ABSTRACT
A gain-of-function mutation in isocitrate dehydrogenase 1 (IDH1) affects immune surveillance in gliomas. As elevated CD47 levels are associated with immune evasion in cancers, its status in gliomas harboring mutant IDH1 (IDH1-MT cells) was investigated. Decreased CD47 expression in IDH1-R132H-overexpressing cells was accompanied by diminished nuclear β-catenin, pyruvate kinase isoform M2 (PKM2), and TCF4 levels compared to those in cells harboring wild-type IDH1 (IDH1-WT cells). The inhibition of β-catenin in IDH1-WT cells abrogated CD47 expression, β-catenin–TCF4 interaction, and the transactivational activity of β-catenin/TCF4. The reverse effect was observed in IDH1-MT cells upon the pharmacological elevation of nuclear β-catenin levels. Genetic and pharmacological manipulation of nuclear PKM2 levels in IDH1-WT and IDH1-MT cells suggested that PKM2 is a positive regulator of the β-catenin–TCF4 interaction. The Cancer Genome Atlas (TCGA) data sets indicated diminished CD47, PKM2, and β-catenin levels in IDH1-MT gliomas compared to IDH1-WT gliomas. Also, elevated BRG1 levels with mutations in the ATP-dependent chromatin-remodeling site were observed in IDH1-MT glioma. The ectopic expression of ATPase-deficient BRG1 diminished CD47 expression as well as TCF4 occupancy on its promoter. Sequential chromatin immunoprecipitation (ChIP–re-ChIP) revealed the recruitment of the PKM2–β-catenin–BRG1–TCF4 complex to the TCF4 site on the CD47 promoter. This occupancy translated into CD47 transcription, as a diminished recruitment of this complex was observed in glioma cells bearing IDH1-R132H. In addition to its involvement in CD47 transcriptional regulation, PKM2–β-catenin–BRG1 cross talk affected the phagocytosis of IDH1-MT cells by microglia.
KEYWORDS: β-catenin, PKM2, IDH1, BRG1, CD47
INTRODUCTION
Somatic mutations in the isocitrate dehydrogenase 1 (IDH1) gene in gliomas have been associated with a better prognosis than in gliomas harboring wild-type IDH1 (1, 2). IDH1 decarboxylates isocitrate to α-ketoglutarate (α-KG), and IDH1 mutations not only result in a loss of the enzymatic wild-type IDH1 function but also are associated with a gain-of-function activity that converts α-KG to d-2-hydroxyglutarate (d-2-HG) (2). The common point mutation R132H in IDH1 (IDH1-R132H) is the most prevalent mutation in gliomas (3). IDH1 is increasingly being recognized as an independent prognostic marker in gliomas, as the occurrence of IDH1 mutations predicts longer survival (4). IDH1 mutations are known to establish CIMP (CpG island methylator phenotype) by remodeling the methylome and transcriptome, with established G-CIMP-positive mutant IDH1-expressing glioblastomas (GBMs) being predictive of improved survival (5). Also, decreased immune cell infiltration in IDH1 mutant gliomas contributes to improved clinical outcomes observed in these patients (6). Not only do IDH1 mutant gliomas escape natural killer cell immune surveillance by the downregulation of NKG2D ligand expression (7), IDH1 mutations also impact CD8+ T cell accumulation and, thereby, immune-evasive responses (8).
The persistent activation of β-catenin is crucial for glioma progression (9), with increased levels of nuclear β-catenin and elevated expression of β-catenin target genes being associated with high-grade GBM (10). Interestingly, the IDH1-R132H mutation in gliomas negatively regulates β-catenin signaling (11). In addition to its well-known role in glycolysis, pyruvate kinase isoform M2 (PKM2) participates in the regulation of gene transcription (12–14). Nuclear PKM2 acts as a coactivator of β-catenin to regulate the Warburg effect (14), and the nuclear expression of PKM2 is upregulated in GBM (15). Also, PKM2-regulated histone modification is essential for its epigenetic regulation of gene expression and glioma tumorigenesis (16). Moreover, BRG1 (SMARCA4), a key subunit of the SWI/SNF chromatin-remodeling complex, binds directly to β-catenin and regulates nucleosome rearrangement at WREs (Wnt-responsive elements) of target genes (17). In addition, meta-analyses of cancer genome sequencing data suggest that SWI/SNF complex subunits are mutated in human cancers with a high frequency (18).
CD47, a transmembrane glycoprotein that mediates a “self/do-not-eat-me” signal on normal cells, communicates with signal regulatory protein α (SIRPα) on macrophages to prevent phagocytosis (19, 20). The upregulation of antiphagocytic CD47 in a variety of cancers renders malignant cells resistant to classical immune surveillance machinery (21). Macrophage-mediated phagocytosis of tumor cells via the blockade of antiphagocytic CD47-SIRPα interactions using anti-CD47 antibodies has shown promise in several solid tumors, including gliomas (20). The enhanced expression of CD47 in malignant tumors facilitates the immunological evasion of tumor cells by rendering tumor cells resistant to immune surveillance. As IDH1 mutation alters the transcriptional program and gene expression in gliomas (5), we examined whether IDH1 mutations affect the status of CD47 in gliomas that exhibit marked difference in effector immune cell functions compared to those of IDH1-WT gliomas (6). Also, since PKM2 regulates β-catenin transactivation to affect gliomagenesis (14) and as β-catenin regulates major histocompatibility complex class (MHC-I) genes associated with immune-evasive responses in gliomas (22), the possible involvement of PKM2 and β-catenin in regulating CD47 associated with immune-evasive responses in IDH1-MT cells was investigated.
RESULTS
Diminished CD47 expression in glioma cells overexpressing IDH1-R132H.
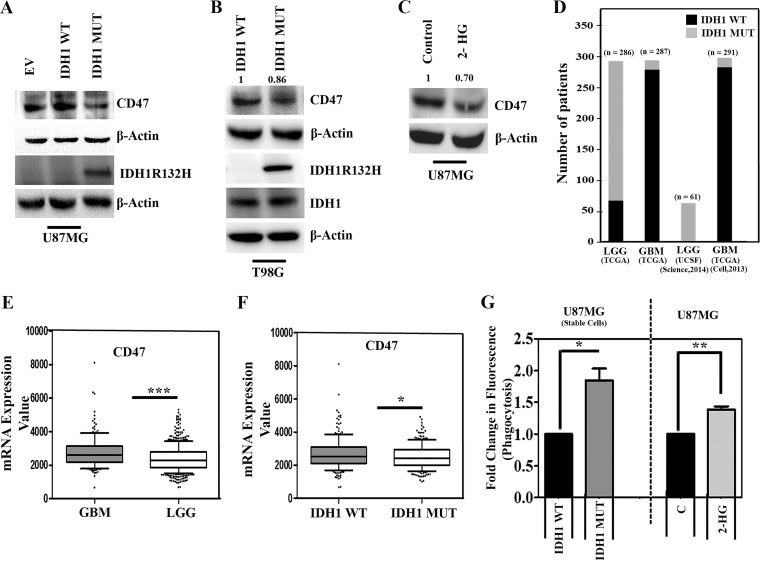
The expression of CD47 on cancer cells allows them to evade innate immune surveillance by serving as an antiphagocytic signal (19, 20). As the IDH1-R132H mutation affects immune-evasive responses (6, 7), we investigated the status of CD47 expression in glioma cells overexpressing IDH1-R132H. A decrease in CD47 expression was observed in glioma cells stably transfected with IDH1-R132H compared to cells transfected with IDH1-WT or the empty vector (Fig. 1A). An increase in the IDH1 mRNA level was observed in stable IDH1-WT cells compared to that in cells transfected with the empty vector (see Fig. S1A in the supplemental material). The oncometabolite 2-HG accumulates to levels of up to 50 mmol/liter in IDH1-R132H-expressing cancer cells (23). The overexpression of IDH1-R132H in U87MG cells induced significant 2-HG accumulation (Fig. S1B). Thus, the stable cell lines recapitulated the IDH1-R132H phenotype in terms of both 2-HG release and IDH1-R132H expression. A decrease in the CD47 level was also observed in T98G those transiently transfected with IDH1-R132H compared to those transfected with IDH1-WT (Fig. 1B). Western blot analysis with a mutation-specific antibody confirmed IDH1-R132H expression in IDH1-MT cells but not in IDH1-WT cells (Fig. 1B). Moreover, similar abundances of IDH1 were observed in both IDH1-WT-expressing and IDH1-R132H-overexpressing cells (Fig. 1B). Upon determining whether 2-HG overproduction contributed to CD47 expression, a reduction in the CD47 level was seen in glioma cells treated with 2-HG (Fig. 1C).
FIG 1.
Ectopic expression of IDH1-MT decreases CD47 expression and enhances phagocytosis of glioma cells. (A and B) Western blot analysis depicting CD47 expression in glioma cells stably (A) or transiently (B) overexpressing IDH1-WT and IDH1-R132H. The transfection efficiency of IDH1-R132H is shown. EV, empty vector. (C) Treatment with 2-HG diminishes CD47 levels in U87MG cells compared to those in untreated controls. The blots in panels A to C are representative of data from three independent experiments with similar results. Blots were stripped and reprobed for β-actin to establish equivalent loading. Densitometric measurements were performed on the immunoblots by using ImageJ. The values indicate fold changes over the values for the controls. Bands were normalized to their corresponding β-actin levels. (D) Graphical representation of the number of patients bearing IDH1-WT and IDH1-MT in LGG and GBM. The data are from multiple LGG and GBM data sets downloaded from the cBioPortal for Cancer Genomics database (http://www.cBioportal.org/). (E) Comparison of CD47 expression levels between LGG and GBM data sets. (F) LGG and GBM data sets were segregated into IDH1-WT and IDH1-MT, and the CD47 expression level was found to be significantly lower in IDH1-MT cells than in IDH1-WT cells. P values were determined by a 2-tailed, unpaired t test using GraphPad Prism. (G) Graph representing increased phagocytosis of IDH1-MT and 2-HG-treated glioma cells by microglia. The results represent averages of data from three different experiments. The P value was determined by a 2-tailed, paired t test using GraphPad Prism. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next analyzed the status of CD47 in glioma cells harboring IDH1-R132H in two public data sets, The Cancer Genome Atlas (TCGA) and UCSF (34, 35, 52, 53). While ∼88% of low-grade glioma (LGG) cells exhibited an IDH1 mutation, ∼5% of GBMs exhibited an IDH1 mutation (Fig. 1D). Upon comparing the CD47 expression levels in GBMs to those in LGGs, a significant decrease in CD47 expression was observed in LGGs (Fig. 1E). Further in silico analysis revealed diminished CD47 expression in glioma patients bearing IDH1-MT, compared to those with the wild-type IDH1 gene (Fig. 1F).
Increased phagocytosis of IDH1-MT glioma cells by microglia.
Microglia are regarded as the professional phagocytes of the central nervous system (CNS), and phagocytosis of glioma cells by microglia leads to a reduction of the number of glioma cells without affecting glioma apoptosis and proliferation (24). Moreover, a significant infiltration of macrophages and microglia occurs in gliomas (25). As CD47 provides an inhibitory signal for macrophage phagocytosis (20), we investigated the ability of microglia to phagocytose IDH1-MT cells expressing diminished CD47 levels. The ability of microglia to phagocytose U87MG cells overexpressing IDH1-MT was greater than the ability of microglia to phagocytose cells expressing IDH1-WT (Fig. 1G). A similar increase in the ability of microglia to phagocytose U87MG cells treated with 2-HG was observed (Fig. 1G).
Decreased nuclear β-catenin and PKM2 accumulation in IDH1-R132H-overexpressing glioma cells.
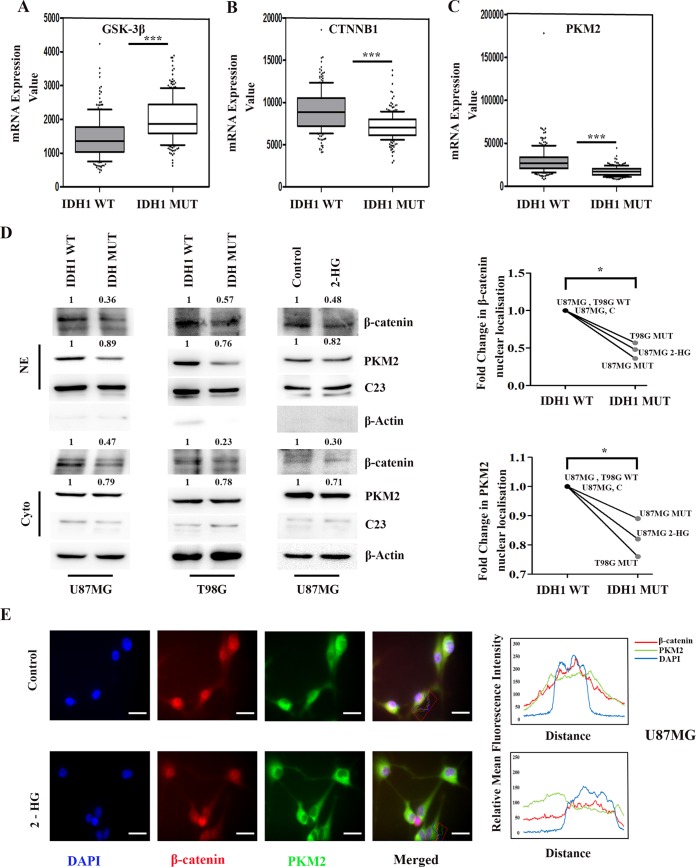
We have previously shown that β-catenin regulates MHC-I expression associated with immune-evasive responses in glioma cells (22). Interestingly, the IDH1 mutation in glioma cells negatively regulates β-catenin signaling (11). To better understand whether β-catenin regulatory mechanisms affect CD47 expression, we used STRING to predict putative interacting partners of β-catenin. This search led to the identification of glycogen synthase kinase 3β (GSK-3β), TCF4, BRG1, and PKM2 as components of the β-catenin interactome (see Fig. S1C in the supplemental material). Importantly, these components were predicted to be involved in transcriptional regulation (Fig. S1C). Decreases in nonphosphorylated β-catenin (involved in the activation of the β-catenin/TCF4 pathway) and total β-catenin levels were accompanied by elevated GSK-3β expression and diminished nuclear TCF4 levels in IDH-MT cells compared to IDH1-WT cells (Fig. S1D). The β-catenin-interacting protein GSK-3β regulates β-catenin phosphorylation and its subsequent degradation (26). The inhibition of this interaction results in β-catenin stabilization followed by its nuclear accumulation to facilitate complex formation with TCF4 (27). As the activation of the β-catenin pathway can occur through alterations in CTNNB1 (encoding β-catenin) expression, analysis of TCGA data for these molecules in IDH1-MT gliomas was performed. In silico analysis revealed a pronounced upregulation of GSK-3β (Fig. 2A) and diminished β-catenin (CTNNB1) levels (Fig. 2B) in IDH1-MT gliomas compared to those in IDH1-WT gliomas. Interestingly, analysis of TCGA data revealed diminished expression levels of the β-catenin-interacting partner PKM2 in glioma patients bearing IDH1-MT (Fig. 2C).
FIG 2.
Decreased nuclear PKM2 levels are accompanied by diminished β-catenin expression in IDH1-MT cells. (A to C) GSK3B (encoding GSK-3β), CTNNB1 (encoding β-catenin), and PKM (encoding PKM2) expression levels (obtained from TCGA) were compared between IDH1-WT and IDH1-MT cells. P values were determined by a 2-tailed, unpaired t test using GraphPad Prism. (D) Western blot analysis showing diminished levels of β-catenin and PKM2, both nuclear and cytosolic, in cells overexpressing IDH1-R132H and in glioma cells treated with 2-HG compared to IDH1-WT and untreated glioma cells, respectively. The blots are representative of data from three independent experiments with similar results. Blots were stripped and reprobed for C23 or β-actin to establish equivalent loading. Densitometric measurements were performed on the immunoblots by using ImageJ. The values indicate fold changes over the values for the controls. Bands were normalized to the levels of their corresponding loading controls. NE, nuclear extract; cyto, cytosolic extract. (E) 2-HG treatment diminishes nuclear localization of β-catenin and PKM2. Cells were immunostained with anti-β-catenin (β-catenin) (red) and anti-PKM2 (PKM2) (green). The nucleus is stained with DAPI (4′,6-diamidino-2-phenylindole) (blue). Merged images are shown. Representative images at a ×63 magnification from three independent experiments are shown. Adjacent line profiles show relative mean fluorescence intensities. *, P < 0.05; ***, P < 0.001.
Upon investigation of the status of β-catenin in cells that either stably or transiently overexpress IDH1-R132H, a significant decrease in nuclear β-catenin levels compared to those in cells harboring IDH1-WT was noted (Fig. 2D). Also, treatment with 2-HG diminished nuclear β-catenin levels in U87MG cells compared to those in untreated control cells (Fig. 2D). As STRING predicted that PKM2 is an interactive partner of β-catenin, and since nuclear PKM2 regulates gene expression (12) by acting as a coactivator of β-catenin (14), we next determined the status of PKM2 in cells that stably or transiently overexpressed IDH1-R132H or those that were treated with 2-HG. A decreased nuclear PKM2 level was observed in cells stably or transiently transfected with the IDH1-MT construct (Fig. 2D) or treated with 2-HG (Fig. 2D). This decrease in the nuclear localization of β-catenin and PKM2 was found to be significant in cells that either stably or transiently expressed IDH1-R132H or cells that were exposed to 2-HG (Fig. 2D).
Fluorescence microscopy revealed a decreased association of nuclear β-catenin and PKM2 in 2-HG-treated cells compared to that in untreated control cells (Fig. 2E), as shown by quantitative image analyses (Fig. 2E).
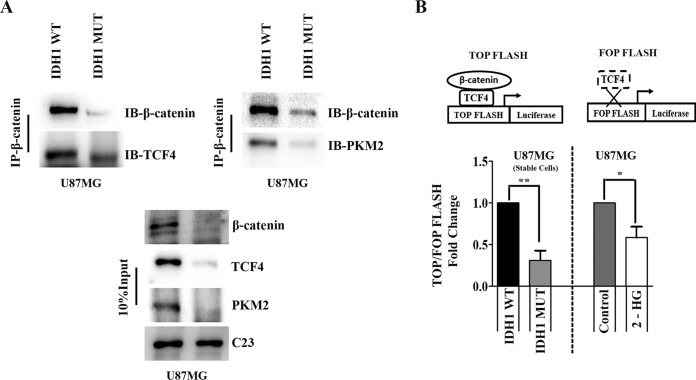
Diminished levels of the PKM2–β-catenin–TCF4 complex in gliomas overexpressing IDH1-R132H.
Given the diminished nuclear recruitment of PKM2 and β-catenin in IDH1-MT cells, immunoprecipitation (IP) studies were performed to reveal their association. The decreases in nuclear levels were concomitant with decreased levels of the PKM2–β-catenin complex (Fig. 3A). TCF4 is a key transcription factor that brings about the activation or repression of genes through the recruitment of β-catenin and associated coactivators/corepressors (28). Since the association of β-catenin with nuclear TCF4 triggers the transcriptional activation of its target genes, we next determined the status of TCF4 in this complex. Decreased TCF4 levels in the PKM2–β-catenin complex were observed in IDH1-MT cells compared to IDH1-WT cells (Fig. 3A). To investigate TCF4 transactivation activity, luciferase systems with a multimerized WT-TCF4 binding site (TOP FLASH) and a mutated TCF4 binding site (FOP FLASH) upstream of the luciferase reporter were used (29). Decreased levels of the PKM2–β-catenin–TCF4 complex in IDH1-MT cells were accompanied by decreased TCF4 luciferase reporter activity, thereby indicating decreased β-catenin transactivation activity (Fig. 3B). A similar decrease in TCF4 activity was observed in U87MG cells treated with 2-HG (Fig. 3B).
FIG 3.
Diminished TCF4/β-catenin transactivation activity in IDH1-MT cells. (A) Coimmunoprecipitation assays with anti-β-catenin antibody show diminished formation of the nuclear β-catenin–PKM2–TCF4 complex in cells overexpressing IDH1-R132H compared to that in cells overexpressing IDH1-WT. IB, immunoblotting. (B) TCF4 transactivation activity in glioma cells was determined by a luciferase reporter assay using TOP Flash and FOP Flash (TOP Flash mutant). The graph depicts TOP/FOP Flash fold changes in relative luciferase units with respect to the control. Values represent the means ± standard errors of the means from three independent experiments. P values were determined by a 2-tailed, paired t test using GraphPad Prism. *, P < 0.05; **, P < 0.01.
CD47 expression is dependent on β-catenin levels.
In the GBM data set of TCGA, CD47 expression was found to be significantly correlated with β-catenin expression (Fig. 4A). Since decreased CD47 expression was concomitant with decreased nuclear β-catenin accumulation, the involvement of β-catenin in CD47 expression was validated by its small interfering RNA (siRNA)-mediated knockdown in stably transfected IDH1-WT cells. Transfection with β-catenin siRNA decreased CD47 expression (Fig. 4B). Treatment of IDH1-WT glioma cells with iCRT14, which prevents the nuclear translocation of β-catenin, also diminished CD47 expression (Fig. 4C). To confirm that CD47 expression is dependent on nuclear β-catenin levels, a complementary experiment was performed to restore nuclear β-catenin levels in stable IDH1-R132H-overexpressing cells. The upregulation of β-catenin nuclear translocation by a small-molecule synergist of the Wnt/β-catenin signaling pathway, QS11, enhanced CD47 expression (Fig. 4C). While the interaction between β-catenin and TCF4 in IDH1-WT cells was abrogated upon iCRT14 treatment (Fig. 4D), an increase in this interaction was noted for IDH1-R132H-expressing cells treated with QS11 (Fig. 4E). As the TCF4–β-catenin interaction and its subsequent binding at the TCF4 binding sites can affect β-catenin transactivation, TCF4/β-catenin transactivation activity was determined in IDH1-WT and IDH1-MT cells upon the inhibition or pharmacological elevation of nuclear β-catenin levels, respectively. The reduction in TCF4/β-catenin transactivation activity upon the inhibition of β-catenin by iCRT14 treatment or siRNA-mediated knockdown (Fig. 4F) in IDH1-WT cells suggested the diminished assembly of TCF4 and β-catenin at the TCF4 binding site. This finding, together with the increased TCF4 promoter activity in IDH1-R132H cells upon QS11 treatment (Fig. 4F), indicated that the association of β-catenin with TCF4 is crucial for the regulation of β-catenin transactivation.
FIG 4.
CD47 expression is β-catenin dependent. (A) Pearson correlation coefficient analysis of CTNNB1 (encoding β-catenin) and CD47 mRNA levels in the GBM data set of TCGA. (B) siRNA-mediated β-catenin knockdown in IDH1-WT cells decreases CD47 levels as demonstrated by Western blotting. The transfection efficiency of β-catenin siRNA is shown. NS, nonspecific. (C) Western blot demonstrating CD47 and β-catenin expression levels in whole-cell lysates (WCL) and nuclear extracts (NE) of IDH1-WT cells treated with the β-catenin inhibitor iCRT14. Treatment with the β-catenin activator QS11 increased CD47 expression in IDH1-MT cells. Blots (B and C) are representative of data from three independent experiments with similar results. DMSO (dimethyl sulfoxide)-treated IDH1-WT and IDH1-MT glioma cells were used as controls for iCRT14 and QS11 treatments, respectively. Blots were stripped and reprobed for C23 or β-actin to establish equivalent loading. (D) Inhibition of β-catenin by iCRT14 diminishes the β-catenin–TCF4 interaction in cells harboring IDH1-WT. (E) Increased β-catenin–TCF4 interactions in IDH1-R132H-overexpressing cells upon treatment with the β-catenin activator QS11. (F and G) β-Catenin regulates TCF4 transactivation activity positively (F) and phagocytosis negatively (G) in glioma cells, as demonstrated by siRNA-mediated knockdown or pharmacological activation/inhibition of β-catenin. The graphs indicate fold changes with respect to the corresponding controls. Values represent the means ± standard errors of the means from 4 or 5 independent experiments. P values were determined by a 2-tailed, paired t test using GraphPad Prism. (H to J) β-Catenin expression (H), β-catenin–TCF4 interactions (I), and TCF4 transactivation activity (J) show positive correlations with CD47 levels and inverse correlations with phagocytosis. Densitometry data for β-catenin and CD47 under different treatment conditions were normalized to the values for the corresponding loading controls. Fold changes of normalized data were analyzed by using a Pearson correlation test in GraphPad Prism. Fold changes in TOP/FOP Flash values are considered for TCF4 transactivation activity, and fold changes in fluorescence are considered for phagocytosis. *, P < 0.05; **, P < 0.01; ***, P < 0.001. r indicates the Pearson correlation coefficient.
Increased phagocytosis of IDH1-MT glioma cells is dependent on β-catenin levels.
Since CD47 expression affects the ability of cells to be phagocytosed, and as β-catenin regulates CD47 expression, we investigated the role of β-catenin in the ability of IDH1-WT and IDH1-MT glioma cells to be phagocytosed. An increase in the phagocytosis of IDH1-WT glioma cells was observed upon the inhibition of β-catenin by iCRT14 treatment (Fig. 4G) or siRNA-mediated knockdown (Fig. 4G). This finding, together with the decreased phagocytosis of QS11-treated IDH1-MT glioma cells (Fig. 4G), indicated that the nuclear β-catenin level affects the phagocytosis of IDH1-WT and IDH1-MT glioma cells, in addition to its ability to regulate CD47 expression. This ability of β-catenin to regulate both CD47 expression and phagocytosis was reflected in data from correlation analyses. While significant positive correlations were found between CD47 levels and (i) β-catenin expression, (ii) β-catenin–TCF4 interactions, and (iii) TCF4/β-catenin transactivation activity (Fig. 4H to J), inverse correlations were observed between phagocytosis and (i) β-catenin expression, (ii) β-catenin–TCF4 interactions, and (iii) TCF4/β-catenin transactivation activity (Fig. 4H to J).
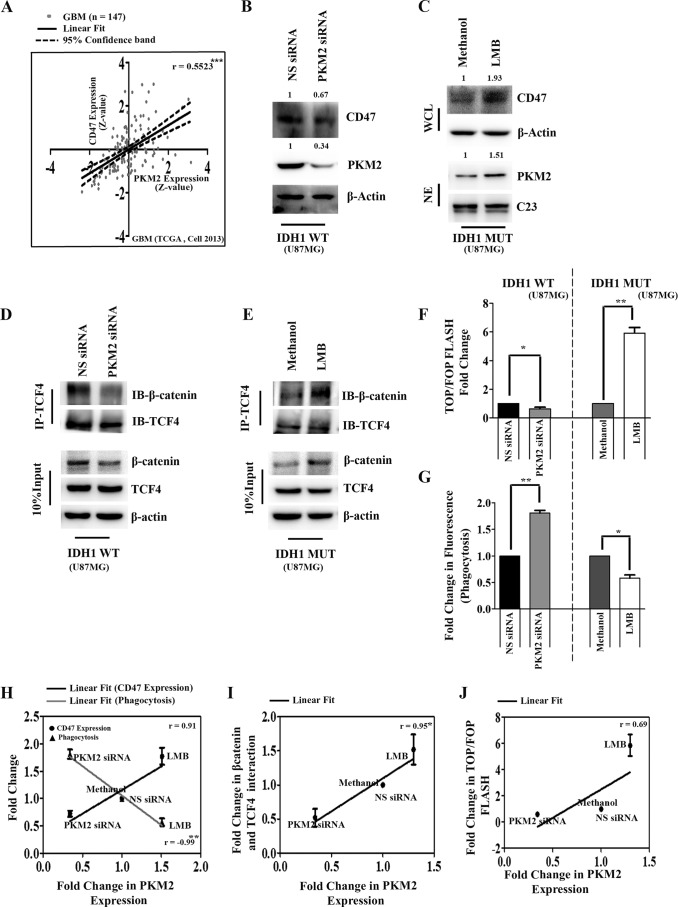
PKM2 serves as a coactivator of β-catenin to regulate CD47 expression.
The GBM data set of TCGA indicated that CD47 expression is significantly correlated with PKM2 expression (Fig. 5A). Based on our observations that β-catenin regulates CD47 expression and PKM2 is crucial for β-catenin transactivation, the possible involvement of nuclear PKM2 in the regulation of CD47 expression was investigated. siRNA-mediated PKM2 knockdown diminished CD47 levels in IDH1-WT cells (Fig. 5B). Treatment with leptomycin B (LMB), which abolishes the export of nuclear proteins, led to the nuclear localization of PKM2 (30). The leptomycin B-mediated increase in nuclear PKM2 accumulation enhanced CD47 levels in cells stably expressing IDH1-R132H (Fig. 5C). The inhibition of PKM2 in IDH1-WT cells or the increase in its nuclear retention in IDH1-MT cells affected the interaction of β-catenin and TCF4 (Fig. 5D and E) as well as TCF4 transactivational activity (Fig. 5F) the and phagocytosis of IDH1-WT and IDH1-MT glioma cells (Fig. 5G). Positive correlations were found between PKM2 levels and (i) CD47 expression, (ii) β-catenin–TCF4 interactions, and (iii) TCF4/β-catenin transactivation activity (Fig. 5H to 5J). Correlational analysis indicated an inverse correlation between PKM2 levels and phagocytosis (Fig. 5H).
FIG 5.
PKM2 functions as a coactivator of β-catenin to regulate CD47 expression. (A) Pearson correlation coefficient analysis of PKM (encoding PKM2) and CD47 mRNA levels in the GBM data set of TCGA. (B) siRNA-mediated knockdown of PKM2 decreases CD47 levels in IDH1-WT cells as determined by Western blotting. The transfection efficiency of PKM2 siRNA is shown. NS, nonspecific. (C) Western blot depicting CD47 and PKM2 expression levels in whole-cell (WCL) and nuclear extracts (NE) of IDH1-R132H cells treated with the nuclear export inhibitor leptomycin B (LMB). Blots in panels B and C are representative of data from three independent experiments with similar results. Blots were stripped and reprobed for C23 or β-actin to establish equivalent loading. Densitometric measurements were performed on the immunoblots by using ImageJ. The values indicate fold changes over values for controls. Bands were normalized to their corresponding β-actin or C23 levels. (D and E) siRNA-mediated knockdown of PKM2 in IDH1-WT cells decreases (D) while leptomycin B treatment increases (E) the β-catenin–TCF4 interaction in IDH1-MT cells. Methanol-treated IDH1-R132H cells were used as a control for LMB treatment. (F and G) PKM2 regulates TCF4 transactivation activity positively (F) and phagocytosis inversely (G) in glioma cells as demonstrated by siRNA-mediated knockdown or upon the nuclear retention of PKM2 by LMB. The graphs indicate fold changes with respect to the values for the corresponding controls. Values represent the means ± standard errors of the means from three independent experiments. P values were determined by a 2-tailed, paired t test using GraphPad Prism. (H to J) PKM2 expression shows positive correlations with CD47 levels (H), β-catenin–TCF4 interactions (I), and TCF4 transactivation activity (J) and inverse correlations with phagocytosis (H). Densitometry data for PKM2 and CD47 under different treatment conditions were normalized to the values for the corresponding loading controls. Fold changes of normalized data were analyzed by a Pearson correlation test using GraphPad Prism. Fold changes in TOP/FOP Flash values are considered for TCF4 transactivation activity, and fold changes in fluorescence are considered for phagocytosis. *, P < 0.05; **, P < 0.01; ***, P < 0.001. r indicates the Pearson correlation coefficient.
BRG1 is mutated in the ATPase domain in IDH1-MT gliomas.
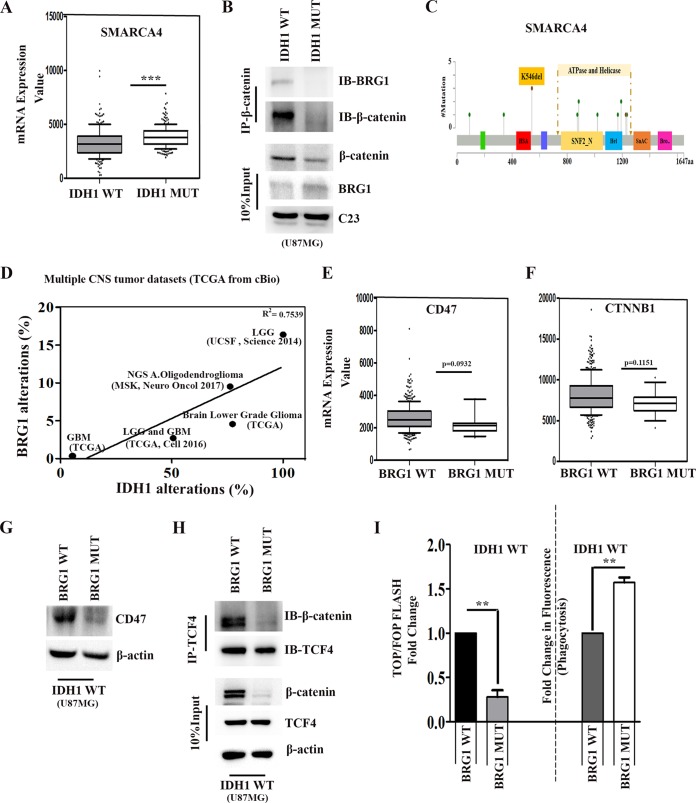
The recruitment of BRG1 to target gene promoters by β-catenin facilitates transcriptional activation through chromatin remodeling (31), and mutations of SMARCA4 (encoding BRG1) have been implicated in many cancer types (18). Upon comparing the statuses of SMARCA4 in IDH1-WT and IDH1-MT gliomas in TCGA data sets, the BRG1 expression level was found to be significantly elevated in IDH1-MT gliomas compared to that in IDH1-WT glioma (Fig. 6A). An increase in the BRG1 level was observed in IDH1-R132H-overexpressing cells compared to that in IDH1-WT cells (see Fig. S1A in the supplemental material). Despite the increase in the level of BRG1, its interaction with β-catenin was diminished in IDH1-MT cells compared to that in IDH1-WT cells (Fig. 6B). This could have resulted from an unavailability of β-catenin for binding with BRG1 in IDH1-MT cells (Fig. 6B).
FIG 6.
BRG1 regulates CD47 expression. (A) TCGA data set analysis of SMARCA4 (encoding BRG1) expression in IDH1-WT and IDH1-MT cells. (B) Coimmunoprecipitation assays with anti-β-catenin antibody show diminished nuclear BRG1–β-catenin complex formation despite increased nuclear BRG1 levels in IDH1-MT cells compared to those in IDH1-WT cells. (C) Mutational profile of BRG1 in LGGs and GBMs (modified from data from cBioPortal) indicating the missense mutation in the ATP binding and helicase activity site of BRG1, which results in the loss of ATPase-dependent BRG1 transcriptional activation. (D) Regression analysis indicates a positive correlation between IDH1 and BRG1 alterations across multiple CNS tumor data sets. R2 values were generated by a linear-fit model using GraphPad Prism. (E and F) Comparison of CD47 and CTNNB1 (β-catenin) expression levels between BRG1-WT and BRG1-MT obtained from TCGA. For panels A, E, and F, P values were determined by a 2-tailed, unpaired t test using GraphPad software. (G) Western blot depicting diminished CD47 expression in IDH1-WT cells transfected with pBJ5-BRG1-K-R. (H) Coimmunoprecipitation showing decreased β-catenin–TCF4 interactions in IDH1-WT cells transfected with pBJ5-BRG1-K-R. Blots in panels B, G, and H are representative of data from two to three independent experiments with similar results. Blots were stripped and reprobed for β-actin and c23 to establish equivalent loading. (I) BRG1 regulates TCF4 transactivation activity positively and phagocytosis inversely in glioma cells. The graphs indicate fold changes with respect to the values for the corresponding controls. Values represent the means ± standard errors of the means from three independent experiments. P values were determined by a 2-tailed, paired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Interestingly, BRG1 is mutated in ∼3% of all gliomas (32), which includes both IDH1-WT and IDH1-MT gliomas. Amino acids 766 to 1246 in BRG1 contain the ATP binding and helicase activity site. Mutations in this region can potentially inhibit the binding of BRG1 to ATP to subsequently inhibit ATP-dependent nucleosome rearrangement at the TCF4 binding site (33) (Fig. 6C). TCGA data suggested the likelihood that BRG1 mutations disrupt gene function and manifest as either early truncating or missense mutations. About >78% of BRG1 mutations occur in the ATP binding and helicase activity site (Fig. 6C). We assessed the correlation between IDH1 mutational status and BRG1 alterations (including mutations, genomic alterations, and gene expression changes) across multiple CNS tumors using cBioPortal data sets (34, 35, 52, 54, 55). This analysis identified a positive correlation between the IDH1 mutational status and Brg1 alterations across CNS tumor types included in the analysis (Fig. 6D). CNS tumors that are prone to IDH1 mutations, such as LGGs, are more likely to have an altered BRG1 mutational status. This indicated the presence of nonfunctional BRG1 in cases bearing IDH1-MT despite its significant increase in expression. In TCGA LGG and GBM data sets, patients were classified as bearing BRG1-WT (n = 381) and BRG1-MT (n = 14). Upon comparison of CD47 and CTNNB1 (β-catenin) expression levels in BRG1-WT and BRG1-MT gliomas, both CD47 and β-catenin levels were found to be lower in BRG1-MT than in BRG1-WT gliomas (Fig. 6E and F).
The ATPase subunit of BRG1 is crucial for TCF4 activity.
The ATPase activity of BRG1 is essential for its chromatin-remodeling activity, and loss-of-function mutations in this domain inhibit transcriptional activation (36). To test the potential involvement of ATP-dependent SWI/SNF chromatin-remodeling complexes in the transcriptional activation of CD47, transient transfections of wild-type or ATPase-defective BRG1 (mutation in ATPase subunit K-R) were carried out in IDH1-WT cells. Inhibition of CD47 expression in IDH1-WT cells upon the ectopic expression of ATPase-deficient BRG1 but not wild-type BRG1 (Fig. 6G) suggested that ATP-dependent chromatin remodeling is required for BRG1 to regulate CD47 expression. A diminished interaction between TCF4 and β-catenin was also observed in IDH1-WT cells upon transfection with ATPase-deficient BRG1 (Fig. 6H). Although ATPase-deficient BRG1 had no effect on TCF4 levels, it decreased β-catenin levels (Fig. 6H). This could possibly account for the diminished levels of β-catenin in IDH1-MT cells harboring ATPase-deficient BRG1 (Fig. 6F). To further demonstrate the role of BRG1 in the TCF4-responsive reporter gene, the effect of ectopic ATPase-deficient BRG1 in IDH1-WT cells was determined. Along with the decrease in TCF4 transactivational activity (Fig. 6I), an increase in the phagocytosis of IDH1-WT cells was observed upon the ectopic expression of ATPase-deficient BRG1 (Fig. 6I).
Recruitment of TCF4 to its binding site on the CD47 promoter is BRG1 dependent.
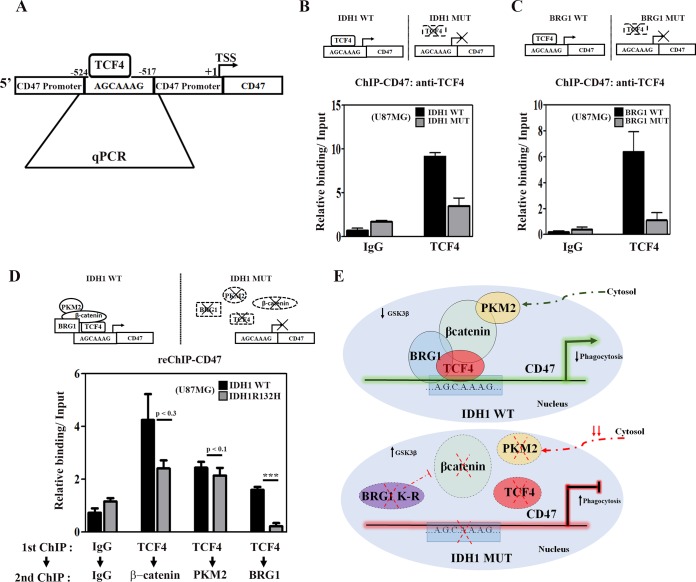
The β-catenin-mediated recruitment of SWI/SNF complexes through its interaction with BRG1 on TCF4 target gene promoters facilitates chromatin remodeling crucial for transcriptional activation (31). A GenBank search for the TCF4 consensus binding sequence (5′-[A/T][A/T]CAAAG-3′) on the CD47 promoter (positions −524 to −517) identified such a site (Fig. 7A). To determine whether TCF4 binding to the defined elements regulates CD47 expression, chromatin immunoprecipitation (ChIP) was performed to determine the enrichment of TCF4 at this site. A significant decrease in TCF4 occupancy at this site was observed in IDH1-MT cells compared to IDH1-WT cells (Fig. 7B). We next investigated whether BRG1 mutants lacking ATPase activity affect TCF4 enrichment on the CD47 promoter. ChIP analysis revealed a decrease in TCF4 occupancy in IDH1-WT cells upon the overexpression of ATPase-deficient BRG1 (Fig. 7C).
FIG 7.
IDH1-MT decreases the occupancy of the PKM2–TCF4–β-catenin–Brg1 complex at the TCF4 site on the CD47 promoter. (A) Schematic representation showing the putative TCF4 binding site on the CD47 promoter (bp −524 to −517). (B) ChIP-qPCR analysis indicating decreased TCF4 occupancy at the TCF4 binding site on the CD47 promoter in IDH1-MT compared to IDH1-WT cells. (C) Ectopic pBJ5-BRG1-K-R expression decreases the occupancy of TCF4 on the CD47 promoter in IDH1-WT cells. (D) ChIP–re-ChIP analyses indicate diminished recruitment of β-catenin, PKM2, and BRG1 at the TCF4 binding site of the CD47 promoter in IDH1-MT compared to IDH1-WT cells. Primary ChIP and secondary ChIP were performed with TCF4 and β-catenin, PKM2, or BRG1 and then analyzed by qPCR. Values represent the means ± standard errors of the means from two independent experiments. (E) Model depicting the role of PKM2 and BRG1 in β-catenin/TCF4-dependent CD47 expression. P values were determined by a 2-tailed, paired t test. ***, P < 0.001.
Corecruitment of PKM2, β-catenin, and BRG1 to the TCF4 site of the CD47 promoter is diminished in IDH1-MT cells.
The involvement of PKM2, BRG1, and β-catenin in TCF4 transactivational activity led us to investigate whether PKM2, β-catenin, and BRG1 are corecruited to the same TCF4 site using a sequential ChIP (ChIP–re-ChIP) assay. IDH1-MT and IDH1-WT cells were subjected to the first ChIP using anti-TCF4. A subsequent re-ChIP with anti-PKM2, anti-β-catenin, or anti-BRG1 was then performed. Results revealed that β-catenin, PKM2, as well as BRG1 are corecruited to the TCF4 site on the CD47 promoter in IDH1-WT cells (Fig. 7D). Although the enrichment of BRG1 on the TCF4 site of the CD47 promoter was significantly reduced in IDH1-MT cells, the levels of recruitment of PKM2 and β-catenin to the same site were comparable between IDH1-WT and IDH1-MT cells (Fig. 7D). Although not significant, a tendency toward diminished recruitment of PKM2 as well as β-catenin to the TCF4 site on the CD47 promoter was observed in IDH1-MT cells. This study provides evidence for a novel transcriptional regulatory network whereby the PKM2–β-catenin complex along with BRG1 are corecruited to the TCF4 site to regulate CD47 expression (Fig. 7E).
DISCUSSION
Macrophage-mediated phagocytosis of tumor cells via blockade of the antiphagocytic CD47-SIRPα interaction has shown promise in preclinical studies of pediatric high-grade gliomas (37). CD47 blockade is also known to promote the T-cell-mediated elimination of immunogenic tumors (21). Previous studies on gene expression data from several solid tumors, including gliomas, suggested that CD47 levels are a clinically relevant prognostic factor (20). The expression of CD47 is a general mechanism through which human solid tumor cells evade phagocytosis (20). Furthermore, using data generated by TCGA, diminished CD47 expression levels were observed in IDH1-MT compared to IDH1-WT gliomas. The ability of the IDH1 mutation to affect the immune component of gliomas has been associated with improved clinical outcomes (6). Our data indicate that the IDH1 mutational context of gliomas can significantly circumvent immune escape mechanisms by facilitating phagocytosis.
The regulation of TCF4/β-catenin transcriptional activity relies on the degradation of β-catenin by the axin–GSK-3β complex (38). Elevated GSK-3β levels coupled with diminished nuclear β-catenin translocation in IDH1-MT cells translated into lower expression levels of CD47. Given that the β-catenin–PKM2 interaction is crucial for glioma growth (14), perhaps the most striking finding in this work is that the nuclear PKM2 and β-catenin levels are positively correlated with CD47 expression. The ability of nuclear PKM2 to regulate the expression of glycolytic genes such as LDHA, which is crucial for the Warburg effect, is known (39). As LDHA is silenced in IDH-MT gliomas (40), it is possible that decreased nuclear PKM2 levels in IDH1-R132H cells also contribute to the altered metabolic profile observed for these tumors (41). Our findings in conjunction with data from a previous study highlight the nonmetabolic function of PKM2 in gene transcription as a transactivator of β-catenin (14). Importantly, our study has highlighted CD47 as a novel target of PKM2.
The SWI/SNF family of ATPases facilitates transcription factor binding to nucleosomal DNA in an ATP-dependent manner (42). The catalytic ATPase subunit in the SWI/SNF complex mediates nucleosome repositioning to regulate the transcription of its targets. Our ChIP and re-ChIP data revealed the recruitment of the PKM2–β-catenin–BRG1–TCF4 complex to the TCF4 site on the CD47 promoter. Importantly, the significant decrease in the recruitment of BRG1 to the TCF4 site in IDH1-MT cells points to the importance of Brg1-facilitated chromatin remodeling as a prerequisite for the transcriptional activation of CD47. Moreover, PKM2 and β-catenin, which are components of the BRG1-TCF4 transcriptional complex, also showed a tendency toward diminished recruitment to the TCF4 site in IDH1-MT cells. The presence of nonfunctional BRG1 with a mutation in the ATPase domain, coupled with diminished PKM2 and β-catenin levels in IDH1-MT gliomas, may have important ramifications for the responsiveness of these tumors to immune surveillance by exhibiting a greater propensity for phagocytosis. Our observations consolidate some unrelated but well-established findings, such as (i) the role of PKM2 as a transcriptional coactivator of β-catenin, (ii) altered immune-evasive responses in IDH1-MT cells, and (iii) BRG1-mediated chromatin remodeling in TCF4 activation.
By remodeling the epigenomic landscape through modulating methylation patterns and changing transcriptional programs, the IDH1 mutation establishes CIMP (5). CIMP, a prevalent molecular signature in low-grade gliomas, confers improved chances of survival chances of in these tumors. The occurrence of IDH1 mutations predicts longer survival and greater sensitivity to chemotherapy in gliomas (43), and CD47 is also known to regulate metabolic pathways that control resistance to ionizing radiation (44). While the inactivation of the BRG1 ATPase domain increases sensitivity to known chemotherapeutic drugs (45), the inhibition of CD47 signaling increases the radiosensitivity of tumors while maintaining the viability of normal tissues (46). Importantly, IDH1 mutations that are common in LGGs are predictive of a better outcome when found in a GBM (47). It is possible that the presence of a mutation in the ATPase domain of BRG1 coupled with diminished CD47 expression in LGGs harboring IDH1-MT cells not only renders glioma cells vulnerable to immune surveillance but also could enhance their responsiveness to therapy. As elevated CD47 levels have a direct bearing on the immune escape mechanism in gliomas, future endeavors to inhibit CD47 by targeting β-catenin–PKM2 cross talk may provide a novel therapeutic approach for modulating immune-evasive responses in glioma. These data provide new insight into the biology of IDH1 by suggesting mechanisms regulating the expression of CD47, a critical regulator of innate immune surveillance. Given the significant contribution of immune cells such as microglia in glioma, the increased ability of these professional phagocytes to engulf IDH1-MT glioma cells may contribute in part to the differences in prognoses associated with mutant IDH1 and wild-type IDH1.
MATERIALS AND METHODS
Cell culture and generation of stable cell lines.
The human glioma cell lines U87MG (ATCC CRL-11268) and T98G (ATCC CRL-1690) and the microglial cell line CHME3 (a gift from Anirban Basu, National Brain Research Centre, India) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS) (Gibco, Gaithersburg, MD) and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Gibco). For the establishment of stable glioma cell lines, U87MG cells transfected with the pEGFP IDH1-WT and pEGFP IDH1-R132H constructs (gifts from Hai Yan, Duke University School of Medicine, USA) were selected in G418 (700 μg/ml)-containing medium for 1 week. The G418-resistant cells were then evaluated for the expression of IDH1-R132H by Western blotting.
Treatment and transfection.
Glioma cells stably harboring IDH1-WT and IDH1-R132H were transfected with 50 nmol/liter duplex PKM2, β-catenin or nonspecific siRNA, or plasmids pBJ5-BRG1 (plasmid 17873; Addgene, Cambridge, MA) and pBJ5-BRG1-K-R (plasmid 17874; Crabtree Lab, Addgene) by using Lipofectamine RNAiMax reagent or Lipofectamine 2000 reagent (Life Technologies-Invitrogen, Carlsbad, CA), respectively, as described previously (22). The control nontargeting siRNA (catalog no. D-001210-03-20) as well as siRNAs targeting β-catenin (catalog no. L-003482-00-0005) and PKM2 (catalog no. L-006781-00) were obtained from Dharmacon (Thermo Fisher Scientific, Waltham, MA). Upon attaining semiconfluence, cells were transferred to serum-free medium (SFM), and after 6 h, cells were treated with 40 mM d-α-hydroxyglutaric acid disodium salt (catalog no. H8378; Sigma, St. Louis, MO) or 50 μM iCRT14 (catalog no. 4299, Tocris Biosciences, Bristol, UK), 10 μM QS11 (catalog no. 3324; Tocris Biosciences), or 15 nM leptomycin B (catalog no. L2913; Sigma) in SFM. For controls, cells were treated with dimethyl sulfoxide (DMSO) or methanol. Similarly, T98G cells were cotransfected with IDH1-WT or IDH1-R132H constructs and different siRNAs or plasmids, as was done for the stable cells. After 24 h or 48 h of treatment, cells were harvested and analyzed by Western blotting.
Western blot analysis.
Western blot analysis was performed with protein from whole-cell extracts, cytosolic extracts, and nuclear extracts of cells transfected with different constructs or siRNA and treated with different inhibitors and activators as described previously (22), using mouse monoclonal anti-CD47 (1:1,000) (catalog no. ab9089; Abcam, Cambridge, UK), rabbit monoclonal anti-PKM2 (1:1,000) (catalog no. 4053; Cell Signaling, Boston, MA), rabbit monoclonal anti-GSK-3β (1:1,000) (catalog no. 9315; Cell Signaling), rabbit polyclonal anti-non-phospho-β-catenin (1:1,000) (catalog no. 4270; Cell Signaling), rabbit monoclonal anti-β-catenin (1:1,000) (catalog no. 8480; Cell Signaling), mouse monoclonal anti-β-catenin (1:1,000) (catalog no. ab19450; Abcam), goat polyclonal anti-TCF4 (1:1,000) (catalog no. sc-8631; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-BRG1 (1:1,000) (catalog no. ab4081; Abcam), mouse monoclonal anti-IDH1-R132H (clone HMab-1; Sigma-Aldrich), and rabbit polyclonal anti-IDH1 (1:1,000) (catalog no. ab135659; Abcam). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Vector Laboratories Inc. (Burlingame, CA). After the addition of a chemiluminescent reagent (Millipore, Billerica, MA), blots were exposed to the Chemigenius Bioimaging system (Syngene, Cambridge, UK), and images were captured with Gene Snap software (Syngene). The blots were stripped and reprobed with anti-β-actin (1:1,000) (catalog no. A3854; Sigma-Aldrich) or anti-C23 (1:1,000) (catalog no. sc-55486; Santa Cruz Biotechnology) to determine equivalent loading as described previously (48).
Coimmunoprecipitation.
For endogenous IP, nuclear extracts from stable IDH1-WT and IDH1-MT cells were incubated with 4 μg of the indicated antibodies at 4°C for 16 h. The lysate was then incubated for 4 h with protein A/G-Sepharose beads (GE Health Care Life Sciences, Marlborough, MA). Beads were washed five times in IP buffer, and immunoprecipitated proteins were resolved by 8 to 10% SDS-PAGE. Briefly, the cells were lysed, and the endogenous β-catenin and TCF4 proteins were immunoprecipitated with anti-β-catenin and anti-TCF4 antibodies. Western blot analysis was performed with the immunoprecipitates by using specific antibodies (48).
Quantitation of d-2-hydroxyglutarate.
The d-2-HG level was measured by using a d-2-HG assay kit (catalog no. K213-100; Bio-Vision Inc., Milpitas, CA) according to the manufacturer's instructions. Briefly, protein extracts from glioma cells stably transfected with IDH1-WT and IDH1-MT were suspended in 100 μl of d-2-HG assay buffer. The lysates were further mixed with a reaction mixture (50 μl/reaction) containing 44 μl of d-2-HG assay buffer, 2 μl of d-2-HG enzyme mix, and 2 μl of the substrate mix and dispensed onto a 96-well microplate. This was followed by incubation at 37°C for 60 min. The optical density (OD) was recorded at 450 nm from both samples and standards by using an enzyme-linked immunosorbent assay (ELISA) plate reader (Tecan pro200; Männedorf, Zürich, Switzerland). Results were calculated as described above and expressed as fold changes over the values for the controls.
Luciferase reporter gene assay.
Luciferase reporter gene assays were performed with cells transfected with different combinations of TCF4 TOP Flash/FOP Flash luciferase, β-catenin-targeting siRNA, PKM2-targeting siRNA, nonspecific siRNA, pBJ5-BRG1, and pBJ5-BRG1-K-R and treated with LMB, iCRT4, and QS11. Briefly, IDH1-WT and IDH1-MT cells (T98G or U87MG stable cells) were transfected with 300 ng of the indicated luciferase reporter constructs and 20 ng of the Renilla luciferase vector (pRL-TK; Promega, Madison, WI) for the normalization of the transfection efficiency. Cell lysates were analyzed with the Dual-Luciferase reporter assay system (Promega) as described previously (22). Plasmid pBJ5-BRG1, plasmid pBJ5-BRG1-K-R, the M50 Super 8× TOP Flash TCF4 reporter plasmid, and the M51 Super 8× FOP Flash TCF4 reporter plasmid (plasmids 12456 and 12457; Moon Laboratory) were obtained from Addgene.
Quantitative real-time PCR.
To analyze the endogenous IDH1 mRNA levels in IDH1-WT and IDH1-MT cells, RNA was isolated by using an RNeasy kit (Qiagen, Hilden, Germany), and cDNA was synthesized by using a High Capacity cDNA reverse transcription kit (Applied Biosystems Inc., Foster City, CA) on a Veriti thermal cycler (Applied Biosystems Inc.). Real-time PCR was performed as described previously (49), using a ViiA7 real-time thermocycler (Applied Biosystems Inc.), and results were plotted as fold changes over the values for the control (empty vector) for the IDH1 mRNA transcript. Values for all samples were normalized to their respective 18S rRNA threshold cycle (CT) values.
The quantitative PCR (qPCR) primers used are as follows: IDH1 forward and reverse primers 5′-GTGCCTGGAGTTTAAAAGGCGA-3′ and 5′-CTACCACAGAACCGCCACTG-3′, respectively, and 18S rRNA forward and reverse primers 5′-CAGCCACCCGAGATTGAGCA-3′ and 5′-TAGTAGCGACGGGCGGTGTG-3′, respectively.
Phagocytosis assay.
Glioma cells under different treatment conditions were fluorescently labeled with carboxyfluorescein succinimidyl ester (CFSE) according to the manufacturer's protocol (Life Technologies-Invitrogen). After 2 h, CFSE-labeled glioma cells were harvested and added to CHME3 microglial cells previously seeded into 24-well plates. Following incubation for 2 h at 37°C in a CO2 incubator, the CFSE-labeled glioma cells were removed, and 100 μl trypan blue was added to the wells for 1 min at room temperature. Upon the removal of trypan blue, microglial cells were washed and subjected to mild lysis. Phagocytosis was then quantified by measuring the fluorescence in a microplate reader (Thermo Electron, Waltham, MA), using 480-nm excitation and 520-nm emission wavelengths (50).
ChIP, re-ChIP, and ChIP-qPCR assays.
ChIP was performed by enzymatic DNA shearing (Chip-IT Enzymatic; Active Motif, Carlsbad, CA) as previously described (22). Cells were fixed in 1% formaldehyde at room temperature for 10 min. Isolated nuclei were lysed, and DNA was enzymatically sheared with the Enzymatic Shearing kit (Active Motif). Anti-TCF4 (catalog no. sc-8631; Santa Cruz Biotechnology) antibody was used for IP, and a nonspecific IgG antibody (Abcam) was used as a control. For re-ChIP, anti-TCF4 (catalog no. sc-8631; Santa Cruz Biotechnology) antibody was used for primary IP, followed by anti-PKM2, anti-β-catenin, or anti-BRG1 for the second IP, and a nonspecific IgG antibody (Abcam) was used as a control. Following reverse cross-linking and DNA purification, DNA from the input (diluted 1:10) and immunoprecipitated DNA recovered by ChIP were analyzed by qPCR using Power SYBR green PCR master mix (Applied Biosystems Inc.) with a ViiA7 real-time thermocycler (Applied Biosystems Inc.) for 40 cycles. Relative fold enrichment with respect to the input was calculated based on the CT as100/2ΔCT, where ΔCT = CT (IP) − (CT input − log2 dilution factor). No target sequences were amplified from the same material as a control for IP in each sample, and results were corrected accordingly. The sequences of the primers used for qPCR analyses of the amplified regions were as follows: CD47 (ChIP) forward and reverse primers 5′-TGGGAGTGAAAGCAAAGAGG-3′ and 5′-CTTCCAGGTCACGTCCTGTC-3′, respectively, and no-target-region forward and reverse primers 5′-AAGTCGGTGAGAAGGGGTTT-3′ and 5′-AAGGAAGAGTTTCCAGCAAGC-3′, respectively.
TCGA analysis.
For studying CD47, β-catenin, PKM2, BRG1, and GSK-3β expression in IDH1 mutants, transcriptome sequencing (RNA-seq) data from TCGA LGG and GBM data sets (51) were downloaded and segregated by IDH mutation status. Patients were classified as bearing IDH1-WT or IDH1-MT tumors. The mutation data for BRG1 were downloaded from cBioPortal (35), and patients were classified as bearing BRG1-WT and BRG1-MT.
Statistical analysis.
All comparisons between groups were performed by using two-tailed paired Student's t test unless otherwise stated. All P values of <0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a research grant from the Department of Biotechnology (DBT) (Government of India grant no. BT/Med/30/SP11016/2015) to E.S.
We acknowledge technical assistance of Shanker Joshi and Rajesh Kumar Kumawat.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00001-18.
REFERENCES
- 1.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. 2008. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 2.Bleeker FE, Atai NA, Lamba S, Jonker A, Rijkeboer D, Bosch KS, Tigchelaar W, Troost D, Vandertop WP, Bardelli A, Van Noorden CJ. 2010. The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol 119:487–494. doi: 10.1007/s00401-010-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amankulor NM, Kim Y, Arora S, Kargl J, Szulzewsky F, Hanke M, Margineantu DH, Rao A, Bolouri H, Delrow J, Hockenbery D, Houghton AM, Holland EC. 2017. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev 31:774–786. doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Rao A, Sette P, Deibert C, Pomerantz A, Kim WJ, Kohanbash G, Chang Y, Park Y, Engh J, Choi J, Chan T, Okada H, Lotze M, Grandi P, Amankulor N. 2016. IDH mutant gliomas escape natural killer cell immune surveillance by downregulation of NKG2D ligand expression. Neuro Oncol 18:1402–1412. doi: 10.1093/neuonc/now061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohanbash G, Carrera DA, Shrivastav S, Ahn BJ, Jahan N, Mazor T, Chheda ZS, Downey KM, Watchmaker PB, Beppler C, Warta R, Amankulor NA, Herold-Mende C, Costello JF, Okada H. 2017. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest 127:1425–1437. doi: 10.1172/JCI90644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, Hu J, Vinjamoori A, Gan B, Sahin E, Chheda MG, Brennan C, Wang YA, Hahn WC, Chin L, DePinho RA. 2010. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sareddy GR, Panigrahi M, Challa S, Mahadevan A, Babu PP. 2009. Activation of Wnt/beta-catenin/Tcf signaling pathway in human astrocytomas. Neurochem Int 55:307–317. doi: 10.1016/j.neuint.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Cui D, Ren J, Shi J, Feng L, Wang K, Zeng T, Jin Y, Gao L. 2016. R132H mutation in IDH1 gene reduces proliferation, cell survival and invasion of human glioma by downregulating Wnt/beta-catenin signaling. Int J Biochem Cell Biol 73:72–81. doi: 10.1016/j.biocel.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. 2012. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. 2011. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. 2011. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee J, Phillips JJ, Zheng S, Wiencke J, Ronen SM, Pieper RO. 2013. Pyruvate kinase M2 expression, but not pyruvate kinase activity, is up-regulated in a grade-specific manner in human glioma. PLoS One 8:e57610. doi: 10.1371/journal.pone.0057610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Yung WKA, Lu Z. 2012. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosimann C, Hausmann G, Basler K. 2009. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 18.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng D, Volkmer JP, Willingham SB, Contreras-Trujillo H, Fathman JW, Fernhoff NB, Seita J, Inlay MA, Weiskopf K, Miyanishi M, Weissman IL. 2013. Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc Natl Acad Sci U S A 110:11103–11108. doi: 10.1073/pnas.1305569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, Lovelace P, Scheeren FA, Chao MP, Weiskopf K, Tang C, Volkmer AK, Naik TJ, Storm TA, Mosley AR, Edris B, Schmid SM, Sun CK, Chua MS, Murillo O, Rajendran P, Cha AC, Chin RK, Kim D, Adorno M, Raveh T, Tseng D, Jaiswal S, Enger PO, Steinberg GK, Li G, So SK, Majeti R, Harsh GR, van de Rijn M, Teng NN, Sunwoo JB, Alizadeh AA, Clarke MF, Weissman IL. 2012. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A 109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Pu Y, Cron K, Deng L, Kline J, Frazier WA, Xu H, Peng H, Fu YX, Xu MM. 2015. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat Med 21:1209–1215. doi: 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh S, Paul A, Sen E. 2013. Tumor necrosis factor alpha-induced hypoxia-inducible factor 1alpha–beta-catenin axis regulates major histocompatibility complex class I gene activation through chromatin remodeling. Mol Cell Biol 33:2718–2731. doi: 10.1128/MCB.01254-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopatz J, Beutner C, Welle K, Bodea LG, Reinhardt J, Claude J, Linnartz-Gerlach B, Neumann H. 2013. Siglec-h on activated microglia for recognition and engulfment of glioma cells. Glia 61:1122–1133. doi: 10.1002/glia.22501. [DOI] [PubMed] [Google Scholar]
- 25.Watters JJ, Schartner JM, Badie B. 2005. Microglia function in brain tumors. J Neurosci Res 81:447–455. doi: 10.1002/jnr.20485. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J 17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H. 2002. Wnt signaling controls the phosphorylation status of beta-catenin. J Biol Chem 277:17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- 28.Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 29.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 13:680–685. doi: 10.1016/S0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 30.Li N, Feng L, Liu H, Wang J, Kasembeli M, Tran MK, Tweardy DJ, Lin SH, Chen J. 2016. PARP inhibition suppresses growth of EGFR-mutant cancers by targeting nuclear PKM2. Cell Rep 15:843–856. doi: 10.1016/j.celrep.2016.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J 20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shain AH, Pollack JR. 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trotter KW, Archer TK. 2008. The BRG1 transcriptional coregulator. Nucl Recept Signal 6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 37.Gholamin S, Mitra SS, Feroze AH, Liu J, Kahn SA, Zhang M, Esparza R, Richard C, Ramaswamy V, Remke M, Volkmer AK, Willingham S, Ponnuswami A, McCarty A, Lovelace P, Storm TA, Schubert S, Hutter G, Narayanan C, Chu P, Raabe EH, Harsh G IV, Taylor MD, Monje M, Cho YJ, Majeti R, Volkmer JP, Fisher PG, Grant G, Steinberg GK, Vogel H, Edwards M, Weissman IL, Cheshier SH. 2017. Disrupting the CD47-SIRPalpha anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci Transl Med 9:eaaf2968. doi: 10.1126/scitranslmed.aaf2968. [DOI] [PubMed] [Google Scholar]
- 38.Nusse R, Clevers H. 2012. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. 2012. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol 14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chesnelong C, Chaumeil MM, Blough MD, Al-Najjar M, Stechishin OD, Chan JA, Pieper RO, Ronen SM, Weiss S, Luchman HA, Cairncross JG. 2014. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro Oncol 16:686–695. doi: 10.1093/neuonc/not243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H. 2011. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A 108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 43.Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, Paris S, Boisselier B, Idbaih A, Laigle-Donadey F, Hoang-Xuan K, Sanson M, Delattre JY. 2010. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 75:1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 44.Miller TW, Soto-Pantoja DR, Schwartz AL, Sipes JM, DeGraff WG, Ridnour LA, Wink DA, Roberts DD. 2015. CD47 receptor globally regulates metabolic pathways that control resistance to ionizing radiation. J Biol Chem 290:24858–24874. doi: 10.1074/jbc.M115.665752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Q, Sharma S, Cui H, LeBlanc SE, Zhang H, Muthuswami R, Nickerson JA, Imbalzano AN. 2016. Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget 7:27158–27175. doi: 10.18632/oncotarget.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maxhimer JB, Soto-Pantoja DR, Ridnour LA, Shih HB, Degraff WG, Tsokos M, Wink DA, Isenberg JS, Roberts DD. 2009. Radioprotection in normal tissue and delayed tumor growth by blockade of CD47 signaling. Sci Transl Med 1:3ra7. doi: 10.1126/scitranslmed.3000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. 2015. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tewari R, Choudhury SR, Ghosh S, Mehta VS, Sen E. 2012. Involvement of TNFalpha-induced TLR4-NF-kappaB and TLR4-HIF-1alpha feed-forward loops in the regulation of inflammatory responses in glioma. J Mol Med (Berl) 90:67–80. doi: 10.1007/s00109-011-0807-6. [DOI] [PubMed] [Google Scholar]
- 49.Ahmad F, Dixit D, Sharma V, Kumar A, Joshi SD, Sarkar C, Sen E. 2016. Nrf2-driven TERT regulates pentose phosphate pathway in glioblastoma. Cell Death Dis 7:e2213. doi: 10.1038/cddis.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uff CR, Pockley AG, Phillips RK. 1993. A rapid microplate-based fluorometric assay for phagocytosis. Immunol Invest 22:407–413. doi: 10.3109/08820139309063419. [DOI] [PubMed] [Google Scholar]
- 51.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, Huse JT, Ciriello G, Poisson LM, Barnholtz-Sloan JS, Berger MS, Brennan C, Colen RR, Colman H, Flanders AE, Giannini C, Grifford M, Iavarone A, Jain R, Joseph I, Kim J, Kasaian K, Mikkelsen T, Murray BA, O'Neill BP, Pachter L, Parsons DW, Sougnez C, Sulman EP, Vandenberg SR, Van Meir EG, von Deimling A, Zhang H, Crain D, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton T, Sherman M, et al. 2015. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov IV, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJM, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF. 2014. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, TCGA Research Network . 2013. The somatic genomic landscape of glioblastoma. Cell 155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas AA, Abrey LE, Terziev R, Raizer J, Martinez NL, Forsyth P, Paleologos N, Matasar M, Sauter CS, Moskowitz C, Nimer SD, DeAngelis LM, Kaley T, Grimm S, Louis DN, Cairncross JG, Panageas KS, Briggs S, Faivre G, Mohile NA, Mehta J, Jonsson P, Chakravarty D, Gao J, Schultz N, Brennan CW, Huse JT, Omuro A. 2017. Multicenter phase II study of temozolomide and myeloablative chemotherapy with autologous stem cell transplant for newly diagnosed anaplastic oligodendroglioma. Neuro Oncol 19:1380–1390. doi: 10.1093/neuonc/nox086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, Anjum S, Wang J, Manyam G, Zoppoli P, Ling S, Rao AA, Grifford M, Cherniack AD, Zhang H, Poisson L, Carlotti CG Jr, Tirapelli DP, Rao A, Mikkelsen T, Lau CC, Yung WK, Rabadan R, Huse J, Brat DJ, Lehman NL, Barnholtz-Sloan JS, Zheng S, Hess K, Rao G, Meyerson M, Beroukhim R, Cooper L, Akbani R, Wrensch M, Haussler D, Aldape KD, Laird PW, Gutmann DH. 2016. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164:55–63. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.