ABSTRACT
Conserved from yeast to humans, the NuA4 histone acetyltransferase is a large multisubunit complex essential for cell viability through the regulation of gene expression, genome maintenance, metabolism, and cell fate during development and stress. How the different NuA4 subunits work in concert with one another to perform these diverse functions remains unclear, and addressing this central question requires a comprehensive understanding of NuA4's molecular architecture and subunit organization. We have determined the structure of fully assembled native yeast NuA4 by single-particle electron microscopy. Our data revealed that NuA4 adopts a trilobal overall architecture, with each of the three lobes constituted by one or two functional modules. By performing cross-linking coupled to mass spectrometry analysis and in vitro protein interaction studies, we further mapped novel intermolecular interfaces within NuA4. Finally, we combined these new data with other known structural information of NuA4 subunits and subassemblies to construct a multiscale model to illustrate how the different NuA4 subunits and modules are spatially arranged. This model shows that the multiple chromatin reader domains are clustered together around the catalytic core, suggesting that NuA4's multimodular architecture enables it to engage in multivalent interactions with its nucleosome substrate.
KEYWORDS: NuA4, cross-linking coupled to mass spectrometry, electron microscopy, histone acetyltransferase, yeast
INTRODUCTION
Eukaryotic genomic DNA exists in the form of chromatin generated through higher-ordered packing of nucleosomes. The nucleosome is composed of approximately 147 bp of DNA wrapped around an octameric protein complex formed by four core histones (H2A, H2B, H3, and H4) (1). A key mechanism for regulating gene expression involves posttranslational modifications (PTMs) of nucleosomal histones, which can lead to changes in chromatin structure or can act as signaling events that regulate the association of factors critical for transcription regulation (2). Acetylation, the covalent linkage of an acetyl group to the epsilon amine of lysine, is a major histone PTM correlated with active transcription. This PTM promotes a more open chromatin structure through neutralizing the positive charges on histones and reducing their affinity toward DNA and neighboring nucleosomes and also creating binding platforms for “reader protein domains” such as bromodomains (3). Although a wealth of high-resolution structural data are now available on the histone acetyltransferase (HAT) enzymes that catalyze histone acetylation (4), most HAT enzymes function in the context of the multiprotein HAT complexes (5). How HAT enzymes work in concert with other noncatalytic subunits within these assemblies is poorly understood.
Conserved from yeast to humans, the nucleosomal acetyltransferase of histone H4 (NuA4) complex is a HAT complex that mediates multiple nuclear processes, ranging from transcription activation to DNA double-strand break repair, being essential for cell viability and development (3, 6–10). NuA4 has been shown to acetylate nonhistone targets, including those that play key roles in metabolism, transcription, cell cycle progression, RNA processing, and autophagy (11–14). The prototypical NuA4 is the one from the model organism Saccharomyces cerevisiae, which is an ∼1-MDa complex composed of 13 unique protein subunits (15). Previous genetics and biochemical studies showed that these yeast NuA4 subunits are organized into four distinct functional modules assembled around the Eaf1 subunit: the Piccolo module, containing the essential catalytic subunit Esa1, and accessory factors Eaf6, Epl1, and Yng2, that mediate nucleosome acetylation (16); the TINTIN module (Eaf3, Eaf5, and Eaf7) that facilitates RNA polymerase (Pol) II binding and transcription elongation (9); the SWR1-C module (Act1, Arp4, Swc4, and Yaf9) that shares subunits with the SWR1 chromatin-remodelling complex (17); and the recruitment module (Tra1) that mediates interaction with transcription activators (18). How these four functionally distinct NuA4 modules cooperate with one another is unclear.
To gain deeper insights into the overall architecture and subunit organization of NuA4, we have developed an improved purification procedure that enabled us to isolate fully assembled native yeast NuA4 while preserving its structural integrity. Our subsequent single-particle electron microscopy (EM) analysis revealed that, contrary to a previous report (14), yeast NuA4 adopts an overall trilobal architecture with the shared and TINTIN modules projecting away from a central core consisting of the recruitment and Piccolo modules. Complementary chemical cross-linking coupled to mass spectrometry (CXMS) analysis combined with in vitro biochemical pulldown further showed that the N terminus of Eaf1 anchors the TINTIN module and associates with the C terminus of Epl1 within NuA4. By combining these new data with other available structural data, including the recent high-resolution structures of Tra1 and the Piccolo module (19, 20), we generated a multiscale structural model of NuA4. This model suggested that its multimodular arrangement enables NuA4 to potentially engage its nucleosome substrate through multivalent interactions.
RESULTS
Yeast NuA4 adopts a trilobal overall architecture.
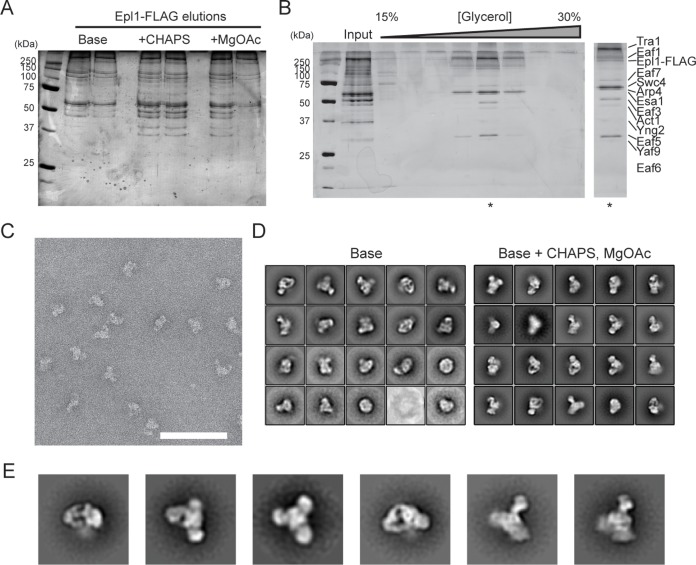
The previously reported yeast NuA4 structure determined by single-particle EM showed a single modular architecture that resembles the “head domain” of the related yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex, which shares a common subunit, the ∼430-kDa subunit Tra1 (14). The authors of that study proposed that the Tra1 subunit likely occupies half of the observed structure. However, our recent structural investigation of yeast SAGA indicated that the “head domain” is occupied predominantly by Tra1 (21), a finding that was substantiated by the very recently reported high-resolution cryo-electron microscopy (cryo-EM) structure of yeast Tra1 (20). To visualize the full yeast NuA4, we first purified this complex from S. cerevisiae expressing C-terminally FLAG-tagged Epl1 using anti-FLAG affinity purification, followed by glycerol gradient centrifugation, an approach that has enabled us previously to preserve the structural integrity of SAGA for structural analysis (21), and then analyzed the purified complex by negative-stain single-particle EM. Our two-dimensional (2D) analysis showed that although we obtained several class averages that resembled the earlier NuA4 structural study, we also detected many class averages that contained a prominent additional density (data not shown). Furthermore, when we analyzed NuA4 that was subject to gradient fixation (GraFix), which involves limited cross-linking using glutaraldehyde (22), the resulting data set showed multiple class averages containing even more than one additional density (Fig. 1D, left panel). The observation that many of these additional densities are blurry is indicative of conformational or compositional heterogeneity of purified NuA4.
FIG 1.
Overall architecture of yeast NuA4. (A) Optimizing buffer conditions for purification of native NuA4 from yeast expressing C-terminally FLAG-tagged Epl1. Elutions from FLAG purification of NuA4 using the base buffer, base plus CHAPS buffer, and base plus CHAPS plus magnesium acetate (MgOAc) buffer were analyzed by SDS-PAGE, followed by silver staining. (B) Glycerol gradient ultracentrifugation purification. Elutions from the FLAG purification in the base plus CHAPS plus MgOAc buffer were subjected to glycerol gradient ultracentrifugation, and individual fractions from the gradient were analyzed by SDS-PAGE. The fraction indicated by an asterisk was used for further EM analysis. (C) Representative segment of a negative-stain EM raw image of purified NuA4. Scale bar, 100 nm. (D) 2D class averages obtained for NuA4 purified from anti-FLAG affinity chromatography and gradient fixation (GraFix) using the base buffer with or without the addition of CHAPS and MgOAc. The two sets of averages (base buffer, modified buffer) were calculated from 9,869 and 23,165 particles, respectively. The box edge length is approximately 467 Å. (E) 2D class averages NuA4 in different orientations. The box edge length is approximately 467 Å.
To obtain a more homogeneous sample for structural analysis, we performed a series of buffer optimization experiments. We found that the addition of magnesium acetate and a small amount of detergent such as CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} to the lysis buffer yielded similar amounts of complex but substantially improved stability of native NuA4 when imaged by negative-stain EM (Fig. 1A to C). 2D analysis of NuA4 purified by the GraFix approach in a buffer containing both magnesium acetate and detergent generated class averages with substantially reduced heterogeneity (Fig. 1D, right panel). In particular, these averages clearly showed that NuA4 adopts a trilobal overall structure of approximately 250 Å in length, with a central core and two additional in-plane lobes projecting away from this core (Fig. 1E).
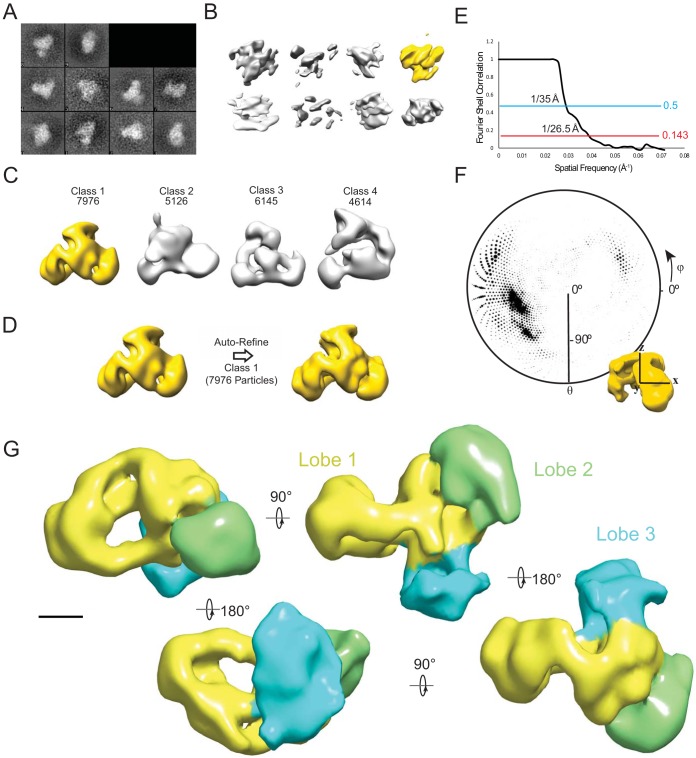
We next generated a three-dimensional (3D) reconstruction of NuA4 by first determining a preliminary reconstruction by the random conical approach and then refining this using the projection data set that generated high-quality 2D averages (Fig. 2A to F). The refined reconstruction recapitulated our observations in 2D by showing three prominent lobes that project into different directions (Fig. 2E). To facilitate discussion, we designate the lobe which encompasses the central core resembling the previously determined EM structure of NuA4 as lobe 1 and the other two densities as lobes 2 and 3, respectively. Lobe 1 shows a hollow cradle-like overall shape characteristic of the family of phosphatidylinositol 3-kinase-related kinase (PIKK) proteins that Tra1 belongs to (20, 23–25), while lobe 2 and lobe 3 are both globular and peripherally attached to lobe 1.
FIG 2.
3D reconstruction of the yeast NuA4 complex. (A) 2D class averages of the 256 untilted particles from tilt pairs used to generate initial random conical tilt (RCT) reconstructions. (B) RCT reconstructions generated from particles gathered at 40°/0° tilt angles. The highlighted model was used for further reconstruction. (C) 3D classification results using the RCT model as a starting model with the number of particles sorted to each class. The highlighted model was used for further refinement. (D) Autorefinement of the most populated 3D model using particles that were classified to class 1 in the 3D classification step. (E) Gold-standard Fourier shell correlation curve indicating the resolution of the model. (F) Angular distribution representation of the input particles to the final model represented as a polar plot. θ and φ correspond to the rotation about the y and z axes, respectively. The radius of the plotted circles represents the number of particles in a particular orientation. (G) 3D reconstruction of NuA4. Different lobes are indicated by color. Scale bar, 50 Å.
The TINTIN and shared SWR1-C modules occupy the two “peripheral” lobes.
The presence of additional densities in our NuA4 reconstruction indicated that the non-Tra1 subunits are spatially arranged according to their functional modules instead of intimately packed against one another adjacent to Tra1. To determine which module constitutes lobes 2 and 3, we performed subunit and module localization using a negative-stain EM-based approach that we have used in our earlier studies on SAGA (21). This approach involves constructing yeast strains containing either green fluorescent protein (GFP)-tagged NuA4 subunits or subunit deletions, analyzing the purified double-tagged or mutant complexes by negative-stain EM, and compared the resulting 2D class averages to those obtained from wild-type NuA4 (Fig. 3A and B).
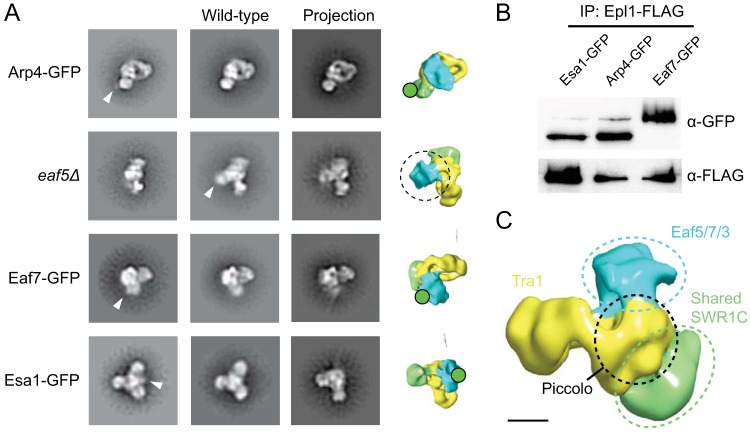
FIG 3.
Localization of yeast NuA4 modules. (A) EM-based analysis of subunit locations. Class averages of purified NuA4 with subunits that are either C-terminally GFP tagged or deleted are compared to wild-type NuA4. Differences in densities between wild-type and mutant NuA4 are indicated by triangles. Projections of the 3D reconstructions corresponding to the class averages are displayed to identify the tagged lobes. The eaf5Δ mutant lacks the entire TINTIN module, and the missing lobe is indicated on the projection by the dotted circle. Class average edge lengths are approximately 600 Å. (B) Western blotting of NuA4 isolated from GFP-tagged strains. Elutions from FLAG purification of NuA4 from Epl1-FLAG-tagged yeast strains expressing GFP fused to Esa1, Arp4, or Eaf7 were analyzed by Western blotting with anti-GFP (top) and anti-FLAG (bottom) antibodies. (C) Schematic of the locations of the identified modules. Scale bar, 50 Å.
We first focused on the shared SWR1-C module since its components Act1, Arp4, Swc4, and Yaf9 are intimately associated with one another within NuA4 (17, 26). We were able to GFP tag the Arp4 subunit of this module and isolate sufficient quantities of intact NuA4 containing GFP-tagged Arp4, as confirmed by Western blotting (Fig. 3B), for our 2D negative stain EM analysis. The class averages obtained from this analysis showed a prominent additional globular density attached to lobe 2, indicating that lobe 2 likely represents the shared SWR1-C module (Fig. 3C). We next turned our attention to the TINTIN module. Previous biochemical studies revealed that this module is anchored to the core of NuA4 through the Eaf5 subunit since the deletion of this protein results in dissociation of the entire module from the full complex (9). We generated an eaf5Δ mutant yeast strain and isolated NuA4 lacking the TINTIN module. Our 2D negative-stain EM analysis showed that this mutant NuA4 lacks lobe 3 (Fig. 3A). Further supportive evidence came from our 2D EM analysis of NuA4 containing GFP-tagged Eaf7 (Fig. 3B), since the resulting class averages showed an additional density tethered to lobe 3. Interestingly, lobe 3 is also the most labile lobe of NuA4, requiring both buffer optimization and gradient fixation to be consistently observed, implicating TINTIN as a metastable component of NuA4 and supporting its independent form and function in vivo (9).
To further confirm these findings, we next attempted to localize the Piccolo module. We isolated NuA4 containing GFP-tagged Esa1 (Fig. 3B) and analyzed this tagged complex by negative-stain EM. Our 2D analysis showed an additional density extending from a region of the central core that corresponds to the proximal half of lobe 1 (Fig. 3A). Collectively, data from these experiments showed that the overall architecture of NuA4 reflects the modular organization that has been implicated from previous genetic and biochemical studies. Notably, the shared and TINTIN modules project outward from the center of the complex, with Piccolo module sandwiched in between (Fig. 3C).
The N terminus of Eaf1 anchors the Eaf5/7/3 and Piccolo modules.
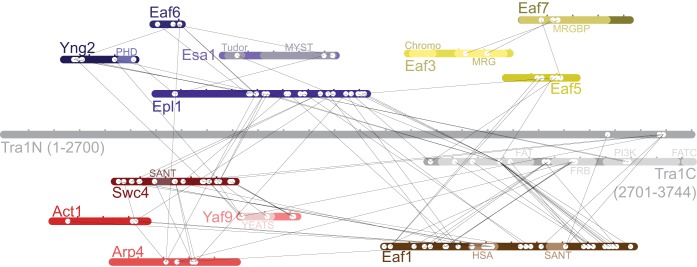
To gain further understanding of how the different subunits are organized with NuA4, we analyzed the purified complex using chemical cross-linking coupled to mass spectrometry (CXMS), a structural proteomics approach capable of generating information on intersubunit proximities within the native assembly (27). We incubated purified NuA4 with the homobifunctional lysine cross-linker disuccinimidyl suberate (DSS) and identified cross-linked subunits through liquid chromatography-tandem mass spectrometry. In total, we identified 108 and 163 intersubunit and intrasubunit cross-links, respectively (Fig. 4; see also the supplemental material). Validation of 57 inter- and intrasubunit cross-links using available high-resolution structures showed that all cross-linked lysine pairs are within the ∼30-Å theorized maximum Cα-Cα cross-linking distance (Table 1) (28). Overall, two general patterns emerged from the CXMS data: (i) subunits within each module are extensively cross-linked with each other, and (ii) the majority of “intermodular” cross-links were found centered on a “hub” containing Tra1, Epl1, and Eaf1.
FIG 4.
CXMS analysis of NuA4. Purified NuA4 was subjected to limited chemical cross-linking with DSS, which targets primary amines, and trypsinized. Peptides connected by DSS were identified by mass spectrometry. The locations of cross-linked intersubunit lysines are shown by two white circles connected by a black line. Subunit representations are marked in 100-residue intervals.
TABLE 1.
Distances measured between Cα and Cα of cross-linked residues identified in this studya
| Cross-link | Protein 1 | Residue 1 | Protein 2 | Residue 1 | Distance (Å) |
|---|---|---|---|---|---|
| 1 | Eaf6 | K104 | Epl1 | K395 | 10.0 |
| 2 | Yng2 | K70 | Epl1 | K374 | 20.1 |
| 3 | Yng2 | K41 | Epl1 | K395 | 18.8 |
| 4 | Yng2 | K70 | Epl1 | K367 | 24.9 |
| 5 | Yng2 | K73 | Epl1 | K367 | 12.1 |
| 6 | Eaf6 | K17 | Epl1 | K367 | 7.5 |
| 7 | Yng2 | K39 | Epl1 | K395 | 8.0 |
| 8 | Yng2 | K41 | Eaf6 | K104 | 15.4 |
| 9 | Yng2 | K40 | Epl1 | K395 | 11.6 |
| 10 | Yng2 | K39 | Eaf6 | K104 | 8.6 |
| 11 | Eaf6 | K104 | Epl1 | K397 | 17.6 |
| 12 | Yng2 | K51 | Epl1 | K395 | 15.8 |
| 13 | Yng2 | K40 | Eaf6 | K104 | 3.6 |
| 14 | Eaf3 | K108 | Eaf3 | K116 | 12.7 |
| 15 | Eaf3 | K109 | Eaf3 | K116 | 13.1 |
| 16 | Eaf3 | K109 | Eaf3 | K119 | 19.8 |
| 17 | Eaf3 | K116 | Eaf3 | K120 | 9.4 |
| 18 | Epl1 | K395 | Epl1 | K397 | 14.6 |
| 19 | Esa1 | K330 | Esa1 | K432 | 8.1 |
| 20 | Yng2 | K34 | Yng2 | K78 | 12.8 |
| 21 | Yng2 | K41 | Yng2 | K51 | 20.9 |
| 22 | Yng2 | K41 | Yng2 | K70 | 14.5 |
| 23 | Yng2 | K41 | Yng2 | K78 | 5.8 |
| 24 | Yng2 | K70 | Yng2 | K78 | 11.7 |
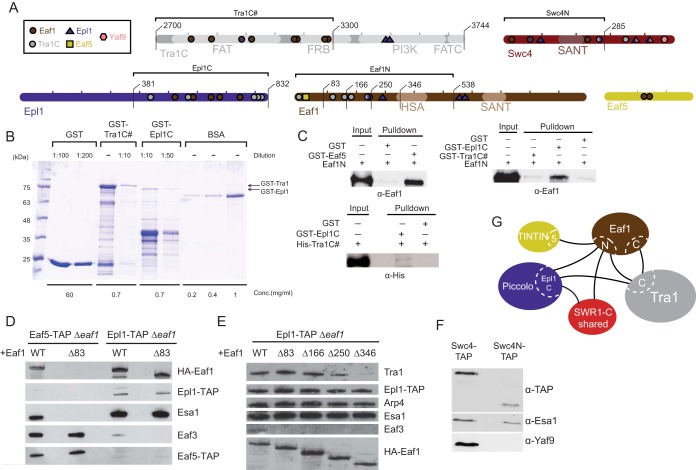
We detected the largest number of intersubunit and intermodular cross-links on Eaf1, a protein that is known to function only within NuA4 and a subunit previously proposed to serve as the platform for coordinating NuA4 assembly (17). A substantial number of intermodular cross-links were mapped between Eaf1 and the C-terminal region of Tra1 encompassing the FAT (FRAP-ATM-TRRAP) and FRB (FKBP12-rapamycin-binding) domains, indicating that these two proteins are likely arranged adjacent to one another. Eaf1 was also found to cross-link “intermodularly” with Eaf5 and Epl1 (Fig. 5A), which have been shown to be anchoring subunits for the TINTIN and Piccolo modules, respectively (9, 16, 17). Indeed, our in vitro glutathione S-transferase (GST) pulldown experiments showed that, in agreement with the CXMS data, the N-terminal region of Eaf1 interacts strongly with Eaf5 (Fig. 5A to C). This was further confirmed by in vivo TAP pulldown analysis, which showed that an N-terminal truncation mutation of Eaf1 causes dissociation of the TINTIN module from NuA4 (Fig. 5D). Collectively, these results pointed to Eaf1 playing a crucial role in anchoring the TINTIN module to the core complex.
FIG 5.
Intersubunit connectivity analysis of NuA4 and its core scaffolding platform. (A) Subunits tested to analyze binding between NuA4 subunits. The locations of cross-linked residues and different construct lengths are indicated. (B) Example of how the amount of recombinant proteins used in GST pulldown assays was estimated. Portions (5 μl) of the indicated dilutions were subjected to SDS-PAGE and stained with Coomassie blue stain. A known BSA concentration was used to estimate the concentration of each fusion protein (full-length band) using ImageJ software. (C) Pulldown assays confirming direct interactions between NuA4 subunits. In vitro GST pulldown assays using the indicated recombinant proteins were performed, and the results were analyzed by Western blotting with anti-Eaf1 or anti-His antibodies. The upper left panel shows that Eaf5 binds directly to the Eaf1 N-terminal region in vitro. rEaf1 (aa 1 to 538) is pulled down by GST-Eaf5. The upper right panel shows that the Eaf1 N-terminal region binds directly to the Epl1 C terminus in vitro. rEaf1N is pulled down by GST-Epl1 (region 381-832) but not by GST-Tra1 (region 2700-3300). In the lower left panel, His-Tra1 (region 2700-3300) shows a very weak interaction with GST-Epl1 (region 381-832) in vitro. The immunoblots shown here and in other panels of this figure are representative images from at least two independent experiments. (D) Eaf1 N-terminal 83 amino acids are required for association of the TINTIN module (Eaf5-Eaf7-Eaf3) to the NuA4 complex in vivo. NuA4 complexes were TAP purified using tagged Epl1 or Eaf5 from yeast strains expressing wild-type or N-terminally truncated Eaf1 and analyzed by Western blotting with anti-HA, anti-TAP, anti-Esa1, and anti-Eaf3 antibodies. (E) The Eaf1 N-terminal deletions identify a region required for the association of Tra1 to the NuA4 complex in vivo, in agreement with the cross-link data. NuA4 complexes were TAP purified using tagged Epl1 as in panel D from yeast strains expressing wild-type or different N-terminally truncated forms of Eaf1 and analyzed by Western blotting. (F) The C terminus of Swc4 is required for the association of Yaf9 to the NuA4 complex in vivo. NuA4 complexes were TAP purified from yeast strains expressing TAP-tagged wild-type Swc4 or its C-terminal truncated form (region 1-285) and analyzed by Western blotting with anti-TAP, anti-Esa1, and anti-Yaf9 antibodies. (G) Schematic of the major interaction regions identified by CXMS and biochemical analysis.
Along the same line, our in vitro GST pulldown experiments showed that the N-terminal region of Eaf1 interacts with the C-terminal region of Epl1, in agreement with the cross-link data (Fig. 5B and C). Since the C terminus of Epl1 anchors Piccolo to NuA4, Eaf1 therefore serves as one of the module's anchoring points. Surprisingly, many putative interaction interfaces inferred from our CXMS data could not be recapitulated using pulldown with recombinant proteins (data not shown). For example, Eaf1 N and C-terminal halves did not bind two Tra1 fragments which had a large number of cross-links (3,201 to 3,744 and 2,700 to 3,300), suggesting more complex physiological interactions and/or the inability of the recombinant fragments to adopt the proper physiological structure/conformation. Nevertheless, progressive N-terminal deletion of Eaf1 in vivo clearly confirmed a critical interaction surface of Tra1 adjacent to the Eaf1 HSA domain, precisely where cross-links were detected (Fig. 5E). In vivo confirmation was also obtained for the detected direct interaction of Yaf9 with Swc4 C terminus within the SWR1-C shared module (Fig. 5F). Altogether, these results lead to a detailed map of the interaction network between the functional modules of NuA4 (Fig. 5G).
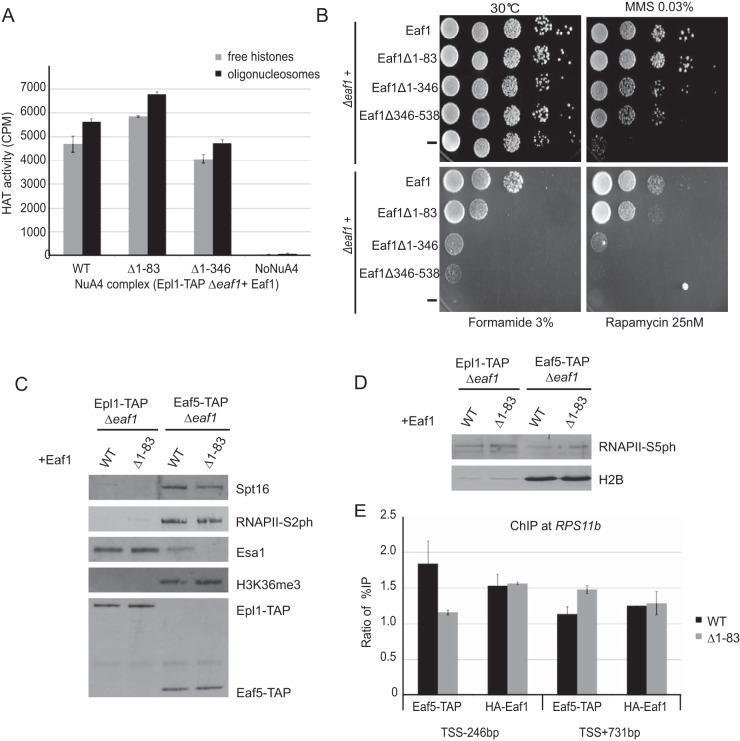
To gain insight on the impact of the intermodular interactions we described in NuA4, we performed a few functional assays. Interestingly, in vitro HAT assays with NuA4 complexes purified from cells expressing Eaf1 deletions leading to loss of TINTIN or Tra1 modules show no obvious defect in HAT activity on free histones or nucleosomes (Fig. 6A). On the other hand, phenotypic analysis of cells carrying Eaf1 deletions shows growth defects on specific media (MMS-DNA repair; formamide-transcription; rapamycin-TOR pathway/ribosomal protein gene expression) reflecting loss of Eaf1 interaction with corresponding functional modules (Δ1-83-TINTIN, Δ1-346-Tra1, Δ346-538-SWR1-C shared and Piccolo; Fig. 6B). Since the TINTIN module has been shown to associate with elongating RNA polymerase II, the FACT histone chaperone, and the H3K36me3 histone mark on the body of active genes (9), we tested the effect of the Eaf1 Δ1-83 truncation that separates TINTIN from NuA4 on these interactions. As shown in Fig. 6C, the interactions of TINTIN (Eaf5-TAP) with the RNA Pol II CTD-Ser2ph isoform, Spt16, and H3K36me3 are clearly not affected by the loss of associated NuA4. In parallel, the interaction of NuA4 (Epl1-TAP) with the early elongating RNA Pol II CTD-Ser5ph isoform located at the 5′ end of active genes is not affected by the loss of TINTIN (Fig. 6D). These results suggest distinct recruitment mechanisms of TINTIN versus the rest of NuA4 at the promoter/transcription start site (TSS) compared to downstream coding region of genes. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) analysis at the NuA4-target gene RPS11B (29). Interestingly, the recruitment of TINTIN (Eaf5-TAP) at the promoter region is fully dependent on its association with NuA4, while this is clearly not the case on the coding region since the TINTIN signal seems to even increase in the absence of NuA4 association (Fig. 6E). In parallel, the loss of TINTIN does not affect NuA4 (HA-Eaf1) recruitment on the RPS11B promoter, an observation in agreement with earlier work using EAF5/7 deletions (8). Altogether, the results obtained with these functional assays support the importance of NuA4 intermodular interaction surfaces that we have mapped in the present study.
FIG 6.
Functional assays of Eaf1 mutants based on mapped molecular interactions. (A) The loss of TINTIN or Tra1 modules does not have an obvious impact on NuA4 HAT activity in vitro. NuA4 complexes purified through Epl1-TAP from cells expressing wild type or Eaf1 truncation mutants (Δ1-83 or Δ1-346) were tested in HAT assays using free histones or oligonucleosomes as the substrate with 3H-acetyl-CoA, followed by spotting on P81 membranes and scintillation counting. Error bars indicate the ranges of duplicate reactions. (B) Phenotypic analysis of yeast strains expressing wild type, different truncation mutants of Eaf1 (Δ1-83, Δ1-346, or Δ346-538) or a mutant expressing no Eaf1. Serial dilution spot assays were performed on rich medium alone or in the presence of formamide, the DNA-damaging agent methylmethane sulfonate (MMS), or the TOR pathway inhibitor rapamycin. Each mutant shows a specific growth phenotype. (C) Loss of association with NuA4 does not affect TINTIN interaction with elongating RNA polymerase on the body of genes (RNAPII-Ser2ph isoform), the FACT histone chaperone (Spt16), and the active coding region-enriched H3K36me3 histone mark. The tobacco etch virus nuclear inclusion a endopeptidase (TEV) elutions of IgG immunoprecipitations from cell extracts as in Fig. 5D (Epl1-TAP or Eaf5-TAP with wild type [WT] or Eaf1-Δ1-83) were analyzed by Western blotting with the indicated antibodies. (D) Loss of TINTIN does not disrupt NuA4 interaction with the early elongating polymerase at the 5′ ends of genes (RNAPII-Ser5ph isoform). Western blots of fractions as in panel C using RNAPII-Ser5ph and H2B antibodies are shown. (E) NuA4-dependent recruitment of TINTIN at the gene promoter but not on the coding region. eaf1 mutant cells (Δ1-83) show a loss of TINTIN (Eaf5-TAP) at the promoter of RPS11B gene but not on its coding region, whereas NuA4 recruitment (Eaf1) on the promoter is not affected by TINTIN. ChIP-qPCR was performed at the promoter region (TSS − 246 bp) and the coding region (TSS + 731 bp) of the NuA4 target gene RPS11B. The data represent the percent immunoprecipitation (IP)/input normalized on signal at the negative-control locus FMP27 (TSS + 5 kb). The error bars indicate the ranges between two biological replicates.
Multiscale structural model of NuA4.
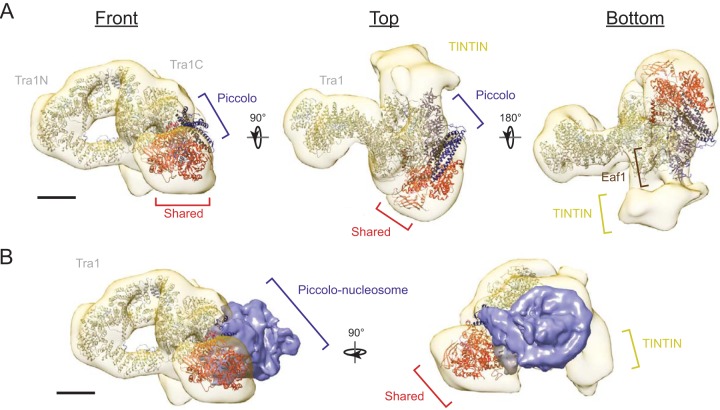
Using our CXMS data as guides, we fitted the high-resolution crystal structures of the core Piccolo module (5J9U) (19), the cryo-EM structure of Tra1 (5OJS) (20), the Esa1 Tudor domain (2RO0) (30), the Arp4-Act1 dimer (5I9E) (31), and the Yaf9 YEATS domain (3FK3) (32), as well as a homology model of Swc4 SANT domain based on the crystal structure of the human ortholog DMAP1 (3HM5), into the refined 3D construction of NuA4. In this multiscale structural model, lobe 1 houses the structural core of NuA4 and contains Tra1, the Piccolo module, and Eaf1 (Fig. 7). Eaf1, predicted to be an intrinsically unstructured protein, occupies the bottom half of lobe1, where it can make direct contacts with multiple modules. The Tra1 N-terminal and C-terminal domains are located distal and proximal to the complex, respectively. The Piccolo module is positioned between lobes 2 and 3, and its nucleosome binding site projects to the space between them. The shared SWR1-C module is contained within lobe 2, which contacts lobe 1 at multiple sites. The Arp4 and Act1 dimer faces lobe 1 based on the presence of cross-links between Act1/Arp4 and Eaf1. We compared our structural model to the recently reported cryo-EM structure of core Piccolo in complex with the nucleosome core particle (NCP) (19). Under the assumption that the structural conformation of isolated Piccolo is the same as in the context of full NuA4, we found that the space cradled by the Piccolo, shared SWR1C, and TINTIN modules can snuggly fit the NCP. Based on this proposed geometry, subunits with chromatin reader domains (Esa1, Yng2, Epl1, Yaf9, and Eaf3) can engage in multivalent interaction with a cognate nucleosome either simultaneously, sequentially, or redundantly to direct NuA4 function (8).
FIG 7.
Multiscale integrated structural model of yeast NuA4. (A) High-resolution structures of Tra1 (5OJS), core Piccolo NuA4 (5J9U), Esa1 Tudor domain (2RO0), Arp4-Act1 dimer (5I9E), and Yaf9 YEATS domain (3FK3), as well as homology models of the Swc4 SANT domain, were fitted to the 3D reconstruction of NuA4 based on EM labeling and cross-linking coupled to mass spectrometry data. The proposed locations of Eaf1 and the TINTIN module are indicated. (B) Overlay of the cryo-EM structure of Piccolo in complex with the NCP (EMD-9536) on the EM density map of full NuA4. Scale bars, 50 Å.
DISCUSSION
Although high-resolution structural information of distinct subunits and even an entire module of NuA4 are now available, the overall architecture of this HAT complex remains obscure due in part to challenges in isolating pure and stable full complexes for structural analysis. The EM-based analysis of yeast NuA4 by Chittuluru et al. represents a major advance by providing the first view on native yeast NuA4 (14). However, the proposed subunit organization from this study is inconsistent with the multimodular nature of NuA4 implicated from other genetics and biochemical studies (17). By developing a procedure that preserved the association of the more labile modules, we were able to obtain a complete structural overview of full NuA4. In addition to having a volume that better reflects the ∼1-MDa overall size of NuA4, our EM reconstruction recapitulates the multimodular arrangement that is known to be a key structural feature of NuA4 and other HAT complexes characterized to date (21). Based on the multiscale structural model we have constructed, we believe the previous earlier EM study was only able to visualize core NuA4 containing Tra1, Eaf1, and the Piccolo module, which according to our studies are intimately associated with one another within lobe 1.
Our CXMS data validated results from previous biochemical and structural analyses on NuA4 subunit connectivity. Eaf1 has previously been shown to serve as a scaffold for the complex, and deletions of its HSA and SANT domain regions result in dissociation of the Piccolo/SWR1-C modules and Tra1, respectively (17). Consistent with these observations, regions directly within or adjacent to these domains cross-linked to Piccolo, SWR1-C module, and Tra1. Furthermore, most of Piccolo's intermodular cross-links center on the C terminus of Epl1, which is required for its association with NuA4 (16). The TINTIN module is anchored to NuA4 through Eaf5, the only subunit of the module with intermodular cross-linking. Specifically, Eaf5 is anchored to NuA4 through the N terminus of Eaf1. We took advantage of this interface to generate an Eaf1 truncation that specifically dissociates TINTIN from NuA4. This mutant allowed us to study NuA4-independent activities of TINTIN, demonstrating independent recruitment and interactions of the respective complexes. Unlike the TINTIN and Piccolo modules, multiple subunits within the shared SWR1-C module were involved in intermodular cross-links, suggesting the presence of multiple anchoring points. Notably, the Swc4-Yaf9 interaction, which is dependent on the C-terminal regions of both proteins (17, 33, 34), was captured by our CXMS analysis and confirmed in vivo (Fig. 5).
Our findings here also potentially provide insights into the molecular assembly of the human NuA4 (TIP60/p400) complex, which is a physical merge of the yeast NuA4 and SWR1 complexes (6). Our 2D and 3D EM analysis of NuA4 indicate that the shared SWR1-C module protrudes outward from the central core, exposing a potential binding surface for other SWR1-C subunits. Interestingly, our CXMS data showed that the regions of Arp4 and Act1 that bind the HSA domain of Swr1 (Act1 regions 130-170 and 345-375 and Arp4 regions 1-35, 145-175, and 440-490) (31) were not cross-linked to other NuA4 subunits, suggesting a distinct mode of interaction within NuA4. This will require further investigation because human NuA4 possesses p400, the lone single HSA-domain-containing scaffold that represents a fusion of Eaf1 and Swr1, the two HSA-domain-containing scaffolding subunits of yeast NuA4 and SWR1-C, respectively (17).
NuA4 possesses a number of characterized chromatin binding domains: the CHD domain of Eaf3 (8, 35), the PHD domain of Yng2 (8), the Tudor/chromobarrel of Esa1 (30, 36), the EPcA basic region of Epl1 (37, 38), and the YEATS domain of Yaf9 (34, 39). The recently reported cryo-EM structure of the core Piccolo module in complex with the nucleosome showed that multiple interactions with one face of the nucleosome defined Piccolo's specificity for its target acetylable lysines (19). Our EM-based labeling experiment and CXMS experiments show that Piccolo resides in the center of the complex, within the proximal half of lobe 1. Combined with the Piccolo-NuA4 structure, our model suggests that nucleosomes are in close proximity to the chromatin binding domain-containing TINTIN and shared SWR1-C modules. This organization raises the possibility that the multiple chromatin binding domains either (i) simultaneously engage with bound nucleosomes, (ii) undergo stages of binding, passing off from one domain to the next, or (iii) redundantly binds nucleosomes, with different domains binding depending on the nucleosome posttranslational modification state. Scenario i is unlikely since NuA4 substrate nucleosomes do not carry modifications recognized by every domain. Based on the observation that the Eaf3 and Yng2 methyllysine binding domains have redundant effects on RNA polymerase II occupancy (8), we favor scenario iii. The multiplicity of chromatin binding domains, then, enables NuA4 to bind and position the various chromatin templates that NuA4 acetylates. This model is consistent with the observation that although loss of the TINTIN module does not affect NuA4 occupancy on the coding regions of active genes, it does alter the acetylation state of histones in this context (8). Since the active site of Esa1 is largely nondiscriminatory for histone tail type, the engagement of other chromatin binding domains is important for defining substrate specificity. The same proliferation of chromatin interacting domains clustering within one region is observed in the SAGA complex (21), suggesting that it may be a feature of other multisubunit histone acetyltransferase complexes. In conclusion, our results allowed us to construct a complete 3D model of the full native NuA4 complex and identify the localization of each of the four functional modules within the structure and the interaction between them. This leads to a better understanding of how this essential large multisubunit assembly works to regulate genome expression and stability, as well as cell fate, in eukaryotes.
MATERIALS AND METHODS
S. cerevisiae strain construction.
Standard S. cerevisiae genetics and culturing methods were used. A list of the strains and plasmids used in this study is provided in Table 2. Serial dilution spot assays for phenotypic analysis on media containing different drugs was performed according to standard procedures.
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Origin | Integration plasmid | Plasmid origin |
|---|---|---|---|---|
| BJ1991 | MATα pep4-3 leu2 trp1 ura3-52 prb1-1122 gal2 | J. Rubinstein | ||
| BJ1991 Epl1-FLAG | BJ1991 EPL1-FLAG::KanMX6 | This study | p3FLAG-KanMX6 | M. Kobor |
| BJ1991 Epl1-FLAG Arp4-GFP | BJ1991 EPL1-FLAG::KanMX6 ARP4-GFP::URA3 | This study | pFA6a-eGFP-URA3 (pKT0209) | K. Thorn (Addgene) |
| BJ1991 Epl1-FLAG Yaf9-GFP | BJ1991 EPL1-FLAG::KanMX6 YAF9-GFP::URA3 | This study | pFA6a-eGFP-URA3 (pKT0209) | K. Thorn (Addgene) |
| BJ1991 Epl1-FLAG Esa1-GFP | BJ1991 EPL1-FLAG::KanMX6 ESA1-GFP::URA3 | This study | pFA6a-eGFP-URA3 (pKT0209) | K. Thorn (Addgene) |
Yeast NuA4 purification.
To isolate native yeast NuA4, 4 to 8 liters of S. cerevisiae strains expressing a C-terminally FLAG-tagged Epl1 were grown to an optical density at 600 nm of ∼4 and harvested by centrifugation. Cell pellets were lysed using the SPEX 6870 freezer mill (SPEX SamplePrep LLC, Metuchen, NJ) under liquid nitrogen temperatures. Cell pellets were resuspended in lysis buffer (40 mM HEPES [pH 7.4], 350 mM NaCl, 0.1% CHAPS, 10% glycerol, 2 mM magnesium acetate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 50 mM NaF, 0.1 mM Na3VO4, 2 mM benzamidine, and cOmplete EDTA-free protease inhibitor cocktail[Roche]) in a 1:2 (wt/vol) ratio. Lysates were precleared by ultracentrifugation at 154,000 × g for 30 min. The cleared lysate was incubated with 500 μl of anti-FLAG M2 resin (Sigma-Aldrich, St. Louis, MO) at 4°C for 1 h. The resin was washed three times with wash buffer (40 mM HEPES [pH 7.4], 350 mM NaCl, 0.1% CHAPS, 10% glycerol, 2 mM magnesium acetate) and then incubated with 2.5 μg/ml RNase A for 30 min at 4°C. The resin was washed another three times with wash buffer, and bound proteins were eluted by incubation with 2× 500 μl elution buffer (wash buffer containing 500 μg/ml 3× FLAG peptide [GenScript, Piscataway, NJ]) for 15 min. Eluted proteins were then subjected to gradient fixation (GraFix) (22). Eluates were loaded onto 15 to 30% glycerol gradients with or without 0 to 0.05% glutaraldehyde and ultracentrifuged at 76,000 × g for 16 h. Gradients were fractionated using the Gradient Station (BioComp, Fredericton, Canada). Fractions from gradients without cross-linker were precipitated using trichloroacetic acid and analyzed by SDS-PAGE. TAP purification of NuA4 complexes from cells expressing wild-type and mutant subunits was performed as previously described (9, 17).
Electron microscopy.
Fractions containing ∼10 μg/ml NuA4 were adsorbed to glow discharged carbon coated grids (Ted Pella, Redding, CA) and stained with uranyl formate as described previously (40). EM specimens were imaged using a Tecnai Spirit G2 (FEI, Hillsboro, OR) operating at an accelerating voltage of 120 kV. Micrographs were acquired at a nominal magnification of 49,000× with an FEI Eagle 4k charge-coupled device camera using a defocus of 1 to 1.5 μm. For collecting tilt pair data for determining an initial 3D model, the same area of the grid was imaged at a 40° tilt and untilted.
Image processing.
To determine the 3D reconstruction of NuA4, 3×3 micrograph pixels were averaged for a final pixel size of 7 Å/pixel and phase-flipped CTF corrected using CTFFIND3 and SPIDER (41, 42). A total of 200 particles were picked manually and then classified, aligned, and used to generate class averages in RELION (43). These class averages are used as the templates for autopicking for a final particle count of 26,402. These particles were subjected to 2D classification, and poor classes were excluded to yield a particle count of 23,165. A separate tilt pair data set containing five micrograph pairs taken at 0 and 40° tilt angles were used to extract 256 tilt pair particles using EMAN2 (44). An initial model was then generated using the tilt pair data and the random conical tilt approach implemented in the EMAN2 software suite (45). The particles were then subjected to 3D classification by RELION using the RCT model as a reference. Then, 7,976 particles from the most highly populated class from 3D classification were selected for autorefinement using RELION. After postprocessing without map sharpening, a final resolution of 26.5 Å was calculated as the 0.143 forward-scatter criterion using the gold-standard method (46).
For subunit localization 2D image analysis, individual particles were selected using RELION. Particle images were then aligned and classified in RELION to generate the class averages. Particles from class averages showing additional GFP tag density or deleted density were subjected to a second round of alignment and classification to further segregate particles with the differing density.
Chemical cross-linking coupled to mass spectrometry.
FLAG eluates of NuA4 were incubated with either 40, 80, 160, or 240 μM DSS for 30 min at room temperature. The cross-linking reaction was quenched by adding 50 mM Tris-HCl (pH 8.0) for 15 min. Cross-linked samples were analyzed on 5 to 20% SDS-PAGE gels (Bio-Rad, Hercules, CA), followed by Coomassie blue G-250 staining. High-molecular-weight bands appearing in the cross-linked samples were excised and processed for mass spectrometry analysis as previously described (47). The mass spectrometry data were analyzed using the pLink software to identify the cross-linked peptides as described previously (28, 47).
Recombinant proteins and pulldown assays.
Standard procedures were used to purify His- and GST-tagged proteins as previously described (16). Briefly, IPTG (isopropyl-β-d-thiogalactopyranoside) induction was performed overnight at 16°C, and the bacterial pellet was lysed with lysozyme, followed by sonication. The extract was incubated for 3 to 4 h at 4°C with glutathione-Sepharose (GE Healthcare) and Ni-nitrilotriacetic acid (NTA)–agarose (Qiagen) beads for GST- and His-tagged proteins, respectively. His-tagged proteins were eluted using imidazole, whereas the GST-tagged proteins immobilized on the beads were stored at 4°C to be used directly for subsequent pulldown assays. The GST pulldown assays was performed using 300 to 600 ng of GST-fused protein and an equivalent amount of His-tagged protein, which was precleared using glutathione-Sepharose beads. Equivalent protein levels were estimated through Coomassie blue-stained SDS-PAGE by comparison with known amounts of bovine serum albumin (BSA) standards. After preclearing, the His-tagged proteins were incubated with GST-immobilized protein in binding buffer (25 mM HEPES [pH 7.5], 100 mM NaCl, 10% glycerol, 100 μg/ml BSA, 1 mM PMSF, 0.5 mM dithiothreitol [DTT], 0.1% Tween 20, and protease inhibitors) at 4°C for 3 h followed by washing the beads three times. To visualize the proteins, the beads were loaded on SDS-PAGE gels, followed by Western blotting with anti-Eaf1 (17), anti-GST (Sigma), and anti-His (Clontech). A 1-μg portion of GST-only protein was used as a control.
HAT assay.
A histone acetyltransferase (HAT) assay was performed as described earlier (8, 48). In brief, 500 ng of free histones or oligonucleosomes purified from HeLa cells was incubated with the NuA4 complex and 0.125 μCi of [3H]acetyl coenzyme A ([3H]acetyl-CoA) in HAT buffer (50 mM Tris-HCl [pH 8], 50 mM KCl plus NaCl, 0.1 mM EDTA, 5% glycerol, 1 mM DTT, 1 mM PMSF, and 20 mM sodium butyrate) at 30°C for 30 min. After this, the reaction mixture was spotted onto p81 filter paper, and scintillation counting was used to determine the incorporation.
ChIP.
ChIP-qPCR was performed as described previously (9). In short, cells were grown in SC-Leu (Formedium) media to an optical density at 600 nm of 0.5 to 1. Cells were cross-linked with formaldehyde and sonicated (Diagenode Bioruptor) to get a chromatin size between 200 and 500 bp. Next, 200 μg of chromatin was used for immunoprecipitation with IgG (Millipore) targeting the protein A part of the TAP tag to follow Eaf5 and anti-HA (Roche) for Eaf1. Dynabeads epoxy M270 (Invitrogen) and Dynabeads protein A (Invitrogen) were used for IgG and hemagglutinin antibodies, respectively. DNA was quantified using LC480 LightCycler (Roche) with primers for the promoter and coding regions of the RPS11B gene, as well as the coding region of the FMP27 gene (nonactive). The percentage of immunoprecipitation on input was measured on RPS11B and is presented as a ratio on the value obtained on FMP27 to show the enrichment level. Primer sequences are available upon request.
Modeling NuA4 architecture.
The 3D reconstruction of NuA4, the high-resolution structures of Act1-Arp4, Yaf9, Tra1, Esa1 chromobarrel, and Piccolo (PDB IDs 5I9E, 3RLS, 5J9Q, 5OJS, 2RO0, and 3TO6), and the homology models of the Swc4 SANT domain based on 3HM5 were used to generate a model of complex architecture using UCSF Chimera (49). Subunits were placed manually based on EM-labeling and CXMS experiments.
Accession number(s).
The refined reconstruction of yeast NuA4 was deposited into the EM Databank under entry number EMD-7131.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a Discovery Grant from Natural Sciences and Engineering Research Council of Canada (418157-2012) to C.K.Y. and by Foundation Grants from the Canadian Institutes of Health Research to C.K.Y. (FDN-143228) and J.C. (FDN-143314).
RELION 3D reconstruction computation was enabled by support provided by the WestGrid (www.westgrid.ca) and Compute Canada Calcul Canada (www.computecanada.ca) Hungabee cluster. Molecular graphics and analyses were performed with the UCSF Chimera package developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California San Francisco. We are grateful to Valerie Côté for technical assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00570-17.
REFERENCES
- 1.McGinty RK, Tan S. 2015. Nucleosome structure and function. Chem Rev 115:2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zentner GE, Henikoff S. 2013. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 3.Steunou AL, Rossetto D, Cote J. 2014. Regulating chromatin by histone acetylation, p 147–212. In Workman JL, Abmayr SM (ed), Fundamentals of chromatin. Springer, New York, NY. [Google Scholar]
- 4.Marmorstein R, Zhou MM. 2014. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalonde ME, Cheng X, Cote J. 2014. Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev 28:1029–1041. doi: 10.1101/gad.236331.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyon Y, Cote J. 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev 14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Voss AK, Thomas T. 2009. MYST family histone acetyltransferases take center stage in stem cells and development. Bioessays 31:1050–1061. doi: 10.1002/bies.200900051. [DOI] [PubMed] [Google Scholar]
- 8.Steunou AL, Cramet M, Rossetto D, Aristizabal MJ, Lacoste N, Drouin S, Cote V, Paquet E, Utley RT, Krogan N, Robert F, Kobor MS, Cote J. 2016. Combined action of histone reader modules regulates NuA4 local acetyltransferase function but not its recruitment on the genome. Mol Cell Biol 36:2768–2781. doi: 10.1128/MCB.00112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossetto D, Cramet M, Wang AY, Steunou AL, Lacoste N, Schulze JM, Cote V, Monnet-Saksouk J, Piquet S, Nourani A, Kobor MS, Cote J. 2014. Eaf5/7/3 form a functionally independent NuA4 submodule linked to RNA polymerase II-coupled nucleosome recycling. EMBO J 33:1397–1415. doi: 10.15252/embj.201386433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC, Pandita TK, Legube G, Doyon Y, Durocher D, Cote J. 2016. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol Cell 62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, Qian J, Zhao Y, Boeke JD, Berger SL, Zhu H. 2009. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, Zhu L, Le Y, Gong X, Yan X, Hong B, Jiang FJ, Xie Z, Miao D, Deng H, Yu L. 2012. Function and molecular mechanism of acetylation in autophagy regulation. Science 336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell L, Huard S, Cotrut M, Pourhanifeh-Lemeri R, Steunou AL, Hamza A, Lambert JP, Zhou H, Ning Z, Basu A, Cote J, Figeys DA, Baetz K. 2013. mChIP-KAT-MS, a method to map protein interactions and acetylation sites for lysine acetyltransferases. Proc Natl Acad Sci U S A 110:E1641–E1650. doi: 10.1073/pnas.1218515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chittuluru JR, Chaban Y, Monnet-Saksouk J, Carrozza MJ, Sapountzi V, Selleck W, Huang J, Utley RT, Cramet M, Allard S, Cai G, Workman JL, Fried MG, Tan S, Cote J, Asturias FJ. 2011. Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes. Nat Struct Mol Biol 18:1196–1203. doi: 10.1038/nsmb.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Cote J. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev 17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Cote J. 2008. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol 28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 19.Xu P, Li C, Chen Z, Jiang S, Fan S, Wang J, Dai J, Zhu P, Chen Z. 2016. The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol Cell 63:965–975. doi: 10.1016/j.molcel.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Diaz-Santin LM, Lukoyanova N, Aciyan E, Cheung AC. 2017. Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 at 3.7 angstrom resolution. Elife 6:e28384. doi: 10.7554/eLife.28384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK. 2015. Conformational flexibility and subunit arrangement of the modular yeast Spt-Ada-Gcn5 acetyltransferase complex. J Biol Chem 290:10057–10070. doi: 10.1074/jbc.M114.624684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, Uchtenhagen H, Urlaub H, Herzog F, Peters JM, Poerschke D, Luhrmann R, Stark H. 2008. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods 5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 23.Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL. 2017. Structures of closed and open conformations of dimeric human ATM. Sci Adv 3:e1700933. doi: 10.1126/sciadv.1700933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. 2016. Architecture of human mTOR complex 1. Science 351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 25.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. 2010. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell 38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu WH, Wu CH, Ladurner A, Mizuguchi G, Wei D, Xiao H, Luk E, Ranjan A, Wu C. 2009. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J Biol Chem 284:6200–6207. doi: 10.1074/jbc.M808830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walzthoeni T, Leitner A, Stengel F, Aebersold R. 2013. Mass spectrometry supported determination of protein complex structure. Curr Opin Struct Biol 23:252–260. doi: 10.1016/j.sbi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. 2012. Identification of cross-linked peptides from complex samples. Nat Methods 9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 29.Reid JL, Iyer VR, Brown PO, Struhl K. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6:1297–1307. doi: 10.1016/S1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 30.Shimojo H, Sano N, Moriwaki Y, Okuda M, Horikoshi M, Nishimura Y. 2008. Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain. J Mol Biol 378:987–1001. doi: 10.1016/j.jmb.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 31.Cao T, Sun L, Jiang Y, Huang S, Wang J, Chen Z. 2016. Crystal structure of a nuclear actin ternary complex. Proc Natl Acad Sci U S A 113:8985–8990. doi: 10.1073/pnas.1602818113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang AY, Schulze JM, Skordalakes E, Gin JW, Berger JM, Rine J, Kobor MS. 2009. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc Natl Acad Sci U S A 106:21573–21578. doi: 10.1073/pnas.0906539106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bittner CB, Zeisig DT, Zeisig BB, Slany RK. 2004. Direct physical and functional interaction of the NuA4 complex components Yaf9p and Swc4p. Eukaryot Cell 3:976–983. doi: 10.1128/EC.3.4.976-983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein BJ, Ahmad S, Vann KR, Andrews FH, Mayo ZA, Bourriquen G, Bridgers JB, Zhang J, Strahl BD, Cote J, Kutateladze TG. 2018. Yaf9 subunit of the NuA4 and SWR1 complexes targets histone H3K27ac through its YEATS domain. Nucleic Acids Res 46:421–430. doi: 10.1093/nar/gkx1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisen A, Utley RT, Nourani A, Allard S, Schmidt P, Lane WS, Lucchesi JC, Cote J. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J Biol Chem 276:3484–3491. doi: 10.1074/jbc.M008159200. [DOI] [PubMed] [Google Scholar]
- 36.Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. 2000. Three yeast proteins related to the human candidate tumor suppressor p33ING1 are associated with histone acetyltransferase activities. Mol Cell Biol 20:3807–3816. doi: 10.1128/MCB.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Tan S. 2013. Piccolo NuA4-catalyzed acetylation of nucleosomal histones: critical roles of an Esa1 Tudor/chromo barrel loop and an Epl1 enhancer of polycomb A (EPcA) basic region. Mol Cell Biol 33:159–169. doi: 10.1128/MCB.01131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selleck W, Fortin I, Sermwittayawong D, Cote J, Tan S. 2005. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol 25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Masson I, Yu DY, Jensen K, Chevalier A, Courbeyrette R, Boulard Y, Smith MM, Mann C. 2003. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol Cell Biol 23:6086–6102. doi: 10.1128/MCB.23.17.6086-6102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohi M, Li Y, Cheng Y, Walz T. 2004. Negative staining and image classification: powerful tools in modern electron microscopy. Biol Proced Online 6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 42.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 43.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 46.Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schroder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL. 2012. Outcome of the first electron microscopy validation task force meeting. Structure 20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Setiaputra DT, Cheng DT, Lu S, Hansen JM, Dalwadi U, Lam CH, To JL, Dong MQ, Yip CK. 2017. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep 18:280–291. doi: 10.15252/embr.201642548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goddard TD, Huang CC, Ferrin TE. 2007. Visualizing density maps with UCSF Chimera. J Struct Biol 157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.