Summary
Insights into the evolution of plant cell walls have important implications for comprehending these diverse and abundant biological structures. In order to understand the evolving structure–function relationships of the plant cell wall, it is imperative to trace the origin of its different components.
The present study is focused on plant 1,4‐β‐xylan, tracing its evolutionary origin by genome and transcriptome mining followed by phylogenetic analysis, utilizing a large selection of plants and algae. It substantiates the findings by heterologous expression and biochemical characterization of a charophyte alga xylan synthase.
Of the 12 known gene classes involved in 1,4‐β‐xylan formation, XYS1/IRX10 in plants, IRX7, IRX8, IRX9, IRX14 and GUX occurred for the first time in charophyte algae. An XYS1/IRX10 ortholog from Klebsormidium flaccidum, designated K. flaccidum XYLAN SYNTHASE‐1 (Kf XYS1), possesses 1,4‐β‐xylan synthase activity, and 1,4‐β‐xylan occurs in the K. flaccidum cell wall.
These data suggest that plant 1,4‐β‐xylan originated in charophytes and shed light on the origin of one of the key cell wall innovations to occur in charophyte algae, facilitating terrestrialization and emergence of polysaccharide‐based plant cell walls.
Keywords: biosynthesis, cell wall, evolution, IRX10, Klebsormidium flaccidum, Klebsormidium nitens, xylan, XYS1
Introduction
A large diversity of cell wall structural variations exist within the plant kingdom (Domozych et al., 2007; Harholt et al., 2012). One important avenue for understanding this diversity is by studying wall evolution and the rise of new architectural solutions and principles. Intriguingly, all plant cell walls are largely based on the same limited number of polysaccharide classes. At the same time, it is clear that some cell wall functions can be fulfilled by different architectural solutions, for example involving different groups of polysaccharides (Carpita & Gibeaut, 1993; Harholt et al., 2012). This suggests that simple carbohydrate composition gives a limited perspective of wall structure, whereas the combinatorial aspects of polymer–polymer interactions are the key players that define architectural complexity. How and why these different interactions evolved, including their specific relationships to wall architecture and function, remain enigmatic. However, before we can answer these questions it is necessary to establish the key routes of cell wall evolution by determining the evolutionary origins of the different cell wall polysaccharide classes to fill in current gaps in our knowledge.
A direct ancestor of land plants (embryophytes) has not been identified, but the Zygnematophyceae (Fig. 1) have been proposed as the closest extant relatives (Wickett et al., 2014). The cell walls of charophytes (streptophyte green algae) vary between taxonomic groups in a similar manner to the taxonomic group variation of land plants (Fangel et al., 2012). Polysaccharide‐based cell walls typical of land plants can be detected in Klebsormidiophyceae (Fig. 1), marking the transition from a wall centered on mineralized organic scales in the basal charophyte Mesostigma viride to one based on polysaccharides (Becker et al., 1991; Domozych et al., 1991; Sørensen et al., 2011). Cell wall complexity is found to increase in the more recently diverged classes, such as Coleochaetophyceae and Zygnematophyceae, seemingly presenting an all‐polysaccharide wall with the full complement of wall glycopolymers typical of plants, that is, cellulose, xylan, xyloglucan, mannan, mixed‐linkage glucan, and pectin. Hence, it has been proposed that what is often conceptualized as a ‘plant cell wall’, based on material composition and organization, emerged in a distant algal ancestor, most likely a common ancestor of Klebsormidiophyceae and plants (Sørensen et al., 2011; Mikkelsen et al., 2014).
Figure 1.
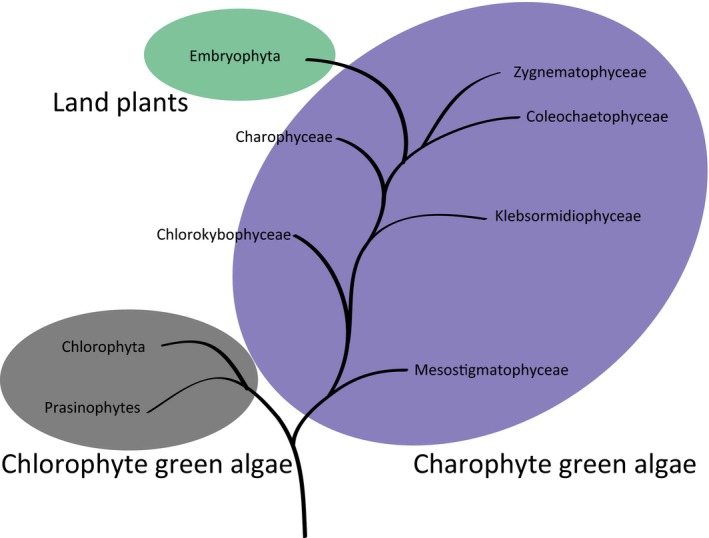
Phylogenetic relationship within Viridiplantae as suggested by Wickett et al. (2014). This clade is defined by chloroplast descendence, the name translating as ‘green plants’. It designates green algae and land plants. The algal species Klebsormidium flaccidum is a member of Klebsormidiophycea, located centrally in the grade of charophyte algae, while Zygnematophyceae is located as the most recently evolved group in the charophyte algae grade.
Cellulose biosynthesis in plants is orthologous to that in prokaryotic organisms, but the origin of the biosynthetic activities for the remaining cell wall polysaccharides in higher plants is less clear. Xyloglucan and pectin putatively originated within charophyte algae (streptophyte green algae), while xylan, mannan, and mixed‐linkage glucan also exist in some noncharophyte algae and bacteria (Painter, 1983; Fangel et al., 2012; Salmeán et al., 2017). Hence, direct proof of functional orthology between algal and plant cell wall biosynthetic activities has been missing. This increases uncertainty as a result of the plausible convergent evolution of the different polymers, exemplified by the multiple occurrences of mixed‐linkage glucans throughout the tree of life. Identification of putative cell wall biosynthetic genes based on a de novo transcriptome assembly in charophytes has previously been published (Mikkelsen et al., 2014); however, this study utilized a limited transcriptome collection of insufficient quality, and carried out no biochemical confirmation of enzyme function, resulting in ambiguous conclusions. The completion of the Klebsormidium flaccidum genome sequence (Hori et al., 2014), in combination with the 1000 Plants (1KP ) Initiative (Matasci et al., 2014), now provides the opportunity to identify full‐length K. flaccidum gene sequences, resolve their phylogeny by comparison with algae and plants across the kingdom, and characterize the biochemical activities of gene products.
Xylan is an abundant and complex cell wall component in plants, particularly in commelinid primary walls and in secondary cell walls of all angiosperms (Scheller & Ulvskov, 2010). It consists of a polymeric backbone of 1,4‐β‐linked d‐xylose (Xyl) decorated mainly with acetyl groups, and is further substituted by l‐arabinofuranose (Araf ) in commelinids or methylated or unmethylated d‐glucuronic acid (GlcA) in noncommelinid angiosperms (Scheller & Ulvskov, 2010; Smith et al., 2017; Fig. 2). The elucidation of the fine structure of xylans found outside angiosperms is less complete and based only on a few species: gymnosperms produce methyl‐glucurono‐arabinoxylan (Busse‐Wicher et al., 2016), lycophytes and pteridophytes contain both methylated and unmethylated GlcA substitutions (Kulkarni et al., 2012), and the bryophyte Physcomitrella patens only contains unmethylated GlcA substituents (Kulkarni et al., 2012).
Figure 2.
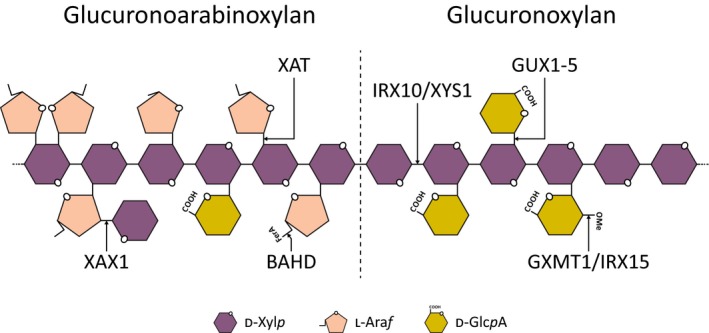
Schematic illustration of xylan structure with known biosynthetic activities represented.
Proteins from eight protein families have been implicated in xylan biosynthesis, and based on their functions, 12 protein classes can be defined (Table 1; Fig. 2). For some of these classes, biochemical function has been established in vitro using recombinant proteins, including IRX10 and IRX10‐L, which are 1,4‐β‐xylan xylosyltransferases producing the 1,4‐β‐xylan backbone polymer (Jensen et al., 2014; Urbanowicz et al., 2014). In the case of IRX10‐L, long 1,4‐β‐xylan oligomers were produced in vitro and the protein was accordingly renamed XYLAN SYNTHASE‐1 (XYS1) (Urbanowicz et al., 2014). P. patens encodes one IRX10/XYS1 ortholog, PpIRX10, which produces 1,4‐β‐xylan polymers that are of a similar size to those made by XYS1 in vitro (Jensen et al., 2014). Other putative glycosyltransferases from families GT8, GT43, and GT47 also affect xylan backbone formation when knocked out in Arabidopsis; however, their specific functions have remained unclear. Enzyme activities that contribute to 1,4‐β‐xylan backbone decorations include acetyl‐, methyl‐, and glycosyltransferases, and genes encoding the main enzyme activities have all been identified and characterized in angiosperms (Table 1).
Table 1.
Phylogenetic origin of glycosyltransferases and accessory activities involved or putatively involved in xylan synthesis in plants
| Xylan protein class | Protein family | Biochemical function | Earliest ortholog | Reference | Fig. |
|---|---|---|---|---|---|
| PARVUS | GT8 | Unknown | Chlorophytaa | Brown et al. (2007) | S2 |
| GXMT1/IRX15 | DUF579 | Glucuronoxylan methyltransferase/unknown | Chlorophytab | Jensen et al. (2014); Urbanowicz et al. (2012) | S3 |
| IRX10/XYS1 | GT47 | β‐1,4‐xylan synthase | Klebsormidiophyceae | Jensen et al. (2014); Urbanowicz et al. (2014) | 3 |
| IRX7 | GT47 | Unknown | Klebsormidiophyceae | Brown et al. (2007) | 3 |
| IRX9 | GT43 | Unknown | Klebsormidiophyceae | Brown et al. (2007) | S4 |
| IRX14 | GT43 | Unknown | Klebsormidiophyceae | Brown et al. (2007) | S4 |
| IRX8 | GT8 | Unknown | Klebsormidiophyceae | Peña et al. (2007) | S5 |
| GUX1‐5 | GT8 | Xylan glucuronsyltransferase | Zygnematophyceae | Rennie et al. (2012) | S6 |
| ESK1 | DUF231 | Acetylation | Unresolved, possibly angiosperms | Urbanowicz et al. (2014) | S7 |
| XAT | GT61 | Xylan arabinosyltransferases | Unresolved, possibly eudicot/monocotc | Anders et al. (2012) | S8 |
| XAX1 | GT61 | Xylan xylosyltransferase | Unresolved, possibly Commelinid monocots | Chiniquy et al. (2012) | S8 |
| BAHD | PF02458 | Coumaric and ferulic acid transferases | Unresolved, possibly Liliales (noncommelinid monocot) | Bartley et al. (2013) | S9 |
Chlorophyta orthologs were identified, but no charophyte algae and the earliest occurrence in plants was in Bryophyta.
Chlorophyta and charophyte algae orthologs were identified ancestral to the split between IRX15(L) and GMXT1/GMXT2 clades.
XAT orthologs have not been identified in Magnoliids or basal angiosperms.
Of the different plant cell wall polysaccharides, three in particular lend themselves to having their emergence in evolution pinpointed, that is, 1,4‐β‐xylan, xyloglucan, and the pectic polysaccharide homogalacturonan. We chose to focus on xylan and its biosynthesis in K. flaccidum, as this polymer was the earliest to emerge of the three and involves a number of biosynthetic activities that have been amenable to biochemical characterization (Sørensen et al., 2011; Rennie et al., 2012; Urbanowicz et al., 2012, 2014; Jensen et al., 2014). Our analysis points to the evolutionary appearance of the various xylan synthesis‐specific genes identified to date, describing innovations of this complex biocatalytic process as they occurred in two major phases over the course of plant evolution. Further, we demonstrated xylan synthase activity for one of the members of the earliest xylan synthesis‐specific gene homologs identified to date: a K. flaccidum IRX10/XYS1 ortholog (KfXYS1). Finally, we identified and characterized the xylan in the cell walls of K. flaccidum, which is probably the product of the KfXYS1. The enzymatic activity of KfXYS1, combined with its evolutionary relatedness to modern IRX10/XYS1 xylan synthases, highlights the functional orthology between algal and plant cell wall biosynthesis.
Materials and Methods
Bioinformatics
The proteome files of the relevant divisions were obtained from the 1000 Plants (1KP) Initiative (Matasci et al., 2014). Relevant sequences for phylogenetic analysis from P. patens, Selaginella moellendorffii, Arabidopsis and rice were obtained using Harholt et al. (2012) as guide. A positive/negative list was made for identification of xylan‐related biosynthetic enzymes and the closest nonxylan related ortholog (Supporting Information Table S1). Sequences from this list were used as a database for blasting proteomes from the 1KP dataset as previously described (Mikkelsen et al., 2014), substituting the CAZy database with our positive list and eliminating false positives by substituting the Arabidopsis GT‐depleted database with our negative list. Phylogenetic analysis was performed as previously described (Ulvskov et al., 2013). Newick format tree files and sequences used in this manuscript are available in Notes S1. Evaluation of evolutionarily conserved protein motifs was performed using the SALAD website with their interactive service (Mihara et al., 2010; http://salad.dna.affrc.go.jp/salad/en/).
Heterologous protein expression in Saccharomyces cerevisiae: purification and activity assays
For heterologous protein expression in S. cerevisiae, YFP was cloned into pESC‐URA by moving it from pPICZ A‐YFP (Jensen et al., 2014) using EcoRI and NotI. KfXYS1 was obtained by de novo DNA synthesis (IDT, Coralville, IA, USA) with codon optimization for S. cerevisiae and cloned into the pESC‐URA‐YFP using EcoRI and PflMI restriction sites, generating pESC‐URA‐KfXYS1‐YFP. The sequence GTAATGGGT was engineered around the start codon for proper initiation of translation.
Saccharomyces cerevisiae YPH499 harboring pESC‐URA‐KfXYS1‐YFP was grown in SC synthetic minimal media (3.4 g Yeast Nitrogen Base l−1 media; ThermoFisher, Waltham, MA, USA), 2.8 g Yeast Synthetic Drop‐out Media (ThermoFisher), 10 g aluminum sulfate, 0.2 g leucine, 0.2 g tryptophan, 0.1 g histidine, and 0.2 g alanine plates with 2% glucose for 24 h at 30°C. Next, scrapes of these plates were used to inoculate 250 ml of liquid SC media with 2% glucose and incubated overnight in baffled flasks at 30°C in an orbital shaker at 180 rpm. At an OD600 of c. 2, the cells were collected by centrifugation (20 min at 2451 g) and resuspended in SC media with 2% galactose, and then incubated at 18°C as before. Expression levels were monitored by fluorescent microscopy and cells were harvested by centrifugation 11 h after induction of recombinant protein expression. At the time of harvest, OD600 was c. 2, corresponding to c. 0.75 g of cells. Cell pellets were stored at −80°C. Protein purification and assays involving the fluorescently labeled xylooligosaccharide acceptor were performed as previously described (Jensen et al., 2014) and were based on c. 0.75 g of cells per batch.
Heterologous protein expression and purification of KfXYS1 in HEK293 cells
KfXYS1 was cloned in a manner similar to that described in Urbanowicz et al. (2014). Briefly, to create Gateway entry clones, the truncated coding region of KfXYS1 (amino acids 27–445) was PCR‐amplified (KfXYS1_27F, 5′‐AACTTGTACTTTCAAGGCAGATCCTCTTTGTTCGT‐3′ and KfXYS1_445F, 5′‐ACAAGAAAGCTGGGTCCTAATTTTCATCATCACCACG‐3′) from pESC‐URA‐KfXYS1‐YFP plasmid DNA. A second set of universal primers (attB_Adapter‐F, 5′‐ GGGGACAAGTTTGTACAAAAAAGCAGGCTCTGAAAACTTGTACTTTCAAGGC‐3′ and attB_Adapter‐R, 5′‐GGGGACCACTTTGTACAAGAAAGCTGGGTC‐3′) was used to complete the attB recombination site and append a tobacco etch virus (TEV) protease cleavage site (Urbanowicz et al., 2014). The attB‐PCR product was cloned into the pDONR221 plasmid vector (Life Technologies, Carlsbad, CA, USA) using Gateway BP Clonase II Enzyme Mix (Life Technologies) to create an entry clone. To generate an expression clone of KfXYS1 (pGEn2‐EXP‐KfXYS1), the entry clone was recombined into a Gateway‐adapted version of the pGEn2 mammalian expression vector (pGEn2‐DEST) (Meng et al., 2013), using Gateway LR Clonase II Enzyme Mix (Life Technologies). The resulting expression construct (His‐GFP‐KfXYS1) encodes a fusion protein comprising an amino‐terminal signal sequence, an 8xHis tag, an AviTag recognition site, the ‘superfolder’ GFP (sfGFP) coding region, the recognition sequence of TEV protease, and residues 27–445 of KfXYS1.
Recombinant expression and purification were performed by transient transfection of suspension culture HEK293‐F cells with pGEn2‐EXP‐KfXYS1 and a HisTap HP 1 ml column (GE Healthcare, Little Chalfont, UK), as previously described (Meng et al., 2013; Urbanowicz et al., 2014). Protein purity was assessed by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (Fig. S1). Purified His‐GFP‐KfXYS1 was concentrated to 0.45 mg ml−1 using an Amicon Ultra Centrifugal Filter Device (30000 MWCO; Merck Millipore; http://www.merckmillipore.com) and dialyzed (3500 MWCO) into HEPES sodium salt‐HCl (75 mM, pH 6.8) or sodium phosphate buffer (75 mM, pH 6.8) and used directly for reactions, or stored at 4 or −80°C in aliquots.
MALDI‐TOF‐MS analysis of KfXYS1 reaction products
Enzyme reactions (20 μl) consisted of 3 mM UDP‐xylose (UDP‐Xyl; Carbosource, Athens, GA, USA), 0.5 mM xylopentaose (Megazyme, Bray, Ireland), labeled at the reducing terminus with 2‐aminobenzamide as previously described (Ishii et al., 2002; Urbanowicz et al., 2014), and 4.5 μg of purified His‐GFP‐KfXYS1 in HEPES sodium salt‐HCl buffer (75 mM, pH 6.8). Reactions were allowed to persist for 4 h before being prepared for analysis by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry analysis on an LT Bruker LT Microflex spectrometer (Bruker, Billerica, MA, USA) as described previously (Urbanowicz et al., 2014). Positive‐ion spectra were recorded with a minimum of 200 laser shots summed to generate each spectrum.
Nuclear magnetic resonance (NMR) analysis of KfXYS1 reaction products
Products of a scaled‐up reaction were structurally characterized by NMR analysis. Reactions (300 μl) were carried out at 25°C in sodium phosphate buffer (75 mM, pH 6.8), containing 5 mM UDP‐Xyl, 1.5 mM xylobiose (Sigma) and 10 μg of purified His‐GFP‐KfXYS1. After 16 h, the reaction mixture was lyophilized, resuspended in 300 μl D2O (99.9%; Cambridge Isotope Laboratories, http://www.isotope.com) and characterized using a Varian 300 MHz NMR spectrometer (Varian, Palo Alto, CA, USA) at 25°C for initial characterization of the full reaction mixture composition. For characterization of higher‐order polysaccharide products only, the reaction mixture was fractionated on a Superdex 75 HR10/30 column and eluted with water to separate oligosaccharides and other reaction components. The fractions were lyophilized and resuspended in 200 μl D2O (99.9%; Cambridge Isotope Laboratories, Tewksbury, MA, USA). The 1D 1H NMR spectra were recorded at 25°C on a Varian Inova NMR spectrometer operating at 600 MHz and equipped with a 5 mm cold probe (Agilent, Santa Clara, CA, USA). Chemical shifts were measured relative to internal DMSO (δ1H 2.721) on both NMR instruments. Data were processed using MestReNova (Mestrelab Research, Santiago Compostela, Spain). Assignments for the UDP‐Xyl, UDP, xylobiose, and xylo‐oligosaccharides synthesized were made based on previously published results (Harper and Bar‐Peled, 2002; Peña et al., 2007; Wishart et al., 2009).
Monosaccharide composition analysis
Cell wall material was prepared by grinding aliquots of 50 mg K. flaccidum cells in a TissueLyser MM 200 (Qiagen, Hilden, Germany) for 2 min at 30 s−1. Each sample was then extracted using 1.5 ml 70% ethanol for 5 d at 55°C, followed by four cycles of 1.5 ml 70% ethanol, one cycle of 1.5 ml acetone, and then air‐dried overnight. Monosaccharide composition was performed on cell wall material as previously described (Øbro et al., 2004) using a Dionex ICS 5000 + DC system (ThermoFisher) equipped with a high‐performance anion exchange chromatograph with pulsed amperometric detection and a 4 μm SA‐10 column (2 × 250 mm and guard column). Run conditions were 40°C column temperature, 0.3 ml min−1 eluent flow rate, 1 mM NaOH for 0–8 min, followed by 100 mM NaOH from 8 to 20 min, and subsequently 10 min equilibration at 1 mM NaOH.
Polysaccharide analysis using carbohydrate gel electrophoresis
Polysaccharide analysis using carbohydrate gel electrophoresis (PACE) was performed as previously described (Goubet et al., 2002) using GH10 xylanase (Brown et al., 2007), GH11 xylanase (Brown et al., 2011), GH115 α‐glucuronosidase (Rogowski et al., 2014), β‐xylosidase (Uniprot: Q92458) and arabinofuranosidase (NS39128). The last two enzymes were kindly provided by Novozymes (Novozymes, Bagsværd, Denmark).
Comprehensive microarray polymer profiling
Samples of 10 mg K. flaccidum cell wall material were extracted with 300 μl of 50 mM diaminocyclohexanetetraacetic acid (CDTA; pH 7.5), and subsequently with 300 μl of 4 M NaOH with 0.1% w/w NaBH4. Extracts were spotted, probed and analyzed as previously described (Pedersen et al., 2012). Antibodies BS‐400‐2 (Meikle et al., 1991; Biosupplies, Bundoora, Australia), and LM10 (McCartney et al., 2005), LM11 (McCartney et al., 2005), LM12 (Pedersen et al., 2012), LM23 (Pedersen et al., 2012), LM27 (Cornuault et al., 2015) and LM28 (Cornuault et al., 2015; PlantProbes, Leeds, UK) were used for the analysis.
Immunohistochemistry
Klebsormidium flaccidum cells were embedded in medium‐grade LR White (Polysciences, Hirschberg an der Bergstrasse, Germany) as previously described (Bell et al., 2013) and sectioned on a Reichert‐Jung/LKB Supernova ultramicrotome (2 μm; Reichert, Depew, NY, USA). Sections were treated with 5% skimmed milk in 100 mM PBS solution for 30 min and labeled using LM11 antibody (PlantProbes). A goat anti‐rat antibody conjugated to Alexa Fluor 555 was used to visualize the binding of LM11. Calcofluor White was applied to stain cell wall β‐glucans. Images were recorded on a Leica CLSM SP5 microscope (Leica, Heidelberg, Germany) using 405 nm excitation and 419–534 nm emission (Calcofluor White), and 543 nm excitation and 582–700 nm emission (Alexa Fluor 555). Negative controls using only the secondary antibody showed no nonspecific binding.
Results
Evolutionary appearance of genes involved in xylan biosynthesis
Utilizing the phylogenetic span of the 1KP dataset (Matasci et al., 2014) and genomic sequences from plants and K. flaccidum (Hori et al., 2014), we obtained phylogenetic trees with ample evolutionary width and resolution. These enabled us to study clade structures of protein families implicated in xylan biosynthesis, and to pinpoint the earliest orthologs for several of them (Table 1; Figs 3, S2–S9). These analyses suggest that xylan evolution occurred in two major phases, one in algae and a second in higher plants.
Figure 3.
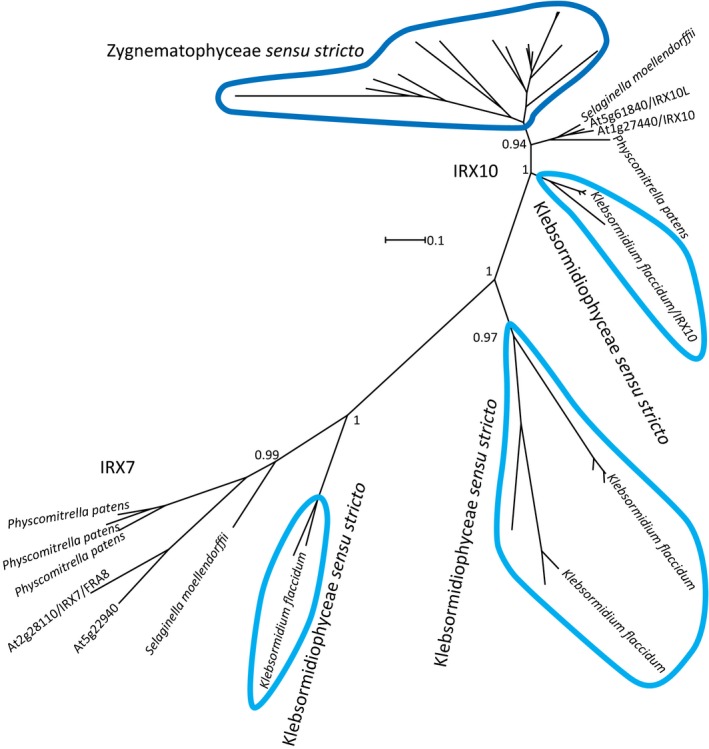
Phylogenetic tree of IRX10/XYS1 and IRX7 orthologs from charophytes, Physcomitrella patens, Selaginella moellendorffii and Arabidopsis. The Klebsormidium flaccidum xylan synthase is designated as Kf XYS1. No algal IRX10/XYS1 or IRX7 orthologs were identified outside Klebsormidiophyceae (light blue) or Zygnematophyceae (dark blue) as indicated by the sensu stricto. Sequences from Klebsormidiophyceae and Zygnematophyceae are ringed in light blue and dark blue, respectively. Branches with no name correspond to charophyte sequences from the 1KP dataset and are probably not full length. All branches with names are sequences from complete genome projects. Branch support was evaluated with an approximate likelihood ratio test and is given as a value between 0 and 1. Supporting Information Notes S1 holds tree files where all sequences used are labeled.
The early algal phase involves charophyte algae and the GT8, GT43, GT47 and DUF579/GXMT protein families. These represent candidate genes implicated in 1,4‐β‐xylan backbone formation, GlcA addition to the backbone, and subsequent GlcA methylation (Fig. 2). The second phase most probably involves flowering plants, specifically basal flowering plants and commelinid monocots, and the DUF231, GT61 and BAHD protein families (Fig. 2). Genes from these families have been implicated in xylan backbone acetylation, addition of arabinosyl and xylosyl substituents to the xylan backbone, and coumaric and ferulic acid transferase activity, respectively. Hence the second phase involves further diversification of xylan structure and probably reflects new functional role(s) of this glycopolymer in wall architecture.
The specific steps of the second phase of xylan evolution appear complex and remained unresolved. The DUF231, GT61 and BAHD protein families are each highly diversified in higher plants, and only a few members from each family have been linked to xylan synthesis. While ancient members of the families exist, none are closely related to the modern xylan‐specific members. Interestingly, we observed a high degree of diversification in basal flowering plants, and then later in commelinid monocots, particularly for GT61 and BAHD. One hypothesis is that xylan‐specific activities developed from different but related catalytic specificities. The time of diversification of these large protein families could in this way indicate the emergence of new specificities towards polymers such as xylan.
A charophyte IRX10/XYS1 ortholog displays β‐1,4‐xylan synthase activity in vitro
Pinpointing early orthologs by sequence analysis primarily defines the oldest possible origin of the individual xylan‐specific activities. However, enzymatic activity of such orthologs is required to unambiguously determine if the modern activities are a result of catalytic conservation or if they evolved from a related enzyme that does not act in the xylan synthesis pathway. IRX10/XYS1 orthologs of the early algal phase are particularly interesting. While other proteins involved in 1,4‐β‐xylan backbone formation exist in plants, the IRX10/XYS1 orthologs are the only group of proteins that have been enzymatically implicated in 1,4‐β‐xylan backbone synthesis, and therefore play a central role among the proteins attributed to this process in plants.
In K. flaccidum, four full‐length sequences showed sequence homology to IRX10/XYS1 and the closely related FRA8 (Table S2). Moreover, one of these proteins, designated K. flaccidum XYLAN SYNTHASE‐1 (KfXYS1), showed a high degree of sequence conservation to IRX10/XYS1. blast protein alignment showed that KfXYS1 and AtXYS1 share a 0.75 identity score (75% amino acid sequence identity across 376 continuous residues of the 415 amino acid AtXYS1 sequence; Table S3). This is a higher score than for the proven 1,4‐β‐xylan xylosyltransferase PoIRX10_4 from Plantago ovata, an herbaceous dicot that is phylogenetically much closer to Arabidopsis than to charophyte algae. In comparison, the K. flaccidum IRX8, IRX9 and IRX14 orthologs were all below 0.50 in identity scores with their respective Arabidopsis orthologs (Table S3), suggesting that these have been less well preserved. Furthermore, protein motif hierarchical clustering showed that all protein motifs shared among the four known 1,4‐β‐xylan xylosyltransferases (with the exception of a protein motif that is unique to PoIRX10_4) are conserved in KfXYS1 (Fig. 4).
Figure 4.
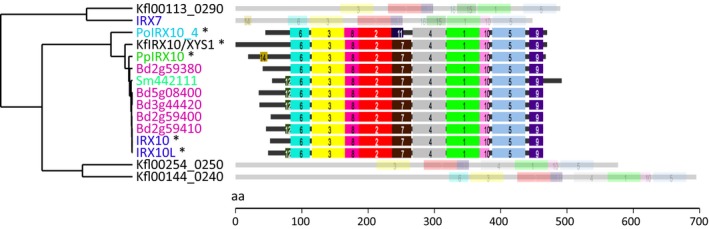
Protein motif analysis of the four IRX10/XYS1 and IRX7 orthologs from Klebsormidium flaccidum. Left part, hierarchical clustering of motif analysis. Right part, graphical representation of identified motifs. Proteins are aligned based on motif 9 (purple). If this motif is not present, the protein is shaded and based on the start codon. IRX10/XYS1 homologs from other plants include Brachypodium distachyon (pink), Arabidopsis (dark blue), Plantago ovata (light blue), Selaginella moellendorffii (light green), and Physcomitrella patens (dark green). Asterisks (*) indicate the five proven xylan xylosyltransferases/xylan synthase activities, including Kf XYS1. Analysis and representation were generated using the SALAD database (Mihara et al., 2010; http://salad.dna.affrc.go.jp/).
A high degree of conservation is supportive of xylan synthase activity; however, to provide biochemical evidence we heterologously expressed KfXYS1 and characterized its enzymatic activity. Initially, we expressed KfXYS1 in S. cerevisiae with a C‐terminal YFP protein tag to facilitate affinity purification. Purified KfXYS1‐YFP was incubated in the presence of UDP‐Xyl and 1,4‐β‐xylotetraose fluorescently labeled with anthranilic acid, and the reaction products were analyzed by normal‐phase high‐performance liquid chromatography. Xylan xylosyltransferase activity was evident by the appearance of multiple new peaks, relative to the control containing only 1,4‐β‐xylotetraose (Fig. 5a). These data suggested that up to nine successive Xyl transfer events occurred under these conditions. Activity levels and peak retention times are comparable to reactions previously reported for AtXYS1 and PpIRX10 (Jensen et al., 2014; Urbanowicz et al., 2014). A partial digestion of the KfXYS1‐YFP reaction products with a 1,4‐β‐xylan‐specific xylanase further supported the idea that the linkages catalyzed by KfXYS1 are indeed β‐1,4‐Xyl linkages (Fig. S10a). No activity was detected when using UDP‐glucose, UDP‐arabinopyranose (UDP‐Arap) or UDP‐arabinofuranose (UDP‐Araf) as donor substrates (Fig. S10b).
Figure 5.
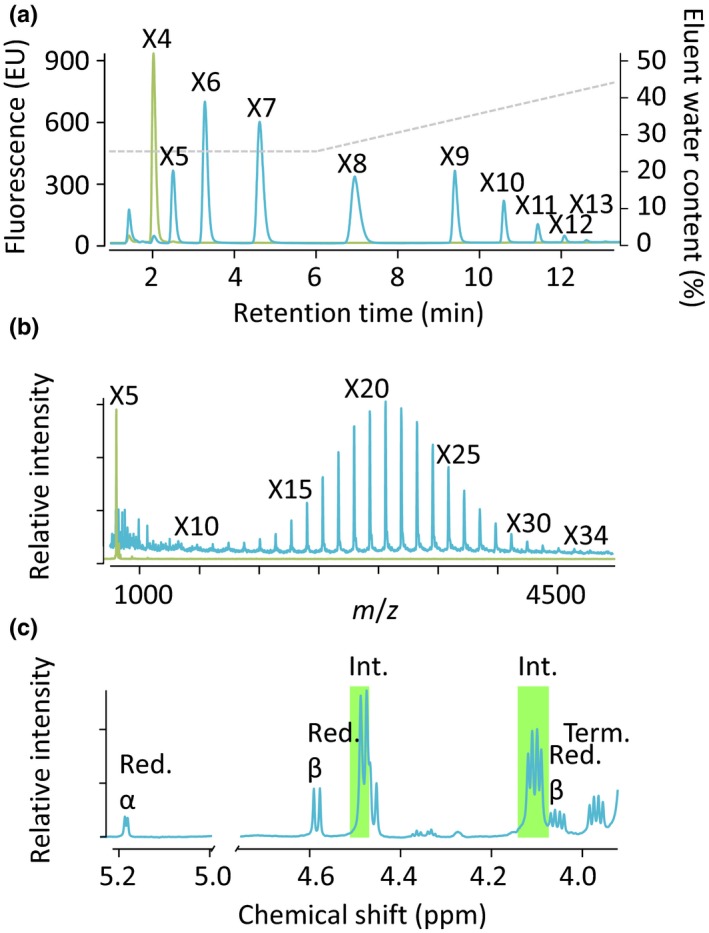
Xylan synthase activity of purified Kf XYS1 expressed in two different heterologous expression systems. (a) Xylan synthase assay using purified Kf XYS1‐YFP produced in Saccharomyces cerevisiae. high‐performance liquid chromatography analysis of reaction mixtures comprising UDP‐xylose and fluorescently labeled xylotetraose (X4) incubated in the presence (blue) or absence (green) of Kf XYS1‐YFP. The gray dashed line represents eluent water content (right y‐axis). X4‐X13 designate progressively longer xylan oligosaccharides. (b) Xylan synthase assay using purified His‐GFP‐Kf XYS1 produced in HEK293 cells. Matrix‐assisted laser desorption ionization‐time of flight mass spectrometry of reaction mixtures after a 4 h reaction in the absence (green) and presence (blue) of UDP‐xylose. The series of annotated [M + H]+ ions are the result of structures with a mass difference of 132 Da, consistent with the sequential addition of xylosyl residues to the 2‐aminobenzamide‐labeled xylopentaose acceptor (X5) generating oligosaccharides with a degree of polymerization (DP) up to 34 (X34). (c) 1D 1H nuclear magnetic resonance (NMR) analysis of reaction products generated by incubation of His‐GFP‐Kf XYS1 with UDP‐xylose as a donor and unlabeled xylobiose as an acceptor substrate. After a 16 h reaction time, xylobiose, UDP‐xylose, and UDP were separated away from higher‐order oligosaccharide products by size exclusion chromatography before NMR analysis. The spectrum shows diagnostic signals of internal β‐(1,4)‐linked xylosyl residues (Int., green shade; Red, Reducing; Term, Terminal).
Xylan polymers consisting solely of 1,3‐β‐Xyl linkages and mixed‐linkage (1,3)(1,4)‐β‐xylans occur in chlorophytes and red algae (Painter, 1983). To further investigate the linkage composition of the KfXYS1 reaction products by NMR spectroscopy, we heterologously expressed the protein in HEK293 cells. A predicted transmembrane domain in the N‐terminus of KfXYS1 was substituted with a His‐GFP protein tag in the HEK293 heterologous expression system, and the recombinant KfXYS1 protein was expressed and purified (His‐GFP‐KfXYS1). Expression and secretion of the His‐GFP‐KfXYS1 fusion protein in transiently transfected HEK293F cells resulted in high amounts of enzyme secretion (c. 100 mg l−1) as determined by measuring the relative fluorescence (GFP fluorescence His‐GFP‐Kf XYS1, 1380) of the recombinant protein secreted into the media (Meng et al., 2013; Urbanowicz et al., 2017), facilitating the large‐scale reactions suitable for detailed product analyses. Expression in HEK293F cells resulted in highly active enzyme preparations capable of adding as many as 29 Xyl residues to a starting 2‐aminobenzamide‐xylopentaose acceptor (Fig. 5b). 1D 1H NMR characterization of reaction mixtures containing xylobiose, UDP‐Xyl and His‐GFP‐KfXYS1 showed depletion of UDP‐Xyl and an increase in internal 1,4‐β‐Xyl linkages, relative to control reactions containing only xylobiose and UDP‐Xyl (Fig. S11). To obtain a less complex spectrum derived from only the higher‐order polysaccharide products, the reaction mixture was subjected to size exclusion chromatography to separate these from other reaction constituents, including UDP and xylobiose. This 1D 1H NMR spectrum revealed prominent diagnostic signals of internal 1,4‐β‐Xyl linkages (Urbanowicz et al., 2014) (Fig. 5c), while 1,3‐β‐Xyl linkages, which would produce a clear signal at 4.68 ppm (Viana et al., 2011), were found to be absent.
These results show that Kf XYS1 expressed in two separate heterologous expression systems using both N‐ and C‐terminal protein tagging, displays 1,4‐β‐xylan synthase activity exclusively, consistent with a high level of protein homology to known plant xylan synthases. Hence, the enzymatic activity of this group of proteins has been conserved as far back as an extinct common ancestor of K. flaccidum and land plants, living c. 700 million yr ago (Douzery et al., 2004).
Klebsormidium flaccidum cell walls contain multiple species of substituted xylan
Having determined that KfXYS1 is a β‐1,4‐xylosyltransferase, we analyzed K. flaccidum cell walls to determine whether they contain 1,4‐β‐xylan. Monosaccharide compositional analysis of K. flaccidum cell wall material showed that they contain 59 μg Xyl mg−1 cell wall material, making it one of the dominant monosaccharide constituents (Fig. 6a). To specifically confirm the presence of 1,4‐β‐xylan and to characterize its structure, we extracted K. flaccidum cell wall material with strong base and subjected these to 1,4‐β‐xylanase digestion. The resulting xylan oligosaccharides were analyzed by gel electrophoresis (PACE). Digestion with GH10 1,4‐β‐xylanase, which requires one unsubstituted Xyl residue for cleavage, resulted in a complex band pattern, indicating the presence of 1,4‐β‐xylan with frequent substitutions. Two bands comigrated with either Xyl or 1,4‐β‐xylobiose (Fig. 6b, lane 3). Subsequent digestion with GH3 β‐xylosidase resulted in a depletion of the xylobiose band and an increase in the Xyl band, confirming their individual identities (Fig. 6b, lane 5). The oligosaccharide migrating near xylotriose was insensitive to β‐xylosidase, indicating that it is not xylotriose, but probably a substituted oligosaccharide. The xylo‐oligosaccharides were insensitive to digestion with GH115 α‐glucuronosidase (Fig. 6b, lanes 6, 8 and 9). By contrast, digestion with diagnostic xylan α‐arabinofuranosidases (GH43 and GH62) resulted in alterations in band patterning, indicating the presence of α‐Araf substituents (Fig. 6b, lane 7–9). The observation that not all the bands were fully digested by these enzymes suggests that there are other kinds of substitutions present in K. flaccidum xylan and possibly linkages other than 1,4‐β‐Xyl in the backbone. In conclusion, K. flaccidum cell wall material shows diagnostic band patterns of 1,4‐β‐xylan with α‐linked arabinosyl substitutions and other additional unidentified modifications or linkages in the backbone. Phylogenetic analysis of the known arabinosyltransferase from GT61 indicated that no orthologs are present in K. flaccidum (Table 1). Therefore, this suggests that another class of glycosyltransferases carries out this function in K. flaccidum, representing a case of convergent evolution with regard to 1,4‐β‐xylan α‐arabinosyl decorations.
Figure 6.
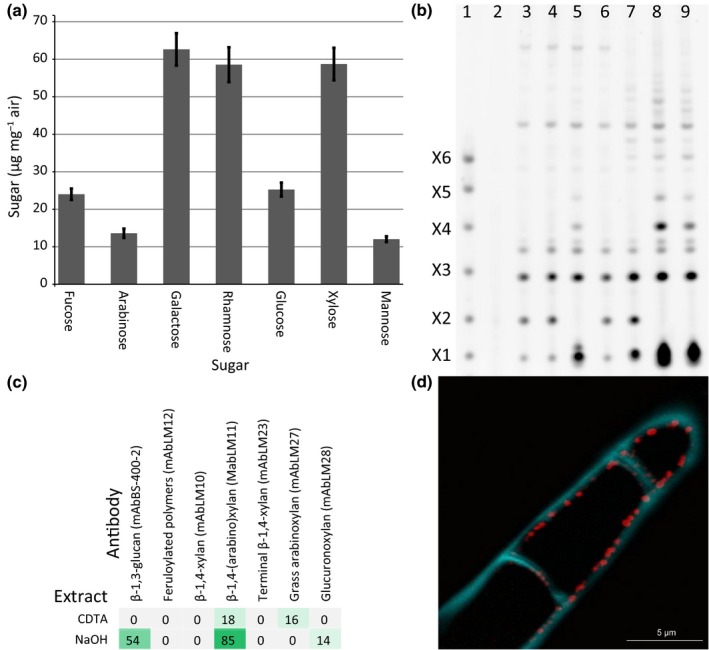
Structural analysis of Klebsormidium flaccidum cell walls. (a) Monosaccharide composition of K. flaccidum cell wall material (n = 3, ± SD). (b) Cell wall material analyzed by polysaccharide analysis using carbohydrate gel electrophoresis using hydrolytic enzymes specific for the 1,4‐β‐xylan backbone or side chain modifications. Lane 1, marker lane, 1,4‐β‐xylan oligosaccharides with degree of polymerization 1 to 6; lane 2, control digestion with no enzyme; lane 3, GH10 1,4‐β‐xylanase; lane 4, GH10 and GH11 1,4‐β‐xylanases; lane 5, GH10 and xylosidase; lane 6, GH10 and GH115 glucuronidase; lane 7, GH10 xylanase plus GH43 and GH62 arabinofuranosidases; lane 8, as lane 7 plus 1,4‐β‐xylosidase and GH115; lane 9, as lane 8 plus GH11. (c) Sequential extraction of K. flaccidum cell wall material and relative quantification of cell wall epitopes by comprehensive microarray polymer profiling (CoMPP). The values in the heat map are mean spot signals from three experiments and the highest signal in the entire dataset was set to 100 and all other data were adjusted accordingly. A cutoff value of 5 was imposed. (d) Immunolocalization of xylan (antibody LM11; red) and β‐glucan (Calcofluor White; blue) in the K. flaccidum cell wall by confocal laser scanning microscopy.
Sequential extraction of K. flaccidum cell wall material with subsequent immobilization to nitrocellulose membranes and probing with xylan‐directed antibodies (comprehensive microarray polymer profiling) resulted in substantial labeling by the 1,4‐β‐xylan‐specific antibody LM11 in both the CDTA and the NaOH‐soluble fractions (Fig. 6c). Additionally, by employing antibodies directed at xylan with complex substitution patterns (LM27, binding to xylan with complex substitution patterns such as grass xylan, and LM28, binding to methylated and unmethylated glucuronosyl substituted xylan) we were able to confirm the complex substitution pattern suggested by the PACE analysis. LM27 and LM28 epitopes were present in separate fractions, suggesting that at least two structurally different pools of xylan are present in K. flaccidum. Immunohistochemical localization with a xylan‐specific antibody (LM11) showed that xylan was present in the K. flaccidum cell wall in a punctate but distinct pattern on the luminal side of the cell, which may be interpreted to be labeling of newly synthesized xylan immediately before integration into the wall matrix (Fig. 6d).
Discussion
The evolution of a polysaccharide‐based cell wall was probably a key event facilitating terrestrialization, making this a defining moment in the evolution of life on land. A growing body of evidence suggests that charophyte algae were the first to inhabit land, and from these algae, land plants subsequently evolved c. 475 million yr ago (Harholt et al., 2016). In the present study, we show enzymatic conservation between 1,4‐β‐xylan synthases separated by c. 700 million yr of evolution. Interestingly, our data‐mining efforts did not identify any additional homologs predating KfXYS1. Using specific antibodies and hydrolytic enzymes, we also showed that K. flaccidum cell walls contain substituted 1,4‐β‐xylans. These xylans are probably the products of a pathway involving KfXYS1. Our study therefore suggests that charophyte algae are the evolutionary origin of the 1,4‐β‐xylans of modern‐day plants.
The xylan synthase activity of KfXYS1 is pivotal evidence in support of the hypothesis that the ‘plant cell wall’ emerged in charophyte algae. For instance, the biochemical orthology between KfXYS1 and embryophyte IRX10/XYS1 makes it likely that other seemingly functionally orthologous relationships in xyloglucan and pectin biosynthesis between charophyte algae and land plants are indeed genuine. With established functional orthology between early diverging charophyte alga and land plant cell wall biosynthetic processes, we can now establish with reasonable certainty that the land plant cell wall originated in an early ancestor in charophyte algae evolution.
Interestingly, IRX9 and IRX14, which are glycosyltransferase‐like proteins required for xylan synthesis in planta, also appear in Klebsormidiophyceae (Figs 2, S4). These proteins do not seem to have essential catalytic functions in higher plants, and most likely have a role in anchoring IRX10 in the Golgi apparatus, as observed in Arabidopsis (Ren et al., 2014) and asparagus (Zeng et al., 2016). The function of IRX9 and IRX14 in Klebsormidiophyceae could be the same as in these higher plants, or they may be unrelated and were recruited later in evolution to participate in xylan synthesis. PARVUS and GXMT1 provide examples of the latter scenario as they are both implicated in xylan biosynthesis in Arabidopsis yet have orthologs in chlorophytes that do not produce xylan.
Klebsormidium flaccidum xylan appears to be highly substituted, but no GT61 or GUX orthologs were identified in K. flaccidum (Table 1; Fig. 2). This suggests that other classes of glycosyltransferases are involved in decorating its xylan backbone. This includes the apparent α‐arabinosyl substituents, which have been associated with GT61 enzymes in grasses (Anders et al., 2012), suggesting a case of convergent evolution. Other examples exist where backbone substitutions are apparently reinvented readily, for example in xyloglucan side chain evolution (Tuomivaara et al., 2015) and possibly in psyllium seed mucilaginous layers that produce a highly arabinosyl‐ and xylosyl‐substituted heteroxylan and encode an unusually high number of GT61 orthologs (Jensen et al., 2013).
While no GUX orthologs were identified in the K. flaccidum genome (consistent with its xylan being insensitive to α‐glucuronosidase and no observable GlcA in the sugar composition analysis), we identified an ortholog in Zygnematophyceae, the most recent group of charophyte algae to have evolved (Fig. 1). The GUX proteins form three phylogenetic subgroups, that is, charophytes, mosses and vascular plants (Fig. S6), with glucuronoxylan isolated and characterized from the latter two subgroups of plants (Kulkarni et al., 2012; Rennie & Scheller, 2014; Busse‐Wicher et al., 2016). This suggests that the GUX proteins function as xylan glucuronosyltransferases in mosses. The vascular plant GUX subgroup further shows division between subgroups of Arabidopsis GUX2/4/5 and GUX1/3 orthologs. In Arabidopsis, even spacing of GlcA residues along the xylan backbone in secondary cell walls is provided by GUX1 (Bromley et al., 2013) and in primary cell walls by GUX3 (Mortimer et al., 2015). The even GlcA spacing facilitates binding to cellulose microfibrils through a twofold helical screw ribbon conformation of the glucuronoxylan, which probably exerts an influence on microfibril aggregation and organization (Simmons et al., 2016). The phylogenetic analysis therefore suggests that this prominent architectural principle arose in basal vascular plants.
As we see the contours of a polysaccharide‐based cell wall evolve in charophytic algae, and its further evolution throughout the plant kingdom, a number of questions become increasingly pressing. Why were these polysaccharide classes (i.e. cellulose, xylan, xyloglucan, mannan, and pectin) selected? Why are some occasionally expendable in some cell walls and cell types, while seemingly indispensable to the organism as a whole and maintained consistently in land plants throughout evolution? Why have so few new polysaccharide classes, such as Rhamnogalacturonan II, emerged during the course of plant evolution? Why does utilization of different polysaccharides change during evolution and what differences in cell wall functionality, architectural principles and solutions drive these evolutionary events? By having a firmer understanding of the evolutionary span and context of the plant cell wall, we are now poised to begin to answer these fundamental questions in plant biology.
In conclusion, using a multifaceted approach involving genome and transcriptome mining and phylogenetic and biochemical analysis, Kf XYS1 was identified and shown to possess 1,4‐β‐xylan synthase activity. Immunolabeling and biochemical analysis of K. flaccidum cell walls identified a cell wall‐localized, highly substituted 1,4‐β‐xylan. These findings, along with an evolutionary analysis of the occurrence of xylan pathway enzymes, link plant xylan evolution to an ancestral charophytic alga with a diverging point c. 700 million yr ago.
Author contributions
J.K.J. and J.H. conceived and initiated the project. J.K.J., M.B‐W., C.P.P., H.J.M., P.J.S., B.R.U. and J.H. designed experiments. J.K.J., M.B‐W., C.P.P., J.U.F., M.H.D., H.J.M., K.W.M., P.J.S., B.R.U., J‐Y.Y., M‐J.P., C.G.W., P.D., H.V.S., P.U. and J.H. performed experiments and interpreted the results. M.M. and G.K‐S.W. provided unpublished sequences. The manuscript was written by J.K.J., B.R.U. and J.H. All authors edited and commented on the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 SDS gel of purified GFP‐KfXYS1 expressed in HEK293 cell culture.
Fig. S2 Phylogenetic tree of PARVUS orthologs.
Fig. S3 Phylogenetic tree of IRX15/GXMT1 orthologs.
Fig. S4 Phylogenetic tree of IRX9 and IRX14 orthologs.
Fig. S5 Phylogenetic tree of IRX8 orthologs.
Fig. S6 Phylogenetic tree of GUX1‐5 orthologs.
Fig. S7 Phylogenetic tree of ESK1 orthologs.
Fig. S8 Phylogenetic tree of GT61 orthologs.
Fig. S9 Phylogenetic tree of OsAT10 orthologs.
Fig. S10 Enzymatic specificity of KfXYS1.
Fig. S11 Characterization of reaction mixtures with or without KfXYS1 by 1D 1H NMR.
Table S1 Positive/negative list used for identification of xylan synthesis‐related sequences
Table S2 Protein sequences of Klebsormidium flaccidum xylan gene orthologs
Table S3 Protein blast analysis of Klebsormidium flaccidum xylan gene orthologs
Acknowledgements
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE‐FC02‐07ER64494) (J.K.J. and C.G.W.). Work by H.V.S. was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE‐AC02‐05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. The Carlsberg Foundation, The Innovation Fund Denmark (IFD) under File no. 5112‐00006B, and the Villum Foundation's Young Investigator Program are gratefully acknowledged for support to M.H.D., C.P.P., J.U.F. and J.H. Immunohistochemistry and confocal microscopy were performed at the Center of Advanced Bioimaging (CAB), SCIENCE KU DK. P.D. and M.B‐W. were supported by the Leverhulme Natural Material Innovation Centre. P.J.S., M‐J.P. and B.R.U. were supported by funding provided by The BioEnergy Science (BESC) and The Center for Bioenergy Innovation (CBI), a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. J‐Y.Y. and K.W.M. were supported by NIH grant P41GM103390 and G.J.B. was supported by NIH grant P01 GM107012. P.U. was supported by the Innovation Fund Denmark. We thank the Center for Plant and Microbial Complex Carbohydrates (DE‐SC0015662) for equipment support. The authors declare no conflict of interest.
References
- Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM et al 2012. Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proceedings of the National Academy of Sciences, USA 109: 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley LE, Peck ML, Kim S‐R, Ebert B, Manisseri C, Chiniquy DM, Sykes R, Gao L, Rautengarten C, Vega‐Sánchez ME et al 2013. Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification. Plant Physiology 161: 1615–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Becker D, Kamerling JP, Melkonian M. 1991. 2‐keto‐sugar acids in green flagellates: a chemical marker for Prasinophycean scales. Journal of Phycology 27: 498–504. [Google Scholar]
- Bell K, Mitchell S, Paultre D, Posch M, Oparka K. 2013. Correlative imaging of fluorescent proteins in resin‐embedded plant material. Plant Physiology 161: 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley JR, Busse‐Wicher M, Tryfona T, Mortimer JC, Zhang Z, Brown DM, Dupree P. 2013. GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant Journal 74: 423–434. [DOI] [PubMed] [Google Scholar]
- Brown D, Wightman R, Zhang Z, Gomez LD, Atanassov I, Bukowski J‐P, Tryfona T, McQueen‐Mason SJ, Dupree P, Turner S. 2011. Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579‐containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant Journal 66: 401–413. [DOI] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. 2007. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant Journal 52: 1154–1168. [DOI] [PubMed] [Google Scholar]
- Busse‐Wicher M, Li A, Silveira RL, Pereira CS, Tryfona T, Gomes TCF, Skaf MS, Dupree P. 2016. Evolution of xylan substitution patterns in gymnosperms and angiosperms: implications for xylan interaction with cellulose. Plant Physiology 171: 2418–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant Journal 3: 1–30. [DOI] [PubMed] [Google Scholar]
- Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, Carroll A, Ulvskov P, Harholt J, Keasling JD et al 2012. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proceedings of the National Academy of Sciences, USA 109: 17117–17122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuault V, Buffetto F, Rydahl MG, Marcus SE, Torode TA, Xue J, Crépeau M‐J, Faria‐Blanc N, Willats WGT, Dupree P et al 2015. Monoclonal antibodies indicate low‐abundance links between heteroxylan and other glycans of plant cell walls. Planta 242: 1321–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domozych DS, Serfis A, Kiemle SN, Gretz MR. 2007. The structure and biochemistry of charophycean cell walls: I. Pectins of Penium margaritaceum . Protoplasma 230: 99–115. [DOI] [PubMed] [Google Scholar]
- Domozych DS, Wells B, Shaw PJ. 1991. Basket scales of the green alga, Mesostigma viride: chemistry and ultrastructure. Journal of Cell Science 100: 397–407. [Google Scholar]
- Douzery EJP, Snell EA, Bapteste E, Delsuc F, Philippe H. 2004. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proceedings of the National Academy of Sciences, USA 101: 15386–15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangel JU, Ulvskov P, Knox JP, Mikkelsen MD, Harholt J, Popper ZA, Willats WGT. 2012. Cell wall evolution and diversity. Frontiers in Plant Science 3: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F, Jackson P, Deery MJ, Dupree P. 2002. Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Analytical Biochemistry 300: 53–68. [DOI] [PubMed] [Google Scholar]
- Harholt J, Moestrup O, Ulvskov P. 2016. Why plants were terrestrial from the beginning. Trends in Plant Science 21: 96–101. [DOI] [PubMed] [Google Scholar]
- Harholt J, Sørensen I, Fangel J, Roberts A, Willats WGT, Scheller HV, Petersen BL, Banks JA, Ulvskov P. 2012. The glycosyltransferase repertoire of the spikemoss Selaginella moellendorffii and a comparative study of its cell wall. PLoS ONE 7: e35846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AD, Bar‐Peled M. 2002. Biosynthesis of UDP‐xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane‐bound UDP‐glucuronic acid decarboxylase isoforms. Plant Physiology 130: 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N et al 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Ichita J, Matsue H, Ono H, Maeda I. 2002. Fluorescent labeling of pectic oligosaccharides with 2‐aminobenzamide and enzyme assay for pectin. Carbohydrate Research 337: 1023–1032. [DOI] [PubMed] [Google Scholar]
- Jensen JK, Johnson N, Wilkerson CG. 2013. Discovery of diversity in xylan biosynthetic genes by transcriptional profiling of a heteroxylan containing mucilaginous tissue. Frontiers in Plant Science 4: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Johnson NR, Wilkerson CG. 2014. Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant Journal 80: 207–215. [DOI] [PubMed] [Google Scholar]
- Jensen JK, Kim H, Cocuron J‐C, Orler R, Ralph J, Wilkerson CG. 2011. The DUF579 domain containing proteins IRX15 and IRX15‐L affect xylan synthesis in Arabidopsis. Plant Journal 66: 387–400. [DOI] [PubMed] [Google Scholar]
- Kulkarni AR, Peña MJ, Avci U, Mazumder K, Urbanowicz BR, Pattathil S, Yin Y, O'Neill MA, Roberts AW, Hahn MG et al 2012. The ability of land plants to synthesize glucuronoxylans predates the evolution of tracheophytes. Glycobiology 22: 439–451. [DOI] [PubMed] [Google Scholar]
- Matasci N, Hung L‐H, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M et al 2014. Data access for the 1,000 plants (1KP) project. GigaScience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP. 2005. Monoclonal antibodies to plant cell wall xylans and arabinoxylans. Journal of Histochemistry and Cytochemistry 53: 543–546. [DOI] [PubMed] [Google Scholar]
- Meikle PJ, Bonig I, Hoogenraad NJ, Clarke AE, Stone BA. 1991. The location of (1→3)‐β‐glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)‐β‐glucan‐specific monoclonal antibody. Planta 185: 1–8. [DOI] [PubMed] [Google Scholar]
- Meng L, Forouhar F, Thieker D, Gao Z, Ramiah A, Moniz H, Xiang Y, Seetharaman J, Milaninia S, Su M et al 2013. Enzymatic basis for N‐glycan sialylation: structure of rat α2,6‐sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. The Journal of Biological Chemistry 288: 34680–34698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T. 2010. SALAD database: a motif‐based database of protein annotations for plant comparative genomics. Nucleic Acids Research 38: D835–D842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Harholt J, Ulvskov P, Johansen IE, Fangel JU, Doblin MS, Bacic A, Willats WGT. 2014. Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae. Annals of Botany 114: 1217–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer JC, Faria‐Blanc N, Yu X, Tryfona T, Sorieul M, Ng YZ, Zhang Z, Stott K, Anders N, Dupree P. 2015. An unusual xylan in Arabidopsis primary cell walls is synthesised by GUX3, IRX9L, IRX10L and IRX14. Plant Journal 83: 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øbro J, Harholt J, Scheller HV, Orfila C. 2004. Rhamnogalacturonan I in Solanum tuberosum tubers contains complex arabinogalactan structures. Phytochemistry 65: 1429–1438. [DOI] [PubMed] [Google Scholar]
- Painter T. 1983. Algal polysaccharides. In: Aspinall G, ed. The polysaccharides. New York, NY, USA: Academic Press, 195–285. [Google Scholar]
- Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet M‐C, Farkas V, von Schantz L, Marcus SE, Andersen MCF et al 2012. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. The Journal of Biological Chemistry 287: 39429–39438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou G‐K, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye Z‐H. 2007. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. The Plant Cell 19: 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Hansen SF, Ebert B, Lau J, Scheller HV. 2014. Site‐directed mutagenesis of IRX9, IRX9L and IRX14 proteins involved in xylan biosynthesis: glycosyltransferase activity is not required for IRX9 function in Arabidopsis. PLoS ONE 9: e105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Hansen SF, Baidoo EEK, Hadi MZ, Keasling JD, Scheller HV. 2012. Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiology 159: 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Scheller HV. 2014. Xylan biosynthesis. Current Opinion in Biotechnology 26: 100–107. [DOI] [PubMed] [Google Scholar]
- Rogowski A, Baslé A, Farinas CS, Solovyova A, Mortimer JC, Dupree P, Gilbert HJ, Bolam DN. 2014. Evidence that GH115 α‐glucuronidase activity, which is required to degrade plant biomass, is dependent on conformational flexibility. The Journal of Biological Chemistry 289: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeán AA, Duffieux D, Harholt J, Qin F, Michel G, Czjzek M, Willats WGT, Hervé C. 2017. Insoluble (1 → 3), (1 → 4)‐β‐d‐glucan is a component of cell walls in brown algae (Phaeophyceae) and is masked by alginates in tissues. Scientific Reports 7: 2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annual Review of Plant Biology 61: 263–289. [DOI] [PubMed] [Google Scholar]
- Simmons TJ, Mortimer JC, Bernardinelli OD, Pöppler A‐C, Brown SP, deAzevedo ER, Dupree R, Dupree P. 2016. Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid‐state NMR. Nature 7: 13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Wang H‐T, York WS, Peña MJ, Urbanowicz BR. 2017. Designer biomass for next‐generation biorefineries: leveraging recent insights into xylan structure and biosynthesis. Biotechnology for Biofuels 10: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen I, Pettolino Fa, Bacic A, Ralph J, Lu F, O'Neill Ma, Fei Z, Rose JKC, Domozych DS, Willats WGT. 2011. The charophyceae green algae provide insights into the early origins of plant cell walls. Plant Journal 68: 201–211. [DOI] [PubMed] [Google Scholar]
- Tuomivaara ST, Yaoi K, O'Neill MA, York WS. 2015. Generation and structural validation of a library of diverse xyloglucan‐derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydrate Research 402: 56–66. [DOI] [PubMed] [Google Scholar]
- Ulvskov P, Paiva DS, Domozych D, Harholt J. 2013. Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes. PLoS ONE 8: e76511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Bharadwaj VS, Alahuhta M, Peña MJ, Lunin VV, Bomble YJ, Wang S, Yang J‐Y, Tuomivaara ST, Himmel ME et al 2017. Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water‐mediated mechanism. Plant Journal 91: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS. 2014. Two Arabidopsis proteins synthesize acetylated xylan in vitro . Plant Journal 80: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, Foston M, Li H, O'Neill MA, Ragauskas AJ et al 2012. 4‐O‐methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proceedings of the National Academy of Sciences, USA 109: 14253–14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana AG, Noseda MD, Gonçalves AG, Duarte MER, Yokoya N, Matulewicz MC, Cerezo AS. 2011. β‐d‐(1→4), β‐d‐(1→3) ‘mixed linkage’ xylans from red seaweeds of the order Nemaliales and Palmariales. Carbohydrate Research 346: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA et al 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S et al 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Research 37: D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Lampugnani ER, Picard KL, Song L, Wu A, Farion IM, Zhao J, Ford K, Doblin M, Bacic A. 2016. Asparagus IRX9, IRX10 and IRX14A are essential for xylan biosynthesis in the Golgi apparatus of asparagus. Plant Physiology 171: 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 SDS gel of purified GFP‐KfXYS1 expressed in HEK293 cell culture.
Fig. S2 Phylogenetic tree of PARVUS orthologs.
Fig. S3 Phylogenetic tree of IRX15/GXMT1 orthologs.
Fig. S4 Phylogenetic tree of IRX9 and IRX14 orthologs.
Fig. S5 Phylogenetic tree of IRX8 orthologs.
Fig. S6 Phylogenetic tree of GUX1‐5 orthologs.
Fig. S7 Phylogenetic tree of ESK1 orthologs.
Fig. S8 Phylogenetic tree of GT61 orthologs.
Fig. S9 Phylogenetic tree of OsAT10 orthologs.
Fig. S10 Enzymatic specificity of KfXYS1.
Fig. S11 Characterization of reaction mixtures with or without KfXYS1 by 1D 1H NMR.
Table S1 Positive/negative list used for identification of xylan synthesis‐related sequences
Table S2 Protein sequences of Klebsormidium flaccidum xylan gene orthologs
Table S3 Protein blast analysis of Klebsormidium flaccidum xylan gene orthologs


