ABSTRACT
One-half of the genes in the human genome contain alternative promoters, some of which generate products with opposing functions. Aberrant silencing or activation of such alternative promoters is associated with multiple diseases, including cancer, but little is known regarding the molecular mechanisms that control alternative promoter choice. The SHC1 gene encodes p46Shc/p52Shc and p66Shc, proteins oppositely regulating anchorage-independent growth that are produced by transcription initiated from the upstream and downstream tandem promoters of SHC1, respectively. Here we demonstrate that activation of these promoters is mutually exclusive on separate alleles in single primary endothelial cells in a heritable fashion, ensuring expression of both transcripts by the cell. Peripheral blood lymphocytes that do not transcribe p66Shc transcribed p52Shc biallelically. This distinct monoallelic transcription pattern is established by allele-specific chromosomal looping between tandem promoters, which silences the upstream promoter. Our results reveal a new mechanism to control alternative promoter usage through higher-order chromatin structure.
KEYWORDS: alternative promoter interaction, gene silencing, SHC1, allele-specific transcription
INTRODUCTION
A large and growing number of mammalian genes have been found to use alternative promoters, which appear to represent a widespread mechanism to expand transcript and protein diversity. Some alternative promoters produce mRNA isoforms with heterogeneous 5′ untranslated regions that can affect the stability or translation efficiency of the mRNA variants, including RUNX1 (1), OTX2 (orthodenticle homeobox 2) (2) and SHOX (short stature homeobox) (3). Some alternative promoters generate mRNA isoforms that encode distinct proteins, sometimes having opposing biological activities, for example LEF1 (4), SHC1 (5, 6), and CTNNA3 (7). Aberrant use of one promoter over another can be associated with various diseases, including cancer (4, 8–10).
One transcriptional process exhibits a suppressive influence on neighboring transcriptional processes through affecting the assembly of preinitiation complexes (11–13). This phenomenon is termed transcriptional interference and has been observed in bacteria, yeast, flies, HIV, and mice (14–19). Thus, long-standing questions include how promoter usage is controlled if multiple promoters are active in one single cell.
SHC1 contains two tandem promoters that generate three proteins, p46Shc, p52Shc, and p66Shc, which differ only in the length of their amino-terminal sequences. Whereas p46Shc and p52Shc arise from alternate start codons on a single transcript, p66Shc is generated from a separate transcript. p52Shc, whose transcript is generated from the upstream promoter and constitutively expressed, binds growth factor receptors and mediates mitogenic Ras signals through recruitment of Grb2 (20). Accordingly, overexpression of p52Shc causes 3T3 cells to become morphologically transformed and tumorigenic (21). In contrast, p66Shc, whose transcript is generated from the downstream promoter, inhibits receptor tyrosine kinase signaling, causes programmed cell death, and promotes differentiation (22–24). More recently, p66Shc was found to localize to focal adhesions and permit anchorage sensing through a RhoA-dependent mechanosensory test of the environment (25). Unlike p52Shc, which permits anchorage-independent growth when overexpressed, p66Shc is required for detachment-induced death, or anoikis (25). Consistent with its role in anoikis, p66Shc is expressed in normal epithelial and endothelial cells but not in hematopoietic cells that survive under unanchored conditions. Therefore, endothelial cells require expression of both p52Shc and p66Shc for proper homeostasis and thus serve as a relevant biological system to explore the coordinate regulation of multipromoter genes.
In the present study, we explored the transcriptional regulation of p52Shc and p66Shc in primary human umbilical vessel endothelial cells (HUVECs) and found that p52Shc and p66Shc are transcribed from separate alleles in a mutually exclusive fashion. This novel monoallelic transcription pattern, which is heritable, is enforced by allele-specific physical interaction between the p66Shc and p52Shc promoters. These data reveal a new mechanism for specification of alternative promoter usage.
RESULTS
p66Shc and p52Shc are generated from separate alleles in single HUVECs.
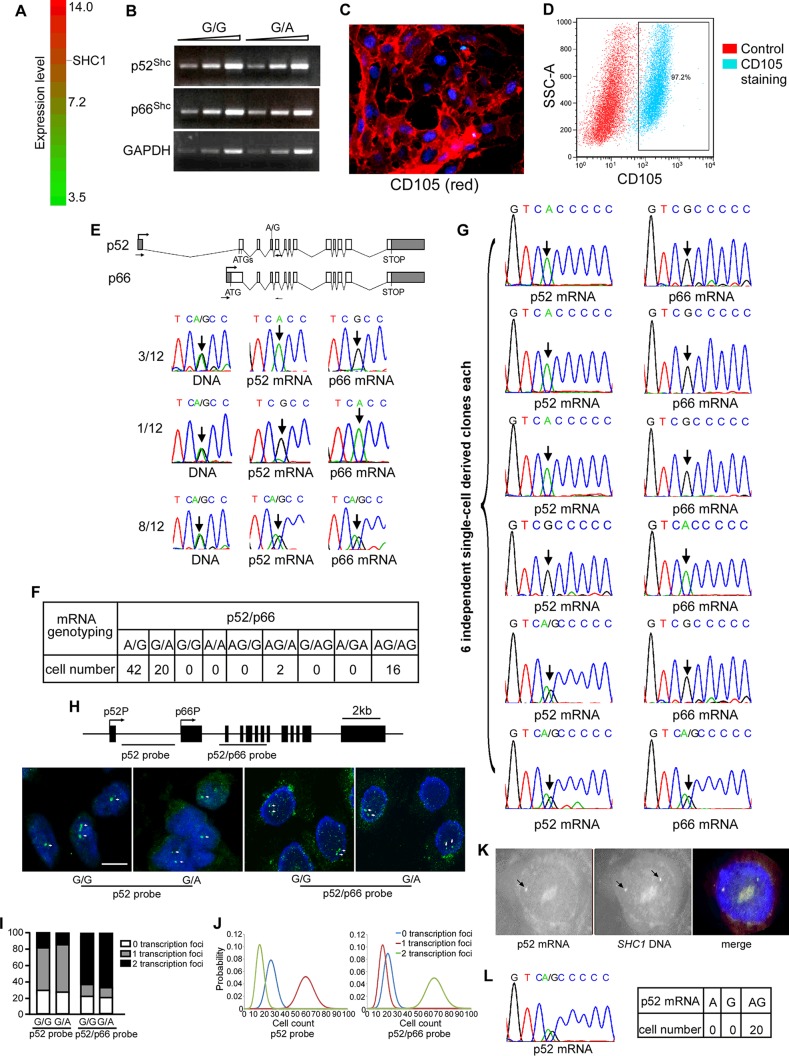
Two tandem promoters of SHC1 are separated by 3.736 kb. Analysis of public microarray data of HUVECs (GSE61989) revealed that SHC1 was highly expressed in HUVECs relative to all transcript signals (Fig. 1A). To avoid transcriptional interference and produce p52Shc and p66Shc efficiently, one possible way is to generate two transcripts from separate alleles. We therefore studied the transcription pattern of p66Shc and p52Shc in HUVECs. Human SHC1 contains an exonic single nucleotide polymorphism (SNP) (rs8191981G/A) that allows us to determine the allele-specific transcription of p66Shc and p52Shc. Semiquantitative reverse transcriptase (RT)-PCR showed that this SNP has no effect on transcription of p66Shc and p52Shc (Fig. 1B). We purified DNA from 40 human umbilical tissues and genotyped the SNP site. Twelve umbilical tissues were heterozygous for the G/A polymorphism and were studied further. We purified and cultured HUVECs from G/A heterozygous umbilical tissues, achieving 97.2% purity as evaluated by CD105 immunofluorescence staining and fluorescence-activated cell sorter (FACS) analysis (Fig. 1C and D). We amplified and genotyped p52Shc and p66Shc transcripts from 106 HUVECs using the indicated primers (Fig. 1E). p52Shc and p66Shc were transcribed separately from two alleles in 4 of 12 umbilical tissues, with three umbilical tissues generating p66Shc from the G allele and p52Shc from the A allele and one umbilical tissue generating p66Shc from the A allele and p52Shc from the G allele (Fig. 1E). Notably, this distinct monoallelic transcription pattern was detected at a population level, indicating that this transcription pattern is heritable across cell division.
FIG 1.
Transcription pattern of p52Shc and p66Shc in HUVECs. (A) Public Affymetrix microarray data of HUVECs (GSE61989) were interrogated for SHC1 transcription. Of 20,305 transcripts with expression levels ranging from 3.5 to 14.0, p52Shc and p66Shc transcripts are quantified as 10.9. (B) Transcription levels of p66Shc, p52Shc, and GAPDH in HUVECs harboring G/G or G/A for the SNP. (C) Representative image of CD105 immunofluorescence staining of HUVECs. Nuclei are stained by DAPI. (D) FACS analysis of percentage of CD105-positive cells. (E, top) SNP site and primers that were used in RT-PCR; (bottom) representative traces of Sanger sequencing of the p52/p66 RT-PCR products containing the informative exonic SNP (arrow). (F) Quantitative analysis of Sanger sequencing data of allele-specific transcription of p52/p66 in single HUVECs. (G) Representative traces of Sanger sequencing of the p52/p66 RT-PCR products containing the informative exonic SNP (arrow) in six single-cell-derived clones. Single cells were purified from biallelic umbilical tissues with undefined transcription pattern. (H, top) Location of probes used in RNA FISH; (bottom) RNA FISH with the p52 probe and p52/p66 probe for examining the nascent transcripts of SHC1 (green) in nuclei stained by DAPI (middle). Bar, 10 μm. (I) Histograms represent the percentages of 0, 1, and 2 transcription sites. (J) Poisson statistics of probability of cell counts per 100 cells. (K) Representative images of RNA-DNA sequential FISH. RNA FISH was performed with the p52 probe (red), and DNA FISH was performed with the p52 probe+p52/p66 probe (green). (L) Representative traces of Sanger sequencing of RT-PCR products of p52Shc containing the informative exonic SNP in single PBL and quantitative analysis of Sanger sequencing data of allele-specific transcription of p52Shc in single PBL.
Given that 8 umbilical tissues showed biallelic transcription of p66Shc and p52Shc, we tested whether the transcription pattern of p66Shc and p52Shc in single cells follows the population average. We amplified and genotyped p52Shc mRNA and p66Shc mRNA from single HUVECs purified from biallelic umbilical tissues. Of 80 single cells, 62 exhibited mutually exclusive transcription of p52Shc and p66Shc from two alleles, with 42 cells generating p66Shc from the G allele and p52Shc from the A allele and 20 cells generating p66Shc from the A allele and p52Shc from the G allele (Fig. 1F), indicating that p66Shc and p52Shc are still transcribed separately from two alleles in single cells, although the population exhibited biallelic transcription. To confirm the inheritance of this monoallelic transcription pattern, we purified HUVECs from biallelic umbilical tissues and established six single-cell-derived clones. Assuming inheritance of monoallelic expression across cell divisions, all cells within each single-cell-derived clone are expected to express the same alleles. Indeed, 4 clones expressed p66Shc from the same allele and p52Shc from the other allele (Fig. 1G). Therefore, generating different isoforms from separate alleles is predominantly used for SHC1 in endothelial cells. This monoallelic transcription pattern is heritable. The biallelic umbilical tissues may result from monoallelic transcription established at a different developmental stage. Because 2 and 1 single-cell clones exhibited biallelic transcription of p52Shc and p66Shc, respectively, SHC1 transcription may not be very tightly controlled by this monoallelic transcription.
To further confirm this result and to test whether the SNP itself affects promoter activity, we performed RNA fluorescence in situ hybridization (FISH) with different HUVEC lines harboring G/G or G/A at the rs8191981. We used a p52 probe that resides within p52Shc native transcripts but is absent from p66Shc transcripts to test p52Shc transcription and a p52/p66 probe located in the region shared by both p66Shc and p52Shc transcripts to examine transcription of both p52Shc and p66Shc. Figure 1H shows the location of the probes used in RNA FISH and a representative image of probe hybridization patterns in HUVEC nuclei. Two hundred cells were counted for each HUVEC line hybridized with each probe, and a Poisson model was used to analyze the probability distributions. Using the p52 probe, 27%, 58%, and 15% of G/G HUVECs and 25%, 60%, and 15% of G/A HUVECs exhibited 0, 1, and 2 transcription sites, respectively (Fig. 1I and J). Using the p52/p66 probe, 21%, 17%, and 62% of G/G HUVECs and 20%, 13%, and 67% of G/A HUVECs showed 0, 1, and 2 transcription sites, respectively (Fig. 1I and J). Transcription foci were not detected in more than 20% of cells in either line, using either the p52 probe or the p52/p66 probe. The reason might be transcription of SHC1 occurring in bursts but not continuously, like globin-β (26). These results indicate that the majority of cells transcribe p52Shc monoallelically regardless of the genotype of the SNP site and that the second allele transcribes p66Shc only. This monoallelic transcription of p52Shc was further confirmed by RNA-DNA sequential FISH (Fig. 1K). The RNA FISH data are consistent with the results of Sanger sequencing of PCR products containing the informative exonic SNP and further demonstrate that the monoallelic transcription is independent of the SNP.
We next sought to determine the transcription pattern of p52Shc in peripheral blood lymphocytes (PBL) that do not express p66Shc (5). We collected PBL from 10 healthy persons, and 2 individuals were G/A heterozygous. Single PBL harboring G/A at rs8191981 from the 2 individuals were analyzed for genotyping of p52Shc transcripts, and all cells exhibited biallelic transcription of p52Shc (Fig. 1L).
Taken together, these data indicate that p52Shc and p66Shc are transcribed from separate alleles in single HUVECs in a heritable fashion.
Two alleles are differentially marked by histone modifications.
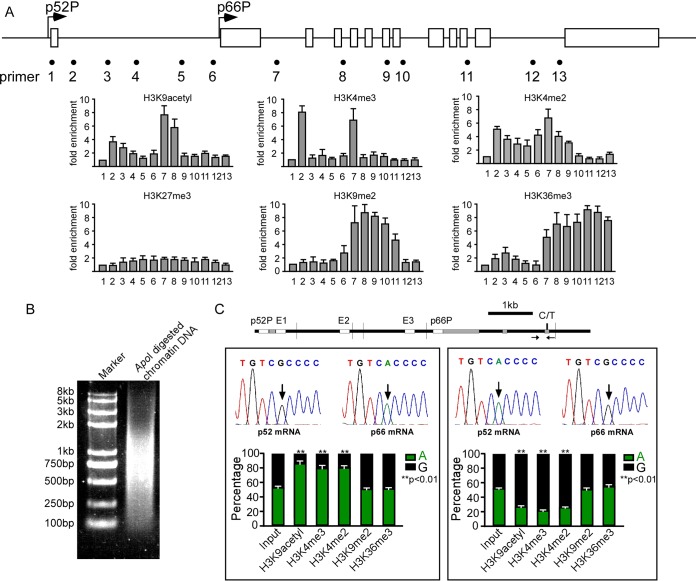
We next explored the mechanisms behind this distinct monoallelic transcription manner. CpG methylation was not detected in two tandem promoters and three enhancers (E1, E2, and E3) of SHC1 in HUVECs (data not shown). We then investigated whether histone modifications may distinguish the two alleles. Chromatin immunoprecipitation (ChIP) analysis showed the association of activating histones (H3K4me3, H3K4me2, and H3K9acetyl) with two tandem promoters (Fig. 2A) and the association of H3K9me2 and H3K36me3 with the gene body region downstream of the p66Shc promoter. H3K27me3, which was reported to control DNA methylation-independent imprinting of multiple genes in preimplantation embryos (27), was not detected at SHC1 (Fig. 2A). To quantify the allele-specific occupancy of these histone marks along SHC1, we performed allele-specific ChIP using two HUVEC lines. One line generates p52Shc from the G allele and p66Shc from the A allele. The other line generates p52Shc from the A allele and p66Shc from the G allele. Chromatin was sheared with ApoI digestion instead of sonication. With this chromatin-shearing method, the p66Shc promoter and the SNP site localize to the same DNA fragment, and therefore allelic association of specified histones with the p66Shc promoter can be measured through genotyping of the SNP in PCR products. The ApoI digestion efficiency is shown in Fig. 2B. Enrichment of H3K4me3, H3K4me2, and H3K9acetyl showed bias toward the active p66Shc promoter, whereas H3K9me2 and H3K36me3 were evenly distributed in the two alleles in both HUVEC lines evaluated by quantitative, allele-specific, and nanofluidic digital PCR (dPCR) (Fig. 2C). This result further confirms the observation that p66Shc was transcribed monoallelically and suggests that the differential alleles might be marked by histone modifications.
FIG 2.
Histone modifications mark differential alleles. (A) ChIP assay in HUVECs shows distributions of modified histones (K9 acetylated H3, K4 di- and trimethylated H3, K27 trimethylated H3, K9 dimethylated H3, and K36 trimethylated H3) along SHC1. Error bars indicate means ± standard deviations (SD) of three different ChIP experiments. (B) ApoI digestion efficiency in allele-specific ChIP assay. (C) Quantitative, nanofluidic, and allele-specific digital PCR was used to evaluate the percentages of genotypes of ChIP products in HUVECs generating p52Shc from the G allele and p66Shc from the A allele (left) and HUVECs generating p52Shc from the A allele and p66Shc from the G allele (right). Error bars indicate means ± SD for three different ChIP experiments.
Changes of chromatin configuration associate with allele-specific transcription of SHC1.
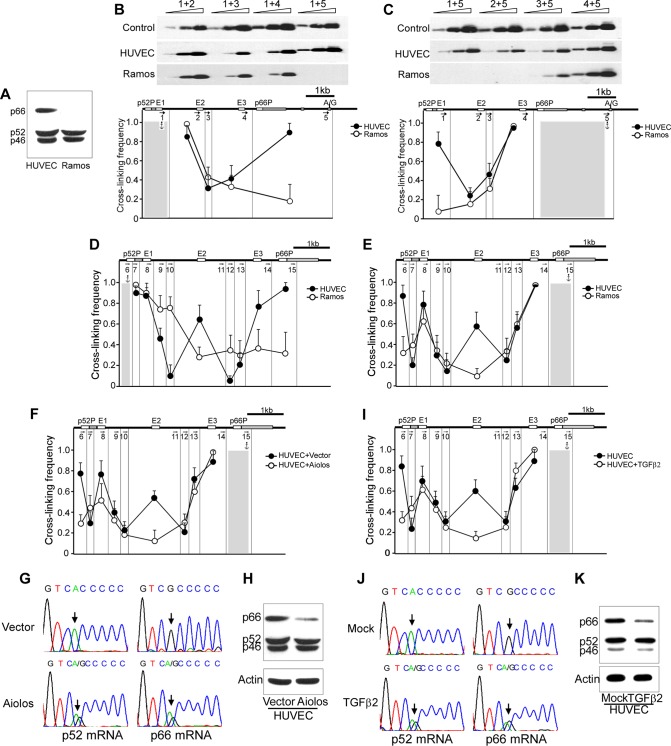
We next explored whether chromatin higher-order organization, which plays a critical role in establishing imprinting of genes such as Igf19/H19 (28–30), is involved. We examined the chromatin configuration of SHC1 in HUVECs and Ramos cells, the latter of which express p52Shc only (Fig. 3A), using chromosome conformation capture (3C) technology. The cross-linked chromatin was digested with ApoI, diluted, and religated. Cross-linking frequencies were examined by semiquantitative PCR with 0.1 μg, 0.3 μg, and 0.9 μg DNA as the template, followed by Southern blotting with a biotin-labeled DNA segment complementary to common sequences adjacent to the anchor primer as a probe. The representative 3C assay results are shown in Fig. 3B and C, top panels. HUVEC-specific promoter-promoter interactions were detected with either the p52Shc promoter as an anchor fragment or the p66Shc promoter as an anchor fragment but were not seen with the hematopoietic-derived Ramos cell line (Fig. 3B and C). Then, we performed 3C again using another restriction enzyme, DpnII, to achieve higher resolution. Consistently, HUVEC-specific promoter-promoter interactions were also detected in DpnII-digested chromatin (Fig. 3D and E). In addition, interactions between two tandem promoters with the three enhancers (E1, E2, and E3) were observed in HUVECs (Fig. 3D and E). Except for the E1-p66Shc promoter, the other interactions did not appear in the Ramos cell line (Fig. 3D and E).
FIG 3.
Changes of higher-order chromatin organization associate with allele-specific transcription. (A) Western blot shows the expression of Shc1 isoforms in HUVECs and Ramos cells. (B and C) The 3C assay was used to calculate the cross-linking frequency between chromatin segments to assess proximity in HUVECs and Ramos cells. Vertical lines represent ApoI restriction sites. Arrows indicate PCR primer sites and directions. Anchor symbols mark the anchoring primer for each data set. Cross-linking frequencies between the p52Shc promoter and other ApoI-defined segments (B) and between the p66Shc promoter and other ApoI-defined segments (C) are shown. Error bars indicate means ± SD for three 3C experiments. (D and E) 3C in HUVECs and Ramos cells using restriction enzyme DpnII to cut the chromatin. Vertical lines represent DpnII restriction sites. Arrows indicate PCR primer sites and direction. Anchor symbols mark the anchoring primer for each data set. Cross-linking frequencies between the p52Shc promoter (D) or the p66Shc promoter (E) and other DpnII-defined segments are shown. Error bars indicate means ± SD for three 3C experiments. (F) Cross-linking frequencies between different DpnII fragments were assessed by 3C assay in Aiolos-overexpressing HUVECs. Error bars indicate means ± SD for three 3C experiments. (G) Representative traces of Sanger sequencing of p52Shc and p66Shc RT-PCR products containing the informative exonic SNP (arrow) in Aiolos-overexpressing HUVECs. (H) Immunoblots of Shc1 and actin in Aiolos-overexpressing HUVECs. (I) Cross-linking frequencies between different DpnII fragments were assessed by 3C assay in TGF-β2-treated HUVECs. Error bars indicate means ± SD for three 3C experiments. (J) Representative traces of Sanger sequencing of RT-PCR products of p52Shc and p66Shc containing the informative exonic SNP (arrow) in TGF-β2-treated HUVECs. (K) Immunoblots of Shc1 and actin in TGF-β2-treated HUVECs.
We next disrupted the physical interactions in HUVECs by two distinct interventions to see whether p52Shc and p66Shc were still transcribed in an allele-specific pattern. First, transient expression of Aiolos, a hematopoietic lineage-restricted transcription factor, in HUVECs purified from monoallelic umbilical tissue (p66Shc from the G allele and p52Shc from the A allele) disrupted the chromatin looping of SHC1 (Fig. 3F), similar to its function in transformed epithelial lung cancer cells (31). We then genotyped p52Shc and p66Shc transcripts in Aiolos-expressing HUVECs. In contrast to the control group, which transcribed p52Shc and p66Shc from separate alleles, the Aiolos-expressing HUVECs transcribed p52Shc and p66Shc from both alleles (Fig. 3G). Accompanying the change of this monoallelic transcription pattern, p66Shc expression was downregulated (Fig. 3H). Second, transforming growth factor β2 (TGF-β2) signaling, which converts endothelial cells to stemlike cells and whose related family member, TGF-β, can downregulate p66Shc in transformed epithelial lung cancer cells (32, 33), also resulted in the disruption of chromatin looping (Fig. 3I), monoallelic-to-biallelic switching of p52Shc and p66Shc transcription (Fig. 3J), and downregulation of p66Shc in HUVECs (Fig. 3K). These results indicate that the mutually exclusive transcription of p52Shc and p66Shc correlates with the chromatin configuration of SHC1.
Physical interaction between two tandem promoters occurs in the p66Shc transcribing allele only.
A promoter-promoter interaction has been found in the INTS1-MAFK locus, in which case the INTS1 promoter functions as an enhancer of the MAFK promoter (34). We then asked whether the tandem promoters that showed strong interaction in HUVECs regulate monoallelic transcription. We examined whether the colocalization of two tandem promoters is allele specific. We performed 3C using ApoI in two HUVEC lines as indicated in the allele-specific ChIP assay. ApoI digestion allows the p66Shc promoter and the SNP site to reside in the same restriction enzyme-cutting DNA fragment. Thus, the allele-specific cross-linking frequency between the p66Shc promoter with upstream fragments can be measured by genotyping the SNP in PCR products. The allele-specific cross-linking frequencies were analyzed by quantitative, allele-specific, nanofluidic digital PCR using the primers indicated in Fig. 4A, top panel. Interestingly, while the adjacent interactions were evenly distributed at the two alleles, interactions between the two tandem promoters and interactions between the p66Shc promoter and the enhancer E2 exhibited bias toward the p66Shc transcribing allele in both HUVEC lines (Fig. 4A and B). Thus, interactions between tandem promoters and between the p66Shc promoter and E2 occur predominantly in the p66Shc transcribing allele. This allele-specific interaction is independent of the genotype of the SNP.
FIG 4.
Physical interaction between two tandem promoters occurs in p66Shc-transcribing allele. (A) Allele-specific 3C shows allele-specific promoter-promoter interactions. The positions of the restriction enzyme cutting sites and primers used in the assay are shown (top panel); also shown are the percentages of genotype of 3C products in HUVECs generating p52Shc from G allele and p66Shc from A allele (middle panel) and HUVECs generating p52Shc from the A allele and p66Shc from the G allele (bottom panel). Quantitative, nanofluidic, and allele-specific digital PCR was used to evaluate the percentages of genotype of 3C products. Error bars indicate means ± SD for three different 3C experiments. (B) Scheme showing the correlation between promoter-promoter interaction and alternative promoter activity.
Deletion of the p66Shc promoter in HUVECs results in biallelic transcription of p52Shc.
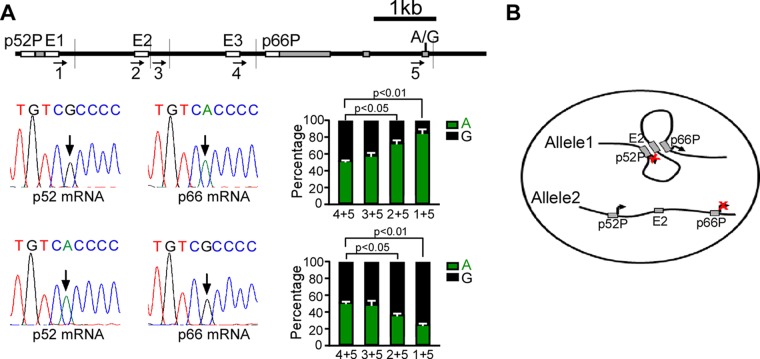
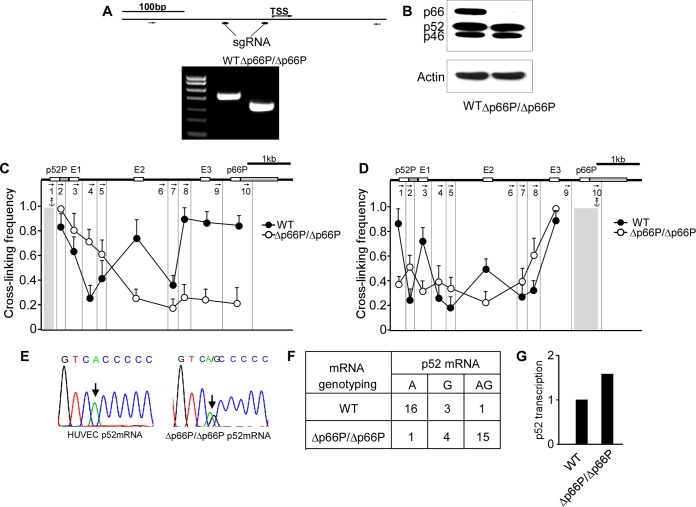
We then utilized the clustered regularly interspaced short palindromic repeat(s) (CRISPR)/Cas9 system to delete the p66Shc promoter in HUVECs, and we evaluated the transcription pattern of p52Shc. Two small guide RNAs (sgRNAs) targeting the flanking sequences located upstream of the TSS of p66Shc and Cas9 cDNA were transfected into HUVECs. After screening, we identified one clone in which the p66Shc promoter was deleted in both alleles (Fig. 5A). This clone exhibited the expected absence of p66Shc expression (Fig. 5B). Notably, deletion of the p66Shc promoter abolished all of the physical interactions among the DNA fragments harboring two tandem promoters and the three enhancers, suggesting that the p66Shc promoter may act as an organizer to establish DNA interactions of SHC1 (Fig. 5C and D). Then, we sequenced the p52Shc mRNA. While control cells monoallelically transcribed p52Shc, HUVECs harboring the p66Shc promoter deletion transcribed p52Shc from both alleles (Fig. 5E). This result was confirmed by a single-cell experiment, which showed biallelic transcription of p52Shc in 15 of 20 single HUVECs (Fig. 5F). Accordingly, a 1.6-fold enhancement of p52Shc transcription was detected by quantitative nanofluidic digital PCR in HUVECs harboring the p66Shc promoter deletion (Fig. 5G). These data indicate that deletion of the p66Shc promoter leads to a monoallelic-to-biallelic transcriptional switch of p52Shc. Thus, the p66Shc promoter can repress the activity of the p52Shc promoter in cis.
FIG 5.
Deletion of the p66Shc promoter results in a monoallelic-to-biallelic transcriptional switch of p52Shc. (A, top) Positions of sgRNAs used in CRISPR/Cas9 and PCR primers used for the PCR assay; (bottom) PCR products generated with the indicated primers. WT, normal HUVECs; Δp66P/Δp66P, HUVECs harboring homozygous p66Shc promoter deletions. (B) Immunoblots of Shc1 and actin in normal HUVECs and p66Shc promoter-deleted HUVECs. (C and D) 3C was performed to assess the change of chromatin configuration of SHC1 when the p66Shc promoter was deleted, with the DNA fragment harboring the p52Shc promoter (C) or the p66Shc promoter (D) as anchor. Error bars indicate means ± SD for three 3C experiments. (E) Representative traces of Sanger sequencing of RT-PCR products of p52Shc containing the informative exonic SNP (arrow) in normal HUVECs and p66Shc promoter-deleted HUVECs. (F) Quantitative analysis of Sanger sequencing of RT-PCR products of p52Shc containing the informative exonic SNP revealing alleles transcribing p52Shc in single HUVECs with the p66Shc promoter deletion. (G) Transcription of p52Shc in HUVECs and HUVECs with p66Shc promoter deletion, evaluated by quantitative and nanofluidic digital PCR assay.
Physical interaction between tandem promoters represses upstream promoter activity.
To examine whether the p66Shc promoter functions as a silencer of the p52Shc promoter, we performed luciferase reporter gene assays, a commonly used method for promoter and cis-regulatory element (enhancer or silencer) characterization (34, 35). The p52Shc promoter was inserted into a construct containing a luciferase reporter gene. The p66Shc promoter and the three enhancers, E1, E2, and E3, were placed side by side upstream of the p52Shc promoter. In contrast to our expectations, the p66Shc promoter in forward or reverse orientation had no effect on the activity of the p52Shc promoter and neither did the three enhancers (Fig. 6A). As a control, the three enhancers increased the p66Shc promoter activity 2- to 3-fold in HUVECs (Fig. 6A). Thus, the p66Shc promoter may not have silencer properties.
FIG 6.
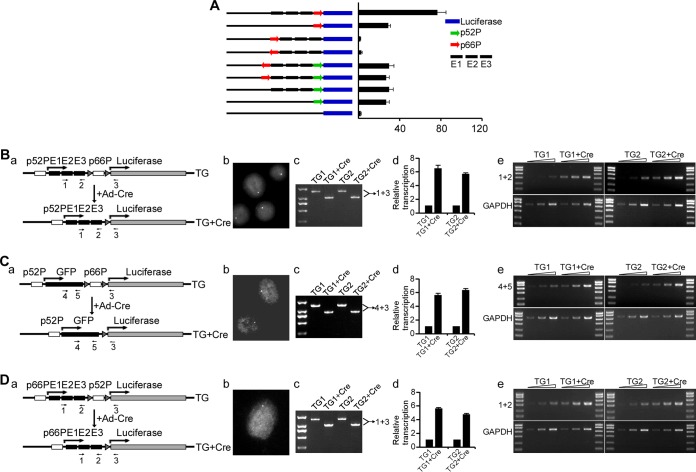
Juxtaposition of two tandem promoters inhibits upstream promoter activity. (A) Luciferase assay shows the effects of three enhancers and the p66Shc promoter on p52Shc promoter activity. Green arrows, p52Shc promoter; red arrows, p66Shc promoter; black boxes, enhancers E1, E2, and E3; blue boxes, luciferase gene. Error bars indicate means ± SD for a representative experiment performed in triplicate. (B to D) (a) Relative positions of the two tandem promoters and the three enhancers in the constructs and the positions of indicated primers used in PCR or RT-PCR assay; (b) DNA FISH showing the copy number of transgene; (c) PCR products (generated with the indicated primers) separated on agarose gels, where TG and TG+Cre refer to the transgene genotypes before and after CMV-Cre transduction, respectively; (d) results from RT-qPCRs showing the relative transcription initiated from the p52Shc promoter (B and C) or initiated from the p66Shc promoter (D) (error bars indicate means ± SD for a representative experiment performed in triplicate); (e) results from semiquantitative RT-PCRs with the indicated primers, in which RT was performed with 1 μg RNA as a template, and 1, 3, or 9 μl of cDNA was used as the template in the following PCRs.
To more carefully evaluate the effect of juxtaposition of two tandem promoters on promoter activity in chromatin environment, we performed stable-transfection experiments. We inserted the p52Shc promoter, E1, E2, and E3 side by side upstream of the p66Shc promoter, which mimics the native chromatin configuration of the p66Shc-expressing allele, in the pGLbasic luciferase vector. We then introduced LoxP sites flanking the p66Shc promoter (Fig. 6Ba). The vectors were transfected into HUVECs. Two clones that contain single-copy transgenes were identified using the TaqMan copy number assay kit and confirmed by DNA FISH (Fig. 6Bb). Then, Cre recombinase adenovirus (Ad-Cre) was added to delete the p66Shc promoter. The efficiency of the p66Shc promoter deletion is shown in Fig. 6Bc. p66Shc promoter deletion results in a striking increase in p52Shc promoter activity, as evaluated by reverse transcriptase quantitative PCR (RT-qPCR) and semiquantitative RT-PCR (Fig. 6Bd and Be), indicating that the p66Shc promoter can impose repressor activity on the p52Shc promoter in cis when juxtaposed to the p52Shc promoter.
To explore whether the suppressive influence of the p66Shc promoter on p52Shc promoter activity requires the three enhancers, we replaced the three enhancers with a nonrelevant sequence, green fluorescent protein (GFP), and performed the same experiments. Similarly, p66Shc promoter deletion enhanced p52Shc promoter activity (Fig. 6C), indicating that the repressor activity of the p66Shc promoter is independent of the three enhancers.
To test whether the repressor activity is promoter specific, we swapped the two promoters and placed the p66Shc promoter upstream of the p52Shc promoter. Interestingly, transcription initiated from the p66Shc promoter was inhibited by the p52Shc promoter (Fig. 6D), suggesting that the repressive activity of the downstream promoter is independent of the specific promoter. Transcription from the downstream promoter can inhibit upstream promoter activity when the two promoters physically interact with each other. These results indicate that physical interaction between the two tandem promoters represses transcription initiation from the upstream promoter and thereby controls promoter choice.
DISCUSSION
Using SHC1 as a model, we studied the mechanism by which alternative promoters are regulated. We found that p66Shc and p52Shc are generated from separate alleles in single cells in a heritable fashion. This distinct monoallelic transcription pattern is established by allele-specific physical interactions between two tandem promoters. Disruption of chromatin looping derepresses the p52Shc promoter in cis and leads to the monoallelic-to-biallelic transcriptional switch of p52Shc.
The SHC1 gene is the first known example of separate alleles generating different isoforms from a single gene in single cells in a heritable fashion. A large number of genes are known to be transcribed monoallelically, such as Igκ in B lymphocytes (36), Albumin in hepatocytes (37), olfactory receptor genes in neurons (38), and interleukin genes in T lymphocytes (39). However, cells accomplish monoallelic expression of these genes through silencing of one allele. In contrast, SHC1 encodes two proteins that are both required for individual endothelial cells to maintain their normal function, raising the problem of how the two promoters are coordinately regulated. Our findings that 70 to 80% of single HUVECs transcribe p52Shc and p66Shc in separate alleles suggest that endothelial cells use this distinct mechanism of allele-specific promoter activation to avoid transcriptional interference and express comparable levels of p66Shc and p52Shc. This transcription pattern is cell type specific because PBL that do not express p66Shc transcribe p52Shc biallelically. In addition to SHC1, a significant number of other genes produce different proteins from alternative promoters in single cells, including WEE1, which encodes Wee1 tyrosine kinase and the kinase domain from tandem promoters in single primary fibroblast IMR90 cells (40). Whether such genes also generate different proteins from separate alleles will be worth investigating.
Notably, this study demonstrates that physical interactions between alternative promoters repress transcription initiation from the upstream promoter. DNA interactions occur at numerous gene loci and reflect a widespread organizing principle of the chromatin fiber (41–46). Although many reports have shown that chromatin loops are highly dynamic and tightly correlate with gene function, whether these long-range interactions are a cause or consequence of dynamic changes in transcription initiation is still an open question. A report by Deng et al. showed that an enforced loop between the promoter and enhancer caused transcriptional activation of β-globin, indicating that chromatin looping causally underlies gene regulation (47). In our study, deletion of the p66Shc promoter in the native position caused derepression of the p52Shc promoter in cis, and ectopic placement of the p66Shc promoter adjacent to but downstream of the p52Shc promoter inhibited the p52Shc promoter activity in a transgene assay. However, two pieces of evidence support the view that the p66Shc promoter is not an intrinsic silencer of the p52Shc promoter. First, the p66Shc promoter had no effect on p52Shc promoter activity when it was placed upstream of the p52Shc promoter in the luciferase assay. Second, when we swapped the two promoters in a transgenic plasmid, the p66Shc promoter lost its silencer activity and, in contrast, the p52Shc promoter inhibited the p66Shc promoter. Therefore, the p52Shc promoter may not be repressed by the p66Shc promoter itself but may be repressed by promoter-promoter interaction, providing another example of chromatin looping underlying transcriptional change. How downstream promoters repress the interacting upstream promoter remains to be determined. Presumably, the colocalization of two tandem promoters sequesters the p52Shc promoter into a small loop, which may impair the association of transcriptional initiation complex with the p52Shc promoter or impede trafficking of the RNA polymerase II during elongation.
The finding that promoter-promoter interaction influences transcription is consistent with previous reports (34, 48). However, the interacting promoters identified in the present study are tandem and intragenic, which differs from the interacting intergenic promoters reported by Ruan and colleagues (34), therefore exhibiting different functions in transcriptional regulation. While the interacting intergenic promoters act as enhancers and positively regulate transcription (34), we found that the interacting tandem and intragenic promoters of SHC1 negatively regulate transcription initiating from the upstream promoter, expanding the functional diversity of promoter-promoter interactions.
CpG methylation and/or histone modification are two well-known epigenetic markers to distinguish differential alleles (49, 50). In this study, we found that three active histone modifications (H3K9acetyl, H3K4me3, H3K4me2) specifically occupied the active p66Shc promoter region. Histone modifications are mitotically heritable and have been reported to mark monoallelically expressed genes, including X-inactivated genes by promoter-restricted H3K4me2 (51) and olfactory receptor genes by H3K9me3 and H4K20me3 (52). Therefore, the two alleles of SHC1 might be specified by differential histone modifications.
In summary, this study demonstrates that intragenic promoter-promoter interactions control promoter usage through inactivation of the upstream promoter, revealing a new mechanism of alternative promoter regulation. This mechanism may allow heritable allelic control of bifunctional eukaryotic genes as a way of preventing asymmetric expression of functionally disparate proteins. Further investigation is warranted to understand how the allele-specific loop interactions are established, whether epithelial cells that also express both p52Shc and p66Shc in single cells use this mechanism to regulate SHC1 transcription, and how generally this mechanism of transcriptional regulation is used to control alternative promoter activity.
MATERIALS AND METHODS
Human tissues.
Human umbilical tissues and blood samples were collected from the Second Hospital, Tianjin Medical University, in accordance with the Internal Review and the Ethics Boards of Tianjin Medical University. Informed consent was obtained from all sample providers.
HUVEC cell purification and single-cell RT-PCR.
HUVECs were purified from umbilical tissues as described in a previous report (53). HUVECs were trypsinized, and the single cell was picked up and transferred into a 0.2-ml thin-wall PCR tube containing 4.9 μl of freshly prepared cell lysis buffer (1× PCR buffer containing 1.35 mM MgCl2, 0.45% NP-40, 4.5 mM dithiothreitol [DTT], 0.36 U RNase inhibitor, and 0.045 mM deoxynucleoside triphosphate [dNTP] mix). To pick up the single cell, we used a mouth tube to control an attached micropipette under a dissection microscope, which permits swift and efficient control of individual cell collection and release. The single cell was incubated at 70°C for 90 s in lysis buffer and then immediately transferred to liquid nitrogen, and all mRNAs were released from the single cell and subjected to subsequent RT-PCR. Then, 15 μl RT mix was added to each tube, so that the total volume for each RT reaction was 20 μl per tube. The tube was incubated at 42°C for 60 min in order to synthesize first-strand cDNAs, and then the reverse transcriptase was inactivated at 70°C for 15 min. A 10-μl volume of cDNA was added to a new PCR tube containing 10 μl of PCR mix to amplify p66Shc, and the other 10 μl of cDNA was added to another tube to amplify p52Shc. The PCR was carried out at 95°C for 3 min followed by 60 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Sequences of primers used are provided in Table 1.
TABLE 1.
Primers used for genotyping SHC1 DNA, p66Shc mRNA, and p52Shc mRNA
| Use and/or name of oligonucleotide | Sequence |
|---|---|
| SHC1-DF | 5′-CATGACAAAGAGCCTGGAGTGCC-3′ |
| SHC1-DR | 5′-TCCTCCTTCCCTACTGGTCTCCT-3′ |
| SNPrs8191981-DF1 | 5′-AATGAGTCTCTGTCATCGCTGGAG-3′ |
| SNPrs8191981-DF2 | 5′-CTGGCAGTGGCGTGATCCGGCAC-3′ |
| SNrs8191981-DR | 5′-GAGACGGTGAGAGTGATTGGCATT-3′ |
| Primers for RNA FISH | |
| p52Shc probe-F | 5′-GGCCCTGGTGAGTCTTGACT-3′ |
| p52Shc probe-R | 5′-AGAAGTCCTGGGGAGGGAGA-3′ |
| p52/p66 probe-F | 5′-TCCTCTGCCTGGGGACGATA-3′ |
| p52/p66 probe-R | 5′-TGGATTAAGCCTGAGGCTGA-3′ |
| Primers for 3C assays | |
| 3C-1 | 5′-GCAACGATCCTCGGCTAACTCCTGTC 3>′ |
| 3C-2 | 5′-CTACACCCAATCACCTGGACTTCCAG-3′ |
| 3C-3 | 5′-TCTCTCTATGCCCGAGAGCTGGCTGG-3′ |
| 3C-4 | 5′-GCTGCTGTAGCAGAGGATGCTAATGGC-3′ |
| 3C-5 | 5′-TGTGTGAGGCTGTGCCGGGTGCTAAG-3′ |
| 3C-ApoI-DNA-F | 5′-CGACACGTTCCAAGCCCTACTACG-3′ |
| 3C-ApoI-DNA-R | 5′-GTCCGTCTGCCCACCTTCACCAAC-3′ |
| ApoI-SouthernProbe1 | 5′-BIOTIN-CCTATCTCCCAAAAGGCAAAACC-3′ |
| ApoI-SouthernProbe2 | 5′BIOTIN-TCCTTGGGGATAGGGATGCTGCTG-3′ |
| 3C-6 | 5′-GAGGGCAGTTCTCACGCTTCTCAAG-3′ |
| 3C-7 | 5′-TAAAGTTTCTCCAGGGAGGCAGGG-3′ |
| 3C-8 | 5′-GACACGTTCCAAGCCCTACTACGAG-3′ |
| 3C-9 | 5′-CGTCAATCCCTGAAACTCCACCTGC-3′ |
| 3C-10 | 5′-CGGAGTTTCGCTCTTATCGTCCAGG-3′ |
| 3C-11 | 5′-TTTGCTCTTGTCACCTAGGCTGGAG-3′ |
| 3C-12 | 5′-CAGGGTTTCACCATGTTGACCAGG-3′ |
| 3C-13 | 5′-TCAACCCATCTTCTCCTGACACCC-3′ |
| 3C-14 | 5′-GTAGCAGAGGATGCTAATGGCCCAG-3′ |
| 3C-15 | 5′-CCACAGAGGTCCAACCAGGCTAAGG-3′ |
| 3C-DpnII-DNA-F | 5′-GGGACACCTCTACTTTCAAGCCTCC-3′ |
| 3C-DpnII-DNA-R | 5′-GGCAGCAGCAACCTTCCATCTACCC-3′ |
| DpnII-SouthernProbe1 | 5′BIOTIN-GGCGTCTTATTGGCCACCACTGCC-3′ |
| DpnII-SouthernProbe2 | 5′BIOTIN-GCAGCAGCCCCTCTTTCACCTCAAC-3′ |
| sgRNAs for p66Shc promoter deletion | |
| sgLacZ | 5′-CGCGATCGTAATCACCCGAGTGT-3′ |
| sgp66P-1 | 5′-CCTGCTCCAGTTCGAGGTTG-3′ |
| sgp66P-2 | 5′-CAGGCTGGACCCAGCGCTTC-3′ |
| sgp66P-Cas9-DF | 5′-GACCCAGACCTCCTATCTCCTTCC-3′ |
| sgp66P-Cas9-DR | 5′-CTTAGCCTGGTTGGACCTCTGTGG-3′ |
| Primers for promoter activity assay in transgene | |
| GAPDH-DF | 5′-GTCAACGGATTTGGTCGTATT-3′ |
| GAPDH-DR | 5′-AGTCTTCTGGGTGGCAGTGAT-3′ |
| Luci-1 | 5′-TCCCCCTAACCAGAGTTCTTG-3′ |
| Luci-2 | 5′-GGAAGAGCAAAGCTGGTGAA-3′ |
| Luci-3 | 5′-GGAACCAGGGCGTATCTC-3′ |
| Luci-4 | 5′-GACCATGTGATCGCGCTTCTC-3′ |
| Luci-5 | 5′-GGCAAGCTGACCCTGAAGTTC-3′ |
| GAPDH-RealTime-F | 5′-GGTGGTCTCCTCTGACTTCAACA-3′ |
| GAPDH-RealTime-R | 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ |
| E123-RealTime-F | 5′-GAGACAGGACAGTGCTTGGC-3′ |
| E123-RealTime-R | 5′-GGAAGAGCAAAGCTGGTGAA-3′ |
| GFP-RealTime-F | 5′-GACCATGTGATCGCGCTTCTC-3′ |
| GFP-RealTime-R | 5′-CGACCACTACCAGCAGAACAC-3′ |
| Primers for copy no. analysis of transgene | |
| Luci-CNV-F | 5′-GTAGCCATCCATCCTTGTC-3′ |
| Luci-CNV-R | 5′-CGCTGGGCGTTAATCAAAG-3′ |
| MGB-probe | 5′-FAM-CTCTCACACACAGTTCG-Q-MGB-3′ |
| Primers for digital PCR | |
| GAPDH-DF | 5′-ACGGATTTGGTCGTATTGGGC-3′ |
| GAPDH-DR | 5′-TTGACGGTGCCATGGAATTTG-3′ |
| GAPDH-probe | 5′-FAM-CCTGGTCACCAGGGCTGCTTTTAA-TAMRA-3′ |
| p52Shc-DF | 5′-GTTAGTGAGGCCGGAAGTG-3′ |
| p52Shc-DR | 5′-GGTTACCGCTCCAACTTTCT-3′ |
| p52Shc-probe | 5′-FAM-AAAGTTTCTCCAGGGAGGCAGGG-Q-MGB-3′ |
| rs8191981-DF | 5′-AGTCTGGTGTGTGAGGCTGTG-3′ |
| rs8191981-DF | 5′-ATTTCCCAGCAGCATCCC-3′ |
| rs8191981-Gprobe | 5′-FAM-CCTCCTTGTCGCCC-Q-MGB-3′ |
| rs8191981-Aprobe | 5′-VIC-CCTCCTTGTCACCC-Q-MGB-3′ |
| Primers for ChIP assays | |
| SHC1-1F | 5′-GAGGCGGGACACCTCTACTTTCAAG-3′ |
| SHC1-1R | 5′-AGCCAATGGCAGCGCGAGAT-3′ |
| SHC1-2F | 5′-GTGACTCACTCTTTCACCCCG-3′ |
| SHC1-2R | 5′-GTGGGGTCGGAGACAAAA-3′ |
| SHC1-3F | 5′-TTTCGCTCTTATCGTCCAGG-3′ |
| SHC1-3R | 5′-CGCCTGTAATCCTAACACTTAAGG-3′ |
| SHC1-4F | 5′-TCCCCCTAACCAGAGTTCTTG-3′ |
| SHC1-4R | 5′-TAGGGGAAAAGCTGTGTGTG-3′ |
| SHC1-5F | 5′-GAGACAGGACAGTGCTTGGC-3′ |
| SHC1-5R | 5′-GGAAGAGCAAAGCTGGTGAA-3′ |
| SHC1-6F | 5′-TGAGTCAGCACTGTCCTCACGATT-3′ |
| SHC1-6R | 5′-AACGTGGAGCCTCTTCTCTGGCT-3′ |
| SHC1-7F | 5′-TTCTAATGCTCACTCCAGCTTGGC-3′ |
| SHC1-7R | 5′-GGGAAGGGATGAGAAAGGTTTAGG-3′ |
| SHC1-8F | 5′-ATCAGTCTGGTGTGTGAGGCTGT-3′ |
| SHC1-8R | 5′-CACATTTCCCAGCAGCATCCCTAT-3′ |
| SHC1-9F | 5′-ACAGCCGAGTATGTCGCCTATGTT-3′ |
| SHC1-9R | 5′-TACTCACCTCCTCAAACAGCCAGA-3′ |
| SHC1-10F | 5′-ACAATACCTCAGGAACCCACCCAA-3′ |
| SHC1-10R | 5′-TCCTAATCCAAGAGCTGCTCCACA-3′ |
| SHC1-11F | 5′-ACCTTTCTACTTGGTTCTGGCCCT-3′ |
| SHC1-11R | 5′-AACACACACCCAAGACCCAGACTT-3′ |
| SHC1-12F | 5′-AGGACTCAGACCACTGTGCTTTCT-3′ |
| SHC1-12R | 5′-TGAACCACCACACCTGGCTAAACA-3′ |
| SHC1-13F | 5′-TGATCAGGATTGACAGGCCAC-3′ |
| SHC1-13R | 5′-TCCAAGGTGGAAGAGGGTGAAACA-3′ |
RNA FISH and RNA-DNA sequential FISH.
RNA probes for RNA FISH were prepared as follows (54). PCR-amplified regions of SHC1 were generated from genomic DNA. The products were subcloned into pGEM vector and were used to provide probes when transcribing from the T7 promoter. The plasmids were linearized by restriction enzyme digestion and labeled using a digoxigenin (DIG)-RNA kit (Roche Molecular Biochemicals, Mannheim, Germany).
RNA FISH was performed as described previously (55). HUVECs were cultured in poly-lysine-coated slides and fixed with 4% formaldehyde. After washing, permeabilization, and dehydration, cells were hybridized overnight with probes.
After RNA FISH, cells were treated with RNase and subsequently denatured and hybridized overnight with probes labeled with green fluorescence. In this analysis, chromosomal DNA was denatured by incubating the slides at 70°C for 5 min. After being counterstained with DAPI (4′,6-diamidino-2-phenylindole), cells were examined with a Leica TCS-4D confocal system.
Chromosome conformation capture.
3C assays were performed as described previously (41). Cells (106) were cross-linked and lysed, and nuclei were digested with 100 U of DpnII or ApoI at 37°C or 50°C for 16 h. After ligation and subsequent DNA purification, the cross-linking frequencies between the anchor and test fragments, as measured by the amount of corresponding ligation product, were estimated by PCRs relative to standards. The PCR product spanning a 6-kb DNA fragment upstream of p66Shc promoter was digested with ApoI or DpnII to completion and ligated at high concentration to generate equimolar mixtures of all possible ligation products and was used to generate standards. The cross-linking and ligation efficiencies between different samples and different experiments were also normalized by setting the highest cross-linking frequency for each experimental series to 1.0.
CRISPR/Cas9-mediated p66Shc promoter deletion.
sgRNAs specific for the p66Shc promoter region were designed from http://crispr.mit.edu, and two sgRNAs with minimized off-target effects were selected and are shown in Table 1. The sgLacZ sequence was used as a control. DNA oligonucleotides were synthesized and ligated into BbsI-digested pX330 (Addgene 42230) after annealing as described previously (56). Then, the two U6-sgRNA cassettes were amplified by PCR and integrated into one pX330 plasmid to subclone a two-in-one U6-sgRNA cassette and a CBH-Cas9 cassette into a lentiviral vector named lenti-sgp66 1-2 Cas9. HUVECs were transfected with lentivirus containing the two-in-one p66Shc-promoter-specific sgRNAs or sgLacZ, and the GFP-positive cells were selected with FACS after 3 days of transfection.
dPCR.
Digital PCR amplifications were carried out on a QuantSudio 3D Digital PCR system (Thermo Fisher Scientific, USA). Reactions were conducted in a 15-μl reaction mixture containing QuantStudio 3D Digital PCR master mix v2 (Thermo Fisher Scientific, USA), 800 nM each primer, 200 nM each TaqMan probe, and template. The primers and TaqMan probes used in this study are listed in Table 1. Positive and negative controls were added in each run. Reaction mixtures of 14.5 μl for each sample were added into QuantStudio 3D Digital PCR 20K Chip (Thermo Fisher Scientific, USA), and the dPCR program of amplifications was developed and carried out according to the manufacturer's user guide. The QuantStudio 3D Digital PCR instrument (Thermo Scientific, USA) was used to determine the number of positive wells among the total number of wells per chip, and the Applied Biosystems QuantStudio 3D AnalysisSuite Cloud software was used to analyze and refine the data derived from the QuantStudio 3D Digital PCR instrument. A Poisson distribution was used to estimate the average number of copies per chip.
Allelle-specific ChIP.
Briefly, 107 HUVECs were cross-linked with 2% formaldehyde for 10 min and quenched with glycine at a final concentration of 0.125 M for 5 min. After washing with ice-cold phosphate-buffered saline (PBS) three times, the cells were lysed with lysis buffer (10 mM Tris-Cl [pH 8.0], 10 mM NaCl, 0.2% NP-40, and 1 mM DTT) on ice for 1 h. The nuclei were harvested and resuspended in ApoI buffer containing 0.3% sodium dodecyl sulfate (SDS) and incubated at 37°C for 1 h while shaking. Triton X-100 was added to 1.8% to sequester the SDS, and samples were incubated at 37°C for another 1 h. Samples were then digested with ApoI at 50°C for 16 h. SDS was added to 1.6%, and the samples were heated at 80°C for 20 min to inactivate ApoI. Triton X-100 was added to 1.8% to sequester the SDS again. Samples were then diluted into NET buffer (50 mM Tris [pH7.4], 150 mM NaCl, 5 mM EDTA, 0.5% NP-40), and the resulting chromatin fragments were immunoprecipitated with anti-acetyl-histone H3 (Lys9), anti-dimethyl-histone H3 (Lys4), anti-trimethyl-histone H3 (Lys4), anti-dimethyl-histone H3 (Lys9), and anti-trimentyl-histone H3 (Lys36) antibodies (Millipore). Purified rabbit IgG (Sigma) was used as a control. We reversed the cross-linking of ChIP samples with proteinase K digestion and subjected the samples to phenol-chloroform extraction before ethanol precipitation with glycogen. Immunoprecipitated DNA and input DNA were quantified using a NanoDrop. The p66Shc promoter and the SNP site rs8191981 belonged to the same DNA fragment; PCR products carrying C/T at SNP rs8191981 were amplified from input or IP. The allelic association of specified histones with the p66Shc promoter and p52Shc promoter was measured through SNP frequency in the PCR products by digital PCR. The primers used in this study are listed in Table 1.
ACKNOWLEDGMENTS
We thank William Garrard at UT Southwestern Medical Center for his critical review.
This work was supported by grants (81773034, 91519331, and 31371295 to Zhe Liu, 81472681 to Xichuan Li, and 31270928 to Yougui Xiang) from the National Natural Science Foundation of China, grant 2014CB910100 to Zhe Liu from the Ministry of Science and Technology of China, grant 15JCZDJC34800 from the Tianjin Municipal Science and Technology Commission to Zhe Liu, Excellent Talent Project to Zhe Liu and grants R01CA208620 from the NIH and RP160307 from the CPRIT to Lance S. Terada.
We declare that we have no potential conflicts of interest.
REFERENCES
- 1.Pozner A, Goldenberg D, Negreanu V, Le SY, Elroy-Stein O, Levanon D, Groner Y. 2000. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol Cell Biol 20:2297–2307. doi: 10.1128/MCB.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courtois V, Chatelain G, Han ZY, Le Novere N, Brun G, Lamonerie T. 2003. New Otx2 mRNA isoforms expressed in the mouse brain. J Neurochem 84:840–853. doi: 10.1046/j.1471-4159.2003.01583.x. [DOI] [PubMed] [Google Scholar]
- 3.Blaschke RJ, Topfer C, Marchini A, Steinbeisser H, Janssen JW, Rappold GA. 2003. Transcriptional and translational regulation of the Leri-Weill and Turner syndrome homeobox gene SHOX. J Biol Chem 278:47820–47826. doi: 10.1074/jbc.M306685200. [DOI] [PubMed] [Google Scholar]
- 4.Arce L, Yokoyama NN, Waterman ML. 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 5.Ventura A, Luzi L, Pacini S, Baldari CT, Pelicci PG. 2002. The p66Shc longevity gene is silenced through epigenetic modifications of an alternative promoter. J Biol Chem 277:22370–22376. doi: 10.1074/jbc.M200280200. [DOI] [PubMed] [Google Scholar]
- 6.Ma Z, Liu Z, Wu RF, Terada LS. 2010. p66(Shc) restrains Ras hyperactivation and suppresses metastatic behavior. Oncogene 29:5559–5567. doi: 10.1038/onc.2010.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goossens S, Janssens B, Vanpoucke G, De Rycke R, van Hengel J, van Roy F. 2007. Truncated isoform of mouse alphaT-catenin is testis-restricted in expression and function. FASEB J 21:647–655. doi: 10.1096/fj.06-6066com. [DOI] [PubMed] [Google Scholar]
- 8.Muller M, Schleithoff ES, Stremmel W, Melino G, Krammer PH, Schilling T. 2006. One, two, three–p53, p63, p73 and chemosensitivity. Drug Resist Updat 9:288–306. doi: 10.1016/j.drup.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Nakanishi T, Bailey-Dell KJ, Hassel BA, Shiozawa K, Sullivan DM, Turner J, Ross DD. 2006. Novel 5′ untranslated region variants of BCRP mRNA are differentially expressed in drug-selected cancer cells and in normal human tissues: implications for drug resistance, tissue-specific expression, and alternative promoter usage. Cancer Res 66:5007–5011. doi: 10.1158/0008-5472.CAN-05-4572. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal VR, Bulun SE, Leitch M, Rohrich R, Simpson ER. 1996. Use of alternative promoters to express the aromatase cytochrome P450 (CYP19) gene in breast adipose tissues of cancer-free and breast cancer patients. J Clin Endocrinol Metab 81:3843–3849. [DOI] [PubMed] [Google Scholar]
- 11.Shearwin KE, Callen BP, Egan JB. 2005. Transcriptional interference—a crash course. Trends Genet 21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. 2009. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell 34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP. 2012. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 14.Callen BP, Shearwin KE, Egan JB. 2004. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell 14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Greger IH, Demarchi F, Giacca M, Proudfoot NJ. 1998. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res 26:1294–1301. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson SL, Ryan K, Sollner-Webb B. 1989. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev 3:212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- 17.Hongay CF, Grisafi PL, Galitski T, Fink GR. 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 18.Martens JA, Laprade L, Winston F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 19.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. 2006. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci PG, Schlessinger J, Pawson T. 1992. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360:689–692. doi: 10.1038/360689a0. [DOI] [PubMed] [Google Scholar]
- 21.Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci PG. 1992. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell 70:93–104. doi: 10.1016/0092-8674(92)90536-L. [DOI] [PubMed] [Google Scholar]
- 22.Xi G, Shen X, Clemmons DR. 2010. p66shc inhibits insulin-like growth factor-I signaling via direct binding to Src through its polyproline and Src homology 2 domains, resulting in impairment of Src kinase activation. J Biol Chem 285:6937–6951. doi: 10.1074/jbc.M109.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, Pelliccia G, Luzi L, Minucci S, Marcaccio M, Pinton P, Rizzuto R, Bernardi P, Paolucci F, Pelicci PG. 2005. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell 122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Natalicchio A, Laviola L, De Tullio C, Renna LA, Montrone C, Perrini S, Valenti G, Procino G, Svelto M, Giorgino F. 2004. Role of the p66Shc isoform in insulin-like growth factor I receptor signaling through MEK/Erk and regulation of actin cytoskeleton in rat myoblasts. J Biol Chem 279:43900–43909. doi: 10.1074/jbc.M403936200. [DOI] [PubMed] [Google Scholar]
- 25.Ma Z, Myers DP, Wu RF, Nwariaku FE, Terada LS. 2007. p66Shc mediates anoikis through RhoA. J Cell Biol 179:23–31. doi: 10.1083/jcb.200706097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartman CR, Hsu SC, Hsiung CC, Raj A, Blobel GA. 2016. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell 62:237–247. doi: 10.1016/j.molcel.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. 2017. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547:419–424. doi: 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 29.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A 103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murrell A, Heeson S, Reik W. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Xu Z, Du W, Zhang Z, Wei Y, Wang H, Zhu Z, Qin L, Wang L, Niu Q, Zhao X, Girard L, Gong Y, Ma Z, Sun B, Yao Z, Minna JD, Terada LS, Liu Z. 2014. Aiolos promotes anchorage independence by silencing p66Shc transcription in cancer cells. Cancer Cell 25:575–589. doi: 10.1016/j.ccr.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medici D, Shore EM, Lounev VY, Kaplan FS, Kalluri R, Olsen BR. 2010. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med 16:1400–1406. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Gao D, Wang H, Li X, Yang J, Yan X, Liu Z, Ma Z. 2015. Negative feedback loop between p66Shc and ZEB1 regulates fibrotic EMT response in lung cancer cells. Cell Death Dis 6:e1708. doi: 10.1038/cddis.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu ZM, George-Raizen JB, Li S, Meyers KC, Chang MY, Garrard WT. 2002. Chromatin structural analyses of the mouse Igkappa gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. J Biol Chem 277:32640–32649. doi: 10.1074/jbc.M204065200. [DOI] [PubMed] [Google Scholar]
- 36.Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. 1998. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev 12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaelson J. 1993. Cellular selection in the genesis of multicellular organization. Lab Invest 69:136–151. [PubMed] [Google Scholar]
- 38.Chess A, Simon I, Cedar H, Axel R. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell 78:823–834. doi: 10.1016/S0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 39.Guo L, Hu-Li J, Paul WE. 2005. Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity 23:89–99. doi: 10.1016/j.immuni.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. 2005. A high-resolution map of active promoters in the human genome. Nature 436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Garrard WT. 2005. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol 25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10:1453–1465. doi: 10.1016/S1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 43.Dean A. 2011. In the loop: long range chromatin interactions and gene regulation. Brief Funct Genomics 10:3–10. doi: 10.1093/bfgp/elq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadauke S, Blobel GA. 2009. Chromatin loops in gene regulation. Biochim Biophys Acta 1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miele A, Dekker J. 2008. Long-range chromosomal interactions and gene regulation. Mol Biosyst 4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoenfelder S, Clay I, Fraser P. 2010. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev 20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoenfelder S, Sugar R, Dimond A, Javierre BM, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, Segonds-Pichon A, Jurkowski W, Wingett SW, Tabbada K, Andrews S, Herman B, LeProust E, Osborne CS, Koseki H, Fraser P, Luscombe NM, Elderkin S. 2015. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet 47:1179–1186. doi: 10.1038/ng.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelsey G, Feil R. 2013. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 368:20110336. doi: 10.1098/rstb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubner MR, Eckersley-Maslin MA, Spector DL. 2013. Chromatin organization and transcriptional regulation. Curr Opin Genet Dev 23:89–95. doi: 10.1016/j.gde.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rougeulle C, Navarro P, Avner P. 2003. Promoter-restricted H3 Lys 4 di-methylation is an epigenetic mark for monoallelic expression. Hum Mol Genet 12:3343–3348. doi: 10.1093/hmg/ddg351. [DOI] [PubMed] [Google Scholar]
- 52.Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, Barnea G, Kellis M, Lomvardas S. 2011. An epigenetic signature for monoallelic olfactory receptor expression. Cell 145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baudin B, Bruneel A, Bosselut N, Vaubourdolle M. 2007. A protocol for isolation and culture of human umbilical vein endothelial cells. Nat Protoc 2:481–485. doi: 10.1038/nprot.2007.54. [DOI] [PubMed] [Google Scholar]
- 54.Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K, Dillon N, Gray D, Perry RP, Merkenschlager M, Fisher AG. 2001. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol 2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 55.Chakalova L, Carter D, Fraser P. 2004. RNA fluorescence in situ hybridization tagging and recovery of associated proteins to analyze in vivo chromatin interactions. Methods Enzymol 375:479–493. doi: 10.1016/S0076-6879(03)75029-0. [DOI] [PubMed] [Google Scholar]
- 56.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]