Abstract
DNA damage has been linked to genomic instability and the progressive breakdown of cellular and organismal homeostasis, leading to the onset of disease and reduced longevity. Insults to DNA from endogenous sources include base deamination, base hydrolysis, base alkylation and metabolism-induced oxidative damage that can lead to single-strand and double-strand DNA breaks. Alternatively, exposure to environmental pollutants, radiation or ultra-violet light, can also contribute to exogenously derived DNA damage. We previously validated a novel, high through-put approach to measure levels of DNA damage in cultured mammalian cells. This new CometChip Platform builds on the classical single cell gel electrophoresis or comet methodology used extensively in environmental toxicology and molecular biology. We asked whether the CometChip Platform could be used to measure DNA damage in samples derived from environmental field studies. To this end, we determined that nucleated erythrocytes from multiple species of turtle could be successfully evaluated in the CometChip Platform to quantify levels of DNA damage. In total, we compared levels of DNA damage in 40 animals from two species: the box turtle (Terrapene carolina) and the red-eared slider (Trachemys scripta elegans). Endogenous levels of DNA damage were identical between the two species, yet we did discover some sex-linked differences and changes in DNA damage accumulation. Based on these results, we confirm that the CometChip Platform allows for the measurement of DNA damage in a large number of samples quickly and accurately, and is particularly adaptable to environmental studies using field-collected samples.
Keywords: CometChip, Erythrocytes, Comet assay, DNA damage, Vertebrates
INTRODUCTION
Global chemical production is increasing exponentially, leading to widespread exposure of many organisms and ecosystems to pollutants (Schwarzenbach et al. 2006). Short- or long-term chemical exposure can cause population decline and even extinction through genetic, physiologic, metabolic, behavioral, or ecological alterations (Bruhl et al. 2013; Goulson et al. 2015), or can lead to potential adaptation of organisms to pollutants; reviewed in (Whitehead 2014). Since chemicals can have diverse impact across organisms and at multiple levels of biological organization (e.g., cells, organs, whole individuals), tools are needed to rapidly assess the nature and magnitude of effects to a given species or system.
Genotoxicity due to DNA damage is an adverse effect that can result from exposure to pollutants. While DNA damage naturally occurs in cells, for example as a result of metabolic processes, chemical exposure has been observed to increase the amount of DNA damage in organisms (Ojha et al. 2013). For this reason, measures of DNA damage have become common markers of environmental genotoxicity (Frenzilli et al. 2009). Genotoxicity can be caused by many types of chemicals including metals (Lacaze et al. 2011), pharmaceuticals (Parrella et al. 2015), and pesticides (Guilherme et al. 2012; Moreno et al. 2014). Studies have linked genotoxicants to effects on growth, reproduction, offspring survival and development, population size and population dynamics (Devaux et al. 2011; Poletta et al. 2011; Sarkar et al. 2017).
Common methods used to detect DNA damage include the presence of micronuclei and sister chromatid exchanges; reviewed in (Frenzilli et al. 2009). However, the classical comet assay (Single Cell Gel Electrophoresis, SCGE) has become one of the most widely applied methods to detect DNA breakage and DNA repair (Azqueta and Collins 2013). Compared to other approaches, the comet assay shows higher sensitivity and reliability in measuring DNA damage in the nucleus of the cell by detecting multiple types of DNA damage (single-strand breaks, double-strand breaks, base damage, DNA cross-links). Further, as a single-cell assay, this approach provides DNA damage quantification for individual cells. SCGE relies on electrophoresis; negatively charged DNA will migrate in an agarose matrix towards the positive anode. When the DNA is undamaged, it remains tightly supercoiled, inhibiting movement through the matrix. However, with increasing DNA damage, the DNA becomes more relaxed due to loss of supercoiling. The less tightly bound DNA allows for further migration in the agarose matrix. The method works for most cell types, requires a small amount of starting material, and can be completed within 24 hours (Olive and Banath 2006; Karbaschi and Cooke 2014).
Recently, we developed and validated a high throughput version (CometChip Platform) of the classical SCGE methodology to increase the number of samples that can be processed at one time, reducing inter/intra assay variability considerably (Sykora et al. 2018). The CometChip Platform is comprised of 96 wells in a standard well configuration of 8×12 wells, with each well consisting of approximately 500 microwells, each able to hold a single cell. Cells are gravity-loaded into the chip, allowing for up to 96 individual experiments to be conducted in a single assay. Extensive testing and damage measurements using the CometChip Platform have been conducted, showing that this method can be used successfully to measure DNA damage in human patient samples as well as cultured adherent and suspension mammalian cells (Sykora et al. 2018). Although the utility of this method has been demonstrated when screening environmental compounds in a laboratory-based setting (Sykora et al. 2018), there is a need for a protocol that can measure DNA damage in a large number of field-derived samples both accurately and reproducibly. Furthermore, while most of the applications using the CometChip Platform have been carried out on mammalian cells, mammalian erythrocytes no longer have a nucleus and hence are incompatible with the comet protocol. Since blood is one of the most accessible tissues to sample for many vertebrates under field conditions, and since the erythrocytes of non-mammalian vertebrates are nucleated, application of the CometChip Platform to screen for genome damage in field-collected samples of nucleated blood would be of great advantage in environmental toxicology. In this work, we consider whether the CometChip Platform can be applied successfully to detect DNA damage in blood samples of turtles sampled under field conditions. Such a technique would find application in many avenues of environmental toxicology requiring field sampling of non-mammalian vertebrates.
MATERIALS AND METHODS
Chemicals
Hydrogen peroxide (H2O2), methyl methanesulfonate (MMS), N-methyl-N′-nitro-N nitrosoguanidine (MNNG), DMSO, glycerol and etoposide were purchased from Sigma-Aldrich (St. Louis, MO). All DNA damaging agents were diluted immediately prior to use.
CometChip supplies
The CometChip Platform is now available from Trevigen, a division of Bio-Techne (Minneapolis, MN). This includes the disposable 30 μM CometChips, the CometChip 96-well magnetically sealed cassettes, the CometChip electrophoresis system and the Comet Analysis Software (CAS).
Animal sampling
Box turtles (Terrapene carolina) and red-eared sliders (Trachemys scripta elegans) were sampled for comparison in this project. This project was conducted under University of South Alabama IACUC permit # 922419. Box turtles were collected by hand, while sliders were collected via baited hoopnet traps in freshwater ponds on the University of South Alabama campus (Mobile, AL). Carapace length and width of each individual were measured using calipers and sex was recorded based on plastron morphology for box turtles (St Clair 1998) and body size, front claw length, and tail length and thickness for sliders (Readel et al. 2008). Additionally, one common snapping turtle (Chelydra serpentina) was sampled for the initial protocol-testing phase of the project. Approximately 200 μl of blood was collected from the sub-carapacial sinus of all individuals using a 23-gauge or a 25-gauge needle and 3 cc syringe and then placed into vacutainer tubes containing lithium heparin (BD biosciences, San Jose, CA). Among the tissues that could potentially be used for the comet assay, blood samples can be considered minimally invasive compared to biopsies from muscles or internal organs (e.g., liver). After sampling, samples were kept on ice until processed for use in the CometChip assay and were transported to the laboratory within two hours of drawing blood.
Blood processing
Cell number was determined using the Countess® II automated cell counter (ThermoFisher Scientific, Waltman, MA). Blood was serial diluted in PBS to reach an erythrocyte concentration of approximately 106 cell per ml. Cell counts were then done in triplicate with the final values averaged. Cellular morphology was imaged on an EVOS XL imaging system (ThermoFisher Scientific).
Freezing protocols
We tested the effect of freezing the samples following common procedures used during field sampling – i.e., blood samples initially stored on ice and then frozen for longer term storage. Before freezing at −80°C, blood aliquots were placed in each of the following storage solutions to test efficacy: (1) Cryostor CS10 (Stem cell technologies, Cambridge, MA), which has 10% DMSO and other unspecified agents; (2) PBS, diluted to 106 cells per ml; (3) 20% glycerol in PBS, where the blood was again diluted to a working concentration before freezing; (4) 10% DMSO in PBS. Blood was also frozen without any dilution or further treatment; these samples are designated as “no treatment”. The frozen samples were stored at −80°C continuously for 6 months before analysis.
General CometChip protocol
The general CometChip protocol was further optimized from the manufacturer’s instructions (Trevigen) and our own previous experiments during the development of the CometChip technology (Sykora et al. 2018). Each well in the 96-well CometChip contains approximately 500 microwells (microwell width of 30 μM). Cells were loaded into the CometChip apparatus at an initial concentration of 50,000 cells per well (Figure 1) which was reduced to 10,000 cells per well for subsequent experiments. Cells were gravity loaded into the 30 μM microwells for 30 min at room temperature (RT). DNA damaging compounds or vehicle controls were diluted in PBS and applied immediately after the cells were loaded into the CometChip. For repair capacity measurements, cells were exposed to DNA damaging agents in the CometChip for 1 hour at RT, washed three times with PBS and allowed to repair in PBS for 1 hour at RT. After treatment (H2O2, MMS or etoposide) in the CometChip apparatus, the chip was washed multiple times with PBS and sealed with low melting point agarose (LMPA) (Topvision, ThermoFisher Scientific) (7 ml; 0.8% LMPA/PBS). The CometChip was then submerged in lysis solution with Triton X-100 detergent (Trevigen) for 40 min at 4°C. The CometChip was run under alkaline (pH>13) conditions (200 mM NaOH, 1 mM EDTA, 0.1% Triton X-100). Electrophoresis was conducted at 22 V for 50 min at 4°C. After electrophoresis, the CometChip was re-equilibrated to neutral pH using Tris buffer (0.4 M Tris·Cl, pH 7.4). Subsequently, the DNA was stained with 1× SYBR Gold (ThermoFisher Scientific) diluted in Tris buffer (20 mM Tris·Cl, pH 7.4) for 30 min and de-stained for 1 h in Tris buffer (20 mM Tris·Cl, pH 7.4).
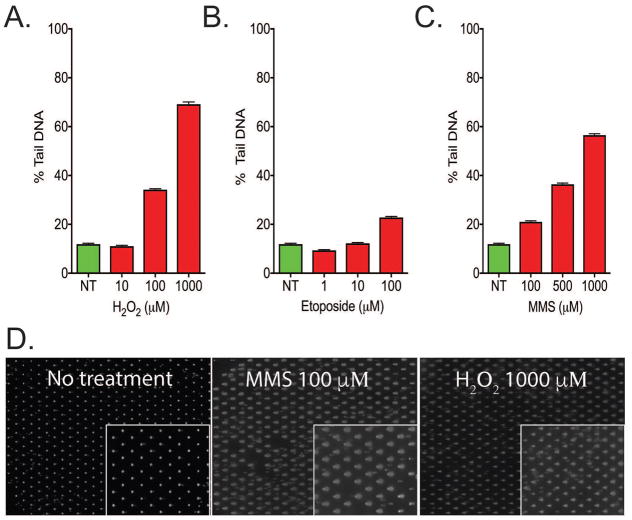
Figure 1. Assessment of nucleated erythrocytes using the CometChip Platform.
Whole blood from a T. scripta was exposed to (A) H2O2, (B) Etoposide or (C) MMS for 30 minutes and measured using the CometChip assay. The CometChip was able to measure DNA damage accumulation, which increased with the concentration of DNA damaging agent. (D) Representative images of the cells after loading into the CometChip and the extent of DNA damage after exposure. (n=3964–4277 from duplicate experiments). Error bars represent mean ± 95% confidence interval.
Automated image acquisition
Image acquisition was conducted on the Celigo S imaging cytometer (Nexcelom Bioscience, Lawrence, MA) at a resolution of 1 micron/pixel with whole plate imaging to avoid imaging variability. Image analysis was conducted using the dedicated Comet Analysis Software (CAS) with the box size set to 220 × 180 pixels which represented a box size that would capture comets from heavily damaged cells without box overlap. Data acquired were exported to Excel (Microsoft) and subsequently to GraphPad (Prism) for statistical analysis.
Statistical Analysis
All statistics and significance analysis was calculated in GraphPad (Prism). Two group comparisons were conducted using unpaired two tailed t-test with Welch’s correction.
RESULTS
Assessment of DNA damage in nucleated erythrocytes using the CometChip Platform
To determine the feasibility of evaluating levels of DNA damage in erythrocyte cells by the CometChip Platform, initial testing was conducted using the whole blood of a single Trachemys scripta elegans (T. scripta) animal. It was determined that the number of cells in the blood sample was between 108 and 109 cells per ml. While validating the CometChip Platform, we found an optimal number of cells to be at a concentration of 1–5 × 105 cells per ml or greater. Therefore, the whole blood was initially loaded at a concentration of 5 × 105 cell per ml (Figure 1), to ensure that all microwells were filled. Subsequent experiments used a loading concentration of 105 per ml. After loading, the cells were exposed to three established DNA damaging agents, each able to induce a different form of DNA damage. Hydrogen peroxide (H2O2) produces a range of oxidative DNA damage and DNA single-strand breaks (SSBs). At high concentrations (greater than 1 mM), H2O2 has been reported to also induce DNA double-strand breaks (DSBs) (Dahm-Daphi et al. 2000). An acute exposure (30 minutes) of the T. scripta erythrocyte cells to H2O2 produced an increase in the amount of DNA in the comet tail compared to the relatively immobile nucleoid, consistent with an increase in DNA damage (Figure 1A). Further, H2O2 at both 100 and 1000 μM but not 10 μM induced DNA damage (>20% tail DNA) in the blood cells. Etoposide, in contrast to H2O2, induces DSBs at low micromolar concentrations in proliferating mammalian cells. In our experiments, T. scripta erythrocyte cells treated with etoposide induced a moderate (>30% tail DNA) level of DNA damage at the highest concentration tested (100 μM) (Figure 1B). MMS is an SN2 alkylating agent, and the erythrocyte cells of T. scripta accumulated greater than 20% tail DNA at all concentrations tested (Figure 1C). Analysis of the comet loading (Figure 1D) showed that the CometChip had greater than 99% loading and that the damage calculated in Figure 1A–C does correspond to visible comet tails. Therefore, the results obtained from tests run on a single animal showed that the CometChip Platform is compatible with measuring DNA damage in cells derived from these blood samples.
Determination of CometChip suitability for field-based research
For the CometChip assay to be adaptable to field collected blood samples, it was necessary to determine that the samples could be handled and stored in a manner that did not induce artificially high levels of DNA damage. We developed a plan (Figure 2A) in which the samples were isolated in the field and then kept on ice for short-term storage while transferred to the laboratory. Once in the laboratory, the samples were separated into three fractions. The first fraction was dedicated to cell profiling to assess physical parameters of the blood including cell number and morphology. The second fraction was used to measure DNA damage and perform a limited analysis of DNA repair capacity. Measurements of DNA repair could not be done on frozen samples. The third fraction of the collected blood was devoted to long-term sample storage. These samples could then be evaluated for endogenous DNA damage at a later stage.
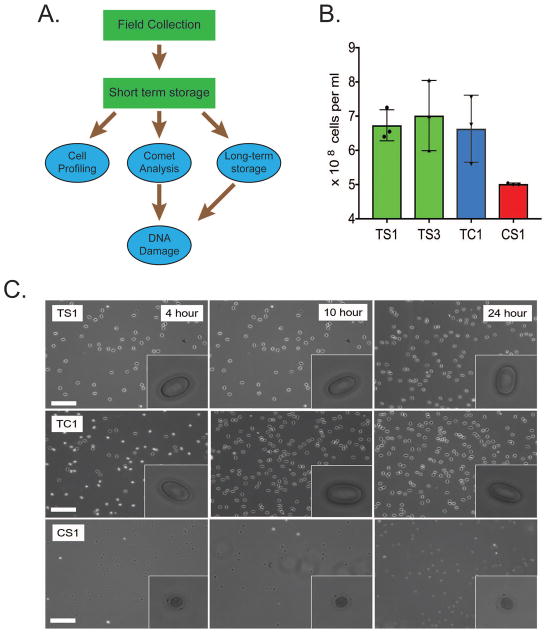
Figure 2. Determination of CometChip suitability for field-based research.
(A) Plan for the collection and utilization of blood samples collected in the field. Steps in green are conducted in the field, the steps in blue are conducted in the laboratory. (B) Cell counts were taken from each of the four turtles: T. scripta (TS), T. carolina (TC) and C. serpentina (CS). All turtle species had a high cell count. Each count was measured in triplicate. (C) Morphology of erythrocytes from the different turtle species. Magnified image of the cells after 4, 10 and 24 hours on ice. Storage on ice did not visibly effect cellular morphology. Erythrocytes from the CS animal had a distinct morphology compared to TS and TC cells. Scale bar represents 200 μm. Inserted box is 20 μm.
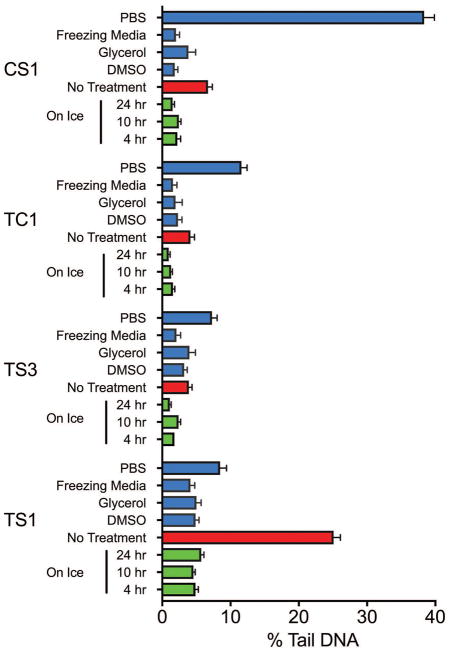
To determine the effectiveness of the work plan, we increased our sample size to four animals, made up of three species: two individuals of T. scripta (TS), one of T. carolina (TC), and one of C. serpentina (CS). The blood was extracted into heparinized tubes and kept on ice until analysis (4 hours). We first assessed blood cell concentration (Figure 2B) and found that all samples were comparable. Remote site fieldwork may also necessitate samples being stored on ice for prolonged periods of time. The samples were inspected for any changes in morphology after 4, 10 and 24 hours on ice (Figure 2C). This analysis allowed us to determine if there were large amounts of cell death or gross changes in morphology. The cells retained morphology on ice up to 24 hours after samples were taken in the field. We next investigated if storage conditions impacted the endogenous levels of DNA damage when measured in the CometChip Platform. Storage on ice for 10 or 24 hours did not cause a significant increase in levels of DNA damage compared to the 4 hour results in any of the samples tested (Figure 3). Finally, because field collection may require the freezing of samples, either due to the remoteness of the location of the research or the duration of sample collection (over many seasons or years), samples need to be stored in such a way as to not promote excessive DNA damage. Five freezing protocols were evaluated (Figure 3). Dilution of the sample in Phosphate Buffered Saline (PBS) or freezing the samples in the heparin tubes in which the sample was originally collected resulted in high levels of DNA damage in many of the samples, suggesting that some further intervention was required to protect the cells from freezing-induced or freeze/thaw-induced DNA damage. Three interventions were tested: (i) 10% DMSO in PBS, (ii) 20% glycerol in PBS or (iii) commercial freezing medium (Cryostor CS10, Stem cell technologies). We found that any of the three intervention protocols were sufficient to protect the DNA when stored at −80°C for up to 6 months. However, the commercial freezing medium offered the most consistently low increase in DNA damage after freezing. These results show that long term storage of blood samples on ice (up to 24 hours) or frozen at −80°C for up to 6 months in freezing medium would both be suitable for studies on DNA damage analysis using the CometChip Platform (Figures 3 and S1).
Figure 3. Effect of various storage conditions on levels of endogenous DNA damage.
Adaptability of erythrocytes to freezing and thawing. Cells were frozen under multiple treatments and the effect on the levels of endogenous DNA damage was evaluated. As expected, PBS alone and no treatment resulted in high levels of DNA damage. The results suggest that any of the three interventions adequately protect the cell. The commercial freezing medium had the greatest protective effect closely followed by samples frozen in 10% DMSO (n= 350–1642). Error bars represent mean ± 95% confidence interval.
Measuring DNA damage in whole blood samples
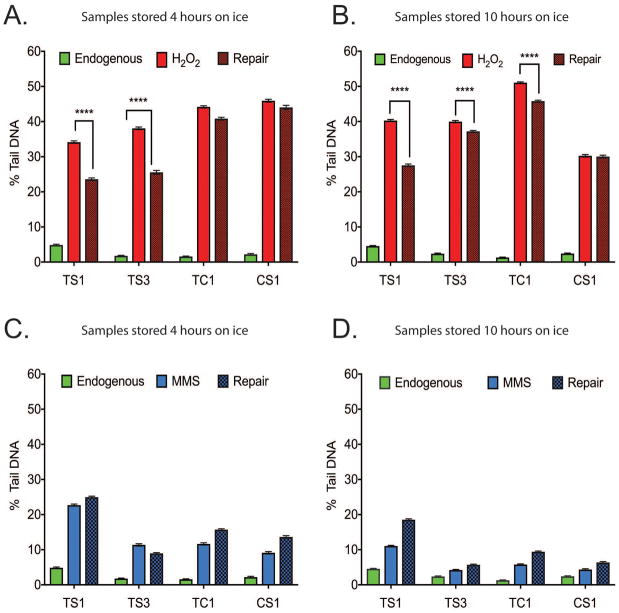
An advantage of the high-throughput CometChip Platform is that it is possible to measure not only levels of endogenous DNA damage but also measure the change in the levels of DNA damage following a repair period (after exposure to DNA damaging agents), providing some limited information on DNA repair capacity. Unlike measuring endogenous DNA damage, measuring DNA repair must be done on live cells, before freezing. We asked whether the cells would be able to repair DNA damage induced by a short exposure (30 minutes) to either H2O2 (100 μM) or MMS (100 μM). Another factor tested was whether the amount of time the samples were stored on ice would influence repair capacity. We compared each species after H2O2 exposure (Figure 4A). The TS samples accumulated less DNA damage than the samples from the other two species. After 30 minutes incubation at room temperature, the TS samples also repaired significantly more of the DNA damage than the other samples. These results were compared to an identical assay conducted on the same samples after a further 6 hours (10 hours total) on ice (Figure 4B). The extended period of time on ice appeared to increase the amount of initial DNA damage accumulated. Overall, results between the two time-points were comparable, with the exception of the CS1 samples that showed substantial variation in DNA damage and repair response. MMS is an SN2 alkylating agent that produces a varied spectrum of DNA damage lesions (Wyatt and Pittman 2006). In contrast to the results obtained with H2O2, only limited DNA damage and repair was measured after exposure to MMS (Figure 4C), consistent with our previous studies evaluating DNA damage in human cells induced by MMS (Sykora et al. 2018). A further difference between the two agents was that there was a decrease in DNA damage after extended storage (10 hours total) on ice (Figure 4D). This data suggests that when performing DNA damage analysis studies for such field-collected cells, all samples should be assayed at the same time.
Figure 4. Measurement of DNA damage levels after exposure to DNA damaging agents when cells are stored on ice.
Levels of DNA damage measured after 4 and 10 hours on ice. The capacity to repair induced DNA damage (exposure period of 30 minutes) was measured following incubation for 30 minutes at RT. (A) Levels of DNA damage in cells kept on ice for 4 hours (from each turtle type) were evaluated by the CometChip Platform. The levels of endogenous DNA damage, DNA damage following a 30-minute exposure to H2O2 (100 μM) and the levels of DNA damage following an additonal 30-minute repair period are shown. (B) Levels of DNA damage in cells kept on ice for 10 hours, as in panel A. (C) Levels of DNA damage in cells kept on ice for 4 hours (from each turtle type) were evaluated by the CometChip Platform. The levels of endogenous DNA damage, DNA damage following a 30-minute exposure to MMS (100 μM) and the levels of DNA damage following an additional 30-minute repair period are shown. (D) Levels of DNA damage in cells kept on ice for 10 hours, as in panel C. (****= p<0.001, n= 488–1272). Error bars represent mean ± SEM. All experiments carried out in unfrozen samples.
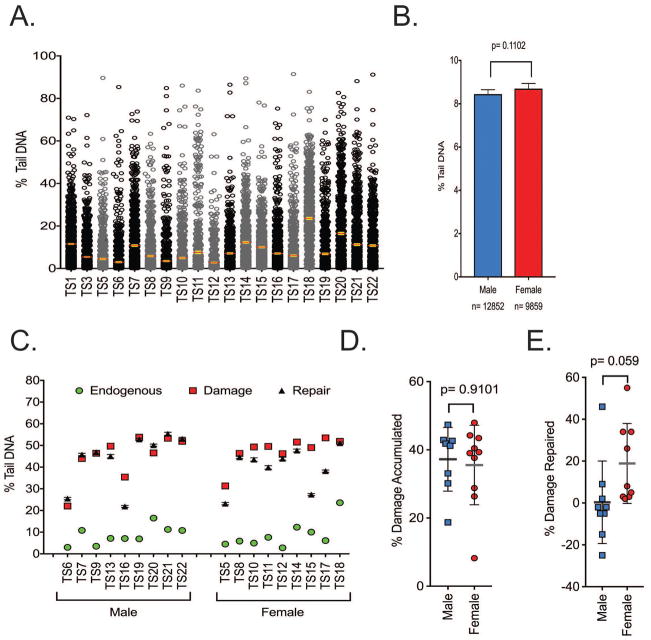
DNA damage analysis in cells from T. scripta
DNA damage parameters were then measured in a larger cohort of 20 T. scripta animals (including the previously used animals TS1 and TS3) (Figure 5A) using un-frozen samples. The mean level of endogenous DNA damage was 8.6% tail DNA with a standard error of ± 0.32%. The data were separated according to the sex of the animal (Figure 5B). The female animals (n=11) showed a trend of more endogenous DNA damage. The samples (TS5-TS22) were then exposed to H2O2 (100 μM) and the initial DNA damage (no repair time) as well as the levels of DNA damage after 30 minutes of repair was measured (Figure 5C). All samples tested had a significant increase in DNA damage after H2O2 exposure. However, repair of the DNA damage was minimal with the exception of three samples (TS15-TS17). We compared any sex-linked changes in either initial DNA damage accumulation or changes in the amount of DNA damage following repair. Levels of DNA damage and repair following treatment with H2O2 (100 μM) were normalized by subtracting the level of endogenous damage (Figure 5D, E). Both sexes accumulated approximately 38% DNA damage after H2O2 exposure. However, samples from the males repaired less of the damage than that from the females after 30 minutes of repair at room temperature (RT).
Figure 5. DNA damage analysis in cells from T. scripta.
(A) Spread of endogenous DNA damage in the cohort. All data points are shown. Error bars are mean ± 95% confidence interval. Female animals are represented in grey, males represented in black. (B) Female animals trended towards having a higher endogenous level of DNA damage, n= 9859–12852 comets analyzed. (C) Samples were exposed to H2O2 and DNA damage measured immediately and after 30 minutes of repair. (D) The amount of DNA damage accumulated was normalized by subtracting the level of endogenous damage. There were no sex-linked differences in damage accumulation. (E) The % of DNA damage that was repaired was also compared. The amount of repaired substrate was also normalized by subtracting the level of endogenous damage. Both sexes repaired the induced DNA damage poorly. (Male n=9, Female n=9). All experiments were technical duplicates using un-frozen samples. Error bars represent mean ± SD.
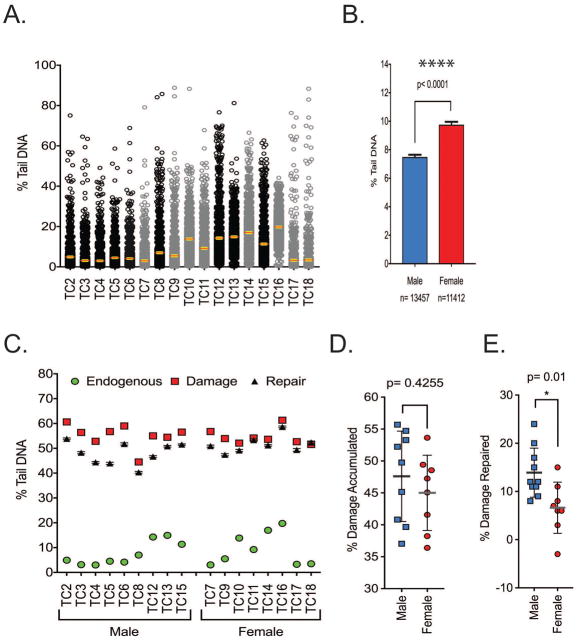
DNA damage analysis in cells from T. carolina
DNA damage levels were also measured in a larger cohort of 17 T. carolina animals (Figure 6A). The mean level of endogenous DNA damage was 8.36%, with a standard error of ±0.21% tail DNA. When comparing sex related differences in the level of endogenous DNA damage, cells isolated from females had statistically higher levels of DNA damage (2.26% increase) (Figure 6B). Samples were exposed to the same H2O2 treatment as the T. scripta cohort and the cells were allowed to repair for 30 minutes (Figure 6C). DNA damage levels and DNA repair capacity did not vary widely within the group. There was also limited difference in the amount of DNA damage accumulated when comparing cells from males and females (Figure 6D). Despite there being only modest difference between the amount of DNA damage accumulated, females repaired approximately half as much of the accumulated DNA damage as compared to the males (Figure 6E).
Figure 6. DNA damage analysis in cells from T. carolina.
(A) Spread of endogenous DNA damage in the cohort, data shows all points. Females are in grey, males in black. Error bars are mean ± 95% confidence interval. (B) Female animals have a statistically higher endogenous level of DNA damage, n=13457–11412 comets analyzed. (C) Samples were exposed to H2O2 and DNA damage measured immediately and after 30 minutes of repair. (D) The amount of DNA damage accumulated was normalized by subtracting the level of endogenous DNA damage. There were no sex-linked differences in damage accumulation but (E) the % of DNA damage that was repaired was statistically lower in the female group. Normalization: the % repair data has had the endogenous DNA damage levels subtracted from the values. (male n=9, Female n=8), ** = p< 0.01. All experiments carried out in unfrozen samples in technical duplicate. Error bars represent mean ± SD.
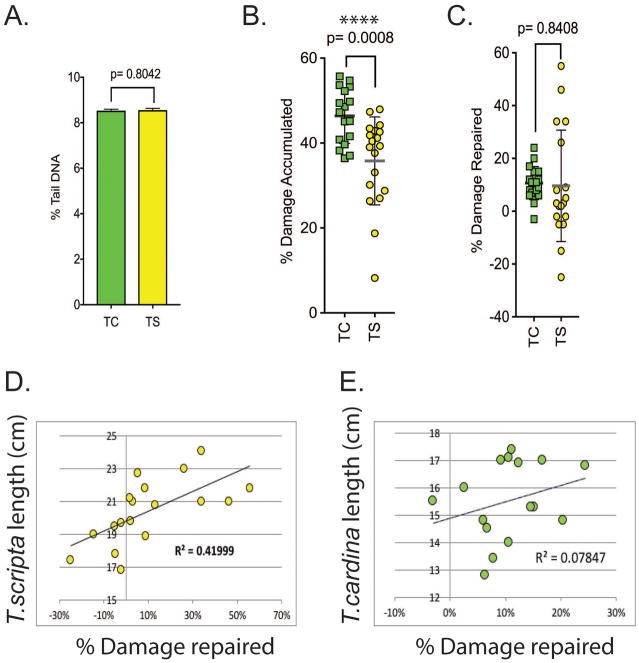
Comparative analysis of T. carolina (TC) and T. scripta (TS)
The data from the two species were combined and the level of endogenous DNA damage was compared (Figure 7A). With % tail DNA values of 8.53 ± 0.06 SEM for the TC and 8.55 ± 0.08 SEM for the TS group, the levels of endogenous DNA damage between the two groups was almost identical. A comparison of the amount of accumulated DNA damage after H2O2 exposure clearly showed that the TC cohort accumulated more DNA damage (Figure 7B), although the amount of DNA damage repaired was comparable between the two species (Figure 7C). Further, when the % damage repaired was compared to the length of the animal, which can be used as a proxy for the age of the animal, it was found that TC had a limited correlation between length and percentage of repair (Figure 7E), whereas this correlation was comparably strong in the TS cohort (Figure 7D).
Figure 7. Comparative analysis of T. carolina and T. scripta.
(A) The endogenous DNA damage levels of the two species T. carolina (TC) and T. scripta (TS) were compared. Data were compared at the single comet level. Individual comets from duplicate assays were evaluated (TC, n= 24869; TS, n=22711). The comparison of the level of accumulated DNA damage shows that (B) the cells from T. scripta animals accumulated less DNA damage than cells from T. carolina animals after H2O2 exposure and (C) the cells from T. scripta animals repaired a greater percentage of the H2O2 damaged DNA than cells from T. carolina animals. (TS n=19, TC n= 17), **** p< 0.001, Error bars represent mean ± SD. (D) The cells from the T. scripta had a correlation (r= 0.419) between the length of the animal and the capacity to repair DNA damage. (E) This correlation was less evident in the cells from the T. carolina cohort (r= 0.078).
DISCUSSION
The ability to measure DNA damage in a large number of samples, quickly and reproducibly, is of undeniable importance in biology. We have shown that it is possible to take field samples and conduct very precise measurements of DNA damage. In this presented research, we measured DNA damage from blood cells in three different species of turtle and predict that any nucleated erythrocyte could be successfully used in the CometChip Platform (Sykora et al. 2018), as long as the size of the cell is considered. We cannot be certain that all the blood cells assessed in the CometChip are erythrocytes. However, from visualization of the blood we can estimate that cells other than erythrocytes make up less than 1% of the total cell count. Hence, the vast majority of these results are from erythrocytes and the influence of any other cell type on the result would be minimal.
The CometChip Platform has the advantage of being able to assess up to 90 samples (considering controls) in each assay, substantially reducing worktime but more pertinently allowing the research to incorporate “on chip” positive and negative controls. These controls can then be used to normalize run conditions and sample variability, as we have described (Sykora et al. 2018). Traditional comet inter\intra assay variability has been a major problem in human studies. Results from experiments measuring the inter laboratory variability of the classic comet assay have proven so large that any actual population differences would be potentially masked (European Standards Committee on Oxidative DNA Damage 2003; Gedik et al. 2005). However, introduction of normalization parameters has improved variability from lab to lab (Collins et al. 2014). An objective of this study was also to create a standardized workflow that allowed a researcher to compare samples taken over extended periods of time (weeks, months, years). This is undoubtedly a strength of the CometChip Platform over current systems. It was successfully shown that samples could be stored long-term with limited preparation and then assessed for DNA damage. Together with the inbuilt controls of each assay run, this will allow field researchers to potentially take samples across an organism’s life span or allow for the measurements of seasonal variations of organismal DNA damage.
In this assessment, we also conducted limited measurements of DNA repair capacity and observed that the amount of DNA damage repaired was less than expected, based on our previous results when evaluating repair in human cells. The low level of DNA damage in the erythrocytes could be due to multiple overlapping factors (Sykora et al. 2018). The erythrocytes are terminally differentiated cells and as such have attenuated DNA repair capabilities compared to actively dividing cells (Sykora et al. 2013). Additional experiments may also be required to determine a more optimal medium for the erythrocytes to incubate while they are repairing. Whereas we have shown here that the CometChip Platform can be used to measure changes in DNA damage levels of the cells from nucleated erythrocytes after a short repair period, we encourage investigators to more completely determine assay conditions for the species of interest before attempting full repair kinetics. Three different DNA damaging agents (H2O2, MMS and etoposide) were assessed for activity in the erythrocytes. Etoposide did not induce high levels of DNA damage in the erythrocytes. Most likely this is due to the non-proliferating nature of the cells. The mechanism of action for etoposide requires actively replicating cells since etoposide forms difficult to resolve DNA/Topoisomerase II-α complexes during replication (Nitiss 2009). The lack of response to etoposide has also been observed in mononuclear cells from human blood (Sykora et al. 2018). Alternatively, it is possible that etoposide does not damage turtle erythrocytes due to differences in the enzyme despite the Topo II family of enzymes having a high level of evolutionary conservation (Forterre et al. 2007).
While genotoxic effects of contaminants may be tissue-specific and therefore some tissues may show more DNA damage than others (Jha 2008; Blanpain et al. 2011; Jackson et al. 2013), obtaining more invasive tissue samples may not be feasible for wild species, especially if endangered. Furthermore, blood samples allow potential monitoring of the same individual at different times (da Silva et al. 2000), a feature that would not be possible if the individual has been sacrificed to obtain other tissues. Finally, while it has been shown that all tissue types can be used for potential biomonitoring (Belpaeme et al. 1998; Frenzilli et al. 2009), tissues other than blood show increased DNA breakage when frozen in liquid nitrogen, a potential requirement of sampling under field conditions (Belpaeme et al. 1998).
We found intra-specific variation in DNA damage levels and DNA repair capacity in both species (T. carolina and T. scripta). Intra-specific variation in DNA damage is commonly used in biomonitoring studies to compare samples from sites experiencing different levels of contamination (Bonisoli-Alquati et al. 2010; Zapata et al. 2016) or different stress levels (Robert and Bronikowski 2010). In this study, individuals were all sampled at the same site and during the same season and therefore the detected differences in levels of DNA damage among individuals are most likely not due to contamination, stress, or seasonal variation among samples; see (Jha 2008) for potential sources of variation. Intra-individual variation in DNA damage has been reported to be affected by age and reproductive status, although comparative data across organisms on the relationship between DNA damage, DNA repair, age and sex differences are lacking (Jha 2008). While published studies have demonstrated increased effects in human and mosquito fish (Gambusia affinis) females vs males in the micronucleus test (Caliani et al. 2009; Fenech and Bonassi 2011), we did not find similar evidence in the literature for the comet assay; this is possibly because the micronucleus test detects a wider array of genotoxic effects (Bhagat 2018). In T. carolina, distinct levels of DNA damage between males and females were observed, with the females showing more damage, probably as a result of lower capacity of DNA repair than males. Furthermore, in T. scripta, we observed a positive relationship between size, used as a proxy for age of the animal, and level of DNA repair. It has been suggested that older individuals may repair more DNA damage (Heuser et al. 2008), but they may also accumulate more damage (Fenech and Bonassi 2011).
Finally, both species showed similar levels of endogenous DNA damage, although their sensitivity to treatment with a damaging agent (H2O2) is different, with T. carolina being more affected than T. scripta. Both species have similar longevity (around 40 years, although records of an individual of T. carolina living up to 138 years have been reported) indicating that similar levels of endogenous DNA damage may reflect comparable life histories. Distinct sensitivity to damaging agents may be an intrinsic characteristic of the species or due to adaptive mechanisms, which cannot be tested with the current data set. However, T. scripta is widespread and successful as an invasive species (Rodder et al. 2009), able to survive and reproduce in contaminated sites (Ferronato et al. 2009). The lower sensitivity to DNA damage may be a species-specific trait conferring more resilience to this species.
The field of environmental toxicology is in need of a new generation of methods that can screen for genotoxic effects across the many chemicals and species co-occurring in the environment. Here, we show that a high-throughput version of the traditional comet assay can be used to measure levels of DNA damage from the blood of many individuals in a single comet run. Moreover, our results report effective methods to preserve samples for short- and long-term storage so that samples can be collected at different time points with minimal artificial damage occurring during the storage process. Although this study was performed in turtles, the approach can be applied to all non-mammalian vertebrates with nucleated erythrocytes. We found evidence for differences in induced DNA damage between the turtle species tested, as well as differences in DNA damage among sexes in T. carolina. These results indicate that the high-throughput CometChip Platform can be used to understand how species and individuals respond differently to genotoxicant exposure.
Supplementary Material
Six months after being frozen samples were measured for endogenous DNA damage. Results generally correlated well with the results measured when the samples were fresh. Samples were frozen in 20% DMSO+PBS as per Figure 3.
Acknowledgments
The authors declare no competing financial interests. RWS is on the scientific advisory board for Bio-Techne/Trevigen.
GRANT SPONSORS
This work was supported by grants from the National Institutes of Health (CA148629 and ES021116) and from the Abraham A. Mitchell Distinguished Investigator fund awarded to RWS. RWS is an Abraham A. Mitchell Distinguished Investigator.
Footnotes
STATEMENT OF AUTHOR CONTRIBUTIONS
PS, SG, AH and NM performed all experiments. PS, SG, YC and RWS participated in the experimental design. PS, YC, SG and RWS wrote the paper.
References
- Azqueta A, Collins AR. The essential comet assay: a comprehensive guide to measuring DNA damage and repair. Arch Toxicol. 2013;87(6):949–968. doi: 10.1007/s00204-013-1070-0. [DOI] [PubMed] [Google Scholar]
- Belpaeme K, Cooreman K, Kirsch-Volders M. Development and validation of the in vivo alkaline comet assay for detecting genomic damage in marine flatfish. Mutat Res. 1998;415(3):167–184. doi: 10.1016/s1383-5718(98)00062-x. [DOI] [PubMed] [Google Scholar]
- Bhagat J. Combinations of genotoxic tests for the evaluation of group 1 IARC carcinogens. J Appl Toxicol. 2018;38(1):81–99. doi: 10.1002/jat.3496. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Mohrin M, Sotiropoulou PA, Passegue E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8(1):16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Bonisoli-Alquati A, Voris A, Mousseau TA, Moller AP, Saino N, Wyatt MD. DNA damage in barn swallows (Hirundo rustica) from the Chernobyl region detected by use of the comet assay. Comp Biochem Physiol C Toxicol Pharmacol. 2010;151(3):271–277. doi: 10.1016/j.cbpc.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Bruhl CA, Schmidt T, Pieper S, Alscher A. Terrestrial pesticide exposure of amphibians: an underestimated cause of global decline? Sci Rep. 2013;3:1135. doi: 10.1038/srep01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliani I, Porcelloni S, Mori G, Frenzilli G, Ferraro M, Marsili L, Casini S, Fossi MC. Genotoxic effects of produced waters in mosquito fish (Gambusia affinis) Ecotoxicology. 2009;18(1):75–80. doi: 10.1007/s10646-008-0259-0. [DOI] [PubMed] [Google Scholar]
- Collins AR, El Yamani N, Lorenzo Y, Shaposhnikov S, Brunborg G, Azqueta A. Controlling variation in the comet assay. Front Genet. 2014;5:359. doi: 10.3389/fgene.2014.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva J, de Freitas TR, Heuser V, Marinho JR, Erdtmann B. Genotoxicity biomonitoring in coal regions using wild rodent Ctenomys torquatus by Comet assay and micronucleus test. Environ Mol Mutagen. 2000;35(4):270–278. [PubMed] [Google Scholar]
- Dahm-Daphi J, Sass C, Alberti W. Comparison of biological effects of DNA damage induced by ionizing radiation and hydrogen peroxide in CHO cells. Int J Radiat Biol. 2000;76(1):67–75. doi: 10.1080/095530000139023. [DOI] [PubMed] [Google Scholar]
- Devaux A, Fiat L, Gillet C, Bony S. Reproduction impairment following paternal genotoxin exposure in brown trout (Salmo trutta) and Arctic charr (Salvelinus alpinus) Aquat Toxicol. 2011;101(2):405–411. doi: 10.1016/j.aquatox.2010.11.017. [DOI] [PubMed] [Google Scholar]
- European Standards Committee on Oxidative DNA Damage. Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic Biol Med. 2003;34(8):1089–1099. doi: 10.1016/s0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- Fenech M, Bonassi S. The effect of age, gender, diet and lifestyle on DNA damage measured using micronucleus frequency in human peripheral blood lymphocytes. Mutagenesis. 2011;26(1):43–49. doi: 10.1093/mutage/geq050. [DOI] [PubMed] [Google Scholar]
- Ferronato BO, Merchant ME, Marques TS, Verdade LM. Characterization of innate immune activity in Phrynops geoffroanus (Testudines: Chelidae) Zoologia (Curitiba) 2009;26:747–752. [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89(4):427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Frenzilli G, Nigro M, Lyons BP. The Comet assay for the evaluation of genotoxic impact in aquatic environments. Mutat Res. 2009;681(1):80–92. doi: 10.1016/j.mrrev.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Gedik CM, Collins A, Escodd Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19(1):82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- Goulson D, Nicholls E, Botias C, Rotheray EL. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science. 2015;347(6229):1255957. doi: 10.1126/science.1255957. [DOI] [PubMed] [Google Scholar]
- Guilherme S, Gaivao I, Santos MA, Pacheco M. DNA damage in fish (Anguilla anguilla) exposed to a glyphosate-based herbicide -- elucidation of organ-specificity and the role of oxidative stress. Mutat Res. 2012;743(1–2):1–9. doi: 10.1016/j.mrgentox.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Heuser VD, de Andrade VM, Peres A, Gomes de Macedo Braga LM, Bogo Chies JA. Influence of age and sex on the spontaneous DNA damage detected by micronucleus test and comet assay in mice peripheral blood cells. Cell Biol Int. 2008;32(10):1223–1229. doi: 10.1016/j.cellbi.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Jackson P, Pedersen LM, Kyjovska ZO, Jacobsen NR, Saber AT, Hougaard KS, Vogel U, Wallin H. Validation of freezing tissues and cells for analysis of DNA strand break levels by comet assay. Mutagenesis. 2013;28(6):699–707. doi: 10.1093/mutage/get049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AN. Ecotoxicological applications and significance of the comet assay. Mutagenesis. 2008;23(3):207–221. doi: 10.1093/mutage/gen014. [DOI] [PubMed] [Google Scholar]
- Karbaschi M, Cooke MS. Novel method for the high-throughput processing of slides for the comet assay. Sci Rep. 2014;4:7200. doi: 10.1038/srep07200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaze E, Devaux A, Mons R, Bony S, Garric J, Geffard A, Geffard O. DNA damage in caged Gammarus fossarum amphipods: a tool for freshwater genotoxicity assessment. Environ Pollut. 2011;159(6):1682–1691. doi: 10.1016/j.envpol.2011.02.038. [DOI] [PubMed] [Google Scholar]
- Moreno NC, Sofia SH, Martinez CB. Genotoxic effects of the herbicide Roundup Transorb and its active ingredient glyphosate on the fish Prochilodus lineatus. Environ Toxicol Pharmacol. 2014;37(1):448–454. doi: 10.1016/j.etap.2013.12.012. [DOI] [PubMed] [Google Scholar]
- Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9(5):338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha A, Yaduvanshi SK, Pant SC, Lomash V, Srivastava N. Evaluation of DNA damage and cytotoxicity induced by three commonly used organophosphate pesticides individually and in mixture, in rat tissues. Environ Toxicol. 2013;28(10):543–552. doi: 10.1002/tox.20748. [DOI] [PubMed] [Google Scholar]
- Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1(1):23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- Parrella A, Lavorgna M, Criscuolo E, Russo C, Isidori M. Eco-genotoxicity of six anticancer drugs using comet assay in daphnids. J Hazard Mater. 2015;286:573–580. doi: 10.1016/j.jhazmat.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Poletta GL, Kleinsorge E, Paonessa A, Mudry MD, Larriera A, Siroski PA. Genetic, enzymatic and developmental alterations observed in Caiman latirostris exposed in ovo to pesticide formulations and mixtures in an experiment simulating environmental exposure. Ecotoxicol Environ Saf. 2011;74(4):852–859. doi: 10.1016/j.ecoenv.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Readel AM, Dreslik MJ, Warner JK, Banning WJ, Phillips CA. A Quantitative Method for Sex Identification in Emydid Turtles Using Secondary Sexual Characters. Copeia. 2008;2008(3):643–647. [Google Scholar]
- Robert KA, Bronikowski AM. Evolution of senescence in nature: physiological evolution in populations of garter snake with divergent life histories. Am Nat. 2010;175(2):147–159. doi: 10.1086/649595. [DOI] [PubMed] [Google Scholar]
- Rodder D, Schmidtlein S, Veith M, Lotters S. Alien invasive slider turtle in unpredicted habitat: a matter of niche shift or of predictors studied? PLoS One. 2009;4(11):e7843. doi: 10.1371/journal.pone.0007843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Bhagat J, Saha Sarker M, Gaitonde DCS, Sarker S. Evaluation of the impact of bioaccumulation of PAH from the marine environment on DNA integrity and oxidative stress in marine rock oyster (Saccostrea cucullata) along the Arabian sea coast. Ecotoxicology. 2017;26(8):1105–1116. doi: 10.1007/s10646-017-1837-9. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach RP, Escher BI, Fenner K, Hofstetter TB, Johnson CA, von Gunten U, Wehrli B. The challenge of micropollutants in aquatic systems. Science. 2006;313(5790):1072–1077. doi: 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- St Clair RC. Patterns of growth and sexual size dimorphism in two species of box turtles with environmental sex determination. Oecologia. 1998;115(4):501–507. doi: 10.1007/s004420050547. [DOI] [PubMed] [Google Scholar]
- Sykora P, Witt KL, Revanna P, Smith-Roe SL, Dismukes J, Lloyd DG, Engelward BP, Sobol RW. Next generation high throughput DNA damage detection platform for genotoxic compound screening. Sci Rep. 2018;8(1):2771. doi: 10.1038/s41598-018-20995-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P, Yang JL, Ferrarelli LK, Tian J, Tadokoro T, Kulkarni A, Weissman L, Keijzers G, Wilson DM, 3rd, Mattson MP, Bohr VA. Modulation of DNA base excision repair during neuronal differentiation. Neurobiol Aging. 2013;34(7):1717–1727. doi: 10.1016/j.neurobiolaging.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A. Evolutionary genomics of environmental pollution. Adv Exp Med Biol. 2014;781:321–337. doi: 10.1007/978-94-007-7347-9_16. [DOI] [PubMed] [Google Scholar]
- Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19(12):1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata LM, Bock BC, Orozco LY, Palacio JA. Application of the micronucleus test and comet assay in Trachemys callirostris erythrocytes as a model for in situ genotoxic monitoring. Ecotoxicol Environ Saf. 2016;127:108–116. doi: 10.1016/j.ecoenv.2016.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Six months after being frozen samples were measured for endogenous DNA damage. Results generally correlated well with the results measured when the samples were fresh. Samples were frozen in 20% DMSO+PBS as per Figure 3.