Abstract

Natural products are well known for their biological relevance, high degree of three-dimensionality, and access to areas of largely unexplored chemical space. To shape our understanding of the interaction between natural products and protein targets in the postgenomic era, we have used native mass spectrometry to investigate 62 potential protein targets for malaria using a natural-product-based fragment library. We reveal here 96 low-molecular-weight natural products identified as binding partners of 32 of the putative malarial targets. Seventy-nine (79) fragments have direct growth inhibition on Plasmodium falciparum at concentrations that are promising for the development of fragment hits against these protein targets. This adds a fragment library to the published HTS active libraries in the public domain.
Keywords: natural products, fragments, native mass spectrometry, target identification, malaria
In 2015, malaria infected an estimated 212 million people globally, leading to 429 000 deaths, most of which were children aged 5 years and under in Africa.1Plasmodium falciparum is the most common species causing malaria in humans. Currently, four major classes of compounds are clinically used to treat malaria: (I) quinolines (e.g., chloroquine, quinine, mefloquine), (II) antifolates (e.g., sulfadoxine, pyrimethamine), (III) artemisinin derivatives (e.g., artesunate, artemether), and (IV) antimicrobials (e.g., doxycycline).2 However, a combination of parasite drug resistance and vector resistance to insecticides remains a huge challenge in combating this disease.3 It is an urgent need that new classes of drugs be developed for the treatment of this pathogen.
Since 2008, almost 6 million compounds have been screened against the blood stages of P. falciparum, and approximately 0.5% of these compounds showed activity consistent with an EC50 of less than 1 μM.4 Chemical structures and associated data have been made public from consortia led by GlaxoSmithKline (GSK), Novartis, and St. Jude Children’s Research Hospital, Memphis (St. Jude) to encourage further research into this disease.5−7 In addition, a subset of 400 compounds has been assembled into the Medicines for Malaria Venture Malaria Box that has catalyzed research into the discovery of new antimalarial clinical candidates.8−13
Finding a ligand for a disease-related protein remains one of the biggest hurdles of the postgenomic era. The identification of synthetic or natural ligands can help decrypt the biochemical functions of proteins with unknown functions.14 Such efforts require a method to detect the interaction between small molecular ligands and macromolecular targets.
Here, we describe a fragment-based approach using native mass spectrometry to detect protein–ligand interactions that exploit proteins produced by structural genomics efforts. The technique relies on nondenaturing electrospray ionization (ESI) to first recognize multicharged proteins in their near-native states. Native screening using high-resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry (ESI-FT-ICR-MS) is a label-free, fast, accurate method that permits the direct observation of noncovalent protein–ligand complexes.15−17 In the resulting mass spectrum, the difference between the mass-to-charge ratio (Δm/z) for the protein–ligand complex and the unbound protein ions multiplied by the charge state (z) directly affords the molecular weight of the bound ligand (MWligand = Δm/z × z). The molecular weight mass information on the ligand allows the identification of the active ligand in a pooled library. The simple mix-and-measure, label-free nature of these experiments makes native ESI-FT-ICR-MS a practical technique in screening fragments.
We previously reported the detection of protein–ligand interactions in complex natural product extracts.18−20 We also demonstrated that ESI-FT-ICR-MS can directly observe weak, noncovalent protein binding of fragment-sized natural products.21 Fragment-based drug discovery (FBDD) is an established alternative approach to high-throughput screening (HTS). Because of the simpler structure and smaller size of fragments, screening of fragment libraries can sample chemical space more efficiently and give higher hit rates in comparison to standard nonfragment chemical libraries.22,23 Although fragment hits generally have lower potency with initial affinities of up to 1 mM,24,25 their high ligand efficiency can offer more efficient and fruitful optimization campaigns, thus reducing attrition rates at all stages.26 FBDD has yielded two marketed drugs, vemurafenib27,28 and venetoclax,29 and there are many other compounds currently in clinical trials,30,31 including verubecestat (MK-8931) for Alzheimer’s disease, where the “intractable” Beta-APP cleaving enzyme (BACE) protein pockets that are long and shallow have proven difficult for traditional drug screenings to identify a hit.32
Natural products, enriched with biosynthetic intermediates and endogenous metabolites resulting from being exposed to a long selection process to interact with biological targets, are excellent resources to search for protein binding partners. Native mass spectrometry provides key advantages for weak binding detection as it has high sensitivity, low sample consumption, does not require modifications or labeling of the protein target, and provides a direct visualization of all species present in solution under binding equilibrium.33,34
Using this assay, we screened a fragment-sized library consisting of low-molecular-weight natural products and identified a number of natural products that bind to putative malaria targets. The hits were further investigated in biological activity assays to identify compounds that inhibit P. falciparum and warrant hit-to-lead development. We have identified 96 fragments that bind to 32 potential targets and have demonstrated that 79 of these fragments can inhibit the growth of malaria parasites in vitro.
Results and Discussion
Protein Selection
The proteins were selected according to a variety of criteria from the analysis of ∼5400 predicted open reading frames in the P. falciparum genome. Initial analysis was restricted to proteins of less than 50 kDa as the 4.7 T FT-ICR-MS detects up to 50 kDa. This produced 2535 genes, of which there was strong evidence that 67 were essential in either the exoerythrocytic stage or intraerythrocytic stages or had demonstrated high expression in gametocytes or liver stages. The evidence for essentiality was based mainly on genetic knockouts35 but also involved pathway-based reasoning. For example, dihydroorotate dehydrogenase (DHODH) has been validated as a drug target,36 so other pyrimidine biosynthesis enzymes were also included. Of these 67 genes, 56 met the criterion of having been validated in an exoerythrocytic stage and/or being highly expressed in gametocyte37 and/or liver stages.38 Proteins that were expected to be difficult to express and/or purify were removed from the analysis, leaving a total of 49 prioritized candidates. From these prioritized genes, 13 proteins were successfully expressed and purified for this study. We attempted to clone and express all 49 proteins but were successful with only 13, broadly consistent with the low success rates previously reported for Plasmodium proteins.39,40 The full list of these proteins is in Table S1. Another 66 proteins were identified from previous selection criteria41 as likely being essential for parasite survival and were amenable to recombinant expression. All proteins were soluble and therefore have a high likelihood of chemotherapeutic accessibility.
Seventy-nine Plasmodium proteins were subjected to mass spectrometry under optimized conditions, yielding protein signals at low charged states between +8 and +18, depending on the molecular weight of the protein. Out of the 79 proteins, 62 proteins had a good signal-to-noise ratio (>100:1), indicating that native mass spectrometry is a powerful technology for recognizing protein in its native-like state (Table S2 and Figure S3).
Fragment Library Assembly and Comparative Analysis of Physicochemical Properties, Shape, and Structural Diversity
The physicochemical properties of most fragment libraries today are compliant with the rule of three (Ro3), initially proposed by Congreve et al. in 2003.42 However, both the structural and shape diversity of a fragment library are important additional factors to consider, and fragment libraries obtained synthetically or purchased from commercial suppliers are predominantly composed of small, flat heterocyclic molecules.31,43 Natural product fragments have different chemotypes and are more populated with three-dimensional (3D) structures, thus offering an opportunity to improve the structural and shape diversity of fragment libraries and consequently to search different regions of biologically relevant chemical space and find structurally different hits.44 We compared natural product fragments present in the Dictionary of Natural Products (DNP, sdf version 211.9) with commercial and synthetic fragments present in six different screening libraries (Asinex, ChemBridge, InterBioscreen (IBS), Molecular Libraries Small Molecule Repository (MLSMR), Life Chemicals, and Otava) using the following filtering criteria: molecular weight (MW) ≤ 250 Da, octanol–water partition coefficient (ALogP) < 4, hydrogen bond donors (HBD) ≤ 4, hydrogen bond acceptors (HBA) ≤ 5, rotatable bonds (RB) ≤ 6, percent polar surface area (%PSA) < 45. After a cleaning process which included salt stripping, structure normalization and standardization, ionization at pH 7.4 and the selection of organic molecules with MW ≥ 100 Da or heavy atom count ≥7 and sulfur atom counts ≤3,45 we found that only 6.7% of the fragments from DNP were considered identical (Tanimoto score of 1) to all of the unique fragments present in the six commercially sourced fragment libraries, as determined by pairwise similarity comparisons using two-dimensional (2D) extended-connectivity fingerprints ECFP_6. In agreement with Hert et al.,46 who noted that 83% of natural product ring scaffolds were absent in commercially available molecules, our substructure search analysis revealed that on average 91% of the scaffolds from the DNP fragments were absent in each of the commercially sourced fragment libraries.
Our natural product fragment library contains 643 molecules with fragment-like physicochemical properties. Of these molecules, 602 (94%) comply strictly with the physicochemical property criteria initially set to guide the selection of these fragments (MW ≤ 250 Da, ALogP < 4, HBD ≤ 4, HBA ≤ 5, RB ≤ 6, %PSA < 45), while the remaining 41 constitute derivatives or structures that despite slightly violating one criterion or another were deemed of interest to screening. For the sake of accurate comparison to other generated fragment libraries, these 41 fragments were left out of the analyses discussed in this section, unless otherwise stated.
The mean distributions among the different fragment libraries for selected physicochemical properties (Figure S1) show that natural-product-based fragments (our 602 natural product fragments and fragments from DNP) have on average the same number of rings but a markedly smaller number of aromatic rings than commercially available fragments (fragments present in Asinex, ChemBridge, IBS, MLSMR, Life Chemicals, and Otava screening libraries). Our natural product fragment library presents itself with the highest average number of HBDs and the lowest average of ALogP and number of RBs (Figure S1).
To characterize the 3D shape of the fragments contained within each library, the fraction of sp3-hybridized carbon atoms (Fsp3) and principal moments of inertia (PMI) were determined for each molecule. The plot of the average of the calculated sum of the normalized PMI ratios (PMIsum) against the average Fsp3 clearly differentiates fragment libraries based on commercial and synthetic compounds from those based on natural products, which occupy a space where the average Fsp3 and average PMIsum are higher (Figure 1a). However, the benefits of populating a fragment library with more 3D-shaped fragments is still controversial.47−52 On one hand, 3D-shaped fragments have been suggested to be capable of sampling a larger chemical space50 and of exploring the pocket space of targets more extensively than 2D-shaped fragments,48 being more likely to deliver biologically active and clinically relevant molecules.51,52 On the other hand, 3D-shaped fragments have raised questions about the implications that their implicit higher molecular complexity can have for hit rates53 and for ease of synthesis and derivatization.54 It has also been shown that three-dimensionality can be introduced, to different degrees depending on the connectivity of the fragment scaffold, during fragment elaboration.47−49 While it is still unclear how important 3D character is for the core scaffold of fragments, we believe that shape diversity should exist in a fragment library. The 643-member natural product fragment library also contains a good representation of flat scaffolds (Bemis-Murcko molecular frameworks including rings, linkers, exocyclic double bonds, and terminal double bonds attached to the linkers), despite containing a higher proportion of 3D-shaped structures than other commercially available fragment libraries.
Figure 1.

Fragment libraries shape (a) and structural (b) diversity. (a) Average sum of normalized PMI ratios (PMIsum) as a function of the average fraction of sp3-hybridized carbon atoms (Fsp3) calculated for each fragment library. The three principal moments of inertia (PMI) were determined for each 3D minimized structure. The normalized PMI ratios (NPR 1 and 2) were calculated by dividing the two lowest PMIs by the highest. NPR1 was added to NPR2 to give the PMIsum. Flat molecules give a PMIsum of 1. A PMIsum higher than 1 indicates a deviation from planarity toward a spherelike shape. (b) Percentage of unique Bemis-Murcko scaffolds as a function of the average number of unique fingerprint features per fragment, calculated as the total number of unique fingerprint features determined using 2D extended-connectivity fingerprints (ECFP_6) divided by the number of fragments in the library.
To assess the structural diversity of each fragment library, the percentage of unique fragment scaffolds (Bemis-Murcko molecular frameworks including rings, linkers, exocyclic double bonds, and terminal double bonds attached to the linkers) present in each library and the average number of unique fingerprint features per fragment (using 2D extended-connectivity fingerprints ECFP_6) were calculated. Figure 1b shows the plot of the former against the latter, demonstrating the high structural diversity achieved with our natural product fragment library.
Prevalence of Fragment Hit Scaffolds among Existing Antimalarial HTS Hits
In order to accelerate research efforts for antimalarial drug discovery, the structures of the hit molecules that resulted from different high-throughput screens (HTS) have been deposited in the public domain (http://www.ebi.ac.uk/chemblntd).
Herein, the structures provided by St. Jude’s Children’s Research Hospital,6 Novartis,55,56 GlaxoSmithKline,5 Harvard Medical School,57 and MMV58 were used to evaluate the representation of the scaffolds derived from our fragment hits within this antimalarial chemical space. Our 96 fragment hits contain 72 unique fragment scaffolds. Through a substructure search, each HTS hit was searched for in the presence of one or more of these fragment scaffolds. Twenty-six (36%) of the scaffolds were found among these libraries. While 8 of the fragment scaffolds are common to all antimalarial HTS hit libraries, some are unique to specific active antimalarial libraries (Figure S2). Table 1 gives examples of some fragment scaffolds represented among the antimalarial HTS hits as well as the originating fragment. The structures of the active drug-sized molecules clearly show that these could have been developed through FBDD by fragment linking, growing, or merging (Table 1). While this provides a validation of our fragment screening approach, it shows that, with 46 (64%) of the fragment hit scaffolds remaining to be mapped within an antimalarial HTS hit molecule, many opportunities exist for fragment elaboration into structurally new active drug-sized molecules. A PMI plot of the unique scaffolds present in our natural product fragment library (325 fragment scaffolds), where those present within fragment hits are discriminated from those absent, is shown in Figure 2. Within our fragment screening campaign, fragment hits were identified across all types of scaffold shapes.
Table 1. Examples of Fragment Hit Scaffolds Present among Antimalarial HTS Hits.

Figure 2.
PMI of the fragment scaffolds (Bemis-Murcko molecular frameworks) contained in our fragment hit library (96 fragment hits, 72 unique fragment scaffolds) and comparison with those not contained in any fragment hit (253 unique fragment scaffolds). NPR1 constitutes the X axis, while NPR2 constitutes the Y axis.
Fragment Screening by Native MS
We performed a native MS screening using the natural product library against each of the 62 proteins. The complexes formed were ranked as strong, medium, or weak according to the ratio between the protein–ligand complex signal intensity and the unbound protein signal intensity (strong, ratio >0.5; medium, ratio 0.15 to 0.5; weak, ratio 0.02 to <0.15) (Figure 3).
Figure 3.
Native proteins and protein–ligand complexes. (a) PF3D7_1457200 thioredoxin and its strong signal complex with 10. (b) PF3D7_1457200 thioredoxin I and its medium signal complex with 16. (c) PF3D7_1457200 thioredoxin I and its weak complex with 8.
This ratio does not provide an absolute measure of the binding affinity but can be used to obtain a relative affinity ranking and provide rough KD estimates. Assuming that the ionization, transmission, and detection efficiencies of the protein–ligand complex and free protein are the same and that relative equilibrium concentrations in solution are preserved upon transfer to the gas phase, the higher the intensity of the protein–ligand complex in the mass spectrum relative to the free protein, the higher the protein occupancy in solution and therefore the higher the ligand affinity for the protein. In our experience17,20 and considering the range of protein and ligand concentrations used throughout this work (1–20 μM protein, 5–400 μM fragment), it is estimated that strong and medium binders have KD < 100 μM. The categorization into strong, medium, and weak binding is particularly useful, together with hit rates, in assessing target ligandability, the ability of a protein target to bind small molecules with high affinity.24 Taking into account the ligandability scoring system proposed by Edfeldt and colleagues,24 we have defined high ligandable targets (with a high ligandability score) as those with ≥2 strong/medium binders and ≥7 hits (greater than 1% of the library); medium ligandability for proteins with at least 1 strong/medium binder and ≥2 hits; and low ligandability for proteins having at least 1 binder (Figure 4). The results clearly identify some targets that are more worthwhile to pursue on the basis of ligandability.
Figure 4.
Summary of 32 proteins that formed complexes with 96 natural products. Number of strong binders (ratio >0.5), medium (ratio 0.15 to 0.5), and weak (ratio 0.02 to <0.15) shown for each protein. Ligandability is based on high (≥2 strong/medium binders and ≥7 hits), medium (at least 1 strong/medium binder and ≥2 hits), and low (at least 1 hit).
Ninety-six compounds (Figure 5) formed complexes with 32 proteins (Figure 6). Of the 96 fragments found to have protein partners, 48 fragments bound 1 protein, 18 bound 2 proteins, 14 bound 3 proteins, 4 bound 4 proteins, 5 bound 5 proteins, and 7 bound 6 proteins. Compounds that formed complexes with greater than 6 proteins were considered promiscuous and were excluded from further analysis. The retained hits are highly selective because they were screened against a total of 62 proteins: 48 fragments did not bind to 61 other proteins, 18 compounds did not bind to 60 other proteins, 14 compounds did not bind to 59 proteins, 4 compounds did not bind to 58 proteins, 5 compounds did not bind to 57 proteins, and 7 compounds did not bind to 56 proteins.
Figure 5.
Structures of 96 low-molecular-weight natural product binders. * indicates a derivative of the indicated natural product. See Table S3 for details.
Figure 6.
Heat map showing hits from the fragment library (643 members) against 32 proteins. Green indicates strong binding (protein–ligand complex/free protein ratio is greater than 50%). Blue indicates medium binding (protein–ligand complex/free protein ratio is greater than 15% and smaller than 50%). Orange indicates weak binding (protein–ligand complex/free protein ratio is from 2 to 15%). White indicates no binding. The proteins are shown in Table S1.
Although it is generally thought that molecules containing an electrophilic warhead (such as Michael acceptors) react indiscriminately and form covalent bonds with a variety of proteins, several studies have shown that such is not the case.59,60 A molecule containing a weak to moderate electrophilic warhead will bind covalently to a nucleophilic protein residue only if it has first bound noncovalently to the target protein and if the reactive electrophile has been properly oriented within the target protein so that a bond can be formed with the nucleophile on the protein.59,60 Many of our fragment hits contain electrophilic groups and are selective for a limited number of proteins (for example, see 15–20). Moreover, our experimental data shows that not all PAINS-containing molecules (such as polyphenols, β-lactams, epoxides, and rhodamine-related compounds) are promiscuous (for example, see 27, 50, 68, and 71).61,62
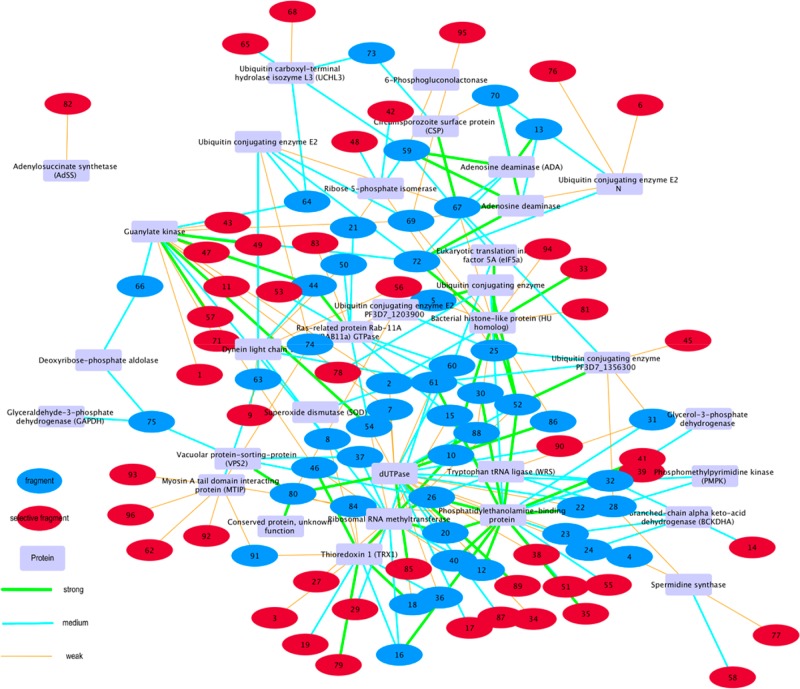
To better display these data obtained from our MS screening, we built a map that shows the relationship between fragments and proteins (Figure 7). Considering hits and proteins as nodes, we connected them to each other based on the binding interaction from Figure 6: the result is a network with 96 fragments. As previously mentioned, there were 48 selective fragments that bound a single protein so that 48 fragments bound more than 1 protein.
Figure 7.
Network visualization of 96 fragment hits against 32 proteins. The network was visualized using the Cytoscape software.63 Circle nodes represent fragment hits, and rectangular nodes represent proteins. The edge represents interaction between fragment hit and protein. The green edge indicates strong binding, blue indicates medium binding, and orange indicates weak binding, according to Figure 3.
Figure 8 shows a limited part of the complete map with 54 fragments and 6 proteins connected to each other through 9 fragments. Twenty-seven hits are binding protein PF3D7_1127100 dUTPase, and six of these hits are joining other four proteins. Fragments 2, 7, and 54 are binding PVX_099895 guanylate kinase, 32 is binding PF3D7_1216200 glycerol-3-phosphate dehydrogenase, and 60 is binding PF3D7_1204300 eukaryotic translation initiation factor 5A (eIF5a). Joint hits most likely identify a similar binding interaction in each protein given the principle that fragments identify single binding sites.
Figure 8.
Enlarged view of Figure 7. Circle nodes represent fragment hits, and rectangular nodes represent proteins. The edge represents the interaction between fragment hit and protein. In this portion of Figure 7, nine fragments are the connecting points among six proteins.
The 96 natural product fragments have different chemotypes and are different from the known antimalarial aminoquinolines, quinolones, or diamidines. An analysis of hits by structural similarity using Tanimoto measurements with a cutoff at 0.75 showed different structure–activity relationships. The two major clusters consist of the sesquiterpene lactones (10, 15–20, 22–24, 27–29, 33, 36, 39–40), which share the same backbone carbon structure as the current most effective antimalarial drug artemisinin. This structural class is known to kill P. falciparum parasites at submicromolar concentration.64 The third largest cluster is the 2,5-disubstituted oxazolones with seven fragments (61, 67–72). There are 6 securinine analogues (80, 81, 86–89) and 40 structural singletons.
Seventy-nine (79) of the 96 fragments had in vitro activity against asexual intraerythrocytic blood stage P. falciparum 3D7 parasites at 100 μM, determined IC50 values, or reported activity in the literature. This corresponded to active compounds against 31 of the 32 proteins (Figure 6, Table S3).
IC50 values were able to be calculated for 24 compounds, with 13 having IC50 < 45 μM. Compounds 5,656,657,669,6727,688569 have previously been reported to have anti-Plasmodial activity. Compound 9 had weak activity against P. falciparum 3D7 (61% inhibition at 100 μM) but had a previously reported IC50 of 9 μM against chloroquine-sensitive (D6, Sierra Leone) and chloroquine-resistant (W2) lines of P. falciparum.(67) The IC50 values of the other compounds correlated with previously reported values. We have previously reported that 80, 81, and 87–89 bind allosterically to recombinant P. falciparum 2′-deoxyuridine 5′-triphosphate nucleotidohydrolase (PfdUTPase, PF3D7_1127100), enhancing enzyme activity and inhibiting the in vitro activity viability of both P. falciparum gametocyte (sexual) and blood (asexual) stage parasites.21
We identified 31 malaria proteins having a fragment–protein complex and in vitro activity against P. falciparum. These proteins warrant further investigation as possible drug targets in Plasmodium. Only one protein had a binder that did not show in vitro activity against P. falciparum: adenylosuccinate synthetase (AdSS) (PF13_0287).
Fragment-based drug discovery is based on the identification of low-affinity ligands. The starting point for vemurafenib27,28 was an IC50 in the mM range with low affinity at ∼200 μM. Similarly, the starting point for venetoclax (ABT-199) was two fragment hits with KD = 400 and 2000 μM that were linked to initially give a compound with an IC50 of 6.9 μM on the way to the development of the approved drug.26,29 In comparison, direct growth inhibition of P. falciparum at 100 μM is significant, and the fact that 14 compounds had IC50 values <45 μM is highly encouraging.
Conclusions
We demonstrated that enriching fragment screening libraries with natural-product-based fragments provides the means not only to increase the structural diversity of the library but also to improve the content of 3D-shaped structures. Increasing the structural diversity of fragment libraries with chemotypes usually absent in commercially available libraries has allowed searching different regions of biologically relevant chemical space and finding structurally different hits.
We have presented a native mass spectrometry method that can recognize native proteins and find a binder concurrently. We have demonstrated the value of native MS screening in fragment-based drug discovery on cloned and purified proteins. We identified 31 proteins worthy of further investigation as anti-plasmodial targets having 79 low-molecular-weight natural product hits with in vitro antimalarial activity out of a total of 96 natural product hits using native MS. This adds a fragment library to the already-published HTS active libraries in the public domain and may facilitate fragment-based drug discovery for malaria.
Material and Methods
Cheminformatics Analysis
Fragment Generation
Fragments from DNP (sdf version 211.9; 166 379 molecules), Asinex (gold and platinum collection; 296 537 molecules), ChemBridge (express-pick collection stock; 459 091 molecules), InterBioscreen (IBS) (synthetic compounds library; 467 251 molecules), the Molecular Libraries Small Molecule Repository (MLSMR) (325 164 molecules), Life Chemicals (general HTS collection; 389 305 molecules), and Otava (collection for prompt delivery; 271 710 molecules) libraries were obtained using a series of filters available in Scitegic Pipeline Pilot (version 9.1.0.1, Accelrys Software, Inc.), which first cleaned up the structures (including salt stripping and selection of the largest fragment, structure normalization and standardization, ionization at pH 7.4, and selection of organic molecules with MW ≥ 100 Da or heavy atom count ≥7 and sulfur atom counts ≤3) and then selected those with fragmentlike physicochemical properties (MW ≤ 250 Da, atomic-based prediction of the octanol–water partition coefficient (A log P) < 4, HBD ≤ 4, HBA ≤ 5, rotatable bonds (RB) ≤ 6, percent polar surface area (%PSA) < 45). After the removal of duplicate molecules using canonical SMILES, 20 510 DNP fragments, 14 484 Asinex fragments, 24 291 ChemBridge fragments, 17 271 IBS fragments, 20 783 MLSMR fragments, 3682 Life Chemicals fragments, and 17 769 Otava fragments were obtained.
Determination of the Fraction of sp3-Hybridized Carbon Atoms (Fsp3) and Principal Moments of Inertia (PMI)
The fraction of sp3-hybridized carbon atoms (Fsp3) of each fragment was determined with Scitegic Pipeline Pilot (version 9.1.0.1, Accelrys Software, Inc.). Principal moments of inertia (PMI) were determined in Maestro (version 10.4.017, Schrodinger Inc., NY) after 3D structure minimization in Scitegic Pipeline Pilot. The normalized PMI ratios (NPR 1 and 2) were calculated by dividing the two lowest PMIs by the highest, as described by Sauer et al.70 NPR1 was added to NPR2 to give the PMIsum.
Generation of Fragment Scaffolds
Within Scitegic Pipeline Pilot, the “generate scaffold tree” component was first used to generate hierarchical ring scaffolds according to the procedure described in ref (71). Then the first-level scaffolds, which correspond to the Bemis-Murcko molecular frameworks (rings, linkers, exocyclic double bonds, and terminal double bonds attached to the linkers), were selected.72 After the removal of duplicate molecules using canonical SMILES, the following number of unique fragment scaffolds were obtained: 3958 DNP, 5163 ChemBridge, 3779 Asinex, 5319 IBS, 1374 Life Chemicals, 6824 MLSMR, 3704 Otava, and 325/72 of our global/hit fragment library. The percentage of unique fragment scaffolds was calculated by dividing the number of unique scaffolds by the number of fragments in the library.
Determination of the Average Number of Unique Fingerprint Features per Fragment
Using Scitegic Pipeline Pilot, each molecule was represented by its extended-connectivity fingerprint, ECFP_6, a 2D circular topological fingerprint where integer identifiers which account for different atom properties (atom type, mass, connection count, etc.) are first assigned to each heavy atom of the molecule and then are combined with identifiers of neighboring atoms in an iterative process until a diameter of six bonds is reached. This iteration process is based on the Morgan Algorithm73 and converts the substructural features around each atom into integer values which constitute the extended-connectivity fingerprint. The average number of unique fingerprint features per fragment was determined by counting the total number of unique ECFP_6 fingerprint features and dividing it by the total number of molecules.
Cleanup of Active Antimalarial HTS Hits
St. Jude’s Children’s Research Hospital (1523 molecules),6 Novartis (5615 molecules),56 GlaxoSmithKline (GSK) (13 467 molecules),5 Harvard Medical School (35 molecules),57 and Medicines for Malaria Venture (MMV) (400 molecules)58 data sets were processed according to the following workflow in Scitegic Pipeline Pilot: salt stripping, selection of the largest fragment in the molecule, structure normalization and standardization, ionization at pH 7.4, selection of organic molecules (molecules containing only organic atoms H, C, N, O, P, S, F, Cl, Br, or I), and removal of duplicate molecules using canonical SMILES. At the end, St. Jude’s Children’s Research Hospital contained 1523 “cleaned” molecules, Novartis 5464, GSK 13 458, Harvard Medical School 35, and MMV 400. These were the structures used for searching for the presence of fragment scaffolds derived from our fragment hit library.
Substructure Search
The fragment scaffolds were used as queries for the substructure search within the molecular structures of interest. Using Scitegic Pipeline Pilot, each query was mapped against all of the molecules to be searched. For each molecule successfully mapped, the number and name of queries mapped were retrieved.
Pairwise Similarity Comparisons
The similarity between two molecules was determined by calculating the similarity between their respective ECFP_6 fingerprints using the Tanimoto function: Tanimoto coefficient = c/(a + b – c), where a is the number of bits present in molecule A but not B, b is the number of bits present in molecule B but not A, and c is the number of bits present in both A and B.
Protein Cloning, Purification, and Purity Analysis
Recombinant histidine-tagged enzymes from P. falciparum and P. vivax were expressed in Escherichia coli. In general, genes were cloned into expression vectors that enabled tagging of the corresponding proteins with an N-terminal 6-histidine tag, as previously described.67,68 In some cases, truncated genes (missing some nucleotides at the 5′ and/or 3′ ends) were cloned to improve the chances of expression. Proteins were purified using a nickel column (immobilized-metal affinity chromatography, IMAC), flash frozen, stored at −80 C, and shipped on dry ice. Residues 61–204 of P. falciparum Myosin A tail domain interacting protein (MTIP) were expressed and purified as published.68
Native MS Screening
Protein Preparation
The proteins were buffer-exchanged into a suitable volatile buffer (ammonium acetate or ammonium bicarbonate) under nearly physiological conditions (pH 7.0) using size exclusion chromatography prior to ESI–MS analysis. Depending on the protein, the buffer and its concentration were chosen to obtain the highest sensitivity in the mass spectrometer. Starting conditions were 10 μM protein in 10 mM volatile buffer.
Instrument Control and Acquisition
Experiments were performed on a Bruker Apex III 4.7T or a Bruker SolariX XR 12T FT-ICR mass spectrometer (Bruker Daltonics Inc., Billerica, MA). An Agilent electrospray tuning mix (G2431A) was used for instrument calibration. Instrument parameters were tuned to avoid the dissociation of noncovalent protein–ligand complexes and achieve the highest signal intensities. As a control, a bovine carbonic anhydrase II (bCAII) sulfanilamide complex was used. Mass spectra were recorded in positive ion and profile modes with a mass range from 50 to 6000 m/z. Each spectrum was a sum of 16 transients (scans) composed of 256 K (Bruker Apex III 4.7T) or 1 M (Bruker SolariX XR 12T) data points. All aspects of pulse sequence control and data acquisition were controlled by Xmass (Bruker Apex III 4.7T) or Solarix control (Bruker SolariX XR 12T) software in a Windows operating system. For fragment library screening, pools of eight fragments (1 μL of each fragment at 5 mM in MeOH) were dried, resuspended in 1 μL MeOH, and incubated with proteins (1–20 μM) for 1 h at room temperature and analyzed by ESI-FT-ICR-MS. Final fragment/protein molar ratios varied between 5:1 and 20:1. When a noncovalent complex was found, the molecular weight of the binding fragment was calculated from the spectrum using the following equation: MWligand = Δm/z × z. The binding of the individual active fragment was confirmed in a separate experiment (Figure 4).
Fragment Library
Approximately half of the compounds were previously isolated in our laboratory and are >90% pure. The other half were purchased. The cited papers (Table S3) are listed to provide easy access to the original publications that reported the compounds.
Asexual Blood Stage Assay
A drug-sensitive (3D7) and -resistant (Dd2) Pf line was to be tested using an established radiometric growth inhibition assay. Briefly, P. falciparum-infected erythrocytes (2.5% hematocrit, 0.5% parasitaemia) were incubated in 96-well plates containing test compounds or DMSO as a control. After 48 h, 3H-hypoxanthine (0.5 microCi/well) was added, and the plates were incubated for another 24 h, followed by an assessment of 3H incorporation.
Acknowledgments
Funding was provided by the Bill and Melinda Gates Foundation (OPP1035218), the Australia Research Council (ARC) (LP120100485, LE20100170, and LE0237908), the National Health and Medical Research Council (NHMRC) (APP1046715), and National Institutes of Health (NIH) Grant 1P50 GM64655-01 “Structural Genomics of Pathogenic Protozoa (SGPP)”. We acknowledge the Australian Red Cross Blood Service for the provision of human blood and sera. We gratefully acknowledge access to the NatureBank biota library housed at the Griffith Institute for Drug Discovery (www.griffith.edu.au/gridd) for isolation of some of the fragments.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.7b00197.
Mean physicochemical properties of different fragment libraries, distribution of fragment hit scaffolds present within different libraries, and proteins studied for cloning and expression. Proteins studied by native mass spectrometry, detected hits and their antimalarial activity, and mass spectra of 62 Plasmodium proteins. (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- WHO. World Malaria Report 2016, pp 1–186.
- Cai S. X.; Risinger A. L.; Nair S.; Peng J. N.; Anderson T. J. C.; Du L.; Powell D. R.; Mooberry S. L.; Cichewicz R. H. (2016) Identification of compounds with efficacy against malaria parasites from common north American plants. J. Nat. Prod. 79, 490–498. 10.1021/acs.jnatprod.5b00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K. K. (2017) Targeting the active sites of malarial proteases for antimalarial drug discovery: approaches, progress and challenges. Int. J. Antimicrob. Agents 50, 287–302. 10.1016/j.ijantimicag.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Spangenberg T.; Burrows J. N.; Kowalczyk P.; McDonald S.; Wells T. N. C.; Willis P. (2013) The open access malaria box: a drug discovery catalyst for neglected diseases. PLoS One 8, e62906. 10.1371/journal.pone.0062906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo F. J.; Sanz L. M.; Vidal J.; de Cozar C.; Alvarez E.; Lavandera J. L.; Vanderwall D. E.; Green D. V. S.; Kumar V.; Hasan S.; Brown J. R.; Peishoff C. E.; Cardon L. R.; Garcia-Bustos J. F. (2010) Thousands of chemical starting points for antimalarial lead identification. Nature 465, 305–310. 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- Guiguemde W. A.; Shelat A. A.; Bouck D.; Duffy S.; Crowther G. J.; Davis P. H.; Smithson D. C.; Connelly M.; Clark J.; Zhu F. Y.; Jimenez-Diaz M. B.; Martinez M. S.; Wilson E. B.; Tripathi A. K.; Gut J.; Sharlow E. R.; Bathurst I.; El Mazouni F.; Fowble J. W.; Forquer I.; McGinley P. L.; Castro S.; Angulo-Barturen I.; Ferrer S.; Rosenthal P. J.; DeRisi J. L.; Sullivan D. J.; Lazo J. S.; Roos D. S.; Riscoe M. K.; Phillips M. A.; Rathod P. K.; Van Voorhis W. C.; Avery V. M.; Guy R. K. (2010) Chemical genetics of Plasmodium falciparum. Nature 465, 311–315. 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S.; Plouffe D. M.; Kuhen K. L.; Bonamy G. M. C.; Wu T.; Barnes S. W.; Bopp S. E.; Borboa R.; Bright A. T.; Che J. W.; Cohen S.; Dharia N. V.; Gagaring K.; Gettayacamin M.; Gordon P.; Groessl T.; Kato N.; Lee M. C. S.; McNamara C. W.; Fidock D. A.; Nagle A.; Nam T. G.; Richmond W.; Roland J.; Rottmann M.; Zhou B.; Froissard P.; Glynne R. J.; Mazier D.; Sattabongkot J.; Schultz P. G.; Tuntland T.; Walker J. R.; Zhou Y. Y.; Chatterjee A.; Diagana T. T.; Winzeler E. A. (2011) Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science 334, 1372–1377. 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessoff K.; Spangenberg T.; Foderaro J. E.; Jumani R. S.; Ward G. E.; Huston C. D. (2014) Identification of Cryptosporidium parvum active chemical series by repurposing the open access malaria box. Antimicrob. Agents Chemother. 58, 2731–2739. 10.1128/AAC.02641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. D.; Merino E. F.; Brooks C. F.; Striepen B.; Carlier P. R.; Cassera M. B. (2014) Antiapicoplast and gametocytocidal screening to identify the mechanisms of action of compounds within the malaria box. Antimicrob. Agents Chemother. 58, 811–819. 10.1128/AAC.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram-Sieber K.; Cowan N.; Panic G.; Vargas M.; Mansour N. R.; Bickle Q. D.; Wells T. N. C.; Spangenberg T.; Keiser J. (2014) Orally active antischistosomal early leads Identified from the open access malaria box. PLoS Neglected Trop. Dis. 8, e2610. 10.1371/journal.pntd.0002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucantoni L.; Duffy S.; Adjalley S. H.; Fidock D. A.; Avery V. M. (2013) Identification of MMV malaria box inhibitors of Plasmodium falciparum early-stage gametocytes using a luciferase-based high-throughput assay. Antimicrob. Agents Chemother. 57, 6050–6062. 10.1128/AAC.00870-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini A.; Bamert R. S.; Kannan-Sivaraman K.; Drinkwater N.; Mistry S. N.; Scammells P. J.; McGowan S. (2015) Screening the Medicines for Malaria Venture “malaria box″ against the Plasmodium falciparum aminopeptidases, M1, M17 and M18. PLoS One 10, e0115859. 10.1371/journal.pone.0115859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhis W. C.; Adams J. H.; Adelfio R.; Ahyong V.; Akabas M. H.; Alano P.; Alday A.; Resto Y. A.; Alsibaee A.; Alzualde A.; Andrews K. T.; Avery S. V.; Avery V. M.; Ayong L.; Baker M.; Baker S.; Ben Mamoun C.; Bhatia S.; Bickle Q.; Bounaadja L.; Bowling T.; Bosch J.; Boucher L. E.; Boyom F. F.; Brea J.; Brennan M.; Burton A.; Caffrey C. R.; Camarda G.; Carrasquilla M.; Carter D.; Cassera M. B.; Cheng K. C. C.; Chindaudomsate W.; Chubb A.; Colon B. L.; Colon-Lopez D. D.; Corbett Y.; Crowther G. J.; Cowan N.; D’Alessandro S.; Le Dang N.; Delves M.; DeRisi J. L.; Du A. Y.; Duffy S.; El-Sayed S. A.; Ferdig M. T.; Robledo J. A. F.; Fidock D. A.; Florent I.; Fokou P. V. T.; Galstian A.; Gamo F. J.; Gokool S.; Gold B.; Golub T.; Goldgof G. M.; Guha R.; Guiguemde W. A.; Gural N.; Guy R. K.; Hansen M. A. E.; Hanson K. K.; Hemphill A.; van Huijsduijnen R. H.; Horii T.; Horrocks P.; Hughes T. B.; Huston C.; Igarashi I.; Ingram-Sieber K.; Itoe M. A.; Jadhav A.; Jensen A. N.; Jensen L. T.; Jiang R. H. Y.; Kaiser A.; Keiser J.; Ketas T.; Kicka S.; Kim S.; Kirk K.; Kumar V. P.; Kyle D. E.; Lafuente M. J.; Landfear S.; Lee N.; Lee S.; Lehane A. M.; Li F. W.; Little D.; Liu L. Q.; Llinas M.; Loza M. I.; Lubar A.; Lucantoni L.; Lucet I.; Maes L.; Mancama D.; Mansour N. R.; March S.; McGowan S.; Vera I. M.; Meister S.; Mercer L.; Mestres J.; Mfopa A. N.; Misra R. N.; Moon S.; Moore J. P.; da Costa F. M. R.; Muller J.; Muriana A.; Hewitt S. N.; Nare B.; Nathan C.; Narraidoo N.; Nawaratna S.; Ojo K. K.; Ortiz D.; Panic G.; Papadatos G.; Parapini S.; Patra K.; Pham N.; Prats S.; Plouffe D. M.; Poulsen S. A.; Pradhan A.; Quevedo C.; Quinn R. J.; Rice C. A.; Rizk M. A.; Ruecker A. St; Onge R.; Ferreira R. S.; Samra J.; Robinett N. G.; Schlecht U.; Schmitt M.; Villela F. S.; Silvestrini F.; Sinden R.; Smith D. A.; Soldati T.; Spitzmuller A.; Stamm S. M.; Sullivan D. J.; Sullivan W.; Suresh S.; Suzuki B. M.; Suzuki Y.; Swamidass S. J.; Taramelli D.; Tchokouaha L. R. Y.; Theron A.; Thomas D.; Tonissen K. F.; Townson S.; Tripathi A. K.; Trofimov V.; Udenze K. O.; Ullah I.; Vallieres C.; Vigil E.; Vinetz J. M.; Vinh P. V.; Vu H.; Watanabe N. A.; Weatherby K.; White P. M.; Wilks A. F.; Winzeler E. A.; Wojcik E.; Wree M.; Wu W.; Yokoyama N.; Zollo P. H. A.; Abla N.; Blasco B.; Burrows J.; Laleu B.; Leroy D.; Spangenberg T.; Wells T.; Willis P. A. (2016) Open source drug discovery with the malaria box compound collection for neglected diseases and beyond. PLoS Pathog. 12, e1005763. 10.1371/journal.ppat.1005763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. E.; Bordeau B.; Cummings M. D.; Petrella E. C.; Pucci M. J.; Zawadzke L. E.; Dougherty B. A.; Tredup J. A.; Bryson J. W.; Yanchunas J. Jr.; Doyle M. L.; Witmer M. R.; Nelen M. I.; DesJarlais R. L.; Jaeger E. P.; Devine H.; Asel E. D.; Springer B. A.; Bone R.; Salemme F. R.; Todd M. J. (2005) Decrypting the biochemical function of an essential gene from Streptococcus pneumoniae using thermofluor technology. J. Biol. Chem. 280, 11704–11712. 10.1074/jbc.M413278200. [DOI] [PubMed] [Google Scholar]
- Vu H.; Quinn R. J.; Nguyen N.-T. (2015) Bioaffinity mass spectrometry screening using droplet-based microfluidics. Micro Nanosyst. 7, 74–79. 10.2174/1876402907666151103210549. [DOI] [Google Scholar]
- Vetting M. W.; Al-Obaidi N.; Zhao S.; San Francisco B.; Kim J.; Wichelecki D. J.; Bouvier J. T.; Solbiati J. O.; Vu H.; Zhang X.; Rodionov D. A.; Love J. D.; Hillerich B. S.; Seidel R. D.; Quinn R. J.; Osterman A. L.; Cronan J. E.; Jacobson M. P.; Gerlt J. A.; Almo S. C. (2015) Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes. Biochemistry 54, 909–931. 10.1021/bi501388y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro L.; Van Voorhis W. C.; Quinn R. J. (2016) Optimization of electrospray ionization by statistical design of experiments and response surface methodology: protein-ligand equilibrium dissociation constant determinations. J. Am. Soc. Mass Spectrom. 27, 1520–1530. 10.1007/s13361-016-1417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca A.; Temperini C.; Vu H.; Pham N. B.; Poulsen S. A.; Scozzafava A.; Quinn R. J.; Supuran C. T. (2009) Non-zinc mediated inhibition of carbonic anhydrases: coumarins are a new class of suicide inhibitors. J. Am. Chem. Soc. 131, 3057–3062. 10.1021/ja809683v. [DOI] [PubMed] [Google Scholar]
- Vu H.; Quinn R. J. (2008) Direct screening of natural product extracts using mass spectrometry. J. Biomol. Screening 13, 265–275. 10.1177/1087057108315739. [DOI] [PubMed] [Google Scholar]
- Yang B.; Feng Y. J.; Vu H.; McCormick B.; Rowley J.; Pedro L.; Crowther G. J.; Van Voorhis W. C.; Forster P. I.; Quinn R. J. (2016) Bioaffinity mass spectrometry screening. J. Biomol. Screening 21, 194–200. 10.1177/1087057115622605. [DOI] [PubMed] [Google Scholar]
- Vu H.; Roullier C.; Campitelli M.; Trenholme K. R.; Gardiner D. L.; Andrews K. T.; Skinner-Adams T.; Crowther G. J.; Van Voorhis W. C.; Quinn R. J. (2013) Plasmodium gametocyte inhibition Identified from a natural-product-based fragment library. ACS Chem. Biol. 8, 2654–2659. 10.1021/cb400582b. [DOI] [PubMed] [Google Scholar]
- Hopkins A. L.; Groom C. R.; Alex A. (2004) Ligand efficiency: a useful metric for lead selection. Drug Discovery Today 9, 430–431. 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D.; Chen K.; Sharp K. A.; Kollman P. A. (1999) The maximal affinity of ligands. Proc. Natl. Acad. Sci. U. S. A. 96, 9997–10002. 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edfeldt F. N. B.; Folmer R. H. A.; Breeze A. L. (2011) Fragment screening to predict druggability (ligandability) and lead discovery success. Drug Discovery Today 16, 284–287. 10.1016/j.drudis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Murray C. W.; Rees D. C. (2009) The rise of fragment-based drug discovery. Nat. Chem. 1, 187–192. 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- Congreve M.; Chessari G.; Tisi D.; Woodhead A. J. (2008) Recent developments in fragment-based drug discovery. J. Med. Chem. 51, 3661–3680. 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- Bollag G.; Hirth P.; Tsai J.; Zhang J. Z.; Ibrahim P. N.; Cho H. N.; Spevak W.; Zhang C.; Zhang Y.; Habets G.; Burton E.; Wong B.; Tsang G.; West B. L.; Powell B.; Shellooe R.; Marimuthu A.; Nguyen H.; Zhang K. Y. J.; Artis D. R.; Schlessinger J.; Su F.; Higgins B.; Iyer R.; D’Andrea K.; Koehler A.; Stumm M.; Lin P. S.; Lee R. J.; Grippo J.; Puzanov I.; Kim K. B.; Ribas A.; McArthur G. A.; Sosman J. A.; Chapman P. B.; Flaherty K. T.; Xu X. W.; Nathanson K. L.; Nolop K. (2010) Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599. 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G.; Tsai J.; Zhang J.; Zhang C.; Ibrahim P.; Nolop K.; Hirth P. (2012) Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat. Rev. Drug Discovery 11, 873–886. 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- Souers A. J.; Leverson J. D.; Boghaert E. R.; Ackler S. L.; Catron N. D.; Chen J.; Dayton B. D.; Ding H.; Enschede S. H.; Fairbrother W. J.; Huang D. C. S.; Hymowitz S. G.; Jin S.; Khaw S. L.; Kovar P. J.; Lam L. T.; Lee J.; Maecker H. L.; Marsh K. C.; Mason K. D.; Mitten M. J.; Nimmer P. M.; Oleksijew A.; Park C. H.; Park C. M.; Phillips D. C.; Roberts A. W.; Sampath D.; Seymour J. F.; Smith M. L.; Sullivan G. M.; Tahir S. K.; Tse C.; Wendt M. D.; Xiao Y.; Xue J. C.; Zhang H. C.; Humerickhouse R. A.; Rosenberg S. H.; Elmore S. W. (2013) ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208. 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Baker M. (2013) Fragment-based lead discovery grows up. Nat. Rev. Drug Discovery 12, 5–10. 10.1038/nrd3926. [DOI] [PubMed] [Google Scholar]
- Erlanson D. A.; Fesik S. W.; Hubbard R. E.; Jahnke W.; Jhoti H. (2016) Twenty years on: the impact of fragments on drug discovery. Nat. Rev. Drug Discovery 15, 605–619. 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- Albert J. S. (2009) Progress in the development of β-secretase inhibitors for Alzheimer’s Disease. Prog. Med. Chem. 48, 133–161. 10.1016/S0079-6468(09)04804-8. [DOI] [PubMed] [Google Scholar]
- Pedro L.; Quinn R. J. (2016) Native mass spectrometry in fragment-based drug discovery. Molecules 21, 984. 10.3390/molecules21080984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. S.-H.; Whitehouse A. J.; Coyne A. G.; Abell C. (2017) Mass spectrometry for fragment screening. Essays Biochem. 61, 465–473. 10.1042/EBC20170071. [DOI] [PubMed] [Google Scholar]
- Khan S. M.; Kroeze H.; Franke-Fayard B.; Janse C. J. (2012) Standardization in generating and reporting genetically modified rodent malaria parasites: the RMgmDB database. Methods Mol. Biol. 923, 139–150. 10.1007/978-1-62703-026-7_9. [DOI] [PubMed] [Google Scholar]
- Phillips M. A.; Lotharius J.; Marsh K.; White J.; Dayan A.; White K. L.; Njoroge J. W.; El Mazouni F.; Lao Y. B.; Kokkonda S.; Tomchick D. R.; Deng X. Y.; Laird T.; Bhatia S. N.; March S.; Ng C. L.; Fidock D. A.; Wittlin S.; Lafuente-Monasterio M.; Benito F. J. G.; Alonso L. M. S.; Martinez M. S.; Jimenez-Diaz M. B.; Bazaga S. F.; Angulo-Barturen I.; Haselden J. N.; Louttit J.; Cui Y.; Sridhar A.; Zeeman A. M.; Kocken C.; Sauerwein R.; Dechering K.; Avery V. M.; Duffy S.; Delves M.; Sinden R.; Ruecker A.; Wickham K. S.; Rochford R.; Gahagen J.; Iyer L.; Riccio E.; Mirsalis J.; Bathhurst I.; Rueckle T.; Ding X.; Campo B.; Leroy D.; Rogers M. J.; Rathod P. K.; Burrows J. N.; Charman S. A. (2015) A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Sci. Transl. Med. 7, 296ra111. 10.1126/scitranslmed.aaa6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. A.; Fivelman Q. L.; Blair P. L.; de la Vega P.; Le Roch K. G.; Zhou Y. Y.; Carucci D. J.; Baker D. A.; Winzeler E. A. (2005) The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143, 67–79. 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Tarun A. S.; Peng X.; Dumpit R. F.; Ogata Y.; Silva-Rivera H.; Camargo N.; Daly T. M.; Bergman L. W.; Kappe S. H. I. (2008) A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. U. S. A. 105, 305–310. 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadi M.; Lew J.; Artz J.; Amani M.; Zhao Y.; Dong A. P.; Wasney G. A.; Gao M.; Hills T.; Brokx S.; Qiu W.; Sharma S.; Diassiti A.; Alam Z.; Melone M.; Mulichak A.; Wernimont A.; Bray J.; Loppnau P.; Plotnikova O.; Newberry K.; Sundararajan E.; Houston S.; Walker J.; Tempel W.; Bochkarev A.; Kozieradzki L.; Edwards A.; Arrowsmith C.; Roos D.; Kain K.; Hui R. (2007) Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 151, 100–110. 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Mehlin C.; Boni E.; Buckner F. S.; Engel L.; Feist T.; Gelb M. H.; Haji L.; Kim D.; Liu C.; Mueller N.; Myler P. J.; Reddy J. T.; Sampson J. N.; Subramanian E.; Van Voorhis W. C.; Worthey E.; Zucker F.; Hol W. G. J. (2006) Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol. Biochem. Parasitol. 148, 144–160. 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Fan E.; Baker D.; Fields S.; Gelb M. H.; Buckner F. S.; Van Voorhis W. C.; Phizicky E.; Dumont M.; Mehlin C.; Grayhack E.; Sullivan M.; Verlinde C.; Detitta G.; Meldrum D. R.; Merritt E. A.; Earnest T.; Soltis M.; Zucker F.; Myler P. J.; Schoenfeld L.; Kim D.; Worthey L.; Lacount D.; Vignali M.; Li J.; Mondal S.; Massey A.; Carroll B.; Gulde S.; Luft J.; Desoto L.; Holl M.; Caruthers J.; Bosch J.; Robien M.; Arakaki T.; Holmes M.; Le Trong I.; Hol W. G. (2008) Structural genomics of pathogenic protozoa: an overview. Methods Mol. Biol. 426, 497–513. 10.1007/978-1-60327-058-8_33. [DOI] [PubMed] [Google Scholar]
- Congreve M.; Carr R.; Murray C.; Jhoti H. (2003) A rule of three for fragment-based lead discovery?. Drug Discovery Today 8, 876–877. 10.1016/S1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- Scott D. E.; Coyne A. G.; Hudson S. A.; Abell C. (2012) Fragment-based approaches in drug discovery and chemical biology. Biochemistry 51, 4990–5003. 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- Over B.; Wetzel S.; Grutter C.; Nakai Y.; Renner S.; Rauh D.; Waldmann H. (2013) Natural-product-derived fragments for fragment-based ligand discovery. Nat. Chem. 5, 21–28. 10.1038/nchem.1506. [DOI] [PubMed] [Google Scholar]
- Pascolutti M.; Campitelli M.; Nguyen B.; Pham N.; Gorse A.-D.; Quinn R. J. (2015) Capturing nature’s diversity. PLoS One 10, e0120942. 10.1371/journal.pone.0120942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hert J.; Irwin J. J.; Laggner C.; Keiser M. J.; Shoichet B. K. (2009) Quantifying biogenic bias in screening libraries. Nat. Chem. Biol. 5, 479–483. 10.1038/nchembio.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J.; Carter M.; Mok N. Y.; Brown N. (2016) On the origins of three-dimensionality in drug-like molecules. Future Med. Chem. 8, 1753–1767. 10.4155/fmc-2016-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller N.; Spadola L.; Cowen S.; Patel J.; Schonherr H.; Cao Q.; McKenzie A.; Edfeldt F.; Rabow A.; Goodnow R. (2016) An improved model for fragment-based lead generation at AstraZeneca. Drug Discovery Today 21, 1272–1283. 10.1016/j.drudis.2016.04.023. [DOI] [PubMed] [Google Scholar]
- Hall R. J.; Mortenson P. N.; Murray C. W. (2014) Efficient exploration of chemical space by fragment-based screening. Prog. Biophys. Mol. Biol. 116, 82–91. 10.1016/j.pbiomolbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Morley A. D.; Pugliese A.; Birchall K.; Bower J.; Brennan P.; Brown N.; Chapman T.; Drysdale M.; Gilbert I. H.; Hoelder S.; Jordan A.; Ley S. V.; Merritt A.; Miller D.; Swarbrick M. E.; Wyatt P. G. (2013) Fragment-based hit identification: thinking in 3D. Drug Discovery Today 18, 1221–1227. 10.1016/j.drudis.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Mok N. Y.; Brenk R.; Brown N. (2014) Increasing the coverage of medicinal chemistry-relevant space in commercial fragments screening. J. Chem. Inf. Model. 54, 79–85. 10.1021/ci400632y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldeghi M.; Malhotra S.; Selwood D. L.; Chan A. W. E. (2014) Two-and three-dimensional rings in drugs. Chem. Biol. Drug Des. 83, 450–461. 10.1111/cbdd.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann M. M.; Leach A. R.; Harper G. (2001) Molecular complexity and its impact on the probability of finding leads for drug discovery. J. Chem. Inf. Comput. Sci. 41, 856–864. 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- Keseru G. M.; Erlanson D. A.; Ferenczy G. G.; Hann M. M.; Murray C. W.; Pickett S. D. (2016) Design principles for fragment libraries: maximizing the value of learnings from pharma fragment-based drug discovery (FBDD) programs for use in Academia. J. Med. Chem. 59, 8189–8206. 10.1021/acs.jmedchem.6b00197. [DOI] [PubMed] [Google Scholar]
- Plouffe D.; Brinker A.; McNamara C.; Henson K.; Kato N.; Kuhen K.; Nagle A.; Adrian F.; Matzen J. T.; Anderson P.; Nam T. G.; Gray N. S.; Chatterjee A.; Janes J.; Yan S. F.; Trager R.; Caldwell J. S.; Schultz P. G.; Zhou Y.; Winzeler E. A. (2008) In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc. Natl. Acad. Sci. U. S. A. 105, 9059–9064. 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagaring K., Borboa R., Francek C., Chen Z., Buenviaje J., Plouffe D., Winzeler E., Brinker A., Diagana T., Taylor J., Glynne R., Chatterjee A., and Kuhen K. Novartis-GNF Malaria Box; Genomics Institute of the Novartis Research Foundation (GNF), 2010; www.ebi.ac.uk/chemblntd.
- Derbyshire E. R.; Prudencio M.; Mota M. M.; Clardy J. (2012) Liver-stage malaria parasites vulnerable to diverse chemical scaffolds. Proc. Natl. Acad. Sci. U. S. A. 109, 8511–8516. 10.1073/pnas.1118370109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery V. M.; Bashyam S.; Burrows J. N.; Duffy S.; Papadatos G.; Puthukkuti S.; Sambandan Y.; Singh S.; Spangenberg T.; Waterson D.; Willis P. (2014) Screening and hit evaluation of a chemical library against blood-stage Plasmodium falciparum. Malar. J. 13, 190. 10.1186/1475-2875-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J.; Petter R. C.; Baillie T. A.; Whitty A. (2011) The resurgence of covalent drugs. Nat. Rev. Drug Discovery 10, 307–317. 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- Mah R.; Thomas J. R.; Shafer C. M. (2014) Drug discovery considerations in the development of covalent inhibitors. Bioorg. Med. Chem. Lett. 24, 33–39. 10.1016/j.bmcl.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. A. (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Baell J. B. (2016) Feeling nature’s PAINS: natural products, natural product drugs, and pan assay interference compounds (PAINS). J. Nat. Prod. 79, 616–628. 10.1021/acs.jnatprod.5b00947. [DOI] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N.; Wang J.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur K.; Jain M.; Kaur T.; Jain R. (2009) Antimalarials from nature. Bioorg. Med. Chem. 17, 3229–3256. 10.1016/j.bmc.2009.02.050. [DOI] [PubMed] [Google Scholar]
- Bajsa J.; Singh K.; Nanayakkara D.; Duke S. O.; Rimando A. M.; Evidente A.; Tekwani B. L. (2007) A survey of synthetic and natural phytotoxic compounds and phytoalexins as potential antimalarial compounds. Biol. Pharm. Bull. 30, 1740–1744. 10.1248/bpb.30.1740. [DOI] [PubMed] [Google Scholar]
- Gengenbacher M.; Fitzpatrick T. B.; Raschle T.; Flicker K.; Sinning I.; Muller S.; Macheroux P.; Tews I.; Kappes B. (2006) Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum - Biochemical and structural insights. J. Biol. Chem. 281, 3633–3641. 10.1074/jbc.M508696200. [DOI] [PubMed] [Google Scholar]
- Bajsa J.; McCluskey A.; Gordon C. P.; Stewart S. G.; Hill T. A.; Sahu R.; Duke S. O.; Tekwani B. L. (2010) The antiplasmodial activity of norcantharidin analogs. Bioorg. Med. Chem. Lett. 20, 6688–6695. 10.1016/j.bmcl.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Suberu J. O.; Gorka A. P.; Jacobs L.; Roepe P. D.; Sullivan N.; Barker G. C.; Lapkin A. A. (2013) Anti-Plasmodial polyvalent interactions in Artemisia annua L. aqueous extract - possible synergistic and resistance mechanisms. PLoS One 8, e80790. 10.1371/journal.pone.0080790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon A. I.; Romero L. I.; Ortega-Barria E.; Brun R.; Correa A. M. D.; Gupta M. P. (2006) Evaluation of larvicidal and in vitro antiparasitic activities of plants in a biodiversity plot in the Altos de Campana National Park, Panama. Pharm. Biol. 44, 487–498. 10.1080/13880200600878361. [DOI] [Google Scholar]
- Sauer W. H. B.; Schwarz M. K. (2003) Molecular shape diversity of combinatorial libraries: a prerequisite for broad bioactivity. J. Chem. Inf. Comput. Sci. 43, 987–1003. 10.1021/ci025599w. [DOI] [PubMed] [Google Scholar]
- Schuffenhauer A.; Ertl P.; Roggo S.; Wetzel S.; Koch M. A.; Waldmann H. (2007) The scaffold tree - visualization of the scaffold universe by hierarchical scaffold classification. J. Chem. Inf. Model. 47, 47–58. 10.1021/ci600338x. [DOI] [PubMed] [Google Scholar]
- Bemis G. W.; Murcko M. A. (1996) The properties of known drugs. 1. molecular frameworks. J. Med. Chem. 39, 2887–2893. 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- Morgan H. L. (1965) Generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service. J. Chem. Doc. 5, 107–113. 10.1021/c160017a018. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Gagaring K., Borboa R., Francek C., Chen Z., Buenviaje J., Plouffe D., Winzeler E., Brinker A., Diagana T., Taylor J., Glynne R., Chatterjee A., and Kuhen K. Novartis-GNF Malaria Box; Genomics Institute of the Novartis Research Foundation (GNF), 2010; www.ebi.ac.uk/chemblntd.