrations between S1 and T4-2 cells (data not shown); however, S-phase-specific aberrations were higher in S1 cells than in T4-2 cells. For the measurement of chromosome aberrations in the S phase of the cell cycle, cells were irradiated with 2 Gy and metaphases were collected 4 to 5 h post-IR. S1 cells showed higher frequencies of chromatid and chromosome aberrations at metaphase post-IR than did T4-2 cells (P < 0.001) (Fig. 1B and C), suggesting a defect in S phase, where HR is the predominant mode of DNA DSB repair. To determine whether the increase in IR-induced chromosomal aberrations in S1 cells was due to a defective DNA damage response, we examined whether IR-induced phosphorylation of H2AX at serine 139 is impaired in S1 cells. There was no significant difference between radioresistant T4-2 cells and S1 cells in their initial levels of IR-induced γ-H2AX foci; however, at 4 h post-IR, S1 cells had higher levels of residual γ-H2AX foci than T4-2 cells (Fig. 1D to F). This suggests that the initial production and detection of DNA damage in S1 cells are similar to those in T4-2 cells but that DNA repair is more efficient in T4-2 cells, a result of elevated β1-integrin levels, which reduces the formation of chromosome aberrations.
FIG 1.
Ionizing radiation (IR)-induced chromosome aberrations and 53BP1/RIF1 cofoci are increased in S1 cells compared to T4-2 cells after IR. (A) Inhibition of β1-integrin in T4-2 cells or ectopic expression of β1-integrin in S1 cells increased or decreased radiosensitivity, respectively. Malignant breast T4-2 cells, derived from the nonmalignant S1 breast epithelial cell line, were left untreated, T4-2 cells were treated with β1-integrin inhibitory antibody AIIB2 (0.1 μg/μl), or S1 cells were transiently transfected with expression vector for β1-integrin (pCMV6-Flag-β1-integrin) or control (pMax-GFP) before exposure to 1, 2, 4, or 8 Gy X rays. Clonogenic survival was measured 14 days after IR. Colonies consisting of more than 50 cells were scored as surviving colonies and normalized against nonirradiated clones. (B and C) Higher frequencies of chromosome aberrations at metaphase post-IR occurred in S1 cells than in T4-2 cells. Metaphase chromosome aberrations were determined in S phase of the cell cycle in cells exposed to 2 Gy X rays. (B) Thick arrow, breaks and gaps; thin arrows, radials. (C) Histogram of S-phase aberrations in T4-2 and S1 cells sham irradiated or exposed to 2 Gy of IR. (D to F) Delayed disappearance of γ-H2AX foci post-IR in S1 cells. Exponentially growing T4-2 and S1 cells were treated with 2 Gy X rays, fixed post-IR, and immunostained for γ-H2AX (histogram of >10 γ-H2AX foci). DAPI, 4′,6-diamidino-2-phenylindole. (G to J) Recruitment of IR-induced 53BP1/RIF1 foci is reduced in T4-2 cells but not in S1 cells. (G to I) Cells were treated with 6 Gy X rays, fixed post-IR, and immunostained for 53BP1 and RIF1. (I and J) Coimmunostaining for 53BP1 and RIF1 was done for fixed cells post-IR. 53BP1/RIF1 foci were counted for 3 sets of 30 cells, and the percentage of colocalized 53BP1/RIF1 foci was calculated relative to the total number of foci, i.e., 53BP1 plus RIF1 foci. (K) Western analysis of 53BP1 and RIF1 in whole-cell lysates prepared from T4-2 and S1 cells sham irradiated or exposed to 6 Gy X rays (GAPDH as a loading control). (A, C, D, G, H, and J) Columns represent the means (n = 3), and bars represent the SDs; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The chromosomal aberration studies suggested impaired S-phase-specific DNA repair in S1 cells. To determine the specific DNA repair pathway usage, several protein components of IR-induced repairosome foci were examined. The p53-binding protein 1 (53BP1) is a key determinant of DSB repair pathway choice (27) that acts as a molecular scaffold for additional DSB-responsive proteins, including RAP1-interacting factor 1 (RIF1), at DNA damage sites. The formation of 53BP1/RIF1 complexes at DSBs blocks the recruitment of DNA resection proteins associated with HR pathway repair and enhances DSB repair by NHEJ (28). To measure 53BP1/RIF1 focus formation at DSBs, T4-2 and S1 cells were exposed to 6 Gy of IR and 53BP1 and RIF1 foci were detected by immunofluorescence at 2 and 4 h post-IR. Exposure to IR induces 53BP1 foci in both cells, with slightly higher levels in S1 cells than in T4-2 cells at 4 h post-IR. There was no significant difference in IR-induced RIF1 foci in T4-2 and S1 cells (Fig. 1G to I), but the level of 53BP1/RIF1 colocalization is much higher in S1 cells and the number of colocalized foci did not decline with time as was observed in T4-2 cells (Fig. 1I and J). As 53BP1 and RIF1 protein levels are similar in both cell lines and did not change post-IR, the focus assays (Fig. 1G to J) indicate that in T4-2 cells with high β1-integrin, the 53BP1/RIF1 complex is displaced more efficiently, which may enhance DSB repair by HR. Conversely, the lower β1-integrin levels in S1 cells favor the persistence of the complex and eventual NHEJ repair that results in increased chromosome aberrations.
IR-induced focus formation is increased by HR-related factors in T4-2 cells.
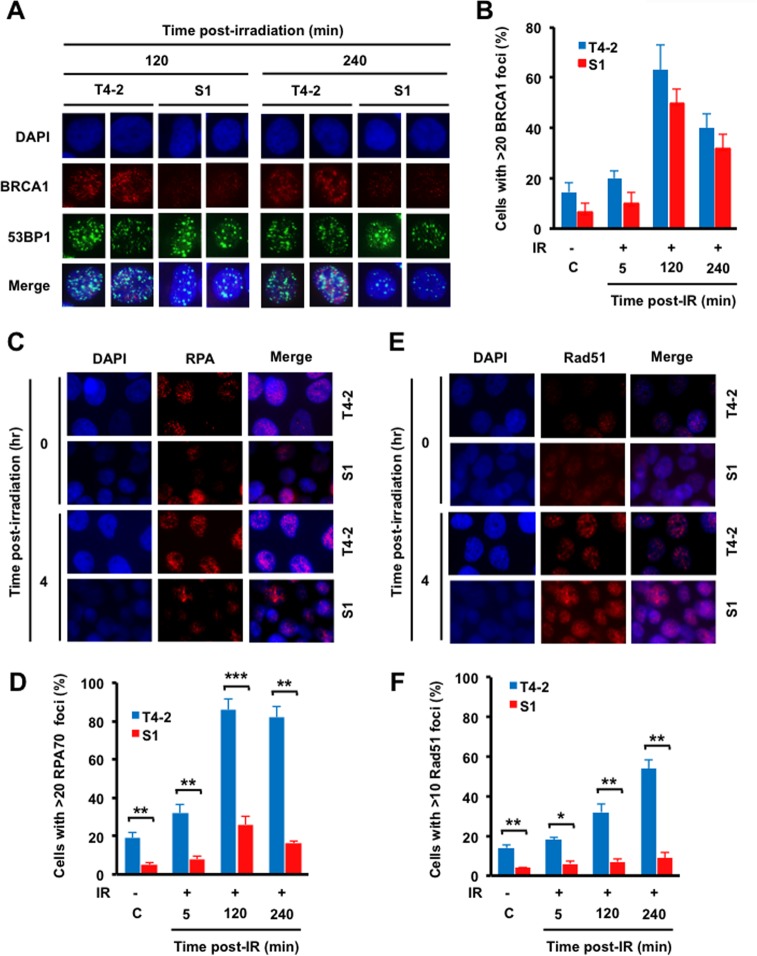
Repair of DSBs by HR is initiated by DNA end resection, which is mediated by MRN complex and facilitated by CtIP (29). This repair process also involves BRCA1 recruitment to replace 53BP1, which is known to inhibit end resection (30). Replication protein A (RPA) then binds to the exposed single-stranded DNA (ssDNA), protecting it from nuclease cleavage and hairpin formation. Subsequent RPA displacement allows for loading and polymerization of Rad51 to initiate the homologous pairing and strand exchange steps of HR (29). Since we observed a decreased number of 53BP1 foci and decreased 53BP1/RIF1 colocalization in T4-2 cells (Fig. 1G, I, and J), we determined whether BRCA1 focus formation was altered (Fig. 2A and B). Although BRCA1 focus formation was increased between 8 and 13% in sham- and IR-treated T4-2 cells compared to that in S1 cells, the difference in focus levels was statistically insignificant. As BRCA1 facilitates the recruitment of RPA and Rad51 post-IR, we also examined RPA focus formation post-IR exposure to 6 Gy of IR. Again, fewer cells were detected with greater than 20 RPA foci in S1 cells than in T4-2 cells (86% versus 26%, 2 h post-IR) (Fig. 2C and D). Finally, the frequency of cells with Rad51 foci was also lower in S1 cells than in the T4-2 cells (32% versus 7%, 2 h post-IR exposure) (Fig. 2E and F). These focus assays all indicate that the HR repair process is more active in radioresistant T4-2 cells than in the nonmalignant S1 cells from which they were derived.
FIG 2.
HR is increased in T4-2 malignant breast epithelial cells compared to that in S1 cells. (A and B) T4-2 and S1 cells were irradiated with 6 Gy X rays, fixed post-IR, and coimmunostained with 53BP1 and BRCA1 antibodies. (A) 53BP1 and BRCA1 staining; (B) histogram of BRCA1 foci. (C and E) Cells were treated with 6 Gy X rays, fixed post-IR, and immunostained for RPA70 (C) and Rad51 (E) antibodies. (D and F) Quantification of focus formation. (D) Histogram of >20 RPA70 foci; (F) histogram of >10 Rad51 foci. (B, D, and F) Columns represent the means (n = 3), and bars represent the SDs; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Increased cellular Rad51 levels contribute to the greater radioresistance of T4-2 cells than of S1 cells.
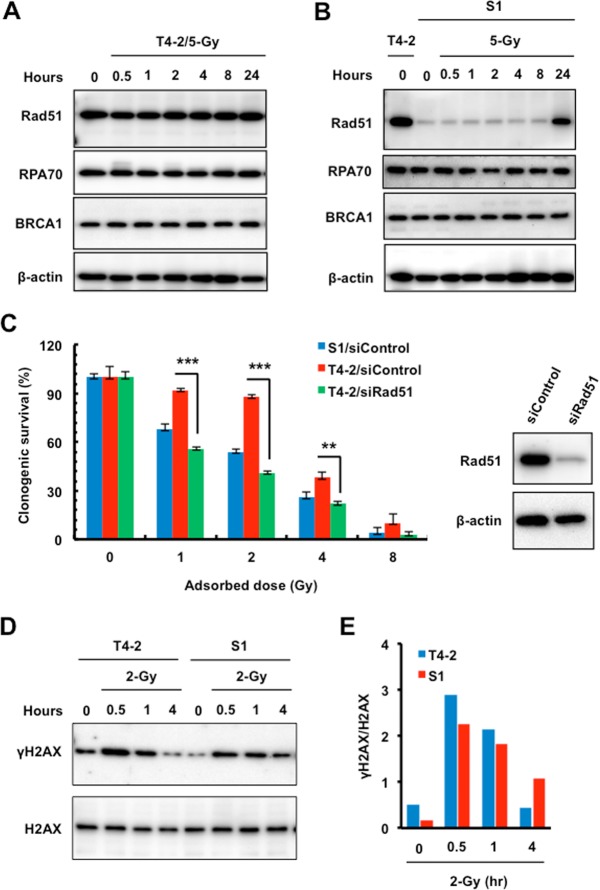
As we observed increased HR-related repairosome focus formation in T4-2 cells, we performed Western analyses to measure Rad51, BRCA1, and RPA70 levels in T4-2 and S1 cells. Interestingly, Rad51, but not BRCA1 or RPA70, was highly upregulated in T4-2 cells compared to S1 cells (Fig. 3A and B). T4-2 cells maintained consistently elevated Rad51 levels before and after IR, while S1 cells had lower basal levels that approached T4-2 levels only at 24 h post-IR (Fig. 3A and B). As Rad51 regulates HR, inhibition of Rad51 should increase radiosensitivity. To test this, Rad51 levels were reduced in T4-2 cells with small interfering RNA (siRNA) and post-IR survival was measured. Rad51 depletion significantly decreased T4-2 cell survival such that it was nearly identical to that of the radiosensitive S1 cell line (Fig. 3C). Western blot analyses of γ-H2AX detected slightly higher protein levels initially in T4-2 cells than in S1 cells until 4 h post-IR, when γ-H2AX was significantly reduced in T4-2 cells compared to S1 cells (Fig. 3D and E). The faster disappearance of γ-H2AX indicates that an efficient DNA repair mechanism exists in T4-2 cells to provide protection from ionizing radiation.
FIG 3.
Inhibition of Rad51 increased malignant breast cancer T4-2 cell radiosensitivity. (A and B) Western analysis of Rad51, BRCA1, and RPA70 using whole-cell lysates prepared from T4-2 and S1 cells sham irradiated or exposed to 5 Gy X rays (β-actin as a loading control). (C) Radiosensitivity assay of T4-2 and S1 cells treated with control siRNA or T4-2 cells treated with Rad51 siRNA before exposure to 1, 2, 4, or 8 Gy X rays. Clonogenic survival was measured 14 days after IR. Colonies consisting of more than 50 cells were scored as surviving colonies and normalized against nonirradiated clones (n = 3, mean ± SD; **, P < 0.01; ***, P < 0.001). Right panel, Western blot analysis of the expression of Rad51 using whole-cell lysates prepared from T4-2 cells electroporated with control or Rad51 siRNA. β-Actin served as an internal loading control. (D and E) Western analysis of γ-H2AX in T4-2 and S1 cells sham irradiated or exposed to 2 Gy X rays. (D) The lysates were also blotted for H2AX as an internal loading control. (E) Relative expression levels of γ-H2AX normalized to the expression levels of H2AX.
β1-Integrin maintains Rad51 protein levels and influences HR-mediated DSB repair.
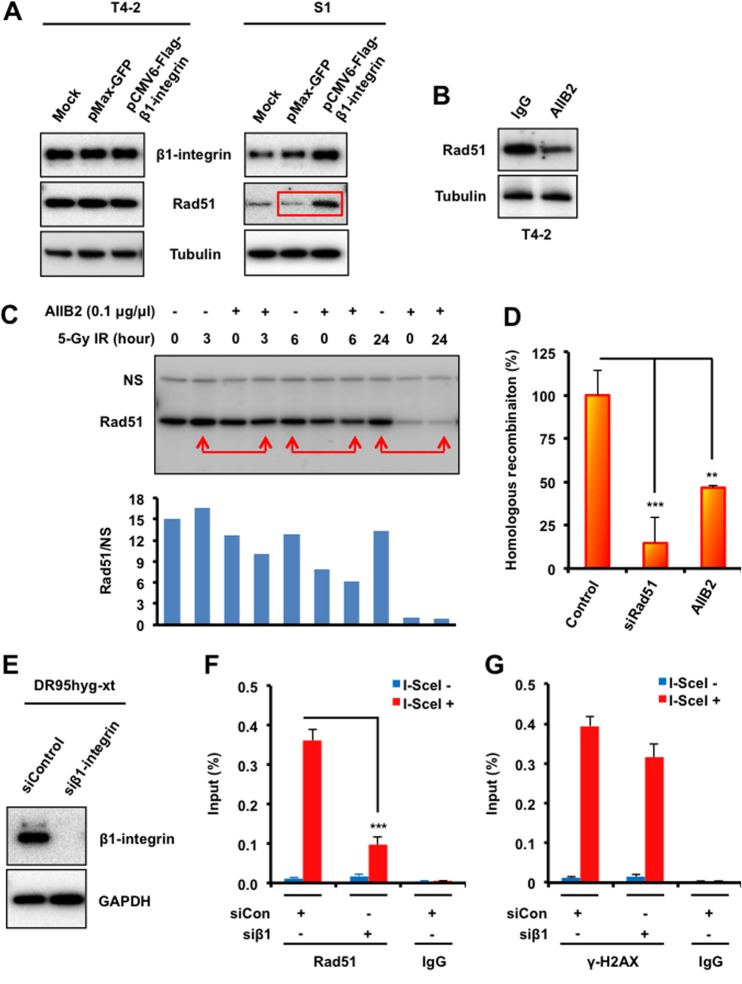
We have shown previously that T4-2 cells are distinguished from S1 cells by having higher levels of β1-integrin (26). Western blot analysis indicates that Rad51 protein levels are also elevated in T4-2 cells (Fig. 3A and B) and that transient overexpression of β1-integrin in S1 cells by transfection increases Rad51 levels (Fig. 4A). Consistent with these results, we found that inhibition of β1-integrin in T4-2 cells reduced Rad51 levels with or without IR (Fig. 4B and C), thus suggesting that Rad51 protein levels may be stabilized by β1-integrin.
FIG 4.
β1-Integrin regulates Rad51 protein levels and recruitment to DSB sites and promotes HR. (A) T4-2 and S1 cells were electroporated with mammalian expression vectors encoding Flag-tagged β1-integrin or with GFP as a control. Cells were lysed 48 h following electroporation and subjected to immunoblotting with β1-integrin and Rad51 antibodies. Tubulin served as an internal loading control. (B and C) Western blot analysis of Rad51 in whole-cell lysates prepared from malignant breast cancer T4-2 cells treated with the β1-integrin inhibitory monoclonal antibody AIIB2 (0.1 μg/μl) or nonspecific rat IgG before exposure to IR (tubulin served as a loading control). (D) HR was assessed using a DR-GFP reporter assay, as described in Materials and Methods, using T4-2 cells electroporated with control siRNA or Rad51 siRNA or treated with AIIB2 or IgG (“control” represents both control siRNA and IgG). (E) DR95hyg-xt cells were electroporated with either control siRNA or β1-integrin siRNA and harvested after 48 h. Cell lysates were immunoblotted for the indicated proteins. (F and G) ChIP analysis of Rad51 (F) and γ-H2AX (G) on a unique DSB induced by I-SceI in vivo in DR95hyg-xt cells treated with control siRNA (siCon) or β1-integrin siRNA (siβ1), before and after DSB induction by I-SceI transfection. Real-time PCR on ChIP samples used primers directed at nucleotides 94 to 378 from the DSB. The enrichment of Rad51 and γ-H2AX after induction of the DSB was compared with that of an IgG control. (D, F, and G) Columns represent the means (n = 3), and bars represent the SDs; **, P < 0.01; ***, P < 0.001.
Inhibition of β1-integrin reduces Rad51, which is a critical component of HR repair, and integrin depletion therefore should affect HR. To test this, a direct repeat-green fluorescent protein (DR-GFP) reporter assay, in which reconstitution of a defective GFP gene is dependent upon HR repair of an introduced DSB, was performed (Fig. 4D). As measured by this assay, the efficiency of HR-mediated DSB repair in T4-2 cells treated with the β1-integrin inhibitor AIIB2 was decreased 53% relative to HR repair efficiency in control cells. This result further indicates that β1-integrin is critical for Rad51-mediated HR repair of DNA DSBs.
Next, chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis was performed to examine Rad51 recruitment at a single DNA DSB site in the β1-integrin-depleted DR95 cell line, which bears a modified GFP gene in which an I-SceI restriction site (18 bp; TAGGGATAACAGGGTAAT) has been engineered (31). Expression of I-SceI creates a specific DSB, and the subsequent recruitment of repair proteins can be detected by ChIP using specific antibodies and site-specific primer sets. We found that Rad51 levels near the I-SceI-induced DSB increased after DSB induction. However, inhibition of β1-integrin highly reduced Rad51 levels (Fig. 4E to G), suggesting that β1-integrin influences Rad51 accumulation at DNA DSBs.
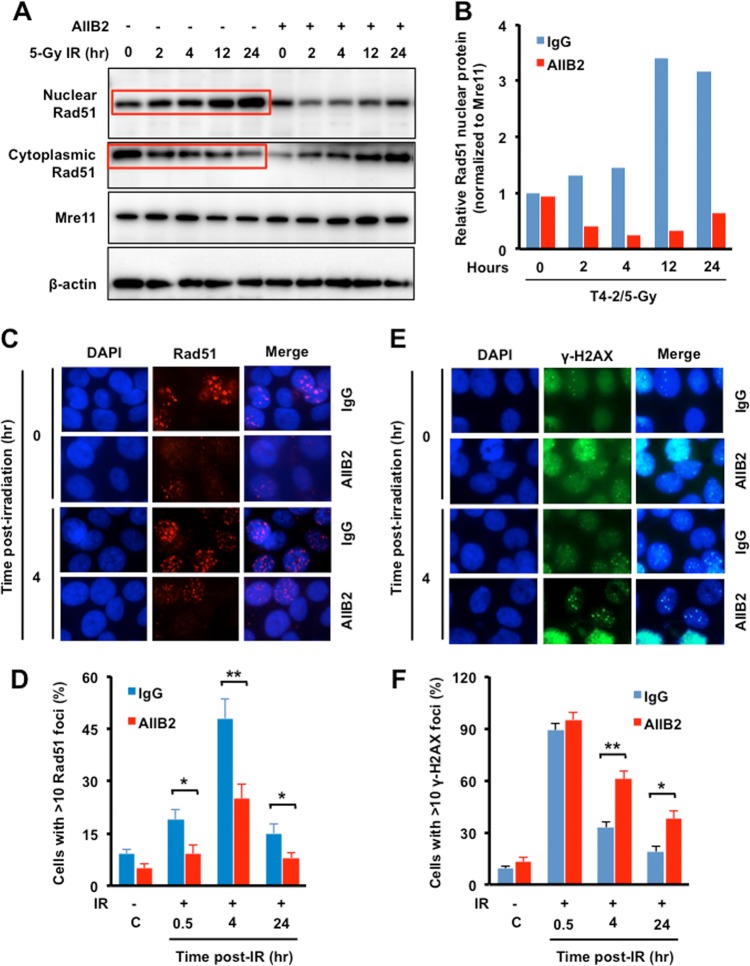
β1-Integrin promotes IR-induced nuclear translocation and focus formation of Rad51.
Several studies have demonstrated that the nuclear translocation of Rad51 after DSB induction is required for its role in repair by HR. Overexpression of Rad51 increases cytoplasmic accumulation, but genotoxic stress triggers cytoplasmic to nuclear translocation (32), which is consistent with the earlier report that Rad51 focus formation requires Rad51 translocation into the nucleus after DSB induction by genotoxic stress (33). However, Rad51 does not have a nuclear localization signal (NLS), so its nuclear entry requires association with other proteins. A role for BRCA2 and Rad51c in the cellular redistribution of Rad51 in response to DNA damage has been reported (34). To determine whether β1-integrin influences nuclear transport of Rad51, nuclear and cytoplasmic fractions were prepared from T4-2 cells that had been treated or not treated with an integrin inhibitor and then irradiated with 5 Gy. Nuclear Rad51 protein levels increased after exposure to IR in a time-dependent manner, but β1-integrin inhibition significantly impaired Rad51 accumulation in the nucleus (Fig. 5A and B). We also examined cytoplasmic Rad51 levels and found that they were increased by IR in β1-integrin-inhibited cells, which is opposite of what occurs in vehicle-treated cells, i.e., Rad51 is reduced in the cytoplasm by IR in a time-dependent manner (Fig. 5A and B). These results suggest that β1-integrin is essential for the nuclear transport of Rad51.
FIG 5.
β1-Integrin promotes IR-induced nuclear accumulation of Rad51 and focus formation in irradiated cells. (A and B) Increased nuclear Rad51 levels in irradiated cells. (A) Western blotting was performed with nuclear and cytosolic Rad51 of T4-2 cells treated with AIIB2 for 16 h before exposure to 5 Gy X rays (Mre11 and β-actin are markers for nuclear and cytosolic fractions). (B) Relative amounts of nuclear Rad51 shown in panel A were estimated by densitometry. (C and E) Decreases in Rad51 foci by AIIB2 treatment were associated with an increase in γ-H2AX foci in irradiated cells. T4-2 cells were treated with AIIB2 before exposure to 6 Gy X rays, fixed post-IR, and immunostained for Rad51 (C) and γ-H2AX (E) antibodies. (D and F) The percentages of cells with more than 10 Rad51 (D) or γ-H2AX (F) foci are represented. (B, D, and F) Columns represent the means (n = 3), and bars represent the SDs; *, P < 0.05; **, P < 0.01.
Since blocking β1-integrin function significantly inhibited nuclear Rad51 levels, we tested whether Rad51 focus formation at DSBs was also affected. Inhibition of β1-integrin significantly reduced IR-induced Rad51 focus formation at all time points post-IR (Fig. 5C and D). The decrease in Rad51 focus frequency by β1-integrin inhibition was associated with delayed resolution of IR-induced γ-H2AX foci (Fig. 5E and F), suggesting impaired DSB repair and consistent with the observed reduction in HR repair (Fig. 4C).
Rad51 protein in S1 cells is a target of both ubiquitin- and calpain-mediated protein degradation.
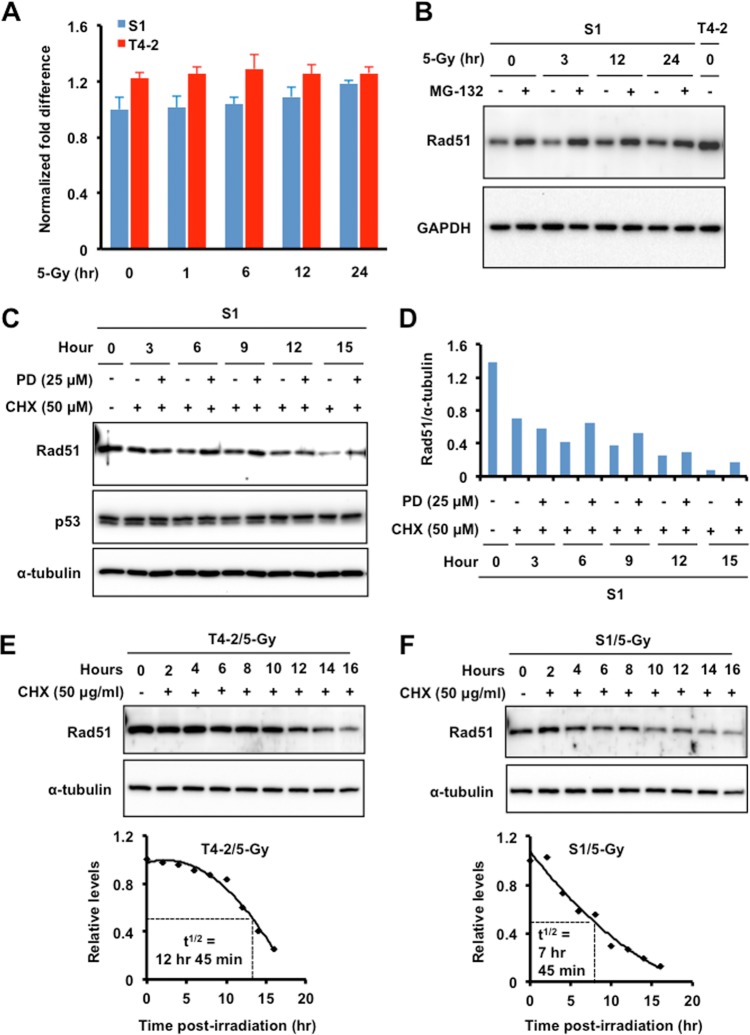
Elevated Rad51 levels have been observed in a wide range of human tumors, where it most likely contributes to tumor drug resistance. Increased Rad51 levels do not arise from gene amplification but rather are more likely due to increased Rad51 gene transcription, altered Rad51 protein stability, or possibly posttranslational modifications. To determine the reasons for lower Rad51 levels in the nonmalignant S1 cells, qPCR was performed to measure Rad51 mRNA transcripts (Fig. 6A). There were only small, statistically insignificant differences between the Rad51 mRNA levels in T4-2 and S1 cells. Next, we investigated whether the low Rad51 levels were due to rapid protein turnover by the ubiquitin proteasomal pathway (UPP). S1 cells were treated with the proteasome inhibitor MG-132 before exposure to 5 Gy of IR (Fig. 6B), and Rad51 levels were measured by Western blotting as a function of post-IR recovery. We found that MG-132 treatment increased Rad51 levels, suggesting that Rad51 in S1 cells is a target for degradation by UPP.
FIG 6.
Regulation of Rad51 protein levels is predominantly through the ubiquitin proteasomal pathway. (A) Quantitative reverse transcription-PCR analysis of β1-integrin mRNA expression in T4-2 and S1 cells exposed to 5 Gy X rays. GAPDH served as an internal control. (B) Proteasomal inhibitor MG-132 (carbobenzoxy-Leu-Leu-leucinal) blocks the degradation of Rad51. Western blot analysis of the expression of Rad51 was done using whole-cell lysate prepared from S1 cells treated with MG-132 (20 μM) or vehicle (dimethyl sulfoxide [DMSO]; GAPDH as a loading control and whole-cell lysate of T4-2 cells as a control for the expression level of Rad51). (C and D) A specific synthetic inhibitor of calpain, PD150606 (PD), blocks the degradation of Rad51. (C) Western blot analysis of Rad51 expression was performed using whole-cell lysate prepared from S1 cells treated with cycloheximide (CHX; 50 μg/ml) to inhibit protein synthesis or not treated with CHX and then treated with PD150606 or vehicle (DMSO; α-tubulin as a loading control and p53 as a negative or system control). (D) Relative expression levels of Rad51 normalized to the expression levels of α-tubulin. (E and F) Half-lives (t1/2) of Rad51 in T4-2 (E) and S1 (F) cells. Western blot analysis of Rad51 expression was performed using whole-cell lysate prepared from S1 cells treated with CHX (50 μg/ml) or not treated with CHX before exposure to 5 Gy X rays (α-tubulin as the loading control). Bottom panels, densitometric analysis of Rad51 normalized with α-tubulin.
Since Rad51 protein levels are much lower in S1 cells than in T4-2 cells, even after the MG-132-induced increase, we investigated whether additional mechanisms might contribute to protein degradation. Proteins are also degraded by calpain-mediated pathways; therefore, new protein synthesis in S1 cells was inhibited with cycloheximide (CHX), and then calpain 1 and calpain 2 activity was blocked with the specific inhibitor PD150606. Higher levels of Rad51 were detected by Western blotting after 6 h of treatment and continued through 15 h posttreatment (Fig. 6C and D). There was no increase over the levels in untreated cells at the 3 h time point, indicating that calpain-mediated degradation starts at a later time than proteasome-mediated degradation (Fig. 6C and D). We utilized p53 levels as a negative or system control because p53 is known to be degraded by the proteasome but is not a substrate for calpain. These results demonstrate that multiple mechanisms are involved in degrading Rad51 in S1 cells (Fig. 6B).
We compared the half-lives of Rad51 protein in T4-2 and S1 cells and found that there was little Rad51 degradation in T4-2 cells up to 10 h after CHX treatment but then significant degradation from 12 h onward, resulting in an overall half-life of 12 h 45 min (Fig. 6E). Compared to T4-2 cells, S1 cells had a nearly linear degradation curve but with a half-life of only 7 h 45 min (Fig. 6F). The significantly shorter half-life of Rad51 protein in S1 cells than in T4-2 cells (5 h) may explain the lower cellular levels.
β1-Integrin regulates Rad51 ubiquitination.
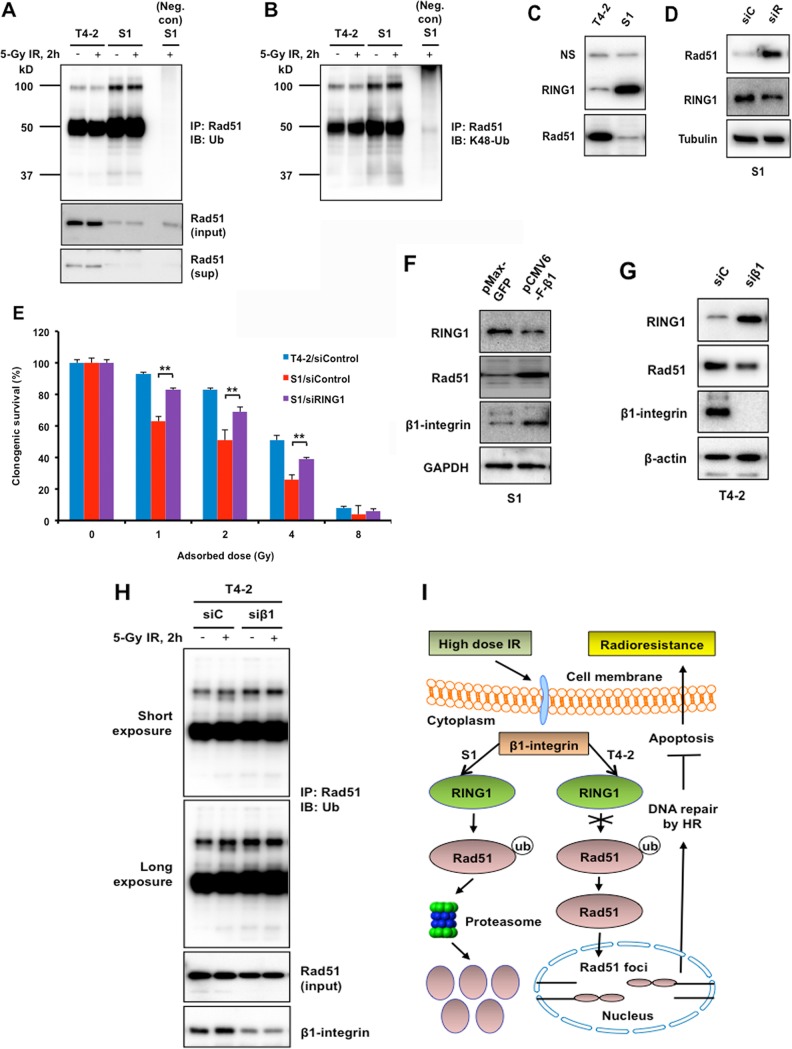
A growing body of research has shown that β1-integrin can affect protein degradation (35); therefore, we examined the potential role of β1-integrin in Rad51 degradation (Fig. 7). The level of Rad51 ubiquitination was determined by immunoprecipitating Rad51 from extracts prepared from cells after IR, followed by Western blotting with a panubiquitin antibody. Rad51 ubiquitination was much higher in S1 cells than in T4-2 cells, with neither showing further induction by IR (Fig. 7A). Moreover, this ubiquitination is K-48 linked, suggesting a degradation role for Rad51 ubiquitination (Fig. 7B).
FIG 7.
β1-Integrin regulates Rad51 ubiquitination and Rad51 protein levels by E3 ubiquitin-protein ligase RING1. (A and B) T4-2 and S1 cells were left untreated or treated with 5 Gy of IR. Cells were lysed under denaturing conditions. Rad51 was immunoprecipitated (IP), and immunoblots (IB) were probed with panubiquitin (A) and K48-Ub (B) antibodies. Positions and sizes in kilodaltons of marker proteins are shown on the left. (A) Lower panels show the Rad51 before immunoprecipitation (input) and Rad51 in the supernatant postimmunoprecipitation (sup). Neg. con, negative control. (C and D) Western blot analysis of RING1 and Rad51 expression using whole-cell lysate prepared from T4-2 and S1 cells (C) and from S1 cells electroporated with control siRNA (siC) or Rad51 siRNA (siR) (D). Nonspecific bands (NS) and α-tubulin served as loading controls. (E) Inhibition of RING1 increased radioresistance in S1 cells. Malignant breast T4-2 cells and the nonmalignant S1 breast epithelial cell counterparts were treated with control siRNA or the S1 cells were treated with RING1 siRNA before exposure to 1, 2, 4, or 8 Gy X rays. Clonogenic survival was measured 14 days post-IR. Colonies consisting of more than 50 cells were scored as surviving colonies and normalized against nonirradiated clones (n = 3, mean ± SD; **, P < 0.01). (F and G) Western blot analysis of RING1, Rad51, and β1-integrin expression using whole-cell lysate prepared from S1 cells electroporated with mammalian expression vector for β1-integrin or GFP (control) (F) and from T4-2 cells electroporated with control siRNA (siC) or β1-integrin siRNA (siβ1) (G). GAPDH and β-actin served as loading controls. (H) T4-2 cells, electroporated with either control siRNA (siC) or β1-integrin siRNA (siβ1), were left untreated or were treated with 5 Gy of IR. Cells were lysed under denaturing conditions. Rad51 was immunoprecipitated, and blots were probed with panubiquitin antibody. Lower panels show the Rad51 (input) and β1-integrin before immunoprecipitation. (I) Model of a novel strategy to enhance the efficacy of radiotherapy in breast cancer patients.
The E3 ubiquitin ligase RING1 ubiquitinates lysine 119 of histone H2A and is ubiquitously expressed in normal tissues, although some variability among different tissues and cell types has been reported (36). Abnormal expression of RING1 has been reported in dividing neoplastic cells; for example, the loss of RING1 expression in 50% of testicular germ cell tumors and some clear-cell renal cell carcinomas has been reported (37). Given the differential expressions of RING1 in normal and cancer cells, we analyzed T4-2 and S1 cells by Western blotting and detected high RING1 levels in S1 cells compared to that in T4-2 cells. This expression pattern of RING1 in T4-2 and S1 cells is exactly opposite the expression levels of Rad51 (Fig. 7C), indicating that increased Rad51 ubiquitination (K48 linked) may be mediated by RING1 (Fig. 7B), resulting in elevated Rad51 degradation in S1 cells.
To test whether the low level of Rad51 in S1 cells is due to elevated RING1 levels, the cells were depleted of RING1 with siRNA and Rad51 levels determined by Western blotting. After RING1 depletion, Rad51 protein levels in S1 cells significantly increased (Fig. 7D) and there was an associated decrease in radiosensitivity (Fig. 7E). In addition, β1-integrin overexpression also reduced RING1 levels and increased Rad51 in S1 cells (Fig. 7F). On the other hand, β1-integrin depletion in T4-2 cells significantly increased RING1 levels and decreased Rad51 levels (Fig. 7G). This induction of RING1 was associated with an increase in Rad51 ubiquitination in T4-2 cells (Fig. 7H). Overall, these results provide evidence that Rad51 protein levels can be regulated via β1-integrin through its effects on RING1.
DISCUSSION
Numerous attempts have been made to overcome the resistance of solid tumors to radiotherapy. Identification of the different factors involved in radioresistance and understanding their mechanism of resistance could be a major step toward improving cancer radiotherapy. Tumor ECM, a major component of the microenvironment, has been shown to increase resistance to cytotoxic cancer therapies, including ionizing radiation (5, 6), and has also been associated with poor outcomes in a subset of breast cancer patients (38). It has been reported that inactivating β1-integrin leads to selective apoptosis and cytostasis in breast cancer cells in vivo without toxicity (7). In addition, a number of studies have shown that β1-integrin regulates IR-induced prosurvival signaling, leading to increased survivability and reproductive capacity of cancer cells exposed to IR (39). However, the underlying mechanism(s) for β1-integrin-mediated resistance to therapy, particularly IR exposure, has not been well studied. Variations in repair of DNA damage can contribute to the molecular mechanisms underlying differences in radioresistance. Although a large number of signaling pathway discrepancies downstream of β1-integrin have been identified, whether β1-integrin participates directly in the regulation of DNA repair is currently unknown. In the present study, we wished to dissect the possible molecular basis of the connection between β1-integrin and the survival of irradiated cells. We discovered that β1-integrin protects Rad51 from proteasome-mediated degradation via regulation of E3 ligase RING1 levels. We also found that β1-integrin impacts Rad51 cytoplasm-to-nucleus translocation and focus formation post-IR exposure (Fig. 5A to D). These mechanisms are associated with increased survival post-IR and sensitization of cells to IR-induced death upon inhibition of Rad51. This pathway is abrogated in S1 human mammary epithelial cells, which have low β1-integrin and Rad51 levels (Fig. 3C). Finally, β1-integrin depletion decreases HR repair (Fig. 4D), suggesting an essential role for β1-integrin in enhancing DNA DSB repair by HR in T4-2 malignant cells but not S1 normal cells, resulting in increased radioresistance of T4-2 cells.
The level and stability of DNA repair proteins are important because they directly impact repair of DNA damage and cellular adaptation to genotoxic conditions, yet little is known regarding the potential repair mechanisms involved in the β1-integrin-dependent survival pathway of cancer cells after IR treatment. Two studies found that β1-integrin-mediated cell adhesion protects cells from bleomycin-induced DNA lesions via PARP-1/ligase III α/XRCC1-dependent base excision repair (24, 25). Recently, Dickreuter and coworkers reported that the FAK/cortactin/JNK signaling axis plays a crucial role in β1-integrin-dependent DNA repair by NHEJ and radioresistance in HNSCC cell lines (23). Here, in line with these observations, we show significantly perturbed DNA DSB repair, which correlates with radiosensitization upon β1-integrin inhibition in malignant breast cancer cells. Specifically, we found that β1-integrin maintains Rad51 protein levels and translocation into the nucleus post-IR with a concomitant increase in binding to a single I-SceI-induced DSB (Fig. 4E to G). The cumulative data all suggest that IR-induced Rad51 activity is mediated by β1-integrin.
As Rad51 is an essential protein in DNA repair, measurement of its expression level in the cell can be a powerful tool to explore its function in tumor biology. In the present study, we observed high Rad51 protein levels in human breast cancer T4-2 cells compared to those of the nonmalignant breast epithelial S1 cells from which they were derived (Fig. 3B). Analyses of Rad51 mRNA expression and protein stability indicate that Rad51 in S1 cells is regulated not at the mRNA level but mainly at the posttranslational level, by β1-integrin-dependent UPP-mediated degradation and, in part, by calpain-mediated degradation. These results suggest that the altered β1-integrin in T4-2 cells leads to increased Rad51 levels, which may enhance HR repair and radioresistance in T4-2 cells.
Another important implication of the current results is the functional role of an E3 ubiquitin-protein ligase RING1 in β1-integrin/Rad51 signaling in breast cancer radioresistance. The expression of RING1 is ubiquitous in every normal tissue analyzed but is altered in many solid and hematopoietic malignancies, including those of testicular germ cell tumors, renal cell carcinomas, Hodgkin's lymphoma, and B-cell lymphoma (37). Deregulation of RING1 expression leads to oncogenic transformation by deregulation of the expression levels of certain oncogenes (40). Our results demonstrate that Rad51 levels are controlled by RING1 and that RING1 knockdown increased Rad51 accumulation and radioresistance in S1 cells (Fig. 7D and E). Interestingly, in T4-2 cells, induction of RING1, via inhibition of β1-integrin, was associated with an increase in ubiquitination of Rad51, followed by a decrease in the protein levels (Fig. 7G and H). While the data suggest that Rad51 could be a direct substrate of RING1, confirmation of this step in the regulatory mechanism requires further study in vitro with purified proteins. RING1 levels themselves are subject to upstream regulation by an ECM receptor, β1-integrin, that ultimately influences Rad51 levels. We propose, therefore, a model in which activation of the β1-integrin/RING1/Rad51 ubiquitylation/degradation pathway represents a novel strategy to enhance the efficacy of radiotherapy in patients with breast cancer (Fig. 7I).
In summary, we describe here a novel finding that Rad51 protein degradation is regulated by β1-integrin through RING1-mediated ubiquitination in breast epithelial cells. Rad51 stability and nuclear translocation regulation by β1-integrin are tightly associated with DNA DSB repair by HR and enhanced clonogenic survival of cells with elevated β1-integrin. Our results suggest that breast cancer therapy may be enhanced by targeting the β1-integrin/RING1/Rad51 axis of radiation-resistant tumors.
MATERIALS AND METHODS
Cell culture.
The isogenic cell lines used, nonmalignant S1 cells and malignant T4-2 cells, from the HMT3522 human breast cancer progression series were maintained as described previously (41). The cell series was established in an attempt to recapitulate the stochastic and prolonged nature of breast cancer progression by continuously culturing S1 cells, derived from reduction mammoplasty, in the absence of serum followed by EGF removal and injection into mice, to give rise to T4-2 cells (26, 42, 43). The S1 cells were propagated as monolayers on plastic in the presence of 10 mg/ml epidermal growth factor (EGF; BD Biosciences, Bedford, MA); T4-2 cells were grown as monolayers on collagen type I (Vitrogen 100; Celtrix Laboratories, Palo Alto, CA)-coated dishes in the absence of EGF.
The H1299 cell line with a stably integrated DR-GFP cassette was grown in Dulbecco's modified Eagle medium (DMEM) (Sigma) and 10% fetal calf serum (Sigma) supplemented with 1% penicillin-streptomycin and 100 μg/ml hygromycin (Invitrogen). The DR95hyg-xt cell line was maintained as previously described (31).
Clonogenic assay.
Standard clonogenic survival assays after exposure to ionizing radiation were performed as described previously (26). The cells were irradiated using a Rad Source RS 2000 X-ray irradiator (Rad Source Technologies, Inc., GA). Different numbers of cells were seeded in 6-well dishes after electroporation (Nucleofector 2b; Lonza) of cells with Rad51 or control siRNA (100 nM; GE Dharmacon) or β1-integrin inhibitory monoclonal antibody AIIB2 (0.1 μg/μl; Argon Biosciences, Morgan Hill, CA) with or without radiation. Each treatment was performed in three wells, and all experiments were repeated in triplicate. The treated and control cells were cultured for 14 days, and colonies with more than 50 cells were scored and normalized to the plating efficiency of each cell line.
DR-GFP assay.
Homologous recombination was assessed using a direct repeat-green fluorescent protein (DR-GFP) cassette, which is stably integrated at a single locus in H1299 cells (44). Briefly, H1299 cells were treated with AIIB2 or siRNA (Rad51 or control) and then transiently transfected with an I-SceI expression plasmid using a Nucleofector 2b device. Cells were trypsinized 48 h after transfection, and the proportion of GFP-positive cells was determined by flow cytometry (FACSCalibur; BD Biosciences) for measurement of HR repair.
Chromosome aberration assay.
Chromosome aberrations at metaphase were examined as previously described (45). G1-type chromosomal aberrations were assessed in cells exposed to 3 Gy of IR. S-phase-specific chromosome aberrations were assessed after exponentially growing cells (pulse-labeled with bromodeoxyuridine [BrdU]) were irradiated with 2 Gy of IR, and metaphases were harvested 4 to 5 h after IR, as previously described (45). For G2-specific aberrations, cells were irradiated with 1 Gy IR and metaphases were collected 1 h posttreatment. After 2 min of hypotonic treatment with 0.56% (wt/vol) KCl at room temperature, cells were fixed with fresh methanol-acetic acid (3:1) and slides were prepared and stained with Giemsa stain (4% in 0.01 M phosphate buffer, pH 6.8) for 10 min. The S-phase-type chromosome aberrations were breaks, gaps, and radials per metaphase.
Western blotting.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer as previously described (46). Equal amounts of protein were loaded onto reducing SDS gels (Invitrogen, Carlsbad, CA). After transfer onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Temecula, CA), blots were blocked with 5% nonfat milk and probed. Primary antibodies against the following were used in this study: from BD Transduction Laboratories, β1-integrin (clone 18); from Cell Signaling, H2AX (2595), γ-H2AX (Ser139; 9718), ATR (2790), pATR (Ser428; 2853), and pATM (Ser1981; 13050); from Santa Cruz, ATM (Sc-53173), 53BP1 (Sc-22760), BRCA1 (Sc-642), and α-tubulin (Sc-5286); from Abcam, Rad51 (ab63801), RPA70 (ab79398), and RING1 (ab32644); from GeneTex, MRE11 (GTX70212); from Bethyl Laboratories, RIF1 (A300-5671); from Proteintech, GAPDH (HRP-60004); from MP Biomedicals, β-actin (691001). Blots were washed, incubated with secondary antibody, and then visualized using the ECL Western blotting detection system (Thermo Scientific, Rockford, IL).
Subcellular fractionation.
T4-2 cells were plated on 10-cm dishes and incubated at 37°C in 5% (vol/vol) CO2. When cells reached ∼90% confluence, selected dishes received β1-integrin inhibitory AIIB2 or control rat IgG antibody (0.1 μg/μl, final concentration). After overnight treatment, a set of plates was exposed to IR (5 Gy X rays). Cells were harvested 2 h, 4 h, 12 h, and 24 h following IR. Control cells were not irradiated. Subcellular fractionation was performed using a subcellular protein fractionation kit (Pierce, USA) according to the manufacturer's instructions. Cytoplasmic and nuclear protein concentrations were determined (Bradford, Bio-Rad), and samples were prepared for gel electrophoresis.
Immunofluorescence microscopy.
Cells grown in chamber slides were fixed and immunostained as previously described (47, 48). To block β1-integrin activity, cells were treated overnight with AIIB2 and then exposed to the indicated doses of X-irradiation. For immunostaining, cells were fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature and then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for γ-H2AX, 53BP1, RIF1, and RPA70 immunofluorescence or with 90% methanol for Rad51 and BRCA1 immunofluorescence assays. Fluorescent focus images were captured by a standard procedure (47). Sections through nuclei were captured, and the images were obtained by projection of the individual sections as previously described (49). The results shown are from three independent experiments.
Real-time PCR analysis.
Total RNA was purified with TRIzol (Invitrogen) according to the manufacturer's instructions, and then 5 μg of each sample was reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen). PCR was carried out with 2 μl of cDNA from each sample using a QuantiTect SYBR green PCR kit (Qiagen) and a QuantStudio 6 Flex real-time PCR detection system (Applied Biosystems by Life Technologies). Primers used to amplify β1-integrin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were purchased from Qiagen. Fragments were amplified using the following protocol: 95°C for 15 min (initial denaturation) and 40 amplification cycles (94°C for 15 s, 55°C for 30 s, and 72°C for 30 s). β1-Integrin mRNA was normalized to the corresponding GAPDH and averaged from three independent experiments. Melting curve analysis verified the presence of a single PCR product.
ChIP assay.
For the chromatin immunoprecipitation assay (ChIP) assay, DR95 cells, carrying an integrated I-SceI site (DR95hyg-xt cells), were electroporated with an I-Scel expression plasmid (31, 50). Protein-DNA cross-linking with 1% formaldehyde for 10 min at room temperature was quenched by the addition of 1.37 M glycine (100 μl/ml) for 5 min. The cells were washed three times with ice-cold PBS containing protease inhibitors (PI), and then the cell pellet was collected and resuspended in 300 μl of sonication buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, 1% SDS, and PI). After shearing with the Diagenode Bioruptor, the chromatin was centrifuged at 20,000 × g for 10 min at 4°C, and the supernatant was collected and diluted (1:10) with ChIP dilution buffer (16.7 mM Tris-HCl [pH 8], 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, and 0.1% SDS). Diluted chromatin was incubated with 2 μg of antibody and Magna ChIP protein A/G beads (Millipore) overnight at 4°C. The beads were washed with wash buffer I (20 mM Tris-HCl [pH 8.0],150 mM NaCl, 1.2 mM EDTA, 1% Triton X-100, 0.1% SDS, and PI), wash buffer II (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1.2 mM EDTA, 1% Triton X-100, 0.1% SDS, and PI), and wash buffer III (10 mM Tris-HCl [pH 8], 1 mM EDTA, 1% sodium deoxycholate, 1% NP-40, and 0.25 M lithium chloride). After washing twice with Tris-EDTA (TE) buffer, ChIP samples were eluted with 200 μl freshly made warm (65°C) ChIP elution buffer (0.1 mM NaHCO3 and 0.01% SDS). Cross-linking was reversed by overnight treatment with 8 μl of 5 M NaCl at 65°C, and the DNA was then purified by using QIAquick Spin columns (Qiagen). Quantitation of the amount of immunoprecipitated DNA was carried out by real-time PCR.
Statistics.
Data were analyzed by Student's t test; the P value was derived to assess the statistical significance. Results are expressed as the mean ± standard deviation (SD) of the results of the number of replicates indicated in the figure legends. The experiments shown are representative of three experiments with similar results.
ACKNOWLEDGMENTS
We thank Mina Bissell (Lawrence Berkeley National Laboratory, Berkeley, CA) and Claus Pietrzik (Universitätsmedizin der Johannes Gutenberg-Universität Mainz, Mainz, Germany) for the gifts of the T4-2 and S1 cells and of the β1-integrin plasmid, respectively. We also thank the members of the Pandita laboratory at the Houston Methodist Research Institute for their support during the execution of this work.
This work was supported by funds from the Houston Methodist Research Institute and MD Anderson Cancer Center, Houston, TX, and National Institutes of Health grants CA129537 and GM109768.
We declare no conflicts of interest.
REFERENCES
- 1.Hynes RO. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Zahir N, Weaver VM. 2004. Death in the third dimension: apoptosis regulation and tissue architecture. Curr Opin Genet Dev 14:71–80. doi: 10.1016/j.gde.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, Wang YA, Look AT, Stratton MR, Chin L, Futreal PA, DePinho RA. 2007. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibue T, Weinberg RA. 2009. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci U S A 106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sethi T, Rintoul RC, Moore SM, MacKinnon AC, Salter D, Choo C, Chilvers ER, Dransfield I, Donnelly SC, Strieter R, Haslett C. 1999. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nat Med 5:662–668. doi: 10.1038/9511. [DOI] [PubMed] [Google Scholar]
- 6.Aoudjit F, Vuori K. 2001. Integrin signaling inhibits paclitaxel-induced apoptosis in breast cancer cells. Oncogene 20:4995–5004. doi: 10.1038/sj.onc.1204554. [DOI] [PubMed] [Google Scholar]
- 7.Park CC, Zhang H, Pallavicini M, Gray JW, Baehner F, Park CJ, Bissell MJ. 2006. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res 66:1526–1535. doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanda R, Kawahara A, Watari K, Murakami Y, Sonoda K, Maeda M, Fujita H, Kage M, Uramoto H, Costa C, Kuwano M, Ono M. 2013. Erlotinib resistance in lung cancer cells mediated by integrin beta1/Src/Akt-driven bypass signaling. Cancer Res 73:6243–6253. doi: 10.1158/0008-5472.CAN-12-4502. [DOI] [PubMed] [Google Scholar]
- 9.Carbonell WS, DeLay M, Jahangiri A, Park CC, Aghi MK. 2013. Beta1 integrin targeting potentiates antiangiogenic therapy and inhibits the growth of bevacizumab-resistant glioblastoma. Cancer Res 73:3145–3154. doi: 10.1158/0008-5472.CAN-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao H, Veine DM, Fay KS, Staszewski ED, Zeng ZZ, Livant DL. 2011. The PHSCN dendrimer as a more potent inhibitor of human breast cancer cell invasion, extravasation, and lung colony formation. Breast Cancer Res Treat 125:363–375. doi: 10.1007/s10549-010-0826-y. [DOI] [PubMed] [Google Scholar]
- 11.Hunt CR, Ramnarain D, Horikoshi N, Iyengar P, Pandita RK, Shay JW, Pandita TK. 2013. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat Res 179:383–392. doi: 10.1667/RR3308.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyer WD, Ehmsen KT, Liu J. 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maacke H, Jost K, Opitz S, Miska S, Yuan Y, Hasselbach L, Luttges J, Kalthoff H, Sturzbecher HW. 2000. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene 19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- 16.Schild D, Wiese C. 2010. Overexpression of RAD51 suppresses recombination defects: a possible mechanism to reverse genomic instability. Nucleic Acids Res 38:1061–1070. doi: 10.1093/nar/gkp1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown ET, Holt JT. 2009. Rad51 overexpression rescues radiation resistance in BRCA2-defective cancer cells. Mol Carcinog 48:105–109. doi: 10.1002/mc.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tennstedt P, Fresow R, Simon R, Marx A, Terracciano L, Petersen C, Sauter G, Dikomey E, Borgmann K. 2013. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. Int J Cancer 132:2118–2126. doi: 10.1002/ijc.27907. [DOI] [PubMed] [Google Scholar]
- 19.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alagpulinsa DA, Ayyadevara S, Shmookler Reis RJ. 2014. A small-molecule inhibitor of RAD51 reduces homologous recombination and sensitizes multiple myeloma cells to doxorubicin. Front Oncol 4:289. doi: 10.3389/fonc.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alshareeda AT, Negm OH, Aleskandarany MA, Green AR, Nolan C, TigHhe PJ, Madhusudan S, Ellis IO, Rakha EA. 2016. Clinical and biological significance of RAD51 expression in breast cancer: a key DNA damage response protein. Breast Cancer Res Treat 159:41–53. doi: 10.1007/s10549-016-3915-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhong X, Luo G, Zhou X, Luo W, Wu X, Zhong R, Wang Y, Xu F, Wang J. 2016. Rad51 in regulating the radiosensitivity of non-small cell lung cancer with different epidermal growth factor receptor mutation status. Thorac Cancer 7:50–60. doi: 10.1111/1759-7714.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickreuter E, Eke I, Krause M, Borgmann K, van Vugt MA, Cordes N. 2016. Targeting of beta1 integrins impairs DNA repair for radiosensitization of head and neck cancer cells. Oncogene 35:1353–1362. doi: 10.1038/onc.2015.212. [DOI] [PubMed] [Google Scholar]
- 24.Jones CB, McIntosh J, Huang H, Graytock A, Hoyt DG. 2001. Regulation of bleomycin-induced DNA breakage and chromatin structure in lung endothelial cells by integrins and poly(ADP-ribose) polymerase. Mol Pharmacol 59:69–75. doi: 10.1124/mol.59.1.69. [DOI] [PubMed] [Google Scholar]
- 25.Rose JL, Reeves KC, Likhotvorik RI, Hoyt DG. 2007. Base excision repair proteins are required for integrin-mediated suppression of bleomycin-induced DNA breakage in murine lung endothelial cells. J Pharmacol Exp Ther 321:318–326. doi: 10.1124/jpet.106.113498. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed KM, Zhang H, Park CC. 2013. NF-kappaB regulates radioresistance mediated by beta1-integrin in three-dimensional culture of breast cancer cells. Cancer Res 73:3737–3748. doi: 10.1158/0008-5472.CAN-12-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun MH, Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daley JM, Sung P. 2014. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol 34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY. 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J 25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mladenov E, Anachkova B, Tsaneva I. 2006. Sub-nuclear localization of Rad51 in response to DNA damage. Genes Cells 11:513–524. doi: 10.1111/j.1365-2443.2006.00958.x. [DOI] [PubMed] [Google Scholar]
- 33.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A 92:2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeyasekharan AD, Liu Y, Hattori H, Pisupati V, Jonsdottir AB, Rajendra E, Lee M, Sundaramoorthy E, Schlachter S, Kaminski CF, Ofir-Rosenfeld Y, Sato K, Savill J, Ayoub N, Venkitaraman AR. 2013. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat Struct Mol Biol 20:1191–1198. doi: 10.1038/nsmb.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazlehurst LA, Argilagos RF, Dalton WS. 2007. Beta1 integrin mediated adhesion increases Bim protein degradation and contributes to drug resistance in leukaemia cells. Br J Haematol 136:269–275. doi: 10.1111/j.1365-2141.2006.06435.x. [DOI] [PubMed] [Google Scholar]
- 36.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Beato M, Sanchez E, Gonzalez-Carrero J, Morente M, Diez A, Sanchez-Verde L, Martin MC, Cigudosa JC, Vidal M, Piris MA. 2006. Variability in the expression of polycomb proteins in different normal and tumoral tissues. A pilot study using tissue microarrays. Mod Pathol 19:684–694. [DOI] [PubMed] [Google Scholar]
- 38.Helleman J, Jansen MP, Ruigrok-Ritstier K, van Staveren IL, Look MP, Meijer-van Gelder ME, Sieuwerts AM, Klijn JG, Sleijfer S, Foekens JA, Berns EM. 2008. Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin Cancer Res 14:5555–5564. doi: 10.1158/1078-0432.CCR-08-0555. [DOI] [PubMed] [Google Scholar]
- 39.Eke I, Storch K, Krause M, Cordes N. 2013. Cetuximab attenuates its cytotoxic and radiosensitizing potential by inducing fibronectin biosynthesis. Cancer Res 73:5869–5879. doi: 10.1158/0008-5472.CAN-13-0344. [DOI] [PubMed] [Google Scholar]
- 40.Satijn DP, Otte AP. 1999. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol 19:57–68. doi: 10.1128/MCB.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. 1997. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briand P, Nielsen KV, Madsen MW, Petersen OW. 1996. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res 56:2039–2044. [PubMed] [Google Scholar]
- 43.Briand P, Petersen OW, Van Deurs B. 1987. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol 23:181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 44.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, Gupta A, Wellinger RJ, Zhang J, Powell SN, Roti Roti JL, Lieberman HB, Pandita TK. 2006. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol 26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed KM, Nantajit D, Fan M, Murley JS, Grdina DJ, Li JJ. 2009. Coactivation of ATM/ERK/NF-kappaB in the low-dose radiation-induced radioadaptive response in human skin keratinocytes. Free Radic Biol Med 46:1543–1550. doi: 10.1016/j.freeradbiomed.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta A, Hunt CR, Chakraborty S, Pandita RK, Yordy J, Ramnarain DB, Horikoshi N, Pandita TK. 2014. Role of 53BP1 in the regulation of DNA double-strand break repair pathway choice. Radiat Res 181:1–8. doi: 10.1667/RR13572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwal M, Pandita S, Hunt CR, Gupta A, Yue X, Khan S, Pandita RK, Pratt D, Shay JW, Taylor JS, Pandita TK. 2008. Inhibition of telomerase activity enhances hyperthermia-mediated radiosensitization. Cancer Res 68:3370–3378. doi: 10.1158/0008-5472.CAN-07-5831. [DOI] [PubMed] [Google Scholar]
- 49.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. 2005. Involvement of human MOF in ATM function. Mol Cell Biol 25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, Takeda S, Hay RT, Gautier J, West SC, Paull TT, Pandita TK, White MF, Khanna KK. 2008. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]