Abstract
Professor Donald Coffey and his laboratory pioneered studies showing the relationships between nuclear shape and cellular function. In doing so, he and his students established the field of nuclear morphometry in prostate cancer. By using perioperative tissues via biopsies and surgical sampling, Dr. Coffey’s team discovered that nuclear shape and other pathologic features correlated with clinical outcome measures. Cancer cells also exist outside of solid tumor masses as they can be shed from both primary and metastatic lesions into the circulatory system. The pool of these circulating tumor cells (CTCs) is heterogeneous. While some of these CTCs are passively shed into the circulation, others are active metastasizers with invasive potential. Advances in nanotechnology now make it possible to study morphologic features such as nuclear shape of CTCs in the bloodstream via liquid biopsy. Compared to traditional tissue sampling, liquid biopsy allows for minimally invasive, repetitive, and systemic disease sampling, which overcomes disease misrepresentation issues due to tumor temporospatial heterogeneity. Our team developed a novel liquid biopsy approach, the NanoVelcro assay, which allows us to identify morphologic heterogeneity in the CTC compartment. By applying classical methods of nuclear morphometry, we identified very small nuclear CTCs (vsnCTCs) in prostate cancer patients. Our initial studies showed that vsnCTCs strongly correlated with unfavorable clinical behaviors including the disposition to visceral metastases. These approaches may continue to yield additional insights into dynamic clinical behaviors, which creates an opportunity for more comprehensive and accurate cancer profiling. Ultimately, these advancements will allow physicians to employ more accurate and personalized treatments, helping the field reach the goal of true precision medicine.
Keywords: Circulating tumor cells, nuclear size, visceral metastases, prostate cancer, NanoVelcro, biomarkers
Introduction
This article is part of a special issue, dedicated to Donald S. Coffey, PhD. The relationship between cell and nuclear shape and cellular function has long been studied and taught by many including Prof. Donald Coffey [1-3]. Particularly, the Coffey lab showed there is an intimate relationship between nuclear shape and nuclear matrix composition [1,4,5]. These findings led to several studies in the area of nuclear morphometry by Coffey and Partin showing relationships between nuclear shape and cellular function [5-10]. The bulk of these studies focused on analysis of perioperative tissue given their accessibility by biopsy or surgical sampling. As the field of cancer biology moved forward, newer means of studying cancer biology arose, including the capability of studying cancer cells that exist within the bloodstream. Even with current advances, the lessons of the past continue to apply.
The need to go beyond tissue analysis in prostate cancer
Histopathologic evaluation of sampled tissue is currently considered the gold standard for cancer diagnosis. The evaluation of tissue samples always includes characterization of cellular features including nuclear size and shape. Additional insight can be gained from immunohistochemistry and profiling of DNA/RNA content. Tissue-based studies have fueled advances in the field of cancer biology. However, this practice relies upon tissue specimen acquisition either by surgical excision or needle biopsy. This requirement presents certain limitations that are particularly difficult in prostate cancer (PCa). The procedures used to obtain tissues samples may be invasive and costly. Tissue collection can be limited by the risk of patient morbidity and psychological stress. In the setting of metastatic disease, the location of tumors can pose technical challenges for acquisition. A common feature of metastatic PCa is the development of osteoblastic metastasis in bone. Sampling of these lesions often requires special equipment such as drills capable of cutting through calcified bone matrix. In addition, the operator must have the technical skills to identify areas where active tumor resides to ensure sampling of viable cells as opposed to calcified bone matrix lacking active tumor cells. Finally, recent studies showing temporospatial heterogeneity [11-17] within a tumor raises concerns regarding the accuracy of using a single biopsy sample as a representative sample of the molecular nature of a spatially heterogonous cancer. The most relevant or most aggressive features may be missed by a lack of sampling.
While these limitations are important, they do not suggest that tissue analysis is not without value. They do, however, point toward a need to find creative solutions to apply these lessons in different settings.
Liquid biopsy and contemporary CTC technology
Liquid biopsies have been proposed as a means of addressing some of the limitations discussed above. Different components in the bloodstream can be utilized for this purpose. These include both CTCs and cell free DNA. In this article, we will focus on the entity of CTCs given feature of this approach.
Since the 1970s, scientists have hypothesized that tumors cells shed by primary and metastatic tumors must be located in the circulation [18-21]. Within this pool of CTCs are those that are passively shed by the shearing forces of blood passing over tumor surfaces (both primary and metastatic). Additionally, this same pool contains cells that have lost contact inhibition and have invaded into the vasculature with the capability of re-invading into distant sites prior to establishing metastatic colonization. These cells are easily obtained through clinical phlebotomy. As such, sampling of CTCs can be more frequent than a tissue biopsy. In fact, it is possible to routinely sample blood for years over the natural course of a cancer, thus providing an opportunity to study the dynamics of an evolving disease in real time [22].
CTCs are very rare cellular events (in comparison to leukocyte or even red blood cells) in the bloodstream. Innovation in the field has led to the development of several platforms capable of identifying and even enumerating CTCs. In 2004, the FDA cleared the CellSearchTM assay [23-25] for clinical use in metastatic breast cancer. The assay was later cleared for PCa in 2008 [25]. While an important step for the field, the approved form of the assay is known to have multiple limitations including the need for cellular fixation and relatively low-resolution image quality that prevents the end user from studying morphologic features.
These limitations have fueled developments of newer technologies and approaches that have improved our ability to study and understand the cells within the circulatory compartment. In particular, advances in materials science engineering, fluid mechanics, chemical engineering, and biomedical engineering have brought the field of nanotechnology to address limitations faced by cancer biologists. This in turn, created a revolution in the field of cancer biology.
A plethora of new technologies has become available for the study of CTCs in the bloodstream. Each system was engineered to address specific problems. As such, we will focus this review on the NanoVelcro Assay, a system that our joint CSMC/UCLA team has developed to address issues of selectively capturing and studying the morphology and biology in CTCs.
Application of NanoVelcro assay in CTC analysis
Over the past decade, collaborative and interdisciplinary efforts have focused on CTC detection, isolation, and characterization [26]. Our group has developed a suite of nanosubstrate microfluidic platforms for capturing and studying CTCs, named the “NanoVelcro” Assay [22,27,28]. The NanoVelcro Assay utilizes a silicon nanowire substrate (SiNS) for capturing CTCs.The mechanism of capture strongly mimics the physical interaction between the complementary faces of a Velcro fastener [29]. In particular, the surface of a CTC is characterized by almost fractal-like structural folds that create hair-like projections with a large surface area suitable for interaction with a nanostructured like NanoVelcro. In this approach, the CTCs are coated with a mixture of biotinylated capture antibodies (such as anti-EpCAM) and run through a microfluidic channel that forces interaction of the CTCs with the SiNS covered in streptavidin. In this fashion, the system utilizes the strength of the biotin-streptavidin interaction to effect capture. The NanoVelcro platform has demonstrated CTC capture efficiency that ranges from 40 to 70% in stationary device settings [30]. Our team simultaneously developed an immunocytochemistry protocol to further verify an immobilized cell as a CTC. Finally, all captured cells are reviewed using single cell image cytometry to verify DAPI staining, CK/CD45 expression, and object size. In the NanoVelcro Assay, an object is called a CTC when the following criteria are met: DAPI+, cytokeratin (CK)+, CD45-, size over 6 µm. Nonspecifically captured white blood cells (WBC) have a different profile: DAPI+, CK-, CD45+, size over 12 µm [22]. Using purposefully engineered variations on the configuration of the NanoVelcro surface, we have developed multiple generations of NanoVelcro chips that are suited for a range of biomedical applications (Figure 1).
Figure 1.
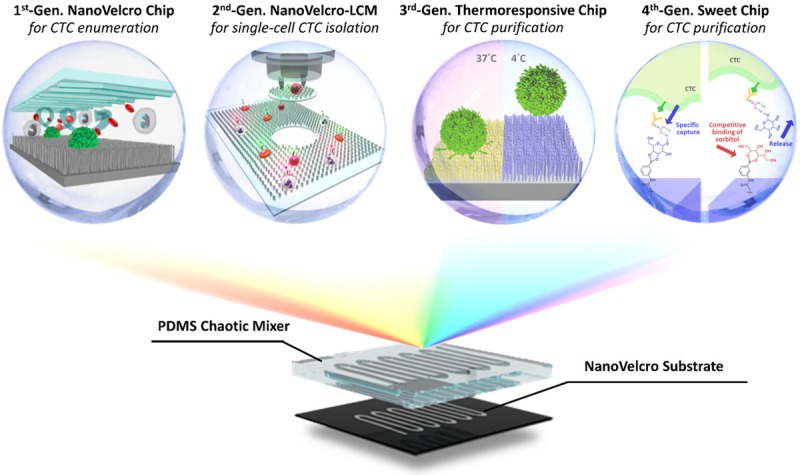
Conceptual illustration of four generations of NanoVelcro Assay developed by UCLA team. The 1st generation NanoVelcro chip [45,46], composed of silicon nanowire substrate (SiNS) in conjunction with an overlaid microfluidic chaotic mixture, was created for CTC capture and enumeration. The 2nd Gen NanoVelcro-LMD (laser microdissection) chip [33-35] was designed for the isolation of captured CTC. The 3rd generation ThermoResponsive NanoVelcro chip [37,38] utilized temperature driven conformation change of polymer brushes to achieve the capture and release of viable CTCs at 37°C and 4°C, respectively. The 4th gen Sweet Chip [39] utilized the competitive binding of sorbitol to phenylboronic acid (PBA) to gently release captured CTC was intact molecular integrity.
1st generation NanoVelcro assay: enumeration and immunocytochemistry (ICC)
The first generation of the NanoVelcro chip is marked by lithographically patterned SiNS with an overlaid polydimethylsiloxane (PDMS) microfluidic chaotic mixer (Figure 2). The PDMS chaotic mixture roof has herringbone structures which induce vertical flow in the microchannels [31], thus increasing the frequency of contact between cells and the nanosubstrates. The captured CTCs are then stained and enumerated using our ICC protocol. Briefly, the NanoVelcro chip is stained with immunofluorescent antibodies for DAPI, cytokeratin (CK), and CD45. Imaging was performed using upright fluorescence microscope (Eclipse 90i, Nikon) [32] under 40× magnification with DAPI, FITC, and TRITC corresponding with nuclear, CK, and CD45 staining, respectively. The chip is scanned using a semi-automated motorized stage and images of individual CTCs are reviewed using the Nikon Elements 2.0 software package. CTC candidates are defined as objects that are DAPI+, CK+, CD45- with a size over 6 µm. The final images are reviewed by a cytopathologist to ensure the calls are morphologically consistent with an epithelial cell. The 1st generation NanoVelcro chip is able to capture CTCs at a remarkable efficiency (>85%) [22]. In a side-by-side comparison study using clinical samples, the NanoVelcro assay performance exceeded that of the CellSearchTM assay [22]. As the CTCs are immobilized onto a small, flat surface, the NanoVelcro platform does not require multiple cross-sectional imaging scans. Instead, chips are reviewed using a single Z-plane, high-resolution, immunofluorescence microscopy [22]. In an early study, blood samples were obtained from 40 PCa patients (8 with localized disease and 32 with metastatic disease) for use with the NanoVelcro assay. CTCs were identified in all patients. Additional blood samples were obtained at intervals between 4-10 weeks of therapy and continued through a follow-up period of over 460 days. CTC counts mirrored the clinical behavior. Declines in counts was associated with a benefit from treatment whereas elevation of counts was associated with disease progression [22]. The 1st generation NanoVelcro chips, along the ICC protocol, have since been used primarily for CTC enumeration.
Figure 2.
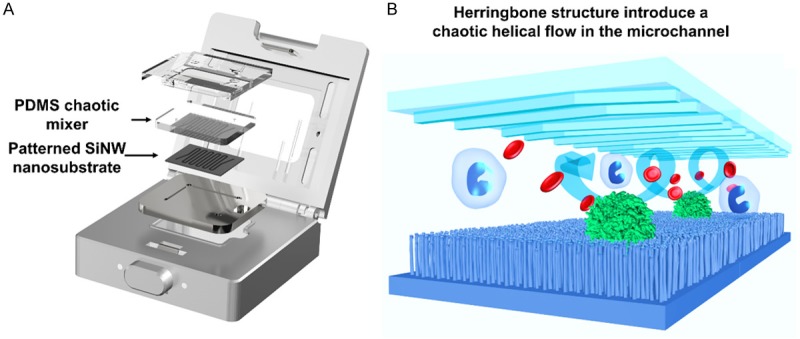
A and B. Configuration of 1st gen NanoVelcro Assay. Composed of silicon nanowire substrate (SiNS) with an overlay of microfluidic chaotic mixture to ensure vertical flow and enhanced cell capture capability.
2nd generation NanoVelcro assay: molecular profiling of single cells
Despite the capability of the 1st generation NanoVelcro chip to perform reproducible detection of CTCs, two challenges needed to be addressed: the limitations of EpCAM and the need to look beyond surface chemistry.
It is well recognized that EpCAM expression varies across the range of solid tumors. In PCa, during certain molecular transitions (e.g. epithelial-to-mesenchymal), EpCAM expression can be suppressed. Thus, we and others identified a need to consider other capture approaches (beyond EpCAM). Moreover, cell surface chemistry is clearly important, but there remains a wealth of information within the cell that goes well beyond cell surface proteins [33-35]. The 2nd generation NanoVelcro assay was designed to address both issues. Thereafter known as the NanoVelcro-LCM (laser capture microdissection) approach, this platform replaced the non-transparent SiNS with a transparent poly (lactic-co-glycolic acid), or PLGA, embedded substrate onto a commercially available laser microdissection (LMD) slide [22]. This was followed by streptavidin binding and conjugation to biotinylated capture agent coated target cancer cells from whole blood samples (Figure 3A). Our team explored anti-CD146, a melanoma specific capturing agent, to test the platform’s efficiency in capturing circulating melanoma cells (CMCs) (Figure 3B) [22]. We also established a new ICC staining protocol based on melanoma specific markers: Mart1 and High Molecular Weight-Melanoma Associated Antigen (HMW-MAA). CMCs are determined by DAPI+, Mart1+, HMW-MAA+, CD45- and 40 mm > cell diameter >10 mm, WBC are determined by DAPI+, Mart1-, HMW-MAA-, CD45+ and cell diameter <10 mm [22]. The 2nd generation platform mirrored the capture efficacy of the 1st generation NanoVelcro chips. Furthermore, the 2nd generation NanoVelcro substrate allowed for integration of LCM technology, where after capture and ICC validation, single CMCs were identified and harvested using laser capture microdissection (Figure 3C) [22]. After isolation, single CMCs underwent whole genome amplification (WGA) prior to sequencing. The team improved upon initial usage of PCR-based WGA by incorporating a multiple displaced amplification (MDA) reaction to amplify CMC DNA in long fragments, which reduced amplification error. This 2nd generation system was then used to study individual CTCs in PCa in a collaboration with the Beijing Genome Institute. We were able to conduct whole genome sequencing (WGS) on 4 CTCs with more than 95% coverage. Using available patient samples, our team showed a striking degree of molecular similarity between CTCs, metastatic tumors, and even the primary tumor [36].
Figure 3.
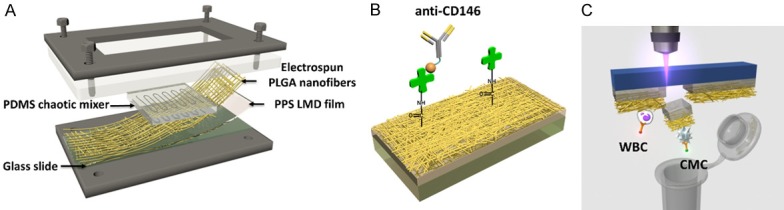
2nd gen NanoVelcro-LCM assay for single-CTC isolation. (A) Configuration of 2nd gen NanoVelcro chip focused on replacing non-transparent SiNS with transparent PLGA nanofibers [35] to allow for laser capture microdissections (LCM). (B) anti-CD146 used as capture agent for Circulating Melanoma Cells (CMC), streptavidin covalently conjugated to PLGA nanofibers mimicking the conjugation between streptavidin and SiNS nanowires of 1st gen NanoVelcro assays (C) Illustration of single cell isolation using LCM technology.
3rd and 4th generation NanoVelcro assay: rapid molecular profiling of viable CTCs
The shift towards molecular characterization and functional analyses drove the development of platforms that would enable CTC isolation at a lower cost and labor point, while still improving the quality of information that could be obtained [22]. The NanoVelcro-LCM platform, despite being able to isolate single CTCs, did not provide viable cells due to the fixation process necessary for ICC. The platform also entailed high labor costs associated with a somewhat complicated platform design [22]. Our group brought forward a 3rd generation NanoVelcro platform to address these concerns, which was named the ThermoResponsive NanoVelcro platform (TR-NanoVelcro) [37,38]. This approached involved grafting thermoresponsive polymer brushes (i.e. poly (N-isopropylacrylamide) PIPAAm) onto the SiNS. Additionally, biotin groups were fixed onto the polymers to allow for anti-EpCAM conjugation. The system takes advantage of PIPAAm conformational changes at 37°C and 4°C (Figure 4). The TR-NanoVelcro system demonstrated efficient capture and release of viable single CTC (>70% recovery and >90% viable at 4°C) [38]. H1975 anti-EpCAM positive non-small cell lung cancer (NSCLC) cells were used as the model system to verify this platform. Two rounds of capture and release on the TR-NanoVelcro platform resulted in improved CTC purification, allowing EGFR point mutation analysis [22]. Sanger sequencing from CTCs from 7 NSCLC patient whole blood samples revealed a molecular signature that strongly correlated with tumor tissues [22]. Ultimately, this assay demonstrated the ability to isolate pure and viable CTC with intact nucleic acid content. This platform can be adapted to create cancer cell lines from individual patients via ex vivo expansion, which can in turn be utilized for individualized drug therapy tests, creating more patient centric and patient specific oncological care [22].
Figure 4.
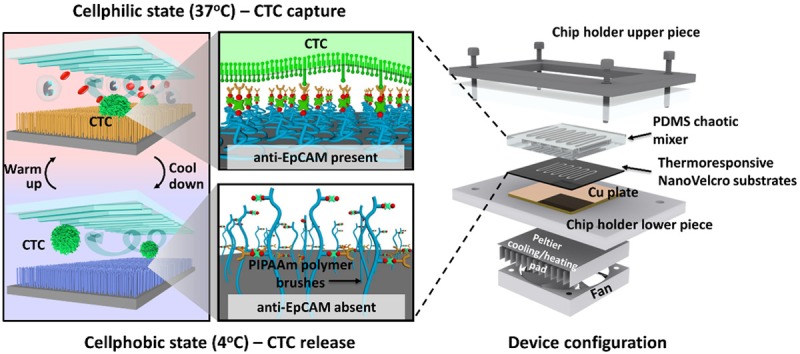
3rd gen ThermoResponsive NanoVelcro Assay. Illustration of temperature dependent conformational change of polymer brushes allowing for capture and release of purified and viable anti-EpCAM coated CTC at 37°C and 4°C respectively.
The 4th generation NanoVelcro assay looked to further improve upon the cell viability and purity. Named the “Sweet Chip”, the 4th generation assay focuses utilization of surface chemistry and competitive binding to enhance CTC capture and release [39]. This platform brings certain features forward: First, the fabrication of poly (3,4-etylene-dioxythiophene) (PEDOT) based nanomaterial on the chip surface provides highly specific CTC capture after antibody conjugation. Second, sorbitol is utilized as a competitive binding agent to PEDOT, allowing for gentle release captured CTCs (Figure 5). The isolated CTCs in this system retain well-preserved RNA transcripts, which allow for analysis of the gene expression of several PCa specific biomarkers (e.g. AR-FL, AR-V7, SchLAP1, FOLH1, KLK3). Along with the 3rd-generation assay, the Sweet Chip has been used in active research to understand the molecular nature of the CTC compartment.
Figure 5.
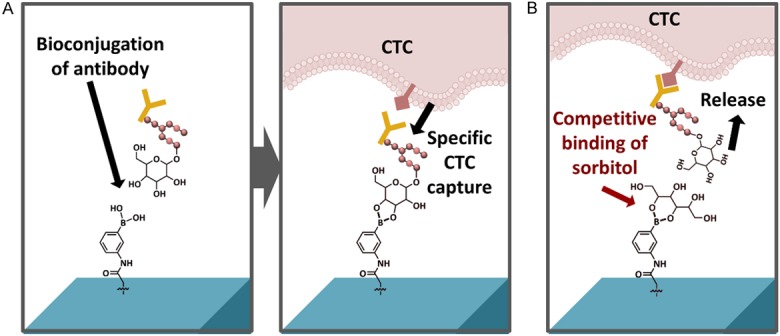
4th gen Sweet Chip NanoVelcro Assay. A. Capture antibody conjugates with phenylboronic acid (PBA) to immobilize and capture target cells. B. Sorbitol has stronger binding with PBA, allowing for gentle release of anti-EpCAM coated CTC with intact RNA integrity.
The existing and forthcoming versions of our assay (and other CTC assays) in the field represent a growing toolkit for biologists and engineers to use for studying the cellular information in the bloodstream. By studying liquid tissue (i.e. blood) in the same manner the field has studied solid tissue (i.e. biopsies and surgically resected material), we may further unravel the intricate biology of PCa and other malignancies. This give us an opportunity to study problems not well addressed by current approaches.
Visceral metastases (VM) in prostate cancer: an important clinical problem
Visceral metastases are important clinical events that, upon detection, evoke concerns from treating oncologists and patients. Their appearance points toward organ failure from metastatic lesions and a short duration of survival for the patient in question. The typical approach of monitoring serum prostate specific antigen (PSA) is not an effective means of monitoring for these important clinical events [32]. Less than 20% of patients who present with visceral metastasis are detected by standard clinical and/or radiographic detection methods [32]. As such, these VM events are typically caught late- i.e. at a time when intervention may not be able to substantially alter clinical course. This problem is likely to become more urgent now that newer, more potent androgen receptor signaling inhibitors are being used early in the natural history of this disease. It is likely that the biology we have associated with late-stage, metastatic, castration-resistant PCa may be altered substantially by the biological pressures on the AR axis. As such, dealing with treatment refractory PCa in the 21st century may no longer look like the “hormone-refractory” prostate cancer studied in the 1980s and 1990s.
Using a liquid biopsy to study VM
During the evolution of the CTC field, our group and others recognized that the pool of CTCs was extremely heterogeneous with respect to the shape of those captured cells from any given patient. This heterogeneity seemed to change over time and in the face of various treatments. With refinements in microscopy, it became readily apparent that classical pathological approaches could be used to describe various subsets of cells within this pool [40,41]. It has been proposed that morphologic analysis may have prognostic value in a fashion akin to what is done with tissue-based analysis.
Liquid biopsies provide a means to gather real time patient status through minimally invasive, serially obtainable, and highly sensitive manner. The 1st generation NanoVelcro ICC assay has proven itself to be highly efficient in the capture and enumeration of CTCs from patient whole blood. This assay also preserves nuclear morphology extremely well through the capture and enumeration processes. As such, it is possible to use this approach to efficiently study nuclear morphology without the use of large volumes of imaging data. Since CTCs on the 1st generation chip sit on a single horizontal plane of focus, the chip may be imaged with a single automated sweep for focused image capture well suited for human review [32]. Using this system, we identified 3 distinct populations of PCa CTCs based on nuclear size that appear to associate with variations in metastatic patterns.
vsnCTCs and visceral metastases
Our group identified and characterized CTCs from annotated specimens from a sample of PCa patients that spanned the range of metastatic burden and sites. Donning a classic pathology mindset, we sought to identify any potential relationships between cellular shape and clinical behavior. In considering various features that could be used, the simplest and most reproducible was nuclear size.
We assembled 148 serial blood samples from 57 PCa patients. Samples were heavily annotated with respect to clinical outcome and disease state. Patients were classified by the location of metastatic states: non-metastatic, non-visceral (osseous and/or nodal), and visceral (hepatic and/or pulmonary) [32]. Between 1 and 11 enumeration studies were completed for each patient. Imaging was performed using upright fluorescence microscope (Eclipse 90i, Nikon) [32] under 40× magnification with DAPI, FITC, and TRITC corresponding with nuclear, CK, and CD45 staining, respectively. Selected CTCs were furthered scanned at 100× or 400× magnification before manual pathological review (Figure 6A) [32]. This analysis identified 3 different populations of CTCs based on nuclear size: large nuclear CTC (lnCTC, nuclear size >15 µm), small nuclear CTC (snCTC, nuclear size 8.5 µm < nuclear size <15 µm), and very small nuclear CTC (vsnCTC, nuclear size <8.5 µm) (Figure 6B) [32]. Patients without metastatic lesions had CTCs classified mostly as lnCTC (62%). Patients with bone and nodal metastases has CTCs classified mostly as snCTC (51%). Strikingly, those patients with visceral metastases had CTCs largely composed of vsnCTCs (65%) [32]. Overall, snCTC + vsnCTC counts correlated strongly with metastatic patients, with vsnCTC counts being able to specifically distinguish between non-visceral and visceral metastatic patient cohorts (Figure 6C) [32]. In the published study, there were patients with metastatic PCa who developed VM. Twelve of the 15 patients blood collections initiated when VM were already present. An additional 3 patients started blood collections when they had only osseous and/or nodal metastasis. These 3 patients later developed VM. vsnCTCs were detected in all 15 of these patients. Furthermore, in the 3 patients who developed VM during the course of their blood collections, vsnCTCs were detected prior to radiographic detection of VM with a lead time of between 104 to 196 days [32]. These particular CTCs show strong correlation with VM in PCa and are being further studied as a prognostic biomarker of disease progression.
Figure 6.
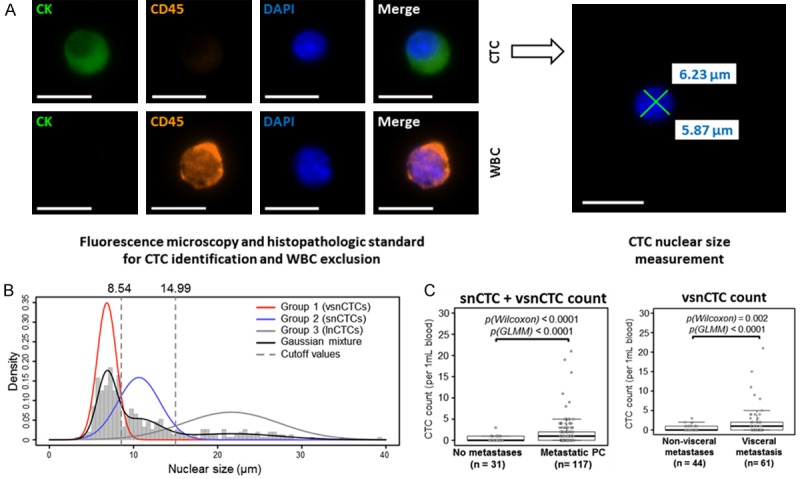
Sub-classification of CTCs based on nucleus size. A. Captured cells from whole blood of 57 prostate cancer patients undergo Immunocytochemistry (ICC) staining before scanning and manual pathological review for CTC confirmation [32]. Confirmed CTCs nuclear dimensions are measured. B. Sub-classification of three groups of CTCs based on nucleus size: large nucleus CTC (lnCTC, nuclear size >15 µm), small nucleus CTC (snCTC, nuclear size 8.5 µm < nuclear size <15 µm), and very small nucleus CTC (vsnCTC, nuclear size <8.5 µm). C. Statistically significant higher snCTC + vsnCTC counts were found in metastatic prostate cancer (PC) patients comparing with non-metastatic PC patients. Also, statistically significant higher vsnCTC counts were found in viceral metastasis patients comparing with non-visceral metastasis patients [32].
Conclusion
The breakthroughs in molecular and cellular biology allowed for tremendous advancement in the field of prostate cancer. While many innovations are focused on understanding genetics and genomics of prostate cancer, the foundation of classical pathology still plays a crucial role in the current oncology practice. Undoubtedly, aberrations at the DNA, RNA, and protein level are important to our understanding of cancer. At the same time, classical shape analysis still provides useful information without the requirement for complicated informatics or technology. By applying classical pathological and histomorphometric analysis to the cells in circulation, we and others have begun to more fully appreciate the value of circulating tumor cells from a “liquid biopsy”. The merging of modern technology and classical analytics makes it possible to obtain large amounts of disease information without incurring great costs or suffering to the patient.
Dr. Coffey spent much of his career speaking with students about the importance of chaos, complexity, and self-organization [42-44]. One central theme of his teachings was the intimate relationship between shape, genomics, and function. This concept runs from the macroscopic organism to organs, cells, and even to an individual chromosome. Our efforts to understand the relationship between circulating tumor cells, their shape, and their respective clinical outcome follow the grand tradition inspired by Dr. Coffey’s doctrines.
We hope by combining emerging technology with classical and novel concepts, we may be able to provide creative solutions to patients with PCa and other malignancies. The frontiers of science in PCa have now been assailed with cutting-edge nanotechnology. We believe that this field will continue to grow at a rapid pace allowing for fruitful collaborations between physicians, biomedical engineers, and cancer biologists. By doing so, we realize a vision set forth by The Chief, himself- a grand merging of engineering, physics, biology, medicine, and ultimately philosophy. While this is a small effort in that spirit, we among the army left behind by Dr. Donald S Coffey continue to heed his battle cry in our efforts to defeat prostate cancer: “Charge!!!”.
References
- 1.Nelson WG, Pienta KJ, Barrack ER, Coffey DS. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem. 1986;15:457–475. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- 2.Mohler JL, Partin AW, Coffey DS. Correlation of prognosis to nuclear roundness and to flow cytometric light scatter. Anal Quant Cytol Histol. 1987;9:156–164. [PubMed] [Google Scholar]
- 3.Pienta KJ, Partin AW, Coffey DS. Cancer as a disease of DNA organization and dynamic cell structure. Cancer Res. 1989;49:2525–2532. [PubMed] [Google Scholar]
- 4.Pienta KJ, Coffey DS. A structural analysis of the role of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J Cell Sci Suppl. 1984;1:123–135. doi: 10.1242/jcs.1984.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- 5.Partin AW, Getzenberg RH, CarMichael MJ, Vindivich D, Yoo J, Epstein JI, Coffey DS. Nuclear matrix protein patterns in human benign prostatic hyperplasia and prostate cancer. Cancer Res. 1993;53:744–746. [PubMed] [Google Scholar]
- 6.Mohler JL, Partin AW, Lohr WD, Coffey DS. Nuclear roundness factor measurement for assessment of prognosis of patients with prostatic carcinoma. I. Testing of a digitization system. J Urol. 1988;139:1080–1084. doi: 10.1016/s0022-5347(17)42791-1. [DOI] [PubMed] [Google Scholar]
- 7.Partin AW, Steinberg GD, Pitcock RV, Wu L, Piantadosi S, Coffey DS, Epstein JI. Use of nuclear morphometry, gleason histologic scoring, clinical stage, and age to predict disease-free survival among patients with prostate cancer. Cancer. 1992;70:161–168. doi: 10.1002/1097-0142(19920701)70:1<161::aid-cncr2820700126>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 8.Partin AW, Walsh AC, Pitcock RV, Mohler JL, Epstein JI, Coffey DS. A comparison of nuclear morphometry and Gleason grade as a predictor of prognosis in stage A2 prostate cancer: a critical analysis. J Urol. 1989;142:1254–1258. doi: 10.1016/s0022-5347(17)39049-3. [DOI] [PubMed] [Google Scholar]
- 9.Veltri RW, Miller MC, Partin AW, Coffey DS, Epstein JI. Ability to predict biochemical progression using Gleason score and a computer-generated quantitative nuclear grade derived from cancer cell nuclei. Urology. 1996;48:685–691. doi: 10.1016/S0090-4295(96)00370-6. [DOI] [PubMed] [Google Scholar]
- 10.Veltri RW, Partin AW, Epstein JE, Marley GM, Miller CM, Singer DS, Patton KP, Criley SR, Coffey DS. Quantitative nuclear morphometry, Markovian texture descriptors, and DNA content captured on a CAS-200 Image analysis system, combined with PCNA and HER-2/neu immunohistochemistry for prediction of prostate cancer progression. J Cell Biochem Suppl. 1994;19:249–258. [PubMed] [Google Scholar]
- 11.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Hognas G, Annala M, Kivinummi K, Goody V, Latimer C, O’Meara S, Dawson KJ, Isaacs W, Emmert-Buck MR, Nykter M, Foster C, Kote-Jarai Z, Easton D, Whitaker HC ICGC Prostate Group. Neal DE, Cooper CS, Eeles RA, Visakorpi T, Campbell PJ, McDermott U, Wedge DC, Bova GS. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O’Donovan M, Malhotra S, di Pietro M, Ivakhno S, He M, Weaver JMJ, Lynch AG, Kingsbury Z, Ross M, Humphray S, Bentley D, Fitzgerald RC Oesophageal Cancer C, Molecular Stratification Study G, Oesophageal Cancer C and Molecular Stratification OSG. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038–1046. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, Zia A, Fox NS, Livingstone J, Shiah YJ, Wang J, Beck TA, Have CL, Chong T, Sam M, Johns J, Timms L, Buchner N, Wong A, Watson JD, Simmons TT, P’ng C, Zafarana G, Nguyen F, Luo X, Chu KC, Prokopec SD, Sykes J, Dal Pra A, Berlin A, Brown A, Chan-Seng-Yue MA, Yousif F, Denroche RE, Chong LC, Chen GM, Jung E, Fung C, Starmans MH, Chen H, Govind SK, Hawley J, D’Costa A, Pintilie M, Waggott D, Hach F, Lambin P, Muthuswamy LB, Cooper C, Eeles R, Neal D, Tetu B, Sahinalp C, Stein LD, Fleshner N, Shah SP, Collins CC, Hudson TJ, McPherson JD, van der Kwast T, Bristow RG. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 16.Kreso A, O’Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, Dunant C, Pollett A, Gallinger S, McPherson J, Mullighan CG, Shibata D, Dick JE. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, Gold KA, Kalhor N, Little L, Mahadeshwar H, Moran C, Protopopov A, Sun H, Tang J, Wu X, Ye Y, William WN, Lee JJ, Heymach JV, Hong WK, Swisher S, Wistuba II, Futreal PA. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donelli MG, Rosso R, Garattini S. Quantitative studies on cancer dissemination. Cancer Res. 1969;29:414–418. [PubMed] [Google Scholar]
- 19.Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38:2651–2660. [PubMed] [Google Scholar]
- 20.Greene HS. A method for determining the presence of tumor cells in blood and organs of experimental animals and Its application to the problems of metastases and retention in organs: a review. Acta Cytol. 1965;9:160–168. [PubMed] [Google Scholar]
- 21.Liotta LA, Kleinerman J, Saidel GM. Quantitative relationships of intravascular tumor cells, tumor vessels, and pulmonary metastases following tumor implantation. Cancer Res. 1974;34:997–1004. [PubMed] [Google Scholar]
- 22.Chen JF, Zhu Y, Lu YT, Hodara E, Hou S, Agopian VG, Tomlinson JS, Posadas EM, Tseng HR. Clinical applications of NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Theranostics. 2016;6:1425–1439. doi: 10.7150/thno.15359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 24.Negin BP, Cohen SJ. Circulating tumor cells in colorectal cancer: past, present, and future challenges. Curr Treat Options Oncol. 2010;11:1–13. doi: 10.1007/s11864-010-0115-3. [DOI] [PubMed] [Google Scholar]
- 25.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 26.Green BJ, Saberi Safaei T, Mepham A, Labib M, Mohamadi RM, Kelley SO. Beyond the capture of circulating tumor cells: next-generation devices and materials. Angew Chem Int Ed Engl. 2016;55:1252–1265. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- 27.Jan YJ, Chen JF, Zhu Y, Lu YT, Chen SH, Chung H, Smalley M, Huang YW, Dong J, Yu HH, Tomlinson JS, Hou S, Agopian VG, Posadas EM, Tseng HR. NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Advanced Drug Delivery Reviews. 2018 doi: 10.1016/j.addr.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc Chem Res. 2014;47:2941–2950. doi: 10.1021/ar5001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer KE, Aleman BJ, Tao SL, Hugh Daniels R, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Biomimetic nanowire coatings for next generation adhesive drug delivery systems. Nano Lett. 2009;9:716–720. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Asghar W, Demirci U, Wan Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today. 2013;8:347–387. doi: 10.1016/j.nantod.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Chaotic mixer for microchannels. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 32.Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA, Chen SF, Liang AJ, Hodara E, Zhau HE, Hou S, Ahmed RS, Luthringer DJ, Huang J, Li KC, Chung LW, Ke Z, Tseng HR, Posadas EM. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer. 2015;121:3240–3251. doi: 10.1002/cncr.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M, Li F, Wu K, Wu H, Lichterman J, Wan H, Lu CL, OuYang W, Ni M, Wang L, Li G, Lee T, Zhang X, Yang J, Rettig M, Chung LW, Yang H, Li KC, Hou Y, Tseng HR, Hou S, Xu X, Wang J, Posadas EM. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015;6:44781–93. doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang WH, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LW, Rettig M, Tseng HR, Posadas EM. High-purity prostate circulating tumor cell isolation by a polymer nanofiber-embedded microchip for whole exome sequencing. Adv Mater. 2013;25:2897–2902. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew Chem Int Ed Engl. 2013;52:3379–3383. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovmar L, Fredriksson M, Liljedahl U, Sigurdsson S, Syvanen AC. Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic Acids Res. 2003;31:e129. doi: 10.1093/nar/gng129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo SC, Tseng HR, Yu HH. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv Mater. 2013;25:1547–1551. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, Liang AJ, Chen SF, Li Q, Fang W, Zhang P, Garcia MA, Lee T, Song M, Lin HA, Zhao H, Luo SC, Hou S, Yu HH, Tseng HR. Programming thermoresponsiveness of NanoVelcro substrates enables effective purification of circulating tumor cells in lung cancer patients. ACS Nano. 2015;9:62–70. doi: 10.1021/nn5056282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen MY, Chen JF, Luo CH, Lee S, Li CH, Yang YL, Tsai YH, Ho BC, Bao L, Jan YJ, Zhu Y, Cheng S, Feng FY, Chen P, Hou S, Agopian V, Hsiao YS, Tseng HR, Posadas EM, Yu HH. Glycan-stimulation enables purification of prostate cancer circulating tumor cells on PEDOT NanoVelcro chips for RNA biomarker detection. Adv Healthc Mater. 2018:7. doi: 10.1002/adhm.201700701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee KB, Wang H, Yu ZT, Lu YT, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff JA, Kornblum HI, Yong WH, Graeber TG, Tseng HR. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamei K, Ohashi M, Gschweng E, Ho Q, Suh J, Tang J, For Yu ZT, Clark AT, Pyle AD, Teitell MA, Lee KB, Witte ON, Tseng HR. Microfluidic image cytometry for quantitative single-cell profiling of human pluripotent stem cells in chemically defined conditions. Lab Chip. 2010;10:1113–1119. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommerfeld HJ, Meeker AK, Piatyszek MA, Bova GS, Shay JW, Coffey DS. Telomerase activity: a prevalent marker of malignant human prostate tissue. Cancer Research. 1996;56:218. [PubMed] [Google Scholar]
- 43.Posadas EM, Criley SR, Coffey DS. Chaotic oscillations in cultured cells: rat prostate cancer. Cancer Research. 1996;56:3682. [PubMed] [Google Scholar]
- 44.Coffey DS. Self-organization, complexity and chaos: the new biology for medicine. Nature Medicine. 1998;4:882. doi: 10.1038/nm0898-882. [DOI] [PubMed] [Google Scholar]
- 45.Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S, Song M, Xu X, Ouyang WH, Ouyang WW, Lichterman J, Luo Z, Xuan X, Huang J, Chung LW, Rettig M, Tseng HR, Shao C, Posadas EM. NanoVelcro Chip for CTC enumeration in prostate cancer patients. Methods. 2013;64:144–152. doi: 10.1016/j.ymeth.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]


