Abstract
Recent studies suggest that biofluid-based metabonomics may identify metabolite markers promising for colorectal cancer (CRC) diagnosis. We report here a follow-up replication study, after a previous CRC metabonomics study, aiming to identify a distinct serum metabolic signature of CRC with diagnostic potential. Serum metabolites from newly diagnosed CRC patients (N = 101) and healthy subjects (N = 102) were profiled using gas chromatography time-of-flight mass spectrometry (GC–TOFMS) and ultraperformance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC–QTOFMS). Differential metabolites were identified with statistical tests of orthogonal partial least-squares-discriminant analysis (VIP > 1) and the Mann–Whitney U test (p < 0.05). With a total of 249 annotated serum metabolites, we were able to differentiate CRC patients from the healthy controls using an orthogonal partial least-squares-discriminant analysis (OPLS-DA) in a learning sample set of 62 CRC patients and 62 matched healthy controls. This established model was able to correctly assign the rest of the samples to the CRC or control groups in a validation set of 39 CRC patients and 40 healthy controls. Consistent with our findings from the previous study, we observed a distinct metabolic signature in CRC patients including tricarboxylic acid (TCA) cycle, urea cycle, glutamine, fatty acids, and gut flora metabolism. Our results demonstrated that a panel of serum metabolite markers is of great potential as a noninvasive diagnostic method for the detection of CRC.
Keywords: colorectal cancer, metabolomics/metabonomics, serum metabolites, diagnostic markers
Graphical Abstract
INTRODUCTION
Colorectal cancer (CRC) is the third most common type of cancer in the world and is a major cause of worldwide cancer morbidity and mortality.1 Due to the lack of early and accurate diagnosis, fewer than 40% of CRC patients were diagnosed at the localized stage with relatively high 5-year survival rate.2 To date, colonoscopy is the gold standard for accurate diagnosis of CRC, but its invasive and unpleasant nature often brings unwanted pain and discomfort to the patient. Although certain tumor biomarkers such as carcinoembryonic antigen (CEA) and fecal occult blood testing (FOBT) are commonly used in clinic, the poor sensitivity and specificity limit their application.3–5 Therefore, development of effective molecular biomarkers for early diagnosis has become increasingly important in the management of CRC patients.
Cancer, as a metabolic disease, is characterized by its metabolic transformations in cells essential to sustain their higher proliferative rates and resist cell death signals with altered flux along key metabolic pathways such as glycolysis and tricarboxylic acid cycle (TCA cycle).6 Metabonomics (or metabolomics) enables the quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification.7 This technology has been extensively used to identify metabolite-based biomarkers in various cancers.8–12 Differentially expressed serum or plasma metabolites have been reported, involving intermediates in glycolysis, TCA cycle, urea cycle, arginine and proline metabolism, fatty acid metabolism, and gut flora metabolism associated with CRC morbidity.13,14 However, cancer metabonomics studies15–20 often generate different metabolite markers due to the different clinical protocols used and the wide dynamic range of metabolites measured by different platforms. As a result, very few metabolic markers in a given cancer type have been consistently discovered, confirmed, and validated by use of this approach. Therefore, it is of central importance to replicate the cancer metabonomics studies and verify the biomarker findings.
Here we present a comprehensive serum metabonomics study designed to replicate a previous CRC study13 by our group and confirm whether a serum based metabonomics approach can be used as diagnostic tool for CRC patients. Serum metabolites from newly diagnosed CRC patients (N = 101) and healthy subjects (N = 102) were measured with gas chromatography time-of-flight mass spectrometry (GC–TOFMS) and ultraperformance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC–QTOFMS). The differentially expressed serum metabolites in CRC were identified using univariate and multivariate statistical tools.
MATERIALS AND METHODS
Clinical Samples
All subjects were recruited in the study with the same sample collection protocol. The patients consisting of 101 CRC patients (aged 24–82 years) and 102 healthy subjects (aged 31–76 years) was recruited and diagnosed at the Ruijin Hospital affiliated with Shanghai Jiao Tong University School of Medicine. CRC and control samples were divided into a learning group and a validation group, with the age (p = 0.364, the Pearson’s chi-squared test) and gender (p = 0.281) matched in the learning group. All patients were not on any medication before sample collection. Any subjects in the healthy control group with inflammatory conditions or gastrointestinal tract disorders were excluded. Blood samples were collected in the morning before breakfast, and sera were prepared within one hour after blood collection and then kept at −80 °C. CRC was staged according to TNM classification of malignant tumors. CEA levels for all CRC patients were also assessed. The demographic information and clinical characteristics of all subjects are provided in Table 1.
Table 1.
Clinical Information and Characteristics of Human Subjectsa
| learning group | validation group | |||
|---|---|---|---|---|
|
|
|
|||
| control (n = 62) | CRC (n = 62) | control (n = 40) | CRC (n = 39) | |
| male/female | 28/34 | 34/28 | 0/40 | 23/16 |
| age (mean, range) | 59.4 (31–75) | 60.1 (24–82) | 55.9 (35–76) | 61.8(36–80) |
| CEA (ng/mL, mean, range)b | 27.9 (0.7–891.2) | 26.4 (0.9–376.4) | ||
| location of tumorc | ||||
| AC | 15 | 6 | ||
| TC | ||||
| DC | 5 | 4 | ||
| SC | 6 | 1 | ||
| R | 35 | 28 | ||
| stage (n, male/female) | ||||
| TNM-I | 16 (11/5) | 10 (9/1) | ||
| TNM-II | 25 (13/12) | 18 (11/7) | ||
| TNM-III | 17 (8/7) | 9 (3/6) | ||
| TNM-IV | 4 (2/2) | 2 (0/2) | ||
One patient of stage IV without a location record in training set.
CEA, carcinoembryonic antigen.
R, rectum; AC, ascending colon; DC, descending colon; SC, sigmoid colon.
All the patients signed a consent form. The study was approved by the institutional ethics committees of the Ruijin Hospital.
Serum Sample Preparation and Analysis by GC–TOFMS
Following our previous procedure,13 each 100 μL of serum sample spiked with two internal standards (10 μL of L-2-chlorophenylalanine in water, 0.3 mg/mL; 10 μL of heptadecanoic acid in methanol, 1 mg/mL) was used for metabolite extraction with 300 μL of methanol:chloroform (3:1) at −20 °C for 10 min. An aliquot of the 300 μL supernatant was used for further analysis after a 12,000 rpm centrifuge for 10 min. The samples were vacuum-dried at room temperature. The residue was subjected to a two-step derivatization procedure with 80 μL of methoxyamine (15 mg/mL in pyridine) for 90 min at 30 °C, and 80 μL of BSTFA (1%TMCS) for 60 min at 70 °C. In addition to the internal standards used for quality control, another quality control sample consisting of multiple reference standards was prepared and run with each of 10 samples (see Supplementary Table 3 in the Supporting Information). This QC sample was vacuum-dried and derivatized using the same procedure along with the samples.
The samples were analyzed by Pegasus HT system (Leco Corporation, St. Joseph, MI, USA) coupled with an Agilent 6890N gas chromatography in the order “control–CRC–control”. A QC sample was run after each 10 serum samples. The injection volume was 1 μL with a splitless mode. The injection was set to 270 °C. A DB-5MS capillary column (30 m × 250 μm i.d., 0.25 μm film thickness; Agilent J&W Scientific, USA) was used to separate the metabolites. Helium was used as the carrier gas, with 1.0 mL/min. The GC oven temperature started at 80 °C for 2 min, then ramped to 180 °C at 10 °C/min, to 230 °C at 6 °C/min, and finally to 295 °C at 40 °C/min. The final temperature of 295 °C was maintained for 8 min. The temperature of the transfer interface and the ion source was set to 270 and 220 °C, respectively. The m/z range was set to 30–600 with electron impact ionization (70 eV). The acquisition rate was set to 20 spectra/second.
Serum Sample Preparation and Analysis by UPLC–QTOFMS
The procedure for serum sample treatment and analysis for UPLC–QTOFMS followed our published report with minor modifications.8,13 Each 80 μL of serum sample was used in UPLC–QTOFMS analysis. After addition of internal standard (10 μL of L-2-chlorophenylalanine in water, 0.3 mg/mL), the samples were combined with 400 μL of a mixture of water, methanol, and acetonitrile (1:2:7). The extraction procedure was performed at −20 °C for 10 min after 2 min vortexing and 1 min ultrasonication. The samples were then centrifuged at 12,000 rpm for 20 min. The supernatant was transferred into the sampling vial for UPLC–QTOFMS analysis. Similar to GC–TOFMS analysis, a QC sample consisting of multiple reference standards was prepared (Supplementary Table 3 in the Supporting Information). This QC sample was run after each 10 serum samples.
The samples were kept at 4 °C during the analysis. A 5 μL aliquot of sample was injected into an ultraperformance liquid chromatography system (Waters, USA) with a 100 mm × 2.1 mm, 1.7 μm BEH C18 column (Waters, USA) in the same order of GC–TOFMS. The column was held at 40 °C. The elution procedure for the column was 1–20% B over 0–1 min, 20–70% B over 1–3 min, 70–85% B over 3–8 min, 85–100% B over 8–9 min, and the composition was held at 100% B for 1 min, where A = water with 0.1% formic acid and B = acetonitrile with 0.1% formic acid for positive mode (ES+), while A = water and B = acetonitrile for negative ion mode (ES−). The flow rate was 0.4 mL/min.
A Waters Q-TOF premier (Manchester, U.K.) was used for data collection, with an electrospray source operating in either positive or negative ion mode. The temperature for the source and desolvation gas was set at 120 and 350 °C, respectively. The gas flow for cone is 50 L/h, and 650 L/h for desolvation gas. The capillary voltage and cone voltage were set to 3.2 kV and 35 V for ES+, and 3 kV and 50 V for ES−, respectively. MassLynx software (Waters) was used to collect the data at a centroid data mode with a mass range of 50 to 1000 m/z. The scan time was set at 0.3 s, and the interscan delay was set at 0.02 s over a 9.5 min analysis time. Leucine-enkephalin was used as the lock mass (m/z 556.2771 in ES+ and 554.2615 in ES−).
Data Analysis
The acquired MS data from GC–TOFMS and UPLC–QTOFMS was analyzed according to our previously published work.13 The GC–TOFMS data was analyzed by ChromaTOF software (v 4.34, LECO, USA). After alignment with Statistic Compare component, the CSV file was obtained with three dimensional data sets including sample information, peak retention time, and peak intensities. The internal standard was used for data normalization. Internal standards and any known pseudo positive peaks, such as peaks caused by noise, column bleed, and BSTFA derivatization procedure, were removed from the data set.
The UPLC–QTOFMS ES+ and ES− raw data was analyzed by the MarkerLynx Applications Manager version 4.1 (Waters, Manchester, U.K.) using the following parameters: the initial and final retention time (RT) was set at be 0, and 9.5 min, respectively. The mass range was set to 50–1000 Da, with mass window of 0.05 Da. The internal standard detection parameters were deselected for peak retention time alignment. The isotopic peaks were excluded from the analysis. The minimum intensity was set to 5% of base peak intensity. The noise elimination level was set at 6 and RT tolerance was set at 0.3 min. A list of the ion intensities of each peak detected was generated, using retention time (RT) and the m/z data pairs as the identifier for each ion. The resulting three-dimensional matrix contains arbitrarily assigned peak index (retention time–m/z pairs), sample names (observations), and ion intensity information (variables). To obtain consistent differential variables, the resulting matrix was further reduced by removing any peaks with missing value (ion intensity = 0) in more than 80% samples. The internal standard was used for data quality control (reproducibility) and data normalization. The ion peaks generated by the internal standard were also removed. The final data was normalized to the peak area of the corresponding internal standard.
For GC–TOFMS generated data, metabolite annotation was processed by comparing the mass fragments with NIST 11 Standard mass spectral databases in ChromaTOF software (v 4.34, LECO, USA) with a similarity of more than 70% and then verified by available reference standards in our lab (~800 mammalian metabolite standards). Metabolites obtained from positive (ES+) and negative (ES−) mode of UPLC–QTOFMS analyses were annotated by means of available reference standards in our lab (by comparing the accurate mass (mass difference <0.02 Da) and retention time (<0.5 min)), in addition to web-based resources such as the Human Metabolome Database (http://www.hmdb.ca/) (by comparing the accurate mass with difference less than 0.005 Da).
All annotated metabolites by GC–TOFMS and UPLC–QTOFMS ES+ and ES− (expressed as G, P, and N, respectively) were combined into a new data set for further investigation (the total list of 249 annotated metabolites is shown in Supplementary Table 1 in the Supporting Information). Principal component analysis (PCA) and orthogonal partial least-squares-discriminant analysis (OPLS-DA) were carried out (SIMCA-P 12.0, Umetrics, Umeå, Sweden) to visualize the metabolic alterations between CRC patients and healthy controls after mean centering and unit variance scaling. The default 7-fold cross-validation was applied, in order to guard against overfitting. The variable importance in the projection (VIP) values of all the metabolites from the 7-fold cross-validated OPLS-DA model was taken as a criterion for differential metabolites selection. Those variables with VIP > 1.0 are considered relevant for group discrimination.21 Additionally, the nonparametric univariate method, Mann–Whitney U test, was applied to measure the significance of each metabolite in discriminating CRC patients from healthy controls. Differential metabolites were selected by consideration of both coefficients (VIP > 1 and p < 0.05). The corresponding up- and downregulated trend (fold change) showed how these selected differential metabolites varied between CRC patients and healthy controls, and was used for subsequent metabolic pathway analysis. Furthermore, we conducted box-plot analysis to show the individual metabolite difference between CRC patients and controls with SPSS software (v19, IBM, USA).
RESULTS
Serum Metabolic Profiles of CRC
We obtained 209, 1293, and 1368 spectral features from each sample analyzed by GC–TOFMS, UPLC–QTOFMS ES+, and ES−, respectively. PCA scores plots showed the separation trend between CRC patients and healthy controls in the learning group (Supplementary Figure 1A,E,I in the Supporting Information). Subsequently, three cross-validated OPLS-DA models were established and demonstrated satisfactory modeling and predictive abilities with 1 predictive component and 2 orthogonal components (R2X = 0.245, R2Ycum = 0.881, Q2cum = 0.767) for GC–TOFMS, 1 predictive component and 1 orthogonal component (R2X = 0.302, R2Ycum = 0.95, Q2cum = 0.938) for UPLC–QTOFMS ES+, and 1 predictive component and 3 orthogonal components (R2X = 0.363, R2Ycum = 0.961, Q2cum = 0.894) for UPLC–QTOFMS ES−, respectively (Supplementary Figure 1B,F,J in the Supporting Information). All these results demonstrated the distinct serum metabolic profiles of CRC patients.
Serum Metabolite Markers of CRC
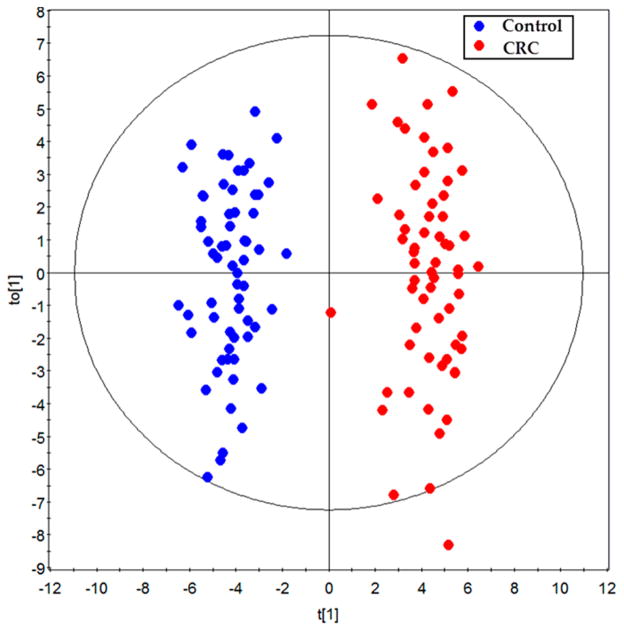
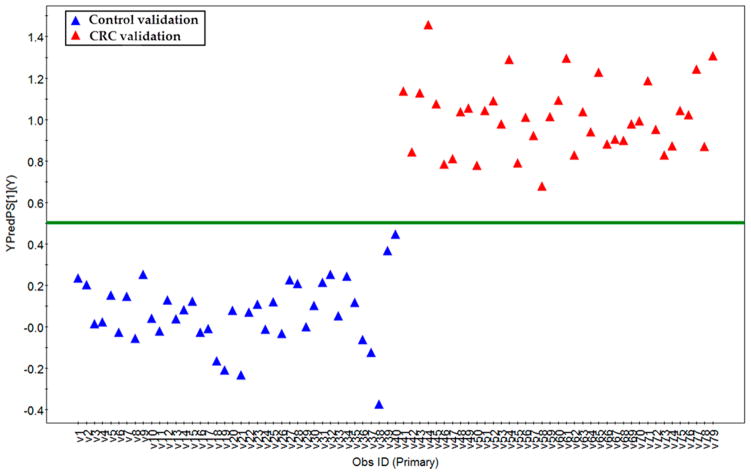
We have annotated 249 metabolites in the sera of each subject (summarized in Supplementary Table 1 in the Supporting Information), which mainly include sugar metabolites (19.2%), amino acid metabolites (14.1%), lipid metabolites (26.9%), short-chain carboxylic acids (12.8%), nucleic acid metabolites (3.6%), gut flora metabolites (10.0%), amines (3.6%), bile acids (3.6%), and others (16.1%). Based on the 249 metabolites in the learning group, an OPLS-DA model was constructed with one predictive component and two orthogonal components with good model parameters (R2X = 0.187, R2Ycum = 0.941, Q2cum = 0.86) (Figure 1). The permutation test assured the validity of the OPLS-DA model with all the R2 (cum) and Q2 (cum) values calculated from the permuted data were lower than the original ones in the validation plot and the Q2 (cum) intercepted the y-axis at −0.309 (Supplementary Figure 2 in the Supporting Information). Furthermore, a set of samples in the validation group (40 control and 39 CRC samples) were used to test the prediction ability of the established OPLS-DA model above. In the Y prediction scores plot, all the samples in the validation groups were correctly assigned to either control or CRC group using a cutoff value of 0.5 (Figure 2, the Y value was set at 1 for CRC, and 0 for controls in the learning group). This result showed the ability of the OPLS-DA model to predict the unknown samples to the right groups with a sensitivity of 100% and a specificity of 100%.
Figure 1.
The scores plot of the OPLS-DA model of the learning group. The OPLS-DA model was constructed using data from 62 CRC patients (red dots) and 62 healthy controls (blue dots).
Figure 2.
Y-predicted scatter plot of samples in validation groups. The erected OPLS-DA model with those samples in the learning group was used to predict the “group membership” (control or CRC) for the samples in a validation data set with 40 healthy controls (blue triangle) and 39 CRC patients (red triangle). The prediction ability of the OPLS-DA model was made with a cutoff of 0.5.
To further test the influence of gender on the quality of the prediction model, two models with males or females only in the training data set were constructed. The samples in the validation set were fed into the models to test the predictive ability of the model. The results showed both models (constructed with only males or females) can correctly assign all the samples in the validation sample set into the right group (CRC or control), suggesting that the gender does not significantly affect the quality of the prediction model (Supplementary Figure 3 in the Supporting Information).
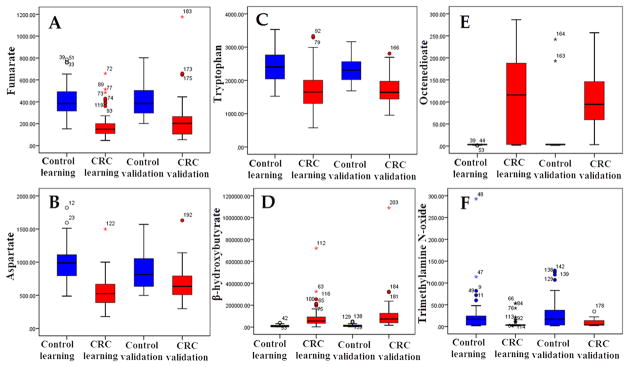
Significantly altered serum metabolites with the VIP threshold (VIP > 1) in the above-mentioned OPLS-DA model, as well as the Mann–Whitney U test (p < 0.05), were selected in CRC patients and are summarized in Table 2. Among these differential metabolites, 36 were confirmed by reference standards, and 10 were identified in both analytical platforms (GC–TOFMS and UPLC–QTOFMS). Variations of these metabolites are expressed in fold change (FC) in CRC subjects from TNM stage I to stage IV (both in learning group and in validation group) in Supplementary Figure 4 in the Supporting Information. Using Kyoto Encyclopedia of Genes and Genomes (KEGG) database,22 several key metabolic pathways that were altered in CRC patients were identified, which involve TCA cycle, urea cycle, tryptophan metabolism, fatty acid metabolism, and gut flora metabolism (Table 2 and Supplementary Figure 4 in the Supporting Information). The box plots of typical metabolites in those metabolic pathways are shown in Figure 3.
Table 2.
Differential Serum Metabolites between CRC Patients and Healthy Controls in the Learning Group
| no. | metabolites | metabolic pathway | FCc | P-valued | VIPe |
|---|---|---|---|---|---|
| 1 | 5-hydroxytryptaminea | tryptophan metabolism | −23.76 | 7.96 × 10−21 | 3.45 |
| 2 | LysoPC(14:0) | phospholipid metabolism | −3.58 | 1.78 × 10−18 | 2.71 |
| 3 | LysoPC(16:1(9Z)) | phospholipid metabolism | −2.65 | 3.77 × 10−18 | 2.60 |
| 4 | 2-oxobutanoic acid | glutathione metabolism | 3.92 | 4.45 × 10−16 | 1.92 |
| 5 | fumaratea | tca cycle | −2.30 | 1.24 × 10−15 | 2.51 |
| 6 | aspartatea | urea cycle | −1.76 | 6.39 × 10−15 | 2.42 |
| 7 | β-hydroxybutyratea | fatty acid metabolism | 8.59 | 9.87 × 10−15 | 1.63 |
| 8 | 4-hydroxystyrene | others | −8.44 | 3.05 × 10−14 | 1.63 |
| 9 | phenylalaninea,b | phenylalanine metabolism | −1.35 | 1.30 × 10−13 | 1.98 |
| 10 | tetrahydrogestrinone | others | 4.18 | 7.16 × 10−13 | 1.84 |
| 11 | LysoPC(20:0) | phospholipid metabolism | −2.32 | 1.33 × 10−12 | 2.23 |
| 12 | glycerola | fatty acid metabolism | 2.61 | 3.14 × 10−12 | 2.22 |
| 13 | glyceric acida | glucose metabolism | −1.55 | 3.37 × 10−12 | 2.30 |
| 14 | cis-aconitatea | TCA cycle | −2.60 | 3.37 × 10−12 | 1.57 |
| 15 | tryptophana,b | tryptophan metabolism | −1.45 | 7.31 × 10−12 | 2.17 |
| 16 | phenol | tyrosine metabolism | −2.85 | 9.33 × 10−12 | 1.59 |
| 17 | octenedioate | fatty acid metabolism | 34.75 | 1.37 × 10−11 | 2.40 |
| 18 | threitola | carbohydrates metabolism | −1.30 | 1.74 × 10−11 | 1.85 |
| 19 | indoleacrylic acid | gut flora metabolism | −1.44 | 3.55 × 10−11 | 2.21 |
| 20 | sphinganine | phospholipid metabolism | 4.08 | 6.29 × 10−11 | 1.35 |
| 21 | 2-hydroxybutyric acida,b | glutathione metabolism | 2.09 | 7.43 × 10−11 | 2.04 |
| 22 | LPA(18:0/0:0) | others | −1.65 | 1.14 × 10−10 | 2.20 |
| 23 | indoxyl sulfate | tryptophan metabolism | −2.85 | 3.46 × 10−10 | 1.79 |
| 24 | histidinea,b | histidine metabolism | −1.28 | 5.82 × 10−9 | 1.91 |
| 25 | 2-aminobutanoic acida | glutathione metabolism | 1.61 | 1.29 × 10−8 | 1.86 |
| 26 | alaninea,b | alanine, aspartate, and glutamate metabolism | −1.39 | 1.68 × 10−8 | 1.66 |
| 27 | 6-phosphogluconic acida,b | pentose phosphate pathway | −1.42 | 6.05 × 10−8 | 1.68 |
| 28 | ribitol | nucleic acid metabolism | −1.49 | 8.45 × 10−8 | 1.62 |
| 29 | 4-hydroxyprolinea | proline metabolism | −1.8 | 3.18 × 10−7 | 1.67 |
| 30 | benzaldehyde | others | −1.38 | 6.12 × 10−7 | 1.33 |
| 31 | hydroquinonea | tyrosine metabolism | −4.87 | 1.25 × 10−6 | 1.55 |
| 32 | ureaa | urea cycle | −1.37 | 1.65 × 10−6 | 1.58 |
| 33 | trihydroxycoprostanoic acid | others | −3.25 | 2.76 × 10−6 | 1.49 |
| 34 | cholic acida | bile acid metabolism | −4.27 | 2.90 × 10−6 | 1.04 |
| 35 | LysoPC(P-18:1(9Z)) | phospholipid metabolism | −3.62 | 3.53 × 10−6 | 1.76 |
| 36 | ubiquinone | oxidative phosphorylation | −1.59 | 4.07 × 10−6 | 1.38 |
| 37 | 3,4,5-trimethoxycinnamic acid | others | −1.57 | 4.59 × 10−6 | 1.20 |
| 38 | ornithinea | urea cycle | −1.27 | 5.43 × 10−6 | 1.20 |
| 39 | indoxyl | tryptophan metabolism | −2.61 | 5.83 × 10−6 | 1.20 |
| 40 | trimethylamine N-oxidea | choline metabolism | −3.39 | 6.71 × 10−6 | 1.01 |
| 41 | oleamidea,b | fatty acid metabolism | 2.55 | 7.91 × 10−6 | 1.68 |
| 42 | oleic acida,b | fatty acid metabolism | 1.55 | 9.31 × 10−6 | 1.58 |
| 43 | inositol | carbohydrates metabolism | −1.28 | 9.31 × 10−6 | 1.40 |
| 44 | chenodeoxycholic acida | bile acid metabolism | −3.54 | 1.17 × 10−5 | 1.10 |
| 45 | serinea | glycine, serine, and threonine metabolism | −1.18 | 1.41 × 10−5 | 1.43 |
| 46 | methioninea,b | cysteine and methionine metabolism | −1.19 | 1.69 × 10−5 | 1.30 |
| 47 | asparaginea | alanine, aspartate, and glutamate metabolism | −1.22 | 2.52 × 10−5 | 1.46 |
| 48 | 2-hydroxy-3-methylpentanoic acid | L-isoleucine metabolism | −8.96 | 3.21 × 10−5 | 1.63 |
| 49 | erythrotetrofuranose | carbohydrates metabolism | −1.32 | 6.67 × 10−5 | 1.32 |
| 50 | carnitine (18:1) | fatty acid metabolism | 1.23 | 7.40 × 10−5 | 1.25 |
| 51 | linolic acida | fatty acid metabolism | 1.43 | 7.56 × 10−5 | 1.34 |
| 52 | creatininea,b | arginine and proline metabolism | 1.30 | 8.75 × 10−5 | 1.43 |
| 53 | β-aspartylserine | glycine, serine, and threonine metabolism | −1.81 | 9.12 × 10−5 | 1.20 |
| 54 | 12a-hydroxy-3-oxocholadienic acid | bile acid metabolism | −2.14 | 1.05 × 10−4 | 1.18 |
| 55 | CPA(18:0/0:0) | phospholipid metabolism | 2.54 | 1.35 × 10−4 | 1.25 |
| 56 | acetyl carnitinea | fatty acid metabolism | 1.30 | 1.82 × 10−4 | 1.11 |
| 57 | allantoic acid | others | −1.20 | 2.05 × 10−4 | 1.07 |
| 58 | elaidic acida | fatty acid metabolism | 1.42 | 2.97 × 10−4 | 1.33 |
| 59 | decanoyl carnitine | fatty acid metabolism | −1.39 | 3.88 × 10−4 | 1.01 |
| 60 | xanthosine | purine nucleotide synthetic | 27.12 | 3.95 × 10−4 | 1.34 |
| 61 | glutamatea,b | glutathione metabolism | −1.29 | 4.95 × 10−4 | 1.02 |
| 62 | adeninea | purine nucleotide synthetic | 1.17 | 5.14 × 10−4 | 1.11 |
| 63 | α-aminoadipic acid | lysine metabolism | −1.93 | 7.70 × 10−4 | 1.20 |
| 64 | pyruvatea | TCA cycle | 1.30 | 1.10 × 10−3 | 1.15 |
| 65 | 3-oxodecanoic acid | fatty acid metabolism | 1.37 | 1.53 × 10−3 | 1.09 |
| 66 | cystinea | cysteine and methionine metabolism | 1.12 | 2.38 × 10−3 | 1.05 |
| 67 | palmitic acida | fatty acid metabolism | 1.24 | 2.46 × 10−3 | 1.09 |
| 68 | allyl isothiocyanate | others | 1.15 | 2.85 × 10−3 | 1.10 |
| 69 | N-acetyl-5-hydroxytryptamine | tryptophan metabolism | −2.49 | 3.87 × 10−3 | 1.01 |
| 70 | proline betaine | others | 4.46 | 7.96 × 10−3 | 1.30 |
| 71 | allisoleucine | valine, leucine, and isoleucine degradation | 1.29 | 1.08 × 10−2 | 1.06 |
| 72 | glycolaldehyde | vitamin B6 metabolism | 2.25 | 2.36 × 10−2 | 1.11 |
Metabolites validated by reference standards.
Metabolites that can be identified by GC–TOFMS and UPLC–QTOFMS.
The fold change (FC) was calculated by the average value of CRC group to that of control group. FC with a value larger than 1.0 indicates a significantly higher level of the serum metabolite in patients while a FC value lower than 1.0 indicates a lower level, compared to healthy controls.
P values were calculated from a nonparametric Mann–Whitney U test.
Variable importance in the projection (VIP) was obtained from OPLS-DA with a threshold of 1.0.
Figure 3.
Box plots of 6 typical differential metabolites concerning 5 metabolic pathways: (A) fumarate, (B) aspartate, (C) tryptophan, (D) β-hydroxybutyrate, (E) octenedioate, (F) trimethylamine N-oxide.
We further compared the differential metabolites identified in this study with those selected from our previous study with a different patient cohort. A panel of 10 metabolites was selected in the two studies between CRC patients and healthy controls with VIP > 1 and p < 0.05 and with the same up and down direction (see first 10 metabolites in Supplementary Table 2 in the Supporting Information). Using these 10 metabolites, an OPLS-DA model was constructed with samples in the training set of the current study (one predictive component and two orthogonal components with R2X = 0.457, R2Ycum = 0.664, Q2cum = 0.600. The samples in the validation data set were fed to the model to test the prediction ability of this metabolite panel. The results showed the model yielded a sensitivity of 83.7% and a specificity of 91.7% (Supplementary Figure 5 in the Supporting Information).
The serum markers identified in this study can also correctly diagnose the CRC patients with low CEA value (<5 ng/mL, n = 58). The samples with high level of CEA and low level of CEA cannot be separated in the PCA scores plot as shown in Supplementary Figure 6 in the Supporting Information. No valid OPLS-DA model can be obtained, suggesting that the CEA values are not associated with changes of serum metabolites. Similar to our previous findings, our attempt to stratify TNM stages (I–IV) of CRC patients using these differential metabolites was not successful. We also found some metabolites that consistently up- and downregulated along with the pathological stages (Supplementary Figure 7 in the Supporting Information). Interestingly, consistently with our previous report, β-hydroxybutyrate was found to continuously increase through stage I to stage IV patients, while two metabolites related to tryptophan metabolism (tryptophan and indoleacrylic acid) were continuously decreasing through stage I to stage IV patients.
DISCUSSION
In this study, we identified 249 serum metabolites of CRC patients with the combination of GC–TOFMS and UPLC–QTOFMS. A robust OPLS-DA model based on these identified metabolites was able to distinguish all of the CRC patients including all the TNM-I stage patients from healthy controls, from which 72 metabolites were found differentially expressed in CRC subjects. Compared with our previous CRC metabonomics findings,13,14 several key metabolic pathways including TCA cycle, urea cycle, glutathione metabolism, fatty acid metabolism, and gut microflora metabolism were consistently altered in association with CRC (Supplementary Table 2 in the Supporting Information). Some previously reported metabolite markers were significantly different between CRC and control subjects with univariate statistics in the current study, but do not meet the criteria in multivariate statistics (Supplementary Table 2 in the Supporting Information). There were also inconsistencies in the differential metabolites compared to our previous study.13 For example, lactate and several amino acids such as tyrosine and leucine were found to be differential metabolites in the serum of CRC patients in our previous study,13 but not identified as biomarkers in the current study. Comparing the two CRC metabonomics studies, among the 32 differential metabolites identified in the previous study, only nervonic acid was not detected in the present study. About 70.1% (22 out of 31) of those detected metabolites are also significantly different between CRC patients and healthy controls in this replication study (Supplementary Table 2 in the Supporting Information). Several possible factors may contribute to such a discrepancy between the two data sets. First, in the current study, 25.7% of the CRC patients were diagnosed at stage I, while only 14.1% of the CRC patients were at stage I in the previous study. Second, different data analysis procedure may also result in differences in the metabolite markers identified. In our previous study,13 the data obtained from GC–TOFMS, UPLC–QTOFMS (ES+), and UPLC–QTOFMS (ES−) were analyzed separately, and then the most statistically significant variables were merged from the three platforms. The metabolite annotation was only performed on those differential variables. However, in this study, with the increased entries in our in-house library and the increased knowledge in metabolite annotation, we were able to annotate the metabolites detected from the three platforms before statistical analysis.
The most significant differential metabolites (based on p value) obtained from our previously published human hepatocellular carcinoma study23 were compared with the CRC metabolite markers. Only two metabolites (oleamide and ornithine) in the most significantly differing 20 metabolites (based on the p value) in the HCC study were found in the 22 differential metabolites (based on p value) in the two CRC studies (Supplementary Table 2 in the Supporting Information), suggesting that the metabolite markers identified in the current study may be specific to CRC phenotype. Consistent with our previous report,13 pyruvate, an important intermediate in glycolysis, was also detected higher in the CRC patients compared with healthy controls. In addition several intermediates in TCA cycle such as fumarate and cis-aconitate were found depleted in CRC subjects. The increased pyruvate and decreased intermediates in TCA cycle suggest an impaired mitochondrial respiration in CRC, which is in line with previous metabonomics studies.14,18 Interestingly, a decreased entry of pyruvate into the TCA cycle in cancer cells was also observed in leukemia cells by Ismael et al.24 These metabolic changes may be indicative of an increased oxidation of non-glucose carbon sources such as fatty acids.
About 10 amino acids detected in our study such as intermediates in urea cycle (aspartate and ornithine) and metabolites related to glutamine and proline metabolism were found decreased significantly in CRC serum compared to healthy controls. While higher concentrations of various amino acids in CRC tissue were reported compared to normal mucosa,17,19,20 the findings with depleted amino acids in CRC serum may suggest the higher absorption of amino acids by the tumor cells to sustain the rapid cell proliferation. One example is that the decreased level of serine in CRC patients may be resulted from the higher consumption of serine in the cancer cells, as evidenced by increased serine 3-phosphoglycerate dehydrogenase and serine hydroxymethyltransferase observed in human colon carcinoma.25 Furthermore, lower level of glutamate in CRC patients was observed in CRC patients in the current study and our previous study.13 As higher consumption of glutamine in tumor cells has been reported to be essential for the production of macromolecules such as fatty acid and nuclear acids,26,27 more circulating glutamate may be transformed to glutamine in CRC patients to compensate the higher consumption in tumor tissue cells.
A notable metabolic feature of CRC subjects was the remarkably disturbed lipid (including fatty acids) metabolism. As shown in Table 2, four lysophosphatidylcholines (LysoPCs) (LysoPC(14:0), LysoPC(16:1), LysoPC(20:0), and LysoPC(P-18:1)) were observed significantly lower in the serum of CRC patients compared with healthy controls. Consistent with our observation, decreased signal of LysoPC in cancers was also observed in previous reports.28,29 The decrease in LysoPCs was reported to be associated with body weight loss and activated inflammatory status in cancer patients.29 Therefore, the observed decreased levels of LysoPCs in CRC patients may be indicative of higher decomposition rate of LysoPCs to support cancer metabolism and activities. Increased degradation of LysoPCs may result in an increased level of the FFAs, which was also observed in our study (for example, oleic acid, linolic acid palmitic acid, and elaidic acid were elevated in CRC patient serum, Table 2). Additionally, increased fatty acid synthesis characterized by increased expression of fatty acid synthase (FAS) and stearoyl-CoA desaturase-1 (SCD 1) was also reported to be an important metabolic characteristic for cancer cells.30,31 The increased levels of fatty acids may also be associated with increased FAS and SCD1 in CRC patients.
Concomitant with the decreased LysoPCs and increased FFAs, there was a significant elevation of glycerol and β-hydroxybutyrate, the most important ketone body of fatty acid β-oxidation, observed in the CRC patient, which was consistent with previous serum metabonomics study of CRC.13 In fact, FFA and glycerol turnover rates were reported to be higher in cancer patients compared with healthy normal subjects. Increased fatty acid β-oxidation may be used as a fuel source in cancer patients.32 Meanwhile, conjugation with carnitine is essential for long chain fatty acids to cross the mitochondrial membranes for β-oxidation. In our study, carnitine (18:1) and acetyl carnitine were both significantly increased, while decanoyl carnitine was found decreased in CRC patients. Fatty acid (18:1) is a long chain fatty acid, and decanoic acid is a medium chain fatty acid. The medium chain fatty acids can freely diffuse into mitochondria and be oxidized, while the long chain fatty acids can cross mitochondrial membrane only after they are conjugated with carnitine. Therefore, the conjugated long chain fatty acid, carnitine (18:1), and its β-oxidation product, acetyl carnitine, but not decanoyl carnitine, are found increased in CRC with increased consumption of energy substrates.
A number of differentially expressed metabolites involved in tryptophan metabolism, phenylalanine and tyrosine metabolism, bile acid metabolism, and choline metabolism (Table 2), which are linked to gut microbial-host cometabolism, were observed in the CRC serum metabolic profile. Consistent with the depleted tryptophan in CRC patients in our previous study,13 tryptophan and its metabolites, 5-hydroxytryptamine (5-HT), N-acetyl-5-HT, indoxyl, and indoxyl sulfate, were found significantly depleted in CRC patients. Particularly, 5-HT was observed to be the most significantly lowered metabolite in the CRC patients compared with healthy controls. Most of the 5-HT production in the human body occurred in the gastrointestinal tract, which is important for the regulation of intestinal activity.33 In fact, a recent report showed that a lower level of serotonin in the rat colon can enhance colonic dysplasia in a high fat diet rat model.34 Our results support the important role of 5-HT metabolism in the CRC carcinogenesis. Intermediates in phenylalanine and tyrosine metabolism, including hippuric acid, phenol, and hydroquinone, were produced in gut microbiota by fermentation of dietary polyphenols and aromatic amino acids.35 Gut microbiota has been suspected to play a key role in the carcinogenesis and progression of CRC.36 These abnormal metabolites related to gut microbiota in our study indicated that the altered gut flora metabolism was closely associated with CRC morbidity, as suggested in our recent study with structural imbalance of gut microbiota in CRC patients.36 We presumed that the abnormalities of gut flora metabolism might be a distinct metabolic signature of CRC.
Significantly elevated levels of 2-hydroxybutyrate, 2-oxobutyrate, and 2-aminobutyrate were observed in our study. As a byproduct of the conversion from cystathione to cysteine (2-oxobutyrate is the intermediate), 2-hydroxybutyrate was considered a biomarker of glutathione status.37 Therefore, the elevation of 2-hydroxybutyrate and 2-oxobutyrate may indicate a higher level of oxidative stress in the CRC patients. Additionally, 2-aminobutyrate is used to synthesize ophthalmate,38 which is indicative of glutathione consumption through activation of γ-glutamyl cysteine synthetase. The elevated level of 2-aminobutyrate further indicates a higher level of oxidative stress in CRC patients compared with healthy controls.
The lower serum level of ubiquinone may be associated with CRC progression along with elevated glutathione metabolism as ubiquinone was reported to suppress fat-induced colon carcinogenesis as an antioxidant.39 In addition, the level of ubiquinone was also reported to be negatively correlated with redox status.40 The lower level of ubiquinone in CRC patients is consistent with the higher levels of metabolites associated with glutathione metabolism such as 2-hydroxybutyrate and 2-aminobutyrate.
In summary, a panel of differentially expressed metabolites was identified, which verified most of the markers discovered in our previous CRC metabonomics study. We confirmed that serum based metabonomics is able to discriminate CRC patients from healthy controls, reassuring us about the feasibility of developing a new diagnostic tool for CRC with a serum metabolite signature.
Supplementary Material
Acknowledgments
This work was financially supported by the National Basic Research Program of China (2007 CB914700, No. 2011CB910603), National Natural Science Foundation of China Program (20875061), and the Natural Science Foundation of Shanghai (10ZR1414800). Shanghai Rising-star Program of the Science and Technology Commission of Shanghai Municipality (No.10QA1401400).
Footnotes
Notes
The authors declare no competing financial interest.
PCA and OPLS-DA scores plots and representative chromatograms; permutation test; Y-predicted scatter plots; heat map; the metabolites with a consistent trend of alteration from stage I to stage IV of CRC patients; a list of the identified serum metabolites; most significant metabolite markers identified in two CRC metabonomics studies; quality control information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet. 2005;365(9454):153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA—Cancer J Clin. 2011;61(4):212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher RH. Carcinoembryonic antigen. Ann Intern Med. 1986;104:66–73. doi: 10.7326/0003-4819-104-1-66. [DOI] [PubMed] [Google Scholar]
- 4.De Noo ME, Tollenaar RA, Deeldar AM, Bouwman LH. Current status and prospects of clinical proteomics studies on detection of colorectal cancer: Hopes and fears. World J Gastroenterol. 2006;12:6594–6601. doi: 10.3748/wjg.v12.i41.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronborg O, Fenger C, Olsen J, Jørgensen OD, Søndergaard O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348(9040):1467–1471. doi: 10.1016/S0140-6736(96)03430-7. [DOI] [PubMed] [Google Scholar]
- 6.Seyfried TN, Shelton LM. Cancer as a metabolic disease. Nutr Metab. 2010;7:1–22. doi: 10.1186/1743-7075-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson JK, Lindon JC, Holmes E. “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, Qi X, Cao Y, Su M, Wang X, Xu LX, Yen Y, Liu P, Jia W. Serum and Urine Metabolite Profiling Reveals Potential Biomarkers of Human Hepatocellular Carcinoma. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Z, Zhao JS, Li JJ, Peng DN, Wang XY, Chen TL, Qiu YP, Chen PP, Li WJ, Xu LY, Li EM, Tam JPM, Qi RZ, Jia W, Xie D. A Combined Proteomics and Metabolomics Profiling of Gastric Cardia Cancer Reveals Characteristic Dysregulations in Glucose Metabolism. Mol Cell Proteomics. 2010;9(12):2617–2628. doi: 10.1074/mcp.M110.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiziani S, Lopes V, Gunther UL. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 2009;11:269–276. doi: 10.1593/neo.81396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine Metabolomics Analysis for Kidney Cancer Detection and Biomarker Discovery. Mol Cell Proteomics. 2009;8(3):558–570. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua Y, Qiu Y, Zhao A, Wang X, Chen T, Zhang Z, Chi Y, Li Q, Sun W, Li G, Cai Z, Zhou Z, Jia W. Dynamic Metabolic Transformation in Tumor Invasion and Metastasis in Mice with LM-8 Osteosarcoma Cell Transplantation. J Proteome Res. 2011;10(8):3513–3521. doi: 10.1021/pr200147g. [DOI] [PubMed] [Google Scholar]
- 13.Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res. 2009;8(10):4844–50. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- 14.Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y, Ni Y, Zhao A, Cai S, Xu LX, Jia W. Urinary metabonomic study on colorectal cancer. J Proteome Res. 2010;9(3):1627–1634. doi: 10.1021/pr901081y. [DOI] [PubMed] [Google Scholar]
- 15.Ma Y, Liu W, Peng J, Huang L, Zhang P, Zhao X, Cheng Y, Qin H. A pilot study of gas chromatograph/mass spectrometry-based serum metabolic profiling of colorectal cancer after operation. Mol Biol Rep. 2010;37(3):1403–1411. doi: 10.1007/s11033-009-9524-4. [DOI] [PubMed] [Google Scholar]
- 16.Ma YL, Qin HL, Liu WJ, Peng JY, Huang L, Zhao XP, Cheng YY. Ultra-High Performance Liquid Chromatography–Mass Spectrometry for the Metabolomic Analysis of Urine in Colorectal Cancer. Dig Dis Sci. 2009;54(12):2655–2662. doi: 10.1007/s10620-008-0665-4. [DOI] [PubMed] [Google Scholar]
- 17.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative Metabolome Profiling of Colon and Stomach Cancer Microenvironment by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 18.Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS) J Proteome Res. 2009;8(1):352–361. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- 19.Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, Fiehn O. Metabolite profiling of human colon carcinoma—deregulation of TCA cycle and amino acid turnover. Mol Cancer. 2008;7:72. doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong ES, Zou L, Li S, Cheah PY, Eu KW, Ong CN. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M900551-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P. Metabolomics Reveals Metabolic Biomarkers of Crohn’s Disease. PLoS ONE. 2009;4(7):e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA—Cancer J Clin. 1999;49(1):33–64. 1. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 23.Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, Qi X, Cao Y, Su M, Xu LX, Yen Y, Liu P, Jia W. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10(7):M110 004945. doi: 10.1074/mcp.M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samudio I, Fiegl M, Andreeff M. Mitochondrial uncoupling and the Warburg effect: molecular basis for the reprogramming of cancer cell metabolism. Cancer Res. 2009;69(6):2163–2166. doi: 10.1158/0008-5472.CAN-08-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57(1):87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuliszkiewicz-Janus M, Janus W, Baczynski S. Application of 31P NMR spectroscopy in clinical analysis of changes of serum phospholipids in leukemia, lymphoma and some other non-haematological cancers. Anticancer Res. 1996;16(3B):1587–1594. [PubMed] [Google Scholar]
- 29.Kriat M, Vion-Dury J, Confort-Gouny S, Favre R, Viout P, Sciaky M, Sari H, Cozzone PJ. Analysis of plasma lipids by NMR spectroscopy: application to modifications induced by malignant tumors. J Lipid Res. 1993;34(6):1009–1019. [PubMed] [Google Scholar]
- 30.Kuhajda FP. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res. 2006;66(12):5977–5980. doi: 10.1158/0008-5472.CAN-05-4673. [DOI] [PubMed] [Google Scholar]
- 31.Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31(9):1509–1515. doi: 10.1093/carcin/bgq131. [DOI] [PubMed] [Google Scholar]
- 32.Legaspi A, Jeevanandam M, Starnes HF, Jr, Brennan MF. Whole body lipid and energy metabolism in the cancer patient. Metabolism. 1987;36(10):958–963. doi: 10.1016/0026-0495(87)90132-6. [DOI] [PubMed] [Google Scholar]
- 33.Berger M, Gray JA, Roth BL. The expanded biology of serotonin. Annu Rev Med. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kannen V, Zanette DL, Fernandes CR, Ferreira FR, Marini T, Carvalho MC, Brandao ML, Elias J, Junior, Mauad FM, Silva WA, Jr, Stopper H, Garcia SB. High-fat diet causes an imbalance in the colonic serotonergic system promoting adipose tissue enlargement and dysplasia in rats. Toxicol Lett. 2012;213(2):135–141. doi: 10.1016/j.toxlet.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 35.Rechner AR, Smith MA, Kuhnle G, Gibson GR, Debnam ES, Srai SKS, Moore KP, Rice-Evans CA. Colonic metabolism of dietary polyphenols: influence of structure on microbial fermentation products. Free Radical Biol Med. 2004;36(2):212–225. doi: 10.1016/j.freeradbiomed.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 36.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2011:1–10. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lord RS, Bralley JA. Clinical applications of urinary organic acids. Part I: Detoxification markers. Altern Med Rev. 2008;13(3):205–15. [PubMed] [Google Scholar]
- 38.Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, Tomita M. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281(24):16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 39.Nohl H, Rohr-Udilova N, Gille L, Bieberschulte W, Jurek D, Marian B, Schulte-Herman R. Suppression of tumour-promoting factors in fat-induced colon carcinogenesis by the antioxidants caroverine and ubiquinone. Anticancer Res. 2005;25(4):2793–2800. [PubMed] [Google Scholar]
- 40.Abdul-Rasheed OF, Farid YY, Al-Nasiri US. Coenzyme Q10 and oxidative stress markers in seminal plasma of Iraqi patients with male infertility. Saudi Med J. 2010;31(5):501–506. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.