INTRODUCTION
The classification of primary nephrotic syndrome (NS) is inadequate because it is based primarily on clinical presentation and histopathology rather than an understanding of the molecular underpinnings of these diseases. From a clinical perspective, the current classification alone does not adequately predict the heterogeneous natural history or response to therapy of individuals within a given glomerular disease category.1 It has been hypothesized that this clinical heterogeneity might be caused by heterogeneity of molecular mechanisms that each present with clinically indistinguishable histopathology. Recent discoveries, including identification of genes associated with familial (Mendelian) focal segmental glomerulosclerosis (FSGS) and treatment-resistant minimal change disease (MCD); the discovery of podocyte autoantigens responsible for primary membranous nephropathy (MN); and the association of APOL1 variants with FSGS histology in African American NS patients, provide indisputable evidence that multiple, specific disease processes can present with indistinguishable NS histopathologic patterns.2–6
Given our poor understanding of MCD, FSGS, and MN biology and our inadequate classification system, it is not surprising that our therapeutic approach to these diseases also is imperfect. No new therapeutic targets have been validated by clinical trials for two decades; current NS therapies rely almost exclusively on immunosuppression, which is used without a clear biological basis, often is not beneficial, and frequently is complicated by significant toxicities.7
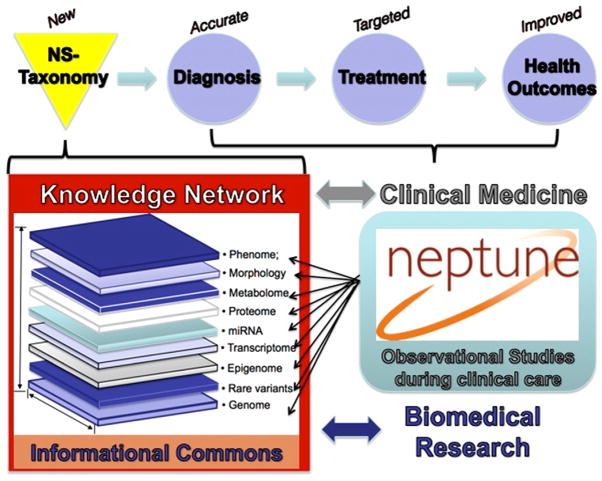
Given these shortcomings, basic science, translational, and clinical studies are needed that address the serious obstacles to providing effective care for our MCD, FSGS, or MN patients. Technologies are in place to obtain comprehensive patient genetic molecular, structural, clinical, environmental, and socioeconomic data at an unprecedented depth, even in multicenter research settings. In-depth knowledge in each of these data domains alone provides valuable information on disease processes. However, disruptive insight can be gained by integrating these multiscalar data sets to define the mechanisms cutting across these knowledge domains, which drive disease initiation and progression. This strategy was coined precision medicine in the Institute of Medicine Report in 2011.8 Charged by the National Research Council of the National Academies to identify a strategy toward a “New Taxonomy of human disease based on molecular biology,”8 the precision medicine strategy proposes the development of new disease definitions to be informed by a multilayered analysis of the disease course under normal clinical care in observational studies. Data from the cohorts of patients along the genotype–phenotype continuum is integrated into an “informational commons”8 and used by the basic and clinical scientists to develop a knowledge network that classifies disease from a molecular mechanism. The new, molecular taxonomy will allow a more accurate diagnosis as a prerequisite for targeted treatments to be used toward improved health outcomes (Fig. 1).
Figure 1.
Precision medicine in NS. Modified with permission from the National Academic Press, 2011.8
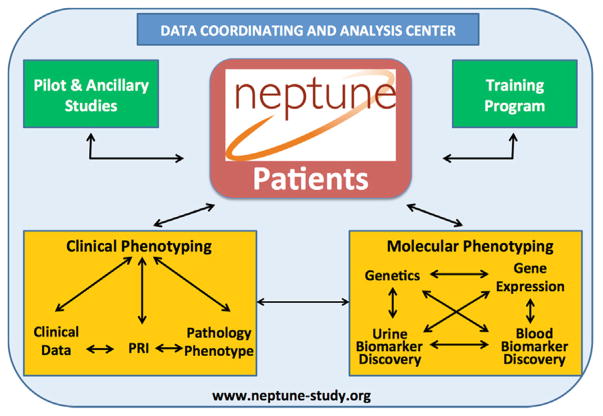
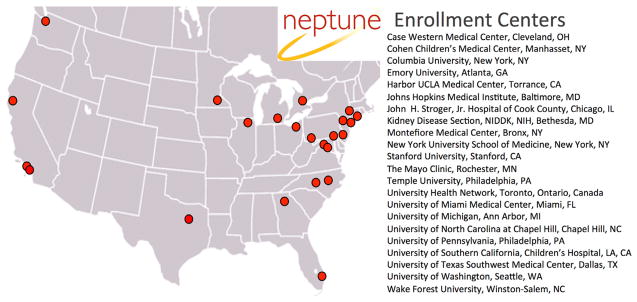
The Nephrotic Syndrome Study Network (NEPTUNE) was created to develop a precision medicine study in patients with NS (Fig. 1). The NEPTUNE cohort studies were designed to generate multilayered data on the disease course encompassing the genotype–phenotype continuum and generating a NS informational commons as recommended by the National Academies (Fig. 2). To date, NEPTUNE has assembled more than 500 NS participants with biosamples for generation of individual catalogues of genome-wide variation, tissue transcriptomes, digital histopathology, longitudinal clinical phenotypes, and patient-reported outcomes across 22 recruitment sites in North America (Fig. 3). These data sets will be used to develop a new taxonomy for discovering disease mechanisms, testing biomarkers, and designing clinical trials that integrate outcomes important to patients.
Figure 2.
NEPTUNE overview. Reprinted with permission from www.neptune-study.org.
Figure 3.
NEPTUNE enrollment centers.
Extensive pilot, ancillary, and training programs will leverage the samples and core data from the cohorts for additional projects that will expand the knowledge base in the NS informational commons. Importantly, NEPTUNE is explicitly inviting investigators from the research community at large and has recruited experts from outside of the core glomerular disease community to enrich its virtual scientific environment.
NEPTUNE is linked closely to EURenOmics, a Framework Program (FP) VII–funded European rare renal disease network (www.eurenomics.eu), in building a human phenotype ontology for rare renal diseases as the base for a new taxonomy of NS. In addition, cohorts of patients with NS are being studied using the precision medicine model in China, the United Kingdom, and Subsaharan Africa. Phenotyping protocols are being harmonized to permit collection of standardized data sets to facilitate discovery and replication across these cohorts.
A strategy to identify and integrate novel biomarkers using genetic messenger RNA, microRNA, protein, digital histology, clinical phenotypes, and self-reported outcomes derived from the cohort study participants are implemented to achieve an accurate diagnosis for NS patients.
Clinical trials of targeted treatments in NS are facilitated using a clinical research network for effective recruitment of rare glomerular disease (ie, Membranous Nephropathy Trial Of Rituximab (MENTOR), clinicaltrials #NCT01180036).
Innovative, web-based, data-sharing and analysis platforms (tranSMART and Nephromine) have been developed to allow the scientific community to access the comprehensive clinical and molecular phenotypes in the informational commons as they emerge from the cohort study analyses.
Longitudinal cohort studies are the main instruments to establish the investigational infrastructure, to establish a molecular definitions of NS, and to facilitate ancillary, pilot, and training studies. To enable deep clinical and molecular phenotyping, a protocol was established9 to longitudinally capture more than 470 parameters for comprehensive analysis. NEPTUNE investigators also have developed a digital archive of whole slide images, which have been annotated with descriptors that characterize patterns of injury.1,10 These agnostic histologic characterizations of glomerular disease processes can be integrated into the informational commons and used to enhance disease mechanism discovery.
Application of precision medicine concepts to the study of primary proteinuric glomerular diseases has necessitated changes in our approach to science. Multi-disciplinary research teams, which bring together scientists who do not traditionally interact and often do not use the same investigative approaches or scientific language, need to find common scientific concepts and communicate effectively.
Studying relatively rare glomerular diseases requires a multicenter approach not only to recruit patients into the cohort, but also to recruit the equally dispersed community of scientific experts necessary to develop the informational commons of precision medicine. As a result, NEPTUNE has collected a cohort of diverse nephrotic syndrome patients from a geographically disparate set of investigative centers, who graciously provided biological samples and clinical and demographic data stored in a central repository managed by a Data Analysis and Coordinating Center, which will be studied by a broad spectrum of core and ancillary investigators from many disciplines.
This issue of Seminars in Nephrology describes the challenges and solutions encountered in developing NEPTUNE as well as some ongoing data integration and analyses. We hope these articles will be instructive for others who implement a precision medicine model to study kidney diseases. For too long we have made marginal progress in the diagnosis, management, and treatment of patients with kidney diseases. The stunning advances in molecular phenotyping technologies (omics), the adoption of electronic records, and the ability to store data on secure servers that allow access to consortium scientists for discovery and study design has allowed NEPTUNE to test the hypothesis that precision medicine will benefit our patients and reinvigorate the study of kidney biology and disease.
Footnotes
Financial disclosure and conflict of interest statements: none.
Contributor Information
Matthias Kretzler, Department of Internal Medicine/Nephrology and Computational Medicine University of Michigan Ann Arbor, Michigan.
John R. Sedor, Departments of Medicine and Physiology and Biophysics Case Western Reserve University Rammelkamp Center for Research and Education The MetroHealth System Cleveland, Ohio.
References
- 1.Schieppati A, Mosconi L, Perna A, Mecca G, Bertani T, Garattini S, et al. Prognosis of untreated patients with idiopathic membranous nephropathy. N Engl J Med. :32985–9. doi: 10.1056/NEJM199307083290203. [DOI] [PubMed] [Google Scholar]
- 2.Hinkes B, Wiggins RC, Gbadegesin R, Vlangos CN, Seelow D, Nürnberg G, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38:1397–405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan J, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–6. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 4.Kestilä M, Lenkkeri U, Männikkö M, Lamerdin J, McCready P, Putaala H, et al. Positionally cloned gene for a novel glomerular protein–nephrin–is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–82. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 5.Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–77. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 6.Reiser J, Polu KR, Möller CC, Kenlan P, Altintas MM, Wei C, et al. TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat Genet. 2005;37:739–44. doi: 10.1038/ng1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cattran D. Management of membranous nephropathy: when and what for treatment. J Am Soc Nephrol. 2005;16:1188–94. doi: 10.1681/ASN.2005010028. [DOI] [PubMed] [Google Scholar]
- 8.Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. Washington, DC: The National Academic Press; 2011. [PubMed] [Google Scholar]
- 9.Gadegbeku CA, Gipson DS, Holzman LB, Ojo AO, Song PX, Barisoni L, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013;83:749–56. doi: 10.1038/ki.2012.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barisoni L, Nast CC, Jennette JC, Hodgin JB, Herzenberg AM, Lemley KV, et al. Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE) Clin J Am Soc Nephrol. 2013;8:1449–59. doi: 10.2215/CJN.08370812. [DOI] [PMC free article] [PubMed] [Google Scholar]