Abstract
CRISPR–Cas9 is a versatile RNA-guided genome editing tool. Here we demonstrate that partial replacement of RNA nucleotides with DNA nucleotides in CRISPR RNA (crRNA) enables efficient gene editing in human cells. This strategy of partial DNA replacement retains on-target activity when used with both crRNA and sgRNA, as well as with multiple guide sequences. Partial DNA replacement also works for crRNA of Cpf1, another CRISPR system. We find that partial DNA replacement in the guide sequence significantly reduces off-target genome editing through focused analysis of off-target cleavage, measurement of mismatch tolerance and genome-wide profiling of off-target sites. Using the structure of the Cas9–sgRNA complex as a guide, the majority of the 3′ end of crRNA can be replaced with DNA nucleotide, and the 5 - and 3′-DNA-replaced crRNA enables efficient genome editing. Cas9 guided by a DNA–RNA chimera may provide a generalized strategy to reduce both the cost and the off-target genome editing in human cells.
The CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 system is a powerful genome editing tool for biology and medicine1–4, and has potential utility for treating a wide range of diseases5. crRNA guides Cas9, a DNA endonuclease, to targeted DNA sequences by forming a two-component RNA structure with transactivating crRNA (tracrRNA)4. Alternatively, crRNA and tracrRNA can be engineered into a single-guide RNA (sgRNA) to guide Cas9 proteins4. The 20 nucleotides at the 5′ end of crRNA or sgRNA hybridize with the complementary DNA sequences through Watson–Crick base pairing between RNA and target DNA1–4. Recognition of the target sequences and the nearby protospacer adjacent motif (PAM) leads to site-specific double-stranded DNA breaks (DSB) produced by Cas9, which can be repaired by nonhomologous end-joining (NHEJ) or homology-directed repair (HDR)3. CRISPR–Cas9 is considered to be an RNA-guided endonuclease1–4. Some members of another family of well-established RNA-guided enzymes, Argonaute (Ago), have been shown to tolerate DNA as a guide6,7. Thus, it is important to understand whether or not CRISPR–Cas9 can use DNA as a guide.
CRISPR–Cas9-mediated genome editing can cause off-target mutations8–13. Multiple strategies for improving its specificity have been developed, including a nickase version of Cas9, structure-guided mutations of the Cas9 protein, fusion of deactivated Cas9 with FokI nuclease, fusion of Cas9 with a programmable DNA-binding domain, and truncated guide RNAs14–21. Most methods to reduce off-target mutations rely on re-engineering the Cas9 protein. Although shortened guide sequences, ranging from 20 to 17 nucleotides, were reported to reduce off-target mutations, they may also decrease the on-target cleavage by Cas9 (ref. 21). Chemically modified crRNA and sgRNA have been developed to enhance efficiency in cells22,23. However, use of chemical modification to reduce off-target effects has not been demonstrated.
Here we report that guide sequences partially composed of DNA nucleotides can direct Cas9 to induce efficient genome editing in human cells. Partial replacement with DNA nucleotides leads to decreased off-target activity compared to the unmodified guide sequence, but similar levels of on-target gene editing activity. Structure guided replacement with DNA nucleotides at both 5′ and 3′ of crRNA maintained its activity in cells. We believe that DNA–RNA chimeric guides may provide a generalized strategy to reduce both the cost and the off-target genome editing by CRISPR–Cas9 in human cells.
RESULTS
DNA–RNA chimeric guides enable efficient genome editing
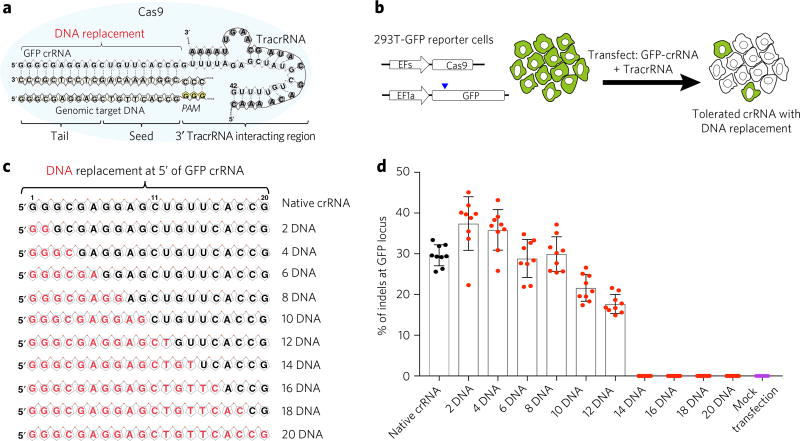
To accelerate the process of guide sequence evaluation, we used a cell reporter system that measures the editing efficiency of various modified crRNAs. HEK293T cells were infected by lentiviruses to constitutively express GFP and Streptococcus pyogenes Cas9 (SpCas9) (Fig. 1a,b)24. Transfecting the cells with a functional 42-nucleotide crRNA targeting GFP and a 75-nucleotide tracrRNA induces frame-shifting indel formations at the GFP site and abrogates the expression of GFP (Fig. 1a,b). Thus, if partial replacement with DNA in the guide sequence is not well tolerated, it will be less efficient than the unmodified RNA at reducing GFP expression in cells.
Figure 1. Partial DNA replacement at the guide region of a GFP crRNA induces gene editing in human cells.
(a) Diagram of the CRISPR system. PAM, protospacer adjacent motif. (b) HEK293T cells stably expressing both elongation factor-1 short (EFs) promoter-SpCas9 and elongation factor 1-α (EF1α) promoter-GFP were transfected with a crRNA targeting GFP and the tracrRNA. Cas9-mediated frame-shift NHEJ yields GFP-negative cells. When replacement of DNA nucleotides in crRNA is tolerated by Cas9, the percent of GFP-negative cells will be retained. The blue arrowhead indicates the cutting site by the Cas9. (c) Illustration of DNA replacement at the guide sequence of GFP crRNAs. The 20-nucleotide (nt) guide region is shown. RNA and DNA are shown in black and red, respectively. (d) HEK293T cells described above were incubated with the tracrRNA and a GFP-targeting crRNA illustrated. TIDE analysis was performed to calculate the percent of indels at GFP locus at day 3. n = 9 biologically independent samples. Error bars show mean ± s.d. Purple color indicates mock-transfection-treated samples. Black dots indicate native crRNA transfected samples. Red dots indicate DNA–RNA chimeric crRNAs-transfected samples.
The crystal structure of Cas9–sgRNA indicates that RNA at the seed region (ten nucleotides at the 3′ end of the guide sequence) is essential for Cas9–sgRNA binding and recognition of targeted DNA25,26. In contrast, the tail region (ten nucleotides at the 5′ end) of the guide sequence interacts with Cas9 less than the seed region25,26. We hypothesized that the guide sequence may tolerate partial DNA replacement based on the structure of Cas9 and guide RNA25,26. To test our hypothesis, we first synthesized unmodified crRNAs (native crRNA) on the basis of a previously identified guide sequence targeting GFP27. A number of crRNAs with the same guide sequence, but varying degrees of DNA substitutions at the 5′ end, were synthesized (Fig. 1c). When evaluated in HEK293T cells, native crRNA targeting GFP generated 27% ± 2% GFP-negative cells (Supplementary Fig. 1a,b). We found that replacing two, four, six, eight or ten RNA nucleotides with DNA nucleotides starting from the 5′ end of the guide sequences also generated similar levels of GFP-negative cells (ranging from 22% to 31%) (Supplementary Fig. 1a,b), indicating that partial replacement of up to ten-nucleotide DNA at the 5′ end of the guide sequence was tolerated. However, replacement with 12-nucleotide DNA generated significantly fewer GFP-negative cells than the native crRNA, and replacement with 14 or more DNA nucleotides reduced GFP-negative cells to background levels (~2%) (Supplementary Fig. 1a,b). The efficiency of genome editing after DNA nucleotide replacement at the guide sequence was confirmed by tracking of indels by decomposition (TIDE) analysis28 of the PCR amplicons from the GFP genomic locus (Fig. 1d).
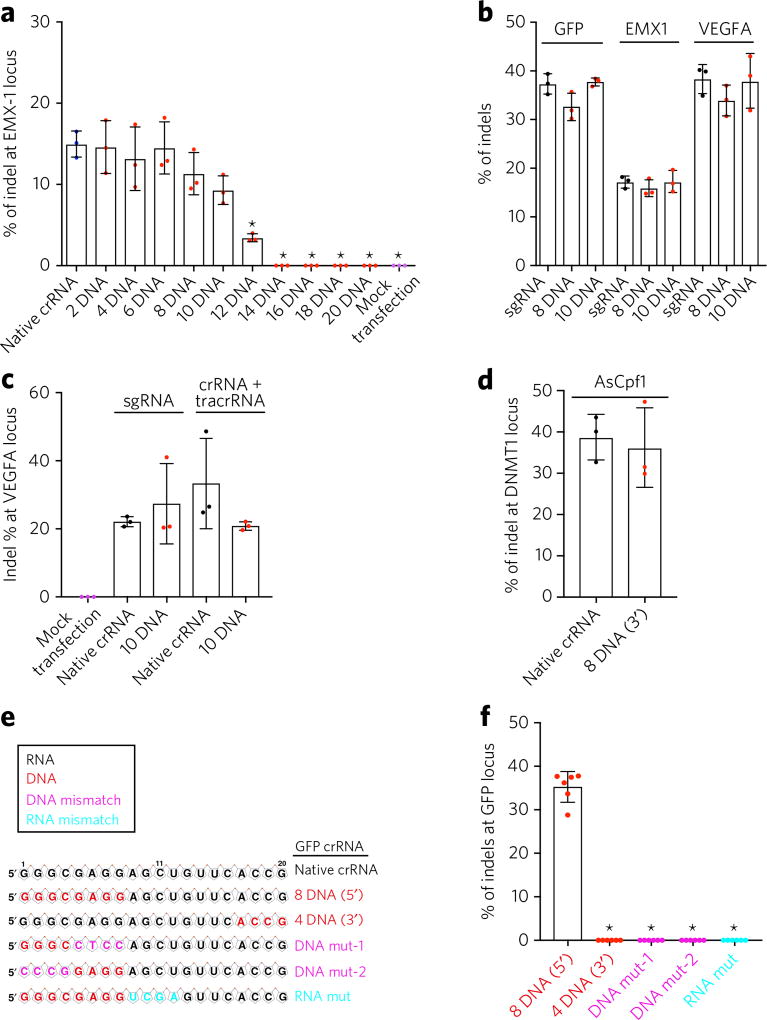
To study the effects of DNA nucleotide replacement on an endogenous gene, we synthesized crRNAs targeting the human gene EMX1 (empty spiracles homeobox 1)29, with various levels of DNA replacement at the 5′ end. After transfecting a crRNA targeting EMX1 and the tracrRNA into HEK293T cells stably expressing SpCas9, we found that replacing two, four, six, eight or ten RNA nucleotides with DNA nucleotides induced frequencies of indel formation comparable to those of the native crRNA (Fig. 2a). In contrast, replacing 12 or more RNA nucleotides with DNA significantly decreased or abolished the editing activity of EMX1 crRNA (Fig. 2a). The efficiency of genome editing at the EMX1 locus was further examined by the Surveyor nuclease assay30, indicating that replacing two to ten RNA nucleotides with DNA induced indels (Supplementary Fig. 2).
Figure 2. Partial DNA replacement at the guide region of crRNA or sgRNA induces efficient gene editing in human cells.
(a) Partial DNA replacement at the guide region of a crRNA targeting EMX1 induced indels in human cells. HEK293T cells described above were transfected with the tracrRNA and an EMX1-targeting crRNA. TIDE analysis was performed to determine indels at EMX1 locus. n = 3 biologically independent samples. *P < 0.01 by one-way ANOV A with Tukey post hoc test. (b) sgRNAs targeting GFP, EMX1 or VE GFA with 8-nt DNA and a 10-nt replacement at the 5′ end (sgRNA-8 DNA and sgRNA-10 DNA) induced indels in HEK293T cells. HEK293T cells described above were transfected with a native sgRNA or sgRNA-8D or sgRNA-10D. TIDE analysis was performed to determine percent of indels. n = 3 biologically independent samples. Black and red dots indicate native crRNA and DNA–RNA chimeric crRNAs transfected samples, respectively. (c) DNA–RNA chimeric crRNAs or sgRNA can mediate efficient genome editing in a RNP setting. VEGFA crRNA + tracrRNA or sgRNA alone were incubated with SpCas9 protein to form RNP complexes and then electroporated into Jurkat T cells by Neon transfection. Genomic DNA was harvested at day 3, and percent of indels was measured by TIDE. n = 3 biologically independent samples. (d) A DNA–RNA chimeric crRNA guides AsCpf1 for efficient genome editing. HEK293T cells were co-transfected with a plasmid expressing AsCpf1 and a native crRNA or crRNA with 8-nt DNA replacement at the 3′ end targeting DNMT1. TIDE analysis was performed to determine percent of indels at the DNMT1 locus 3 d after transfection. n = 3 biologically independent samples. (e,f) 4-nt DNA replacement at the 3′ end of the guide sequence (red), 4-nt mutation of DNA (DNA mut-1 or 2, pink) at the 5′ end, or 4-nt mutation of RNA (RNA mut, cyan) abolished activity of CRISPR in human cells. (e) Illustration of DNA replacement at the 3′ end and DNA or RNAmutations. (f) HEK293T cells described above were incubated with the tracrRNA and a GFP-targeting crRNA as in e. *P < 0.01; n = 6 biologically independent samples. Error bars represent mean ± s.d.
To investigate whether partial DNA replacement can be tolerated by sgRNA, we synthesized sgRNAs targeting GFP, EMX1 and vascular endothelial growth factor A (VEGFA), with and without DNA replacement24. After transfecting the sgRNAs in HEK293T reporter cells, we found that the native sgRNAs and sgRNAs with substitution of eight DNA and ten DNA nucleotides in the 5′ end generated similar levels of indels in cells for all three guide sequences (Fig. 2b). These data show that partial DNA replacement of both crRNA and sgRNA retains on-target genome editing.
To investigate whether DNA–RNA chimeric crRNAs or sgRNA can be applied in therapeutic delivery, we incubated VEGFA 10 DNA (replacing ten RNA nucleotides with DNA nucleotides in the 5’ end) crRNA and a tracrRNA with Cas9 protein or VEGFA 10 DNA sgRNA with Cas9 protein to form ribonucleoprotein (RNP). We found that they each induced levels of indels similar to those of native crRNA or sgRNA after RNP electroporation in Jurkat T cells (Fig. 2c).
To test whether DNA replacement works with other Cas enzymes, we synthesized Acidaminococcus sp. Cpf1 (AsCpf1) native crRNA targeting DNMT1 (ref. 31) or crRNA with an eight-nucleotide DNA replacement at the 3′ end (as the guide sequence of Cpf1 crRNA is at 3′ end). HEK293T cells were transfected with a plasmid expressing AsCpf1 protein followed by crRNAs after 24 h. The Cpf1 crRNA with partial DNA replacement fully maintains its activity in human cells (Fig. 2d).
To investigate whether the DNA replacement could be introduced at the 3′ end of the guide sequence, four RNA nucleotides were replaced with DNA at the 3′ end of the guide sequences (Fig. 2e,f). This modified crRNA did not induce indels of GFP (Fig. 2f), indicating that replacement of RNA with DNA nucleotides at the 3′ end of a guide sequence is not well tolerated.
To further evaluate the potential of DNA nucleotide substitution in the guide sequence, we synthesized GFP crRNAs with eight DNA nucleotides at the 5′ end (GFP-8DNA), but mutated four nucleotides of either the DNA or RNA sequence (Fig. 2e). As expected, mutation of four RNA nucleotides (positions 9–12 from the 5′ end) fully abolished its genome-editing activity (Fig. 2f). We also found that mutations of four DNA nucleotides (positions 1–4 or 5–8 from the 5′ end) eliminated the activity of chimera crRNA, indicating that complementarity of DNA sequences in the guide region plays an essential role in recognizing genomic DNA sequences.
DNA–RNA chimeric guides reduce off-target editing
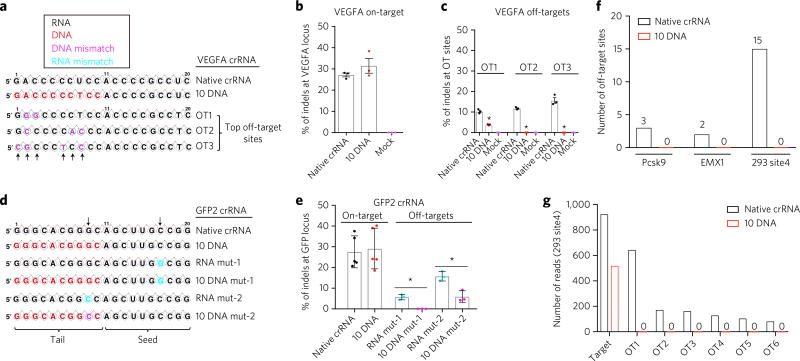
Previous studies have reported that truncation of the RNA guide sequence to 17 RNA nucleotides instead of 20 RNA nucleotides decreases off-target activity of CRISPR–Cas9 (ref. 21). It was also reported that some truncated guide sequences reduce on-target editing21. Because binding of duplexes of DNA–DNA are less thermodynamically stable than those of DNA–RNA duplexes32,33, a partially DNA-substituted crRNA guide sequence is less likely to tolerate mismatches when undergoing base pairing with genomic DNA. We therefore hypothesized that DNA–DNA binding between the crRNA, with a 5′ end DNA replacement and a target genomic DNA strand, might reduce off-target editing. We synthesized crRNAs targeting a site within VEGFA (Fig. 3a) that is known to have high off-target frequencies in the human genome24.
Figure 3. Partial DNA replacement at the guide region reduces off-target effects in human cells.
(a) Illustration of DNA replacement at the guide sequence of VEGFA crRNA. Arrows denote mismatches between target and off-target sites. (b,c) HEK293T cells described in Figure 1 were transfected with the tracrRNA and a VEGFA-targeting crRNA with 10 DNA nt replacement at the 5′ end of the guide sequence (10 DNA) or with native crRNA. Surveyor assay were performed to determine indels at the VEGFA locus (b) and 3 top off-target (OT) sites of the VEGFA guide sequence (c). n = 3 biologically independent samples. *P < 0.01 by one-way ANOV A with Tukey post hoc test. (d,e) Partial DNA replacement in GFP crRNA reduces off-target activity. (d) Illustration of mismatch mutations of GFP2 sequences. Arrows denote point mutations. (e) TIDE analysis was performed to determine percent of indels. n = 3–5 biologically independent samples, as indicated. *P < 0.05 by unpaired, two-tailed Student’s t-tests. (f) GUIDE-seq genome-wide off-target analysis of native and 10 DNA crRNAs from three endogenous genes. The chart indicates the number of off-target peaks detected by GUIDE-seq for each type of crRNA. Six total mismatches are allowed in the guide and PAM. (g) Number of GUIDE-seq reads of 293 site 4. Target is the crRNA target site. OT 1–OT 6 are top off-target sites in the native crRNA data set. Error bars represent mean ± s.d.
We found that the native VEGFA crRNA and the 10 DNA crRNA generated similar levels of indels at the on-target genomic locus (Fig. 3b), further demonstrating that replacement of RNA with DNA was tolerated by various guide sequences. Interestingly, VEGFA 10 DNA crRNA generated no detectable indels at two out of three of the top off-target sites and produced less than 4% indels at another site, whereas native crRNA generated 10–15% off-target indels (Fig. 3c and Supplementary Fig. 3). To compare the off-target effects of a crRNA with partial DNA replacement to those of a crRNA with a truncated 17-nucleotide guide sequence, which has been shown to reduce off-target effects21, we synthesized a VEGFA crRNA with a 17-nucleotide guide sequence. We found that the crRNA that had ten DNA nucleotides replaced showed levels of off-target site indel formation comparable to those of the truncated guide RNA (Supplementary Fig. 4).
We hypothesized that if DNA replacement at the guide sequence provides higher specificity than native crRNA, the chimeric crRNAs would be less tolerant to mismatches than native crRNA. When both native GFP crRNA and GFP-8DNA were mutated at 2–4 nucleotides of the tail region of guide sequences, their genome-editing activities were abolished (Fig. 2e,f, and Supplementary Fig. 5). Importantly, native GFP crRNA, but not GFP-8DNA, tolerated single-nucleotide mismatch in the tail region, as demonstrated by the slight reduction of indels by native GFP crRNA with one nucleotide mutation in the tail region of crRNA compared to the lack of indels produced by GFP-8DNA with one nucleotide mutation (Supplementary Fig. 5). To investigate whether DNA replacement could enhance the specificity of other guide sequences, we studied another GFP guide sequence named GFP2 (Fig. 3d)24. One nucleotide mismatch in the GFP2 guide sequence has also been shown to mediate off-target genome editing24. GFP2 native crRNA and crRNA with ten DNA nucleotides replaced at the 5′ end (GFP2-10 DNA) generated similar levels of indels at the GFP site (Fig. 3d,e). To compare ‘simulating’ off-target editing of GFP2 native crRNA and GFP2-10 DNA, we introduced a point mutation at either the seed or the tail region of GFP2 crRNA and GFP2-10 DNA to create a mismatch between the guide sequence and the target GFP sequence (Fig. 3d). If DNA replacement can enhance specificity and reduce off-target editing, then GFP2-10 DNA would be less tolerant of mismatches than the native GFP crRNA. We found that mutations on GFP2 in native crRNAs induced substantial indels of GFP in human cells (Fig. 3e). In contrast, mutated GFP2-10 DNA showed reduced levels of indels (Fig. 3e), further indicating that partial DNA replacement can reduce off-target effects.
We performed GUIDE-seq10 to systematically compare the genome-wide off-target activity of native and 10 DNA crRNA. Three guide sequences were chosen: mouse Pcsk9 (ref. 34), human EMX1, and human 293 site 4 (ref. 10). Mouse Hepa1-6 liver cells or human HEK293 cells stably expressing SpCas9 were transfected with Guide-seq oligos, tracrRNA, and (1) native crRNA or (2) 10 DNA crRNA. Analysis of the off-target peaks revealed that all three 10 DNA crRNAs had no detectable off-target sites using the threshold of six or fewer total mismatches between the guide sequence and PAM (Fig. 3f,g, and Supplementary Data Set 1), although the depth of the GUIDE-seq data for the EMX1 target site is suboptimal to make definitive conclusions for this sequence. Consistent with the VEGFA off-target analysis, these data collected via an alternate approach indicate that 10 DNA crRNA substantially reduces off-target editing.
Structure-guided DNA substitution at the 3′ of crRNA
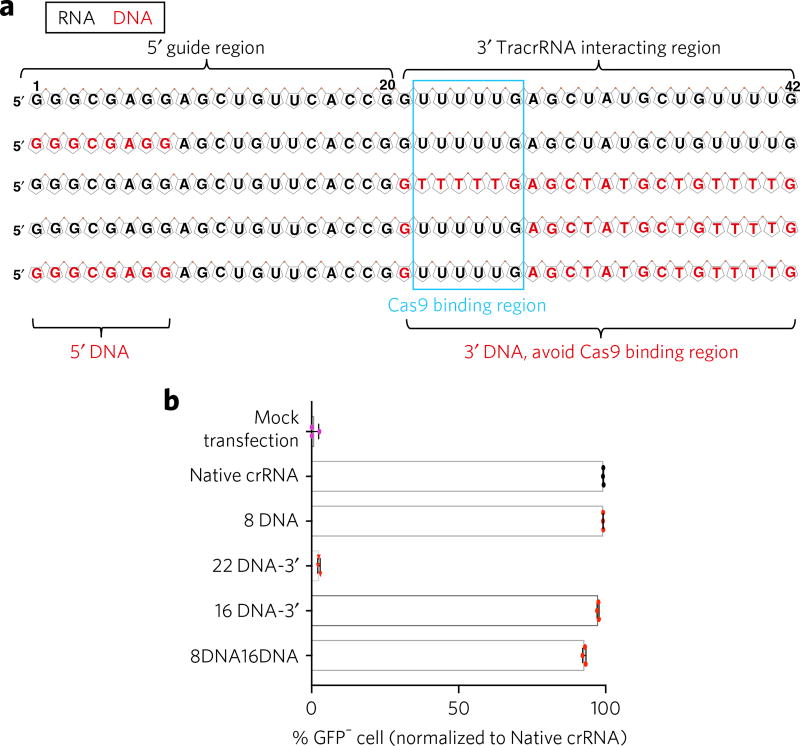
To explore whether the 3′ tracrRNA-interacting region of crRNA can tolerate DNA replacement, we synthesized a series of chimeric crRNAs targeting GFP (Fig. 4a). U2OS-GFP-PEST cells21 stably expressing Cas9 were transfected with crRNAs and tracrRNA. GFP-negative cells caused by Cas9-mediated frameshift NHEJ were measured by FACS at day 3 to report genome editing efficiency. As shown in Figure 4b, replacing all 22 RNA nucleotides in the 3′ region with DNA (22DNA) abolished genome editing. These data are consistent with our recent finding that 2′-O-methyl (2′OMe) chemical RNA modification of the entire 3′ region abolished sgRNA activity34. Within the structure of the Cas9–sgRNA complex25,26, six nucleotides in the 3′ end of crRNA–sgRNA have been shown to interact with the Cas9 protein. We hypothesized that avoiding substitution of RNA bases with DNA bases in those six nucleotides would help preserve crRNA activity. As expected, this 16DNA design (16-nucleotide DNA substitution in the 3′ region, avoiding the Cas9 binding region) efficiently generated GFP− cells, comparable to native crRNA or 8DNA crRNA (Fig. 4b). We subsequently tested whether crRNA functionality can be maintained with 5′ and 3′ DNA substitution within one crRNA. crRNA with both 8DNA and 16DNA design (8DNA16DNA, 8-nucelotide DNA in 5′ and 16-nucleotide DNA in 3′) induced efficient genome editing (Fig. 4b). Of note, the majority of the 8DNA16DNA crRNA is comprised of DNA bases (57%) (Supplementary Fig. 6a). Because DNA bases are more than ten-fold cheaper than RNA bases in oligo synthesis, the cost of synthesizing 8DNA16DNA crRNA is ~60% less than that for native crRNA (Supplementary Fig. 6b). Together, these data present an optimized DNA–RNA chimeric crRNA design that enables efficient genome editing in human cells and has potential to substantially reduce the cost for certain CRISPR–Cas9 applications.
Figure 4. An optimized DNA–RNA chimeric crRNA enables efficient genome editing in human cells.
(a) Illustration of DNA substitution of GFP targeting crRNAs. RNA and DNA are shown in black and red, respectively. The Cas9 binding region is shown by a blue box. (b) U2OS-GFP-PEST cells stably expressing Cas9 were transfected with GFP crRNAs and the tracrRNA. GFP negative cells caused by Cas9-mediated frame shift NHEJ were measured by FACS at day 3. 8 DNA16 DNA design (8-nt DNA in 5′ and 16-nt DNA in 3′, avoiding the Cas9 binding region) mediates efficient genome editing. 3′ all DNA (22DNA) abolished genome editing. n = 3 biologically independent samples. Error bars show mean ± s.d.
DISCUSSION
In this study, we demonstrate that partially replacing guide RNA with DNA can retain on-target genome editing by CRISPR–Cas9. Importantly, replacement of RNA with DNA can significantly reduce off-target activity. Our study indicates that partial DNA-guided genome editing of Cas9 is feasible in mammalian cells, extending the toolkit of CRISPR from an RNA-guided nuclease platform to a partial DNA-guided nuclease system. Moreover, our study indicates a simple and effective way to decrease off-target effects that may be combined with modified Cas9 protein or improved delivery technologies to further minimize off-target activity14–20,35.
Our study indicates that partial DNA replacement is feasible for crRNA of spCas9 and Cpf1 (Fig. 2d). It is possible that this approach has utility for guide sequences of other Cas9 proteins, such as Staphylococcus aureus (Sa) Cas9 (ref. 36) or Neisseria meningitidis (Nm) Cas9 (ref. 37). Future work is required to study other types of chemical modifications or incorporation of DNA aptamers at the 5′ end of crRNA and sgRNA38. In addition, whether DNA Cpf1 crRNA can decrease off-target specificity has yet to be tested. Because crRNA alone is sufficient to mediate genome editing by Cpf1, DNA–RNA chimera crRNA may be particularly attractive for generating array or plate of crRNA libraries with reduced cost.
Our study indicates that the tail region of the guide sequence is more amenable to DNA replacement than the seed region. Replacement of the 5′ end of the guide sequence maintains the genome-editing activity. However, Cas9 loses its genome-editing capability in cells with crRNA harboring >12-nucleotide DNA at the 5′ end or 4-nucleotide DNA at the 3′ end of the guide region. The crystal structure of the Cas9–sgRNA complex indicates that the seed region of the guide sequence is essential for Cas9–sgRNA binding and recognition of targeted DNA25,26; however, the tail region is less interactive with Cas9 than the seed region25,26. This likely explains why the tail region, but not the seed region, can tolerate DNA replacement (Figs. 1 and 2). This is consistent with previously studies reporting that the seed region is sensitive to chemical modifications23. In our recent study, we demonstrated that by using the structure of Cas9–sgRNA as a guide, sgRNA and crRNA can be chemically modified to enhance their activity in cells and animals34. We identified regions of sgRNA and crRNA that can tolerate modifications of the 2′OH and avoided modifying the 2′OH that interact with the Cas9 protein34. We also showed that modification of even one ‘interacting’ 2′OH with 2′OMe at the guide region of crRNA abolishes its activity in mammalian cells34. This finding is consistent with the data showing that replacing RNA with DNA at the tail region, but not the seed region, can maintain the activity of crRNA (Figs. 1 and 2). Furthermore, the helix geometry of DNA and RNA are different32,33. It is possible that the variation of helix geometry of DNA–RNA chimeras may, in part, explain their activities in cells.
We noticed that in the GUIDE-seq assay, the number of sequencing reads for crRNAs targeting the human EMX1 and 293 site 4 in HEK293T cells were lower than the crRNA targeting mouse Pcsk9 in Hepa1-6 cells (Fig. 3f,g and Supplementary Data Set 1). The numbers of our GUIDE-seq reads for the on-target locus are similar to those of the published study10. Furthermore, the different forms of guides and the delivery methods applied may also affect the outcome of the GUIDE-seq assay10,34. The higher reads in Hepa1-6 cells relative to HEK293T cells may be explained by the higher concentration of GUIDE-seq oligonucleotide used in Hepa1-6 cells, because those cells are more tolerant of DNA oligo transfection. We noticed that in the GUIDE-seq of EMX1, three off-target sites have around 10% reads of the on-target site for the DNA-substituted crRNA (Supplementary Data Set 1). However, to rule out unspecific reads, we applied GUIDE-seq criteria using the threshold of ≤6 mismatches between the guide and the PAM sequence in total10. These three off-target sites contain more than six mismatches, so they are not likely to be bona fide off-target cleavage sites.
Here we demonstrated, for the first time, the use of chemical modification to significantly reduce off-target activities of CRISPR–Cas9 (Fig. 3). We learned from RNA therapeutics, in particular siRNA therapy, that other chemical modifications also hold the potential to reduce off-target activities39. For example, nucleobase modifications of siRNA have been shown to reduce miRNA-like off-target effects40. Position-dependent 2′OMe modification in the guide strand of siRNAs decreases ‘off-target’ transcript silencing41. It is worthy to investigate other chemical modifications to reduce off-target activities of CRISPR systems.
High-fidelity and enhanced specificity Cas9 variants demonstrated reduced off-target activities in mammalian cells18,19. A recent study indicated that tuning the natural conformational threshold of the Cas9–sgRNA–substrate DNA complex can improve the specificity of Cas9, and a new hyper-accurate Cas9 variant was designed and developed accordingly42. It is possible that the DNA–RNA chimera guides, along with truncate guides and Cas9 variants, share the similar principles of reducing off-target activities. It is worthwhile to further study the underlining mechanisms of how chemically modified guides are able to reduce off-target cleavages.
Our data suggest that the DNA nucleotides play a functional role in the recognition of the genomic DNA and support the endonuclease activity of Cas9. Interestingly, truncation of the guide sequence to 14 RNA nucleotides, combined with catalytically active Cas9 fused with a functional domain such as a transcription activator or inhibitor, can modulate gene expression without inducing double-stranded breaks43,44. Our data indicate that guide sequences with 14 RNA nucleotides and 6 DNA nucleotides are fully capable of inducing double-stranded breaks in the human genome (Fig. 1b,c and Fig. 2a). Indeed, mutation of 1–4 DNA nucleotides in the guide region abolishes the editing effect (Fig. 2e,f and Supplementary Fig. 5), demonstrating that DNA sequence substitutions can play a role in recognizing the complementary genomic sequences. Considering the weaker binding of DNA–DNA compared to DNA–RNA32,33, it will be interesting to test whether some DNA–RNA chimeras may guide catalytically active Cas9 to bind a genomic locus and modulate gene expression but not to cause indels.
Because the synthetic cost of sgRNA is high, a recent study conjugated a 65-nucleotide 5′-hexyne tracrRNA and a 34-nucleotide 3′-azide crRNA component45. The synthetic conjugated sgRNA showed efficient genome-editing activity in cells45. It is feasible to replace about half of the RNA with DNA nucleotides in the 3′-azide crRNA component, which has the guide sequence. Such a strategy may further reduce the cost of synthesis and increase the specificity of guide sequences. It also allows generation of single synthetic sgRNA and libraries of synthetic sgRNA that are more practical for research and development purposes.
Our data showed that DNA–RNA chimeric guides can induce efficient genome editing in human cells with reduced off-target effects, highlighting their possible usage for biomedical research and therapeutic genome editing. Such partial DNA crRNAs or sgRNA can be easily synthesized for genome editing in cells at reduced cost, and can potentially be delivered to animals using lipid or polymer nanoparticles for research and therapeutic applications34,35. It is worth investigating whether DNA–RNA chimeric guides can reduce off-target effects in living animals and understanding the associated immune responses. Our data suggests that incorporation of other nucleotides or chemical modifications into guide-RNA sequences may have the potential to further decrease off-target effects of CRISPR–Cas systems.
ONLINE METHODS
Oligonucleotides synthesis
The oligonucleotides were synthesized by Integrated DNA Technologies (IDT) using the solid-phase synthesis and phosphoroamidite chemistry22. The sequences of all oligonucleotide are shown in Supplementary Table 1. The guide sequences were published elsewhere22,24,27. The oligonucleotides were dissolved in sodium citrate buffer (pH 4.5), aliquoted and stored in −80 °C.
Cell culture and FACS
HEK293T cells were infected by lentiviral particles to stably express EF1a-GFP-PGK-Puro (Addgene; 26777)46 and EFs-spCas9-Blast (Addgene; 52962)47. Functional titer was used to ensure low multiplicity of infection (MOI, as the number of virus particles on average infecting one cell). Cells were infected with a limiting dilution of lentivirus, and wells with <40% GFP signal or cells surviving Blast selection (MOI < 1) were chosen as described34. U2OS-GFP-PEST cells (kindly provided by the K. Joung lab, Massachusetts General Hospital21) were transfected with lentivirus to allow stable expression of Cas9. HEK293T Cells were transfected with a crRNA targeting GFP and the tracrRNA (26 nM each, final concentration) using Lipofectamine (Thermo Fisher Scientific). U2OS-GFP-PEST cells were transfected with the same concentration of the crRNA and the tracrRNA by electroporation (Neon Transfection System, Thermo Fisher Scientific; see section below for details of electroporation). GFP-negative cells were counted by FACS 7 d for HEK293T cells and 3 d for U2OS-GFP-PEST after transfection. FACS was performed using MoFlo cell sorter (Beckman) or LSR (BD Biosciences) as described35. Flowjo was used to perform data analysis. The axis labels indicate the fluorochrome used (Supplementary Fig. 1b). The GFP-negative cells were gated according to untreated GFP-positive cells.
Determining allele modification frequencies using TIDE analysis and Surveyor assay
Genomic DNA was extracted using QuickExtract DNA Extraction Solution (Epicentre). PCR (initiate heating and 25–30 cycles of 15 s at 94 °C, 15 s at 55–62 °C and 1 min at 72 °C) was performed with 50 ng genomic DNA to yield the amplicons of the CRISPR target sites. The sequences of primer pairs are presented in Supplementary Table 2. For the tracking of indels by decomposition (TIDE)28, the PCR products were purified using PCR purification kit (Qiagen), sequenced by Sanger method (Quintarabio) and analyzed by the online software (http://tide.nki.nl). For the surveyor assay, purified PCR amplicons were denatured, re-annealed and subsequently digested with Surveyor nuclease (IDT). Digested DNA was resolved by electrophoresis in a 4–20% TBE gel (Thermo Fisher Scientific), stained briefly with ethidium bromide, and visualized by UV light. The gels are representative of three experiments. Off-target sites of VEGFA were published elsewhere24. Indel percentage was measured as 100 × (1 − (1 − (b + c)/(a + b + c))1/2), where a is the intensity of the uncut PCR product, and b and c are the intensities of cleavage PCR products29.
GUIDE-seq off-target analysis
We performed GUIDE-seq10 with some modifications as described previously20. Briefly, Hepa1-6 or HEK293T cells stably expressing SpCas9 were transfected using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s suggested protocol. 26 nM of crRNA/tracrRNA and annealed GUIDE-seq oligonucleotide (7.5 pmol for HEK293T cells and 100 pmol for Hepa1-6 cells) were transfected into cells in each well of a 24 well plate. 48 h after transfection, genomic DNA was extracted with a PureLink Genomic DNA Mini Kit (Invitrogen) according to the manufacturer’s suggested protocol. Library preparations were done with original adaptors according to protocols described by Joung et al.10. Each library was barcoded within the P5 and P7 adaptors for pooled sequencing. The barcoded, purified libraries were deep-sequenced as a pool using a paired-end 150-bp Illumina MiSeq run. Data were analyzed using the GUIDE-seq Bioconductor package48 using the default settings except that min. reads is set to 2 and distance. Threshold to 70 with the identified sites filtered to those sequences with six or fewer mismatches within the input guide and PAM (NGG) sequences.
Neon transfection for Jurkat T cells
We prepared a 5-µl mixture containing crRNA, tracrRNA and spCas9 protein (2 µM final for each) and kept them at room temperature (20–25 °C) for 15 min to form the Cas9 RNP complex. Jurkat T cells were harvested, washed and resuspended in Resuspension Buffer (included in Neon Kits) at a final density of 2 × 107 cells/ml. Cells were mixed with the 5 µl RNP complex. Transfection was performed according to the manufacturer’s suggested protocol in a 10-µl tip with the following parameters: 1,325 v /10 ms /4 pulses. After transfection, cells were transferred into 500 µl RPMI 1640 media with l-glutamine and 10% FBS, but without antibiotics, and incubated at 37 °C in a humidified CO2 incubator. Three days after transfection, genomic DNA was harvested for the analysis of indel% by TIDE.
Statistics
Student’s t-tests or one-way ANOVA with Tukey post hoc test were used to calculate P values by Prism 7 (GraphPad). No adjustments were made for multiple comparisons. P < 0.05 is considered significant. The definition of error bars is s.d. N indicates biological replicates as number of cell cultures. No inclusion and exclusion criteria of samples were used.
Life Sciences Reporting Summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. The original deep sequencing data is available using the BioProject accession code PRJNA420401.
Supplementary Material
Acknowledgments
We thank T. Jacks, P. Sharp, Z. Weng, C. Mello, and E. Sontheimer for discussions, and Y. Li for technical assistance. We thank K. Joung (Massachusetts General Hospital and Harvard Medical School) for sharing U2OS-GFP-PEST cells and J. Smith and A. Sheel for proofreading. This work is supported by grants from the National Institutes of Health (NIH), 5R00CA169512, DP2HL137167 and P01HL131471 (to W.X.). H.Y. is supported by 5-U54-CA151884-04 (NIH) and Skoltech Center. W.X. was supported by the Lung Cancer Research Foundation, Hyundai Hope on Wheels, and ALS Association. This work is supported in part by Cancer Center Support (core) grant P30-CA14051 from the NIH. We thank the Swanson Biotechnology Center at MIT for technical support.
Footnotes
Author contributions
H.Y. conceived of and designed the study and directed the project. H.Y., C.-Q.S, S.S., S.W., Q.W., J.D., S.-Y.K., L.J.Z., and S.A.W. performed experiments and analyzed data. H.Y. made the figures with C.-Q.S. V.K., W.X., and R.L.B., and R.L. provided conceptual advice. H.Y. wrote the manuscript with comments from all authors. W.X., D.G.A. and R.L. supervised the project.
Competing financial interests
The authors declare competing financial interests: details accompany the online version of the paper.
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
References
- 1.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 4.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swarts DC, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 9.Sander JD, et al. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acids Res. 2013;41:e181. doi: 10.1093/nar/gkt716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR–Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 14.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolukbasi MF, et al. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat. Methods. 2015;12:1150–1156. doi: 10.1038/nmeth.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendel A, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahdar M, et al. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. USA. 2015;112:E7110–E7117. doi: 10.1073/pnas.1520883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 26.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesnik EA, Freier SM. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34:10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- 33.Gyi JI, Lane AN, Conn GL, Brown T. The orientation and dynamics of the C2′-OH and hydration of RNA and DNA.RNA hybrids. Nucleic Acids Res. 1998;26:3104–3110. doi: 10.1093/nar/26.13.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin H, et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee K, et al. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. eLife. 2017;6:e25312. doi: 10.7554/eLife.25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Suter SR, et al. Controlling miRNA-like off-target effects of an siRNA with nucleobase modifications. Org. Biomol. Chem. 2017;15:10029–10036. doi: 10.1039/c7ob02654d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson AL, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiani S, et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahlman JE, et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He K, Chou ET, Begay S, Anderson EM, van Brabant Smith A. Conjugation and evaluation of triazole-linked single guide RNA for CRISPR-Cas9 gene editing. ChemBioChem. 2016;17:1809–1812. doi: 10.1002/cbic.201600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou J, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu LJ, et al. GUIDEseq: a bioconductor package to analyze GUIDE-seq datasets for CRISPR-Cas nucleases. BMC Genomics. 2017;18:379. doi: 10.1186/s12864-017-3746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. The original deep sequencing data is available using the BioProject accession code PRJNA420401.