Abstract
The PI3K–AKT kinase signaling pathway is frequently deregulated in human cancers, particularly breast cancer where amplification and somatic mutations of the PIK3CA gene occur with high frequency in patients. Numerous small molecule inhibitors targeting both PI3K and AKT are under clinical evaluation, but dose-limiting toxicities and the emergence of resistance limit therapeutic efficacy. Various resistance mechanisms to PI3K inhibitors have been identified, including de novo mutations, feedback activation of the AKT or cross-talk pathways. Here, we found a previously unknown resistance mechanism to PI3K pathway inhibition that results in AKT rebound activation. In a subset of triple-negative breast cancer cell lines, treatment with PI3K inhibitor or depletion of PIK3CA expression ultimately promoted AKT reactivation in a manner dependent on the E3 ubiquitin ligase Skp2 but independent of PI3K activity or PIP3 production. Resistance to PI3K inhibitors correlated with increased abundance of Skp2, ubiquitylation of AKT, cell proliferation in culture, and xenograft tumor growth in mice. The findings reveal a ubiquitin signaling feedback mechanism by which PI3K inhibitor resistance may emerge in aggressive breast cancer cells.
Introduction
The phosphoinositide 3-kinase (PI3K) signaling pathway is frequently deregulated in virtually all human solid tumors including breast cancer as well as hematological malignancies (1). Amplifications, somatic mutations and other genetic lesions in genes encoding proteins in PI3K signal relay play critical roles in breast cancer by regulating phenotypes such as cell proliferation, survival, metastasis and metabolic reprogramming (2). The three class IA PI3K catalytic subunits p110α, p110β and p110δ are encoded by the PIK3CA, PIK3CB and PIK3CD genes, respectively, and all are activated downstream of receptor tyrosine kinases (RTKs), although PIK3CB can also be activated by G protein-coupled receptor (GPCR) signaling (3). Activated PI3K phosphorylates phosphatidylinositol (4,5)-bisphosphate (PI4,5P2) to generate the second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3). Consequently, synthesis of PIP3 leads to activation of several downstream effector proteins, including the serine and threonine protein kinase AKT, also known as protein kinase B (PKB) (2).
AKT is activated by interaction of the Pleckstrin Homology (PH) domain with either PI3,4P2 or PIP3 (4–7). Maximal AKT activation is achieved by phosphorylation at both threonine 308 (Thr308) and serine 473 (Ser473) mediated by the Phosphoinositide-Dependent Kinase-1 (PDK-1) and Mammalian or Mechanistic Target of Rapamycin Complex 2 (mTORC2), respectively (8–10). Signal termination of PI3K and AKT signaling, on the other hand, is mediated primarily by the tumor suppressors Phosphatase and Tensin homolog (PTEN), which dephosphorylates PIP3 back to PI4,5P2 (11, 12) and Inositol Polyphosphate-4-Phosphatases type I and II (INPP4A and INPP4B) that dephosphorylate PI3,4P2 resulting in PI3P (13). Moreover, protein phosphatase 2A (PP2A) and PH domain leucine-rich repeat-containing protein phosphatase (PHLPP) dephosphorylate AKT at Thr308 and Ser473 respectively, and thereby also participate in signal termination of AKT signaling (14, 15). Once fully activated, AKT mediates downstream signal relay by phosphorylating a myriad of substrates whose phosphorylation is causally implicated in multiple phenotypes associated with malignancies (2).
Amplification and gain-of-function somatic mutations of the PIK3CA gene occur in many human carcinomas, but are most prevalent in breast tumors, particularly in estrogen receptor (ER)-positive breast cancer patients where approximately 40% of cases harbor one of the two most frequent activating PIK3CA hotspot mutations, H1047R and E545K (16, 17). Loss-of-function mutations, deletions and loss of heterozygosity (LOH) in the PTEN gene are also frequently observed in cancers and result in PIP3 accumulation and hyperactivation of AKT signaling (18). Additional genetic lesions in the PI3K and AKT pathway include mutations of PIK3R1 that lead to misregulation of p110α, LOH of INPP4B, activating mutations or copy number gain of the AKT1, AKT2 and AKT3 genes, amplification of PDPK1, as well as a plethora of genetic alterations of upstream RTKs such as HER2 (2).
The frequency of genetic lesions in the PI3K and AKT pathway has made it an attractive target for the development of small molecule inhibitors for therapeutic benefit (19). Pan-, isoform-specific (PIK3CA, PIK3CB and PIK3CD) and dual PI3K inhibitors, mTOR inhibitors, and catalytic and allosteric AKT inhibitors are being evaluated in preclinical as well as phase I and II clinical trials; however, aside from the PIK3CD inhibitor Idelalisib, to date they have shown only relatively modest responses in patients (19). The development of drug resistance, including incomplete inhibition of PI3K, reactivation of PI3K and AKT and activation of compensatory pro-survival pathways is often cited as a major obstacle in achieving complete clinical responses (20, 21). For example, in HER2-amplified tumors, AKT inhibition induces the expression of HER3, such that the combination of HER-targeted kinase inhibitors with AKT inhibitors improve anti-tumor efficacy (22). An adaptive resistance mechanism to PI3K inhibition in PIK3CA-mutant breast cancer cells, increased phosphorylation of the tumor suppressor retinoblastoma (RB) has been observed, and is reversed using a combination of PI3K and cyclin-dependent kinase 4 and 6 (CDK4/6) inhibitors (23). In addition, prolonged treatment with the p110α inhibitor BYL-719 in patients harboring activating PIK3CA mutations leads to loss of PTEN, providing an alternative mechanism of PI3K activation (24). Additional mechanisms of PI3K inhibitor resistance have previously been uncovered, including enhanced ER function mediated by the epigenetic regulator lysine methyltransferase 2D (KMT2D) (25, 26). A systematic analysis of genes that promote PI3K inhibitor resistance also identified proto-oncogene proviral integration site for moloney murine leukemia virus (PIM) kinase overexpression as an AKT-independent mechanism of resistance (27). Other mechanisms of PI3K inhibitor resistance including signaling through ribosomal S6 kinases 3 and 4 (RSK3/4) and AXL have also been uncovered (28, 29).
In the context of additional potential mechanisms of PI3K inhibitor resistance, non-canonical AKT reactivation mechanisms have been described. Non-proteolytic ubiquitination by the S-phase kinase-associated protein 2 (Skp2) promotes the activation of AKT (30), and Skp2 overexpression correlates with AKT activation in cancer and is associated with poor prognosis (31–33), Here, we further investigated this pathway of AKT activation in the context of PI3K inhibitor-treated triple negative breast cancer (TNBC) cells.
Results
PI3K Inhibition Induces Reactivation of AKT Signaling
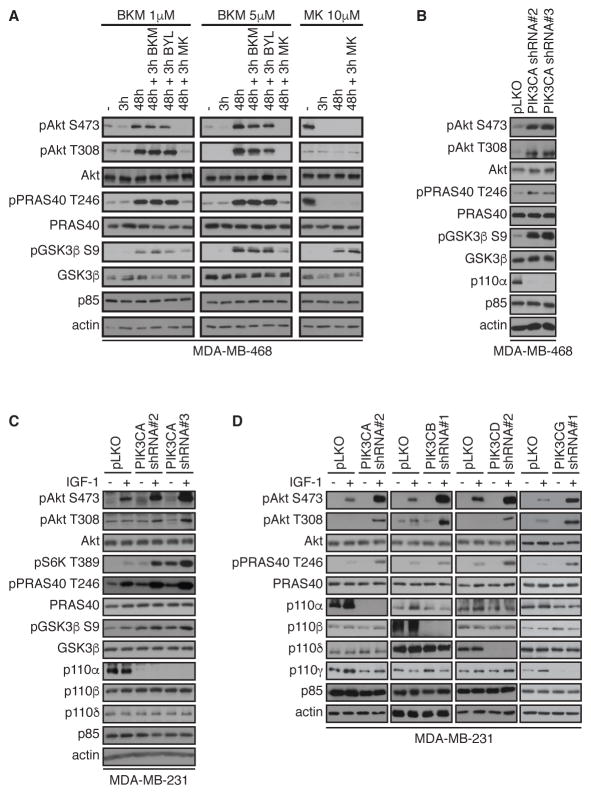
Numerous studies have shown that AKT signaling is reactivated in various cancer cell lines after treatment with PI3K inhibitors. To examine the effect of PI3K inhibitors on AKT signaling, MDA-MB-468 breast cancer cells were treated with two different concentrations of BKM-120 (a pan class I PI3K inhibitor, trade name buparlisib) and the AKT-specific inhibitor, MK-2206. As expected, AKT phosphorylation at Ser473 and Thr308 was blocked after a 3-hour treatment with both PI3K and AKT inhibitors (Figure 1A). However, when cells were treated for 48 hours with BKM-120, inhibition of AKT phosphorylation was no longer observed, and reactivation of AKT signaling was evidenced by enhanced AKT phosphorylation at Ser473 and Thr308 as well as phosphorylation of the AKT substrates PRAS40 and GSK3β.
Figure 1. PI 3-K inhibition and depletion lead to AKT reactivation.
(A) MDA-MB-468 cells were treated with BKM-120 (1 or 5 μM), MK-2206 (10 μM) for 3 or 48 hours, alone or with 1 μM of BKM-120, BYL-719, or MK-2206 for an additional 3 hours, as indicated. Cells were then harvested and lysates were immunoblotted for the indicated total and phosphorylated proteins. T, Thr; S, Ser.
(B) MDA-MB-468 cells were infected with shRNA lentivirus targeting PIK3CA or control vector (pLKO). After selection, cells were harvested, and lysates were immunoblotted for the indicated total and phosphorylated proteins.
(C) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO. Cells were serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+), harvested, and lysates were immunoblotted for the indicated antibodies.
(D) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA, PIK3CB, PIK3CD, PIK3CG or control pLKO. Cells were serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+), harvested and lysates were immunoblotted with the indicated antibodies. Blots are representative of at least 3 independent experiments. See also fig. S1A–C for additional experiments with BKM-120 time course, depletion of PIK3CA with multiple shRNA hairpins and analysis of pAKT1 pS473 and pS474 with PIK3CA depletion.
AKT reactivation in response to PI3K inhibition was observed in MDA-MB-468 cells as long 120 hours after treatment with BKM-120 (fig. S1A). AKT reactivation was not affected by the addition of fresh BKM-120 or BYL-719 (a selective p110α inhibitor also known as alpelisib), for an additional 3 hours or 48 hours after the initial treatment (Figure 1A). Re-addition of PI3K inhibitors did not reduce AKT phosphorylation at Ser473 and Thr308, nor did the treatment affect the phosphorylation of PRAS40 and GSK3β, suggesting that AKT reactivation was independent of PI3K activity. However, AKT phosphorylation was detectably eliminated by the addition of MK-2206 alone or in addition to BKM-120 (Figure 1A).
Depletion of PIK3CA with two distinct shRNA hairpins in MDA-MB-468 cells also induced AKT reactivation evidenced by increased AKT phosphorylation at Ser473, Thr308 and phosphorylation of PRAS40 and GSK-3β (Figure 1B). Similarly, AKT reactivation was induced by PIK3CA depletion in MDA-MB-231 breast cancer cells in response to IGF-1 stimulation (Figure 1C). Moreover, AKT reactivation was also observed with 4 additional and distinct shRNA hairpin sequences, indicating that AKT reactivation is unlikely resulting from shRNA off-target effects (fig. S1B). Furthermore, both AKT1 and AKT2 showed reactivation in PIK3CA-depleted cells (fig. S1C), and AKT reactivation was not due to compensatory expression of PIK3CB or PIK3CD (Figure 1C). Finally, depletion of both PIK3CA, PIK3CB, PIK3CD and PIK3CG in MDA-MB-231 cells induced AKT reactivation in response to IGF-1 (Figure 1D), indicating that AKT reactivation is a common event upon PI3K pathway inhibition.
AKT Reactivation Upon PI3K Pathway Inhibition is Restricted to a Subset of TNBC Cell Lines
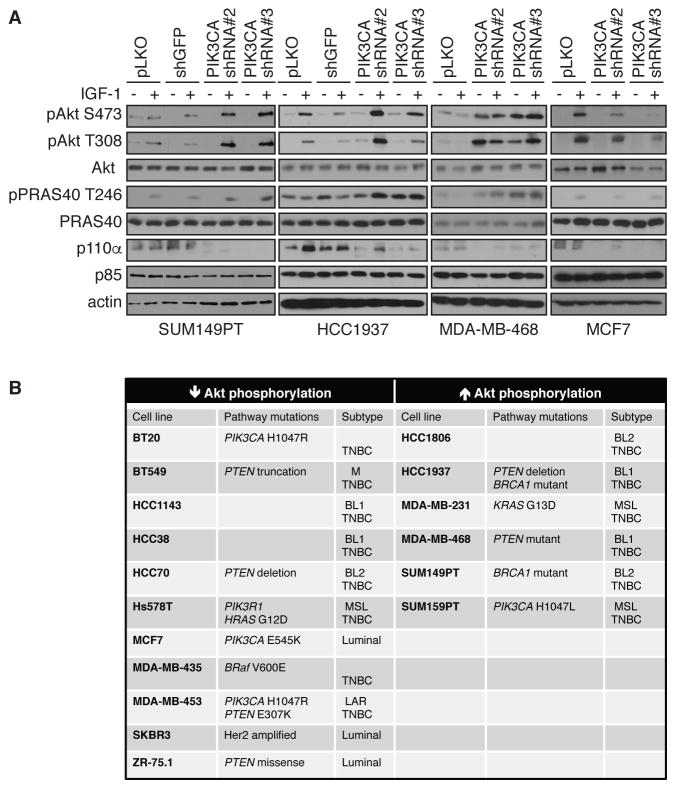
We next tested whether AKT reactivation upon PI3K pathway inhibition is a common event in breast cancer cells. Surprisingly to us, whereas AKT reactivation was observed in SUM149PT, HCC1937 or MDA-MB-468 cells, in the ER-positive breast cancer cell line MCF7, PIK3CA depletion showed a marked inhibition of AKT phosphorylation and activity (Figure 2A). We then tested a larger panel of breast cancer cell lines using the same PIK3CA shRNA approach and evaluated AKT Ser473 and Thr308 phosphorylation and activity (pPRAS40 and pGSK3β). The results (Figure 2B) revealed that of the total 17 breast cancer cell lines tested here, 6 showed AKT reactivation and other cell lines in this panel showed inhibition of AKT. All 6 cell lines that showed AKT reactivation upon PIK3CA depletion are TNBC, although this was not a defining characteristic of the reactivation phenotype, since several TNBC lines showed AKT inhibition and not reactivation upon PI3K pathway inhibition (Figure 2B).
Figure 2. PI3K depletion does not lead to AKT reactivation in all breast cancer cell lines.
(A) SUM149PT, HCC1937, MDA-MB-468 and MCF7 cells were infected with shRNA lentivirus targeting PIK3CA, control pLKO or shGFP as additional negative control. Cells were serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+), harvested and lysates were immunoblotted with the indicated antibodies. Blots are representative of at least 3 independent experiments.
(B) Summary table showing the different breast cancer cell lines tested in this study for AKT reactivation. TNBC, triple negative breast cancer. TNBC classification according to Lehmann et al (51), BL1, Basal-like 1; BL2, basal-like 2; M, mesenchymal; MSL, mesenchymal stem-like; LAR, luminal androgen receptor. Note that BT20 and MDA-MB-435 are only classified as TNBC.
AKT Reactivation is Dependent on mTORC2 and PDK-1
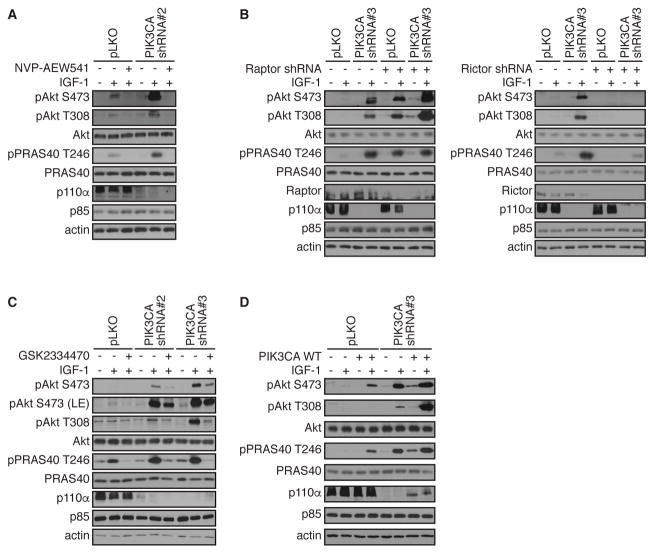
Given that AKT reactivation was observed upon IGF-1 stimulation (Figure 1C), we next treated PIK3CA-depleted MDA-MB-231 cells with the IGF-1R inhibitor NVP-AEW541. AKT reactivation in PIK3CA-depleted cells was completely blocked upon IGF-1R inhibition (Figure 3A). Similarly, AKT reactivation in PIK3CA-depleted cells was attenuated in Rictor-depleted cells, indicative of an mTORC2 requirement for Ser473 phosphorylation as part of the AKT reactivation mechanism (Figure 3B). By contrast, depletion of the mTORC1 component Raptor (RPTOR) actually further promoted AKT reactivation compared to PIK3CA depletion alone (Figure 3B), consistent with previous studies demonstrating that Raptor depletion induces AKT activation (9). Therefore, AKT reactivation upon PI3K pathway inhibition is mTORC2 dependent.
Figure 3. AKT reactivation is dependent on upstream regulators.
(A) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO. Cells were serum-starved overnight (−), treated with IGF-1R inhibitor, NVP-AEW541 for 15 min before stimulation with IGF-1 for 20 min (+). Cells were harvested and lysates were immunoblotted with the indicated antibodies.
(B) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO alone or with the indicated combinations of shRNA specific for Raptor or Rictor, respectively. Cells were serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+). Cells were harvested and lysates were immunoblotted with the indicated antibodies.
(C) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO. Cells were serum-starved overnight (−), treated with PDK-1 inhibitor GSK2334470 for 15 min before stimulation with IGF-1 for 20 min (+). Cells were harvested and lysates were immunoblotted with the indicated antibodies. Data are representative of at least 3 independent experiments.
(D) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO, in the absence or presence of cDNA directing expression of p110α. Cells were serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+). Cells were harvested and lysates were immunoblotted with the indicated antibodies.
AKT reactivation in PIK3CA-depleted cells was also blocked by pre-treatment with the PDK-1 inhibitor GSK2334470 (34) (Figure 3C). Finally, wild type p110α was expressed in control cells, and as expected, this promoted AKT phosphorylation at Ser473 and Thr308 and PRAS40 and GSK-3β phosphorylation (Figure 3D). Notably, enhanced AKT activity was also observed in PIK3CA-depleted cells. Together, these data show that AKT reactivation in PI3K pathway-inhibited cells is dependent on the canonical IGF-1R/PDK-1/mTORC2 pathway.
AKT Reactivation is PI3K-independent
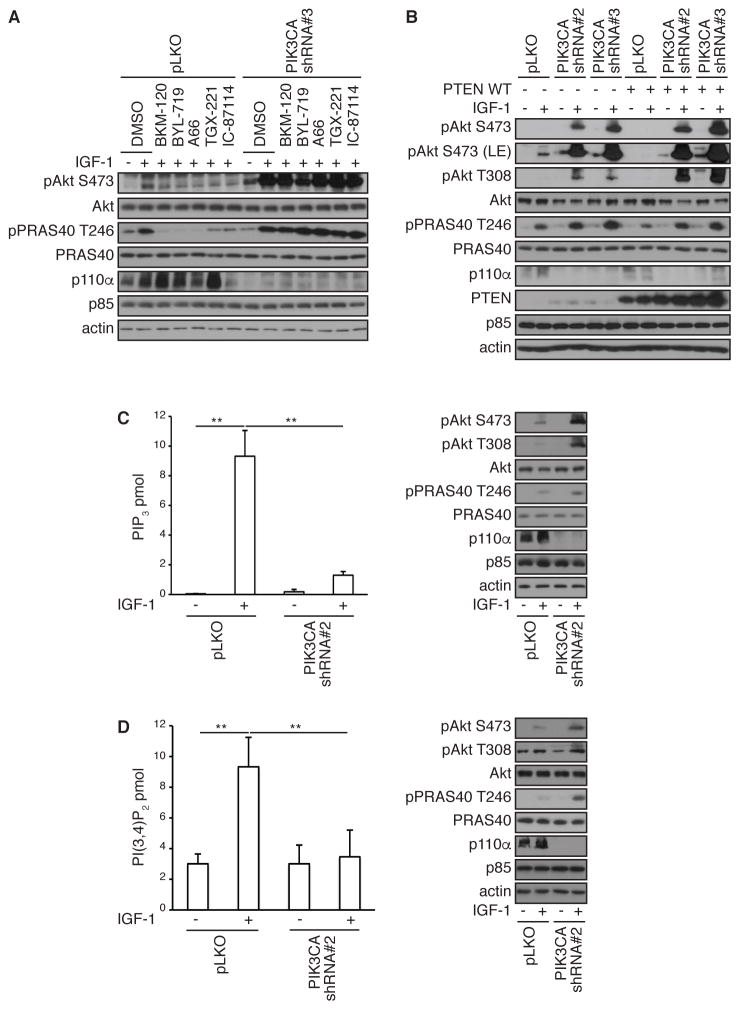
We next determined whether AKT reactivation in PI3K pathway-inhibited cells required any form of PI3K activity. Control and PIK3CA-depleted cells were treated with BKM-120, BYL-719, A66, TGX-221 or IC-87114, inhibitors that are specific to p110α (PIK3CA), p110β (PIK3CB) and p110δ (PIK3CD), respectively. Control cells showed AKT phosphorylation and activity in response to IGF-1, and this was robustly attenuated by all PI3K inhibitors. However, AKT reactivation in PIK3CA-depleted cells was unaffected by either pan or specific PI3K inhibitors, indicating that AKT reactivation is largely independent of PI3K activity.
To provide further evidence for PI3K-independent AKT reactivation, PTEN was expressed in control and PIK3CA-depleted cells. Whereas expression of PTEN robustly attenuated AKT and PRAS40 phosphorylation in control cells, as expected, there was no effect on AKT reactivation in PIK3CA-depleted cells (Figure 4B). We also measured the production of PIP3 and PI3,4P2 in control and PIK3CA-depleted cells in response to IGF-1. Depletion of PIK3CA in MDA-MB-231 cells promoted AKT and PRAS40 phosphorylation (Figure 4C). In control cells, we detected a significant increase in PIP3 also in response to IGF-1. However, depletion of PIK3CA profoundly reduced PIP3 production (Figure 4C), yet under these conditions, robust AKT activity is observed. Similar responses were detected with PI3,4P2 (Figure 4D). Thus, class I PI3K and PIP3/PI3,4P2 do not prominently contribute to AKT reactivation in MDA-MB-231 breast cancer cells in which the PI3K pathway is inhibited.
Figure 4. AKT reactivation is PI3K-independent.
(A) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO. Cells were serum-starved overnight (−), treated with indicated inhibitors for 15 min before stimulation with IGF-1 for 20 min (+). Cells were harvested and lysates were immunoblotted with the indicated antibodies.
(B) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO and then transfected with PTEN WT or control pcDNA vector. Cells were serum-starved overnight (−), or stimulated with IGF-1 for 20 min (+). Cells were harvested and lysates were immunoblotted with the indicated antibodies.
(C and D) MDA-MB-231 cells were infected with shRNA lentivirus targeting PIK3CA or control pLKO. Cells were serum-starved overnight (−), or stimulated with IGF-1 for 20 min (+). Phospholipids were isolated and PIP3 (C) or PI3,4P2 (D) were quantified by ELISA or cells lysates were immunoblotted with the indicated antibodies. Data are means ± S.E.M. from three independent experiments, each carried out in triplicate. ** P < 0.01 by a two-sided Student’s t-test.
AKT Reactivation is Skp2-dependent
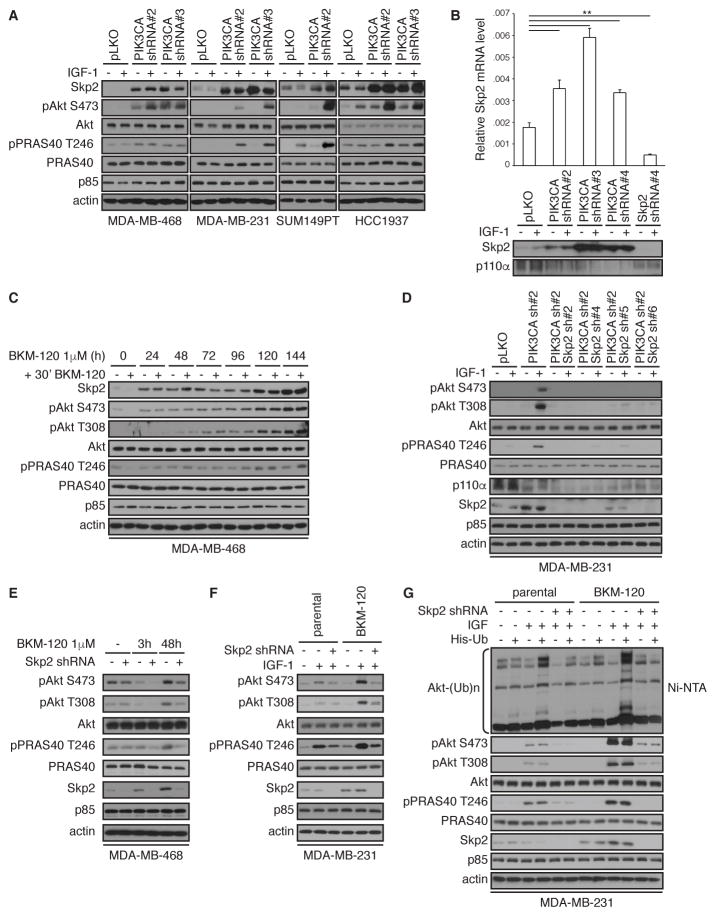
To identify the molecular mechanism by which AKT is reactivated in a PI3K-independent manner, we investigated the role of the ubiquitin E3 ligase Skp2, which has been shown to contribute to AKT activation through ubiquitination (30, 35). Strikingly, we detected increased abundance of total Skp2 protein in breast cancer cell lines depleted with PIK3CA and which show AKT reactivation (Figure 5A). This was also evident at the mRNA level with 3 distinct PIK3CA shRNA hairpins (Figure 5B).
Figure 5. AKT reactivation is Skp2-dependent.
(A) The indicated cell lines were infected with shRNA lentivirus targeting PIK3CA or control pLKO. Cells were serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+), harvested and lysates were immunoblotted with the indicated antibodies.
(B) MDA-MB-231cells infected with PIK3CA and/or SKP2 shRNA lentiviral vectors or control pLKO. SKP2 mRNA expression was measured by real-time RT-PCR and corresponding lysates were immunoblotted with indicated antibodies. Data are means ± S.E.M. from three independent experiments. ** P < 0.01 by a two-sided Student’s t-test.
(C) Immunblotting in lysates from MDA-MB-468 cells treated with BKM-120 (1 μM) for up to 144 hours, then treated again with BKM-120 (1 μM) for an additional 30 min.
(D) Immunblotting in lysates from MDA-MB-231cells infected with PIK3CA, SKP2, or PIK3CA in combination with SKP2 shRNA lentiviral vectors or control vector (pLKO) and serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+).
(E) Immunblotting in lysates from MDA-MB-468 cells infected with SKP2 shRNA lentiviral vector or control pLKO and then treated with BKM-120 (1 μM) for 3 or 48 hours.
(F) Immunblotting in lysates from MDA-MB-231 cells, parental and resistant to 1 μM BKM-120 (BKM-120), that were infected with SKP2 shRNA lentiviral vector or control pLKO vector and serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+).
(G) MDA-MB-231 cells, parental and BKM-120–resistant, were infected with SKP2 shRNA lentiviral vector or control pLKO. After transfection with pcDNA3 or His-Ub, His-ubiquitin complexes were isolated, followed by immunoblotting. Blots are representative of at least 3 independent experiments. See also fig. S1D for additional time course with BKM-120 in cells expressing cDNA for SKP2.
Similar to PIK3CA depletion, PI3K inhibition (by BKM-120) also promoted Skp2 expression (Figure 5C). In this experiment, to confirm that AKT reactivation was PI3K independent, cells were treated with BKM-120 for an additional 30 min after the initial drug administration, and this had no substantially apparent effect on AKT phosphorylation. AKT reactivation required Skp2 in MDA-MB-231 cells given that SKP2 depletion with each of 4 distinct shRNA hairpins in the context of PIK3CA depletion attenuated AKT reactivation, when compared to PIK3CA depletion alone (Figure 5D). Depletion of SKP2 in MDA-MB-468 cells control-treated attenuated AKT phosphorylation, as previously documented (30). AKT reactivation in cells treated with BKM-120 for 48 hours was also attenuated with SKP2 depletion (Figure 5E). We also generated MDA-MB-231 cells resistant to 1μM BKM-120 using a dose escalation protocol. In agreement with the above data, BKM-120-resistant cells show AKT reactivation and Skp2 expression, when compared to parental cells (Figure 5F) and SKP2 depletion attenuated AKT reactivation in IGF-1-stimulated cells. Furthermore, expression of Skp2 promoted AKT phosphorylation and activation, and this was further promoted upon chronic treatment with BKM-120 (fig. S1D). Finally, we used an established cell-based ubiquitination assay to evaluate AKT ubiquitination in the reactivation mechanism. Notably, AKT ubiquitination was induced in parental cells in response to IGF-1 stimulation (Figure 5G). As expected, depletion of SKP2 suppressed AKT ubiquitination as well as phosphorylation and activity. Robust ubiquitination of AKT in response to IGF-1was also detected in MDA-MB-231 cells that were resistant to BKM-120, and this was prominently attenuated by SKP2 depletion (Figure 5G). Together, these data demonstrate that AKT reactivation upon PI3K pathway inhibition depends, at least in part, on Skp2 activity.
AKT Reactivation Promotes Cell Proliferation
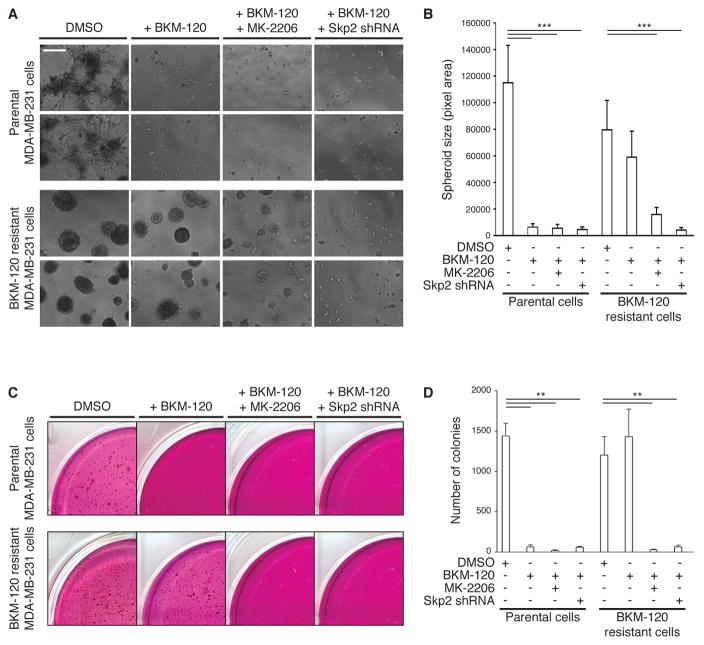
We next explored the functional significance of AKT reactivation by Skp2. The spheroid growth of MDA-MB-231 parental cells in 3D in Matrigel was significantly reduced by treatment with BKM-120 (Figures 6, A and B). However, spheroid growth in BKM-120-resistant MDA-MB-231 cells was not significantly affected by BKM-120 (Figure 6B), although spheroid growth was significantly inhibited upon shRNA-mediated depletion of SKP2 or by treatment with the AKT inhibitor MK-2206. Similar results were obtained with anchorage independence of growth assays; MDA-MB-231 parental cells formed colonies in soft agar, but treatment with BKM-120 significantly inhibited the number of colonies growing in this anchorage-independent manner (Figure 6, C and D). Moreover, additional treatment with MK-2206 or depletion of SKP2 expression of shRNA-resistant SKP2 cDNA did not rescue the defect in anchorage-independent growth. On the other hand, in BKM-120 resistant cells, which also formed a substantial number of colonies, treatment with BKM-120 did not reduce colony formation, but additional treatment with MK-2206 or depletion of SKP2 in BKM-120 resistant cells did abrogate colony formation (Figure 6, C and D). These results show that AKT reactivation in the context of PI3K inhibitor resistance is primarily driven by Skp2.
Figure 6. Skp2 promotes 3D culture and anchorage independence of growth in BKM-120 resistant cells.
(A) Representative images of MDA-MB-231 spheroids, from parental or BKM-120–resistant cells, that were infected with SKP2 shRNA lentiviral vector or control pLKO, and grown for 14 days in a 3-dimensional Matrigel/growth media mixture and treated with BKM-120 and/or MK-2206 (each 1 μM). Scale bar, 500 μm (4x magnification).
(B) Quantitation of spheroid sizes from (A) was performed using ImageJ software. Data are means ± S.E.M. ***P < 0.001 by a two-sided Student’s t-test.
(C) Representative images of MDA-MB-231 colony growth, from parental or BKM-120–resistant cells, that were infected with SKP2 shRNA lentiviral vector or control pLKO, and grown in an agar/growth media mixture for 28 days in the presence of DMSO, BKM-120 and/or MK-2206 (each 1 μM).
(D) Number of colonies in soft agar obtained in (C) was quantitated using MatLab software. Data are means ± S.E.M. **P < 0.01 by a two-sided Student’s t-test. All data and blots are representative of at least 3 independent experiments.
Depletion of Skp2 Reduces Progression of BKM-120-resistant Breast Cancer Xenografts
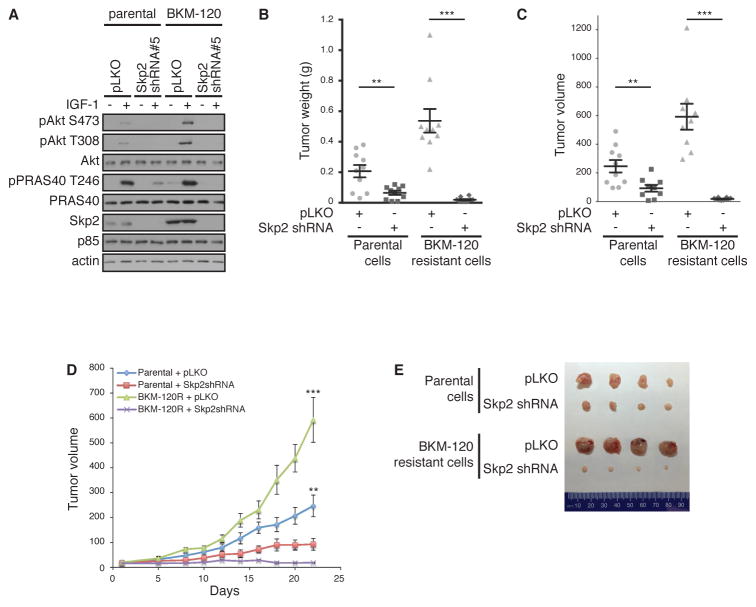
Because SKP2 depletion as well as inhibition of AKT significantly retarded both anchorage-dependent and -independent cell growth, we performed xenograft experiments utilizing parental and BKM-120 resistant cells with SKP2 depletion. We first confirmed that BKM-120-resistant cells display AKT reactivation that is reversed with SKP2 depletion (Figure 7A). Cells were then subcutaneously injected into mice and tumorigenicity assessed in xenograft growth. Tumors derived from xenografted BKM-120 resistant cells grew larger than those derived from parental MDA-MB-231 cells (Figure 7, B–E). Moreover, consistent with in vitro assays (Figure 6), depletion of SKP2 resulted in significantly retarded tumorigenesis in both parental and BKM-120-resistant cells. Finally, BKM-120 resistant cells displayed more pronounced effects upon SKP2 depletion with respect to delay in tumor growth (Figure 7, D and E). Together, these results indicate that Skp2 is a major mechanism for tumor growth of BKM-120-resistant tumors.
Figure 7. Skp2 depletion attenuates tumor growth in vivo.
(A) Immunoblotting of lysates from MDA-MB-231, parental and BKM-120–resistant, cells that were infected with SKP2 shRNA lentiviral vector or control pLKO. Cells were serum-starved overnight (−) or stimulated with IGF-1 for 20 min (+).
(B and C) Tumor weight (B) and volume (C) of xenografts formed from cells described in (A) inoculated subcutaneously into nude mice (n=10). 1×106 cells were injected in each flank. Tumor measurements were taken 22 days after injection. Data are means ± S.E.M. from 10 mice each condition. **P < 0.01, ***P < 0.001 by a two-sided Student’s t-test.
(C) Tumor xenograft volumes taken 22 days after injection.
(D) Growth curves of tumors described in (B). Tumor volume was measured starting day 1 after injection, then on days 5, 8, and every 2 days up to day 22.
(E) Representative pictures of tumors described in (B), surgically removed at day 22.
Discussion
The aberrant activation of the PI3K/AKT/mTOR signaling pathway is frequently observed in virtually all cancers types, including breast cancer. Somatic mutations and amplification of PIK3CA, the gene that encodes the p110α catalytic subunit of class I PI3K, occur with high frequency in both hormone receptor-positive and triple negative breast cancers (1, 36). Although a large number of potent and highly selective PI3K inhibitors have been developed and are under clinical evaluation, the emergence of resistance mechanisms that result in pathway bypass or downstream effector reactivation limits therapeutic efficacy in patients (19, 20). Therefore, elucidating the molecular mechanisms underlying acquired resistance to PI3K-targeted therapies may afford a critical diagnostic assessment of which subtype of breast cancers will develop drug resistance and inform the development of combination therapies to limit resistance. In this study, we identified a previously unidentified mechanism of resistance to PI3K inhibition in breast cancer cells. We show that inhibition of PI3K (with either shRNA-mediated depletion of p110α, or administration of PI3K inhibitors) promotes AKT reactivation by the E3 ubiquitin ligase Skp2 and the canonical upstream AKT-kinases PDK1 and mTORC2, resulting in enhanced growth of breast cancer cells both in vitro and in mouse xenografts. We further identified that AKT reactivation was independent of PI3K and PIP3. Skp2 is implicated in the development and progression of various cancers, including breast cancer (37, 38). We propose that this Skp2–AKT mechanism may represent a central node for a subset of breast cancers that develop resistance to PI3K inhibitors.
The precise molecular mechanism(s) leading to increased Skp2 expression in PI3K inhibitor-treated (and resistant) cells is not yet clear. A number of pathways are likely to contribute. Activation of Notch1, a transcriptional activator of SKP2 (49), is frequently observed in PI3K inhibitor-resistant cells (48); potentially, mutations that enhance Notch signaling could be a source of the resistance and, therefore, a genetic biomarker for patients likely to relapse after BKM-120 treatment. Alternatively, inactivation of FOXO3-mediated suppression of SKP2 may also contribute to the increase in Skp2 in PI3K inhibitor-treated cells (50). Notably, Skp2 expression and AKT kinase activation positively correlates in breast cancer cells(39, 40). AKT itself is a positive regulator of Skp2 function through both transcriptional, translational and post-translational mechanisms. Specifically, AKT1 induces SKP2 transcription in an E2F1-dependent manner (41) and antagonizes the transcriptional repressor function of FOXO3 that binds the SKP2 promoter (42); AKT1/2 reportedly enhance the translation of SKP2 mRNA through activation of the translation initiation protein eIF4E (43);, and AKT1 promotes Skp2 protein stability in part through direct phosphorylation of Skp2 at Ser72 (40, 44). Collectively, these findings indicate that AKT and Skp2 reciprocally enhance their functional activation through a positive feedback loop.
Whether this mechanism of AKT reactivation mediated by Skp2 in the absence of productive PI3K activity occurs in patients treated with inhibitors, such as BKM-120 or BYL-719, remains to be determined. Likewise, it is presently unknown what genetic or epigenetic characteristics define the ability of breast cancer cells to coopt Skp2-mediated AKT activation in response to PI3K inhibition. This is further compounded by the current lack of any predictive biomarkers that would allow a classification of cells that display AKT rebound activation as part of PI3K inhibitor resistance. Our observation that AKT reactivation upon depletion of any PI3K isoform was surprising, given that under conditions of depletion of an individual p110 isoform, other isoforms should compensate and retain intact PI3K signaling. That depletion of PIK3CA alone was sufficient to quantitatively eliminate PIP3 and PI3,4P2 in IGF-1-stimulated cells suggests that inhibition of individual p110 isoforms cannot be efficiently compensated for, at least under these conditions. It is also worth speculating that other PI3K effectors (such as SGK isoforms) could also display rebound activation in the context of resistance to PI3K inhibitors. SGK3 may be particularly relevant, given that SGK3, and not AKT, mediates the PI3K oncogenic signal in certain PI3KCA-mutant cells (46), and in a manner that also requires the PI3,4P2 phosphatase INPP4B (47).
Further work with mouse models of PIK3CA-driven breast cancer treated with combination regimens of PI3K and Skp2 inhibitors will be required to support the model proposed by these data. However, this concept is supported by studies that have revealed therapeutic efficacy of Skp2 inhibitors in mouse models (35, 52, 53). Nonetheless, the findings uncover a mechanism of PI3K inhibitor resistance in breast cancer cells in which the Skp2 ubiquitin ligase enables PI3K-independent AKT signaling.
Materials and Methods
Cell culture and transfection
BT20, Hs578T, MCF7, MDA-MB-231, MDA-MB-435 and MDA-MB-468 cells were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Cellgro) supplemented with 10% Fetal Bovine Serum (FBS; Gibco). BT549, HCC1143, HCC1806, HCC1937, HCC38, HCC70 and ZR-75.1 cells obtained from ATCC were grown in RPMI 1640 medium (Cellgro) supplemented with 10% FBS. MDA-MB-453 and SKBR3 cells obtained from ATCC were grown in McCoy’s 5A medium (Cellgro) supplemented with 10% FBS. SUM149PT and SUM159PT were grown in HAM’s F12 medium (Cellgro) with 5% FBS and 5 μg/ml insulin (Sigma-Aldrich) and 500 ng/ml hydrocortisone (Sigma-Aldrich). Cells were passaged for no more than 6 months and routinely assayed for mycoplasma contamination. Transient transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. To generate MDA-MB-231 resistant cells, the parental cells were treated with increasing concentration of BKM-120 until a target concentration of 1μM BKM-120 was achieved.
Growth factors and inhibitors
Cells were stimulated for 20 min with recombinant human IGF-1 (R&D systems) at a final concentration of 100 ng/ml. PI3K inhibitors BKM-120 (Selleck Chemicals), BYL-719 (Selleck Chemicals), A66 (Selleck Chemicals), TGX-221 and IC87114 [gifts from Peter Shepherd, University of Auckland, New Zealand; (54)] were added to cells 15 min prior to IGF-1 stimulation at final concentration of 1 μM. AKT inhibitor MK-2206 (Active Biochem) was added to cells 15 min prior to IGF-1 stimulation at final concentration of 1 μM. NVP-AEW541 (Cayman Chemical) and GSK2334470 (Tocris) were added to cells 15 min prior to growth factor stimulation at final concentration of 1 μM.
Antibodies
Antibodies recognizing AKT (4685), phospho-AKT Ser473 (4060), phospho-AKT Thr308 (2965), PRAS40 (2691), phospho-PRAS40 (2997), GSK3β (9315), phospho-GSK3β (9336), p110α (4249), phospho-S6K T389 (9205), PTEN (9188), actin (4970) and p110γ (5405) were purchased from Cell Signaling Technologies. Antibodies recognizing p110β (sc-602), p110δ (sc-7176) and raptor (sc-81537) were from Santa Cruz. Antibody recognizing rictor (A300-459A) was from Bethyl Laboratories. Antibody recognizing Skp2 (32-3300) was from Invitrogen. Antibody recognizing p85 was generated in house and has been described (55). Horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse immunoglobulin G antibodies were from Millipore.
Plasmids
(His)6-ubiquitin plasmid was gift from Hui-Kuan Lin (The University of Texas M. D. Anderson Cancer Center, Houston). PTEN-WT plasmid was obtained from Addgene (Cambridge, MA).
RNA interference
For shRNA-mediating silencing of PIK3CA, PIK3CB, PIK3CD and PIK3CG, a set of oligonucleotides composed of a target shRNA sequence and its complement were synthesized. The hairpin sequences were generated as follows:
PIK3CAshRNA#2, sense, 5′-CCGGGCACAATCCATGAACAGCATTCTCGAGAATGCTGTTCATGGATTGTGCTTTTTTG-3′; PIK3CAshRNA#2, antisense, 5′-AATTCAAAAACACAATCCATGAACAGCATTCTCGAGAATGCTGTTCATGGATTGTG-3′
PIK3CAshRNA#3, sense, 5′-CCGGGCATTAGAATTTACAGCAAGACTCGAGTCTTGCTGTAAATTCTAATGCTTTTTTG-3′; PIK3CAshRNA#3, antisense, 5′-AATTCAAAAAGCATTAGAATTTACAGCAAGACTCGAGTCTTGCTGTAAATTCTAATGC-3′
PIK3CBshRNA#1, sense, 5′-CCGGGCAACAGCTTTGCATGTTAAACTCGAGTTTAACATGCAAAGCTGTTGCTTTTTG-3′; PIK3CBshRNA#1, antisense, 5′-AATTCAAAAAGCAACAGCTTTGCATGTTAAACTCGAGTTTAACATGCAAAGCTGTTGC-3′
PIK3CDshRNA#2, sense, 5′-CCGGGATCTTTCTCTCTGACTATACCTCGAGGTATAGTCAGAGAGAAAGATCTTTTTG-3′; PIK3CDshRNA#2, antisense, 5′-AATTCAAAAAGATCTTTCTCTCTGACTATACCTCGAGGTATAGTCAGAGAGAAAGATC-3′
SKP2shRNA#2, sense, 5′-CCGGGAAGGCCTGCGGCTTTCGGATCCTCGAGGATCCGAAAGCCGCAGGCCTTCTTTTTTG-3′; SKP2shRNA#2, antisense, 5′-AATTCAAAAAAAGGCCTGCGGCTTTCGGATCCTCGAGGATCCGAAAGCCGCAGGCC TT-3′
The oligonucleotide pairs were each annealed and inserted into pLKO.1. For shRNA-mediated knockdown of SKP2 and PIK3CG, simple hairpin shRNAs in the pLKO.1 lentiviral vector designed by The RNAi Consortium (TRC) were ordered from Thermo Fisher Scientific: 5′-GCCTAAGCTAAATCGAGAGAA-3′ (SKP2shRNA#4; clone TRCN0000007530), 5′-CCACGATCATTTATGGACCAA-3′ (SKP2shRNA#5; TRCN0000007531), 5′-CCATTGTCAATACTCTCGCAA-3′ (SKP2shRNA#6; TRCN0000007532), 5′-TAATTCCCAAGAATGTGCCCG-3′ (PIK3CGshRNA#1; TRCN0000033282)
To produce lentiviral supernatants, 293T cells were co-transfected with VSVG, psPAX2 and control-pLKO, PIK3CA, PIK3CB, PIK3CD, PIK3CG or SKP2 shRNA-containing vectors for 48 hrs.
Immunoblotting
Cells were washed with ice-cold PBS and lysed in RIPA buffer (150 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 20 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 50 nM calyculin A, proteinase inhibitor cocktail) for 15 min at 4°C. Cell extracts were centrifuged at 13,000 rpm for 10 min at 4°C and protein concentration was measured with the Bio-Rad protein assay reagent using a Beckman Coulter DU-800 machine. Lysates were then resolved on 8% acrylamide gels by SDS-PAGE and transferred electrophoretically to nitrocellulose membrane (Bio-Rad) at 100 V for 60 min. The blots were blocked in TBST buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 0.2% Tween 20) containing 5% nonfat dry milk for 30 min and then incubated with the specific primary antibody at 4°C overnight. Membranes were washed 3 times in TBST and incubated with HRP-conjugated secondary antibody for 1 hr at room temperature. Membranes were washed 3 times and developed using chemiluminescent HRP substrate (Millipore).
Quantitative RT-PCR analysis
RNA was extracted using the Qiagen RNeasy mini kit and the reverse transcription reaction was carried out with ABI Taqman Reverse Transcriptional Reagent. After mixing the resulting template with Skp2 (Mm00449925_m1) or glyceraldehyde-3-phospahte dehydrogenase (GAPDH; Mm99999915_g1) primers and ABI Taqman Fast Universal PCR Master Mix, the RT-PCR reaction was performed with the ABI-7500 Fast Real-time PCR system.
PIP3 and PI3,4P2 assay
PIP3 and PI3,4P2 were extracted as described in the manufacturer’s protocol (Echelon, K-2500s and K-3800). Briefly, after IGF-1 stimulation, media was removed from the plates and 10 mL ice cold 0.5 M TCA was immediately added. Cells were scraped and centrifuged at 3000 rpm for 5 min. The pellet was resuspended in 3 mL 5% TCA/1 mM EDTA, vortexed and centrifuged at 1500 rpm for 5 min. To extract neutral lipids, 3 mL MeOH:CHCl3 (2:1) was added to the pellet, vortexed 3 times over 10 min and centrifuged at 1500 rpm for 5 min. Acidic lipids were extracted by adding 2.25 mL MeOH:CHCl3:12 M HCl (80:40:1), vortexed 4 times over 15 min and centrifuged at 1500 rpm for 5 min. The supernatant was transferred to a new 15 mL tube and 0.75 mL CHCl3 and 1.35 mL of 0.1 M HCl were added to the supernatant, vortexed and centrifuged at 1500 rpm for 5 min to separate organic and aqueous phases. The organic (lower) phase was collected and dried in vacuum dryer for 1 hr. Once isolated from cells, PIP3 and PI3,4P2 were measured using ELISA kits (Echelon, K-2500s and K-3800) according to the manufacturer’s instructions.
3D Culture Assay
3D cultures were performed as described (56). Chamber slides were coated with growth factor-reduced Matrigel (BD Biosciences) and allowed to solidify for 30 min. Subsequently, 5000 MDA-MB-231 cells were seeded to each chamber slide in assay media (DMEM supplemented with 2% FBS and 5% Matrigel). Assay medium was replaced 4 days after seeding. ImageJ was used to quantify 2D surface area on day 14.
Soft Agar Colony Formation Assay
Soft agar assays were performed by coating 6-well plates with 2 mL of 0.8% Noble agar/growth media (10% FBS/DMEM) with indicated inhibitors and allowed to solidify at 20°C. 2×104 cells were plated in 1 mL top layer 0.4% Noble agar/growth media with indicated inhibitors. Growth media with inhibitors was replaced every 4 days and cells were counted and measured 28 days after seeding. Quantitation was performed using MatLab software (MathWorks).
Ubiquitination Assay
Cell based in vivo ubiquitination assays was performed as described (30). MDA-MB-231parental and BKM-120 resistant cells were transfected with His-Ubiquitin or pcDNA3 as control. Subsequently, cells were infected with Skp2 shRNA lentiviral vector or control pLKO. Cells were serum-starved overnight or stimulated with IGF-1 for 20 min and lysed in denatured buffer (6M guanidine-HCl, 0.1M Na2HPO4/NaH2PO4, 10 mM imidazole). The cell lysates were incubated with nickel beads for 3 hr, washed, and subjected to immunoblotting analysis.
Tumor Xenografts
6- to 8-week-old female nude mice were purchased from Taconic and maintained in a pathogen-free environment. All procedures were carried out under the approval of the Institutional Animal Care and Use Committee at the Beth Israel Deaconess Medical Center and comply with the federal guidelines for maintenance and care of laboratory animals. 10 mice for each experimental condition were injected subcutaneously in the flank with 1.0 × 106 MDA-MB-231 cells in PBS with 50% Matrigel. Tumor volume was measured by volume = 0.52 x length x width2.
Statistics and Reproducibility
Sample sizes and reproducibility for each figure are denoted in the figure legends. Unless otherwise noted, all immunoblots are representative of at least three biologically independent experiments. Statistical significance between conditions was assessed by two-tailed Student t-tests. All error bars represent s.e.m., and significance between conditions is denoted as *P < 0.05; **P < 0.01; and ***P < 0.001.
Supplementary Material
Fig. S1. Reactivation of AKT in response to PIK3CA depletion and inhibition.
Acknowledgments
We thank the members of the Toker and Wei laboratories for productive discussions, Peter Shepherd (University of Auckland) and Hui-Kuan Lin (The University of Texas M. D. Anderson Cancer Center, Houston) for providing reagents, and Rohit Arora (Beth Israel Deaconess Medical Center) for evaluation of statistical analyses.
Funding: This study was supported in part by Susan G. Komen for the Cure postdoctoral fellowship (KG111139 to E.C.), the National Institutes of Health (CA177910 and CA200671 to A.T.; GM089763 and GM094777 to W.W.) and the Ludwig Center at Harvard (to A.T.).
Footnotes
Author Contributions: E.C., A.T. and W.W. designed the study and interpreted the results. E.C. performed all experiments unless otherwise noted. H. I. performed the ubiquitination experiments and mouse xenograft studies. N.T.N. performed a subset of the shRNA and Skp2 expression assays. E.C., A.T., H.I. and W.W. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 2.Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 4.Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 5.James SR, Downes CP, Gigg R, Grove SJA, Holmes AB, Alessi DR. Specific Binding of the Akt-1 protein kinase to PIP3 without subsequent activation. Biochem J. 1996;315:709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frech M, Andjelkovic M, Ingley E, RKK, Falck JR, Hemmings BA. High Affinity Binding of Inositol Phosphates and Phosphoinositides to the Pleckstrin Homology Domain of RAC/Protein Kinase B and Their Influence on Kinase Activity. The Journal of biological chemistry. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 7.Klippel A, Kavanaugh WM, Pot D, Williams LT. A specific product of PI 3-K directly activates the protein kinase Akt through its pleckstrin homology domain. Molecular and cellular biology. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 10.Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, Painter GF, Holmes AB, McCormick F, Hawkins PT. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer [see comments] Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 12.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 13.Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, Pandolfi PP, Cantley LC. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andjelkovic M, Jakubowicz T, Cron P, Ming XF, Han JW, Hemmings BA. Activation and phosphorylation of a pleckstrin homology domain containing protein kinase (RAC-PK/PKB) promoted by serum and protein phosphatase inhibitors. Proc Natl Acad Sci USA. 1996;93:5699–5704. doi: 10.1073/pnas.93.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Molecular cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nature reviews Drug discovery. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 17.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 18.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 19.Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med. 2016;67:11–28. doi: 10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- 20.Brown KK, Toker A. The phosphoinositide 3-kinase pathway and therapy resistance in cancer. F1000Prime Rep. 2015;7:13. doi: 10.12703/P7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero-Zotano A, Mayer IA, Arteaga CL. PI3K/AKT/mTOR: role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 2016;35:515–524. doi: 10.1007/s10555-016-9637-x. [DOI] [PubMed] [Google Scholar]
- 22.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M, Li X, Lehar J, Wiesmann M, Wartmann M, Chen Y, Cao ZA, Pinzon-Ortiz M, Kim S, Schlegel R, Huang A, Engelman JA. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, Shah RH, Huynh T, Mino-Kenudson M, Sgroi D, Isakoff S, Thabet A, Elamine L, Solit DB, Lowe SW, Quadt C, Peters M, Derti A, Schegel R, Huang A, Mardis ER, Berger MF, Baselga J, Scaltriti M. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature. 2015;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, Tao JJ, Spratt DE, Viola-Villegas NT, Castel P, Minuesa G, Morse N, Rodon J, Ibrahim Y, Cortes J, Perez-Garcia J, Galvan P, Grueso J, Guzman M, Katzenellenbogen JA, Kharas M, Lewis JS, Dickler M, Serra V, Rosen N, Chandarlapaty S, Scaltriti M, Baselga J. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med. 2015;7:283ra251. doi: 10.1126/scitranslmed.aaa4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, Dickler MN, Scaltriti M, Leslie CS, Armstrong SA, Baselga J. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355:1324–1330. doi: 10.1126/science.aah6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le X, Antony R, Razavi P, Treacy DJ, Luo F, Ghandi M, Castel P, Scaltriti M, Baselga J, Garraway LA. Systematic Functional Characterization of Resistance to PI3K Inhibition in Breast Cancer. Cancer Discov. 2016;6:1134–1147. doi: 10.1158/2159-8290.CD-16-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serra V, Eichhorn PJ, Garcia-Garcia C, Ibrahim YH, Prudkin L, Sanchez G, Rodriguez O, Anton P, Parra JL, Marlow S, Scaltriti M, Perez-Garcia J, Prat A, Arribas J, Hahn WC, Kim SY, Baselga J. RSK3/4 mediate resistance to PI3K pathway inhibitors in breast cancer. J Clin Invest. 2013;123:2551–2563. doi: 10.1172/JCI66343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof RA, Brook S, Benzaken AO, Rodon J, Morse N, Yan JJ, Liu M, Das R, Chen Y, Tam A, Wang H, Liang J, Gurski JM, Kerr DA, Rosell R, Teixido C, Huang A, Ghossein RA, Rosen N, Bivona TG, Scaltriti M, Baselga J. AXL mediates resistance to PI3Kalpha inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27:533–546. doi: 10.1016/j.ccell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Invest. 2002;110:633–641. doi: 10.1172/JCI15795. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Radke S, Pirkmaier A, Germain D. Differential expression of the F-box proteins Skp2 and Skp2B in breast cancer. Oncogene. 2005;24:3448–3458. doi: 10.1038/sj.onc.1208328. [DOI] [PubMed] [Google Scholar]
- 33.Fujita T, Liu W, Doihara H, Date H, Wan Y. Dissection of the APCCdh1-Skp2 cascade in breast cancer. Clin Cancer Res. 2008;14:1966–1975. doi: 10.1158/1078-0432.CCR-07-1585. [DOI] [PubMed] [Google Scholar]
- 34.Najafov A, Sommer EM, Axten JM, Deyoung MP, Alessi DR. Characterization of GSK2334470, a novel and highly specific inhibitor of PDK1. Biochem J. 2011;433:357–369. doi: 10.1042/BJ20101732. [DOI] [PubMed] [Google Scholar]
- 35.Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, Logothetis CJ, Hung MC, Zhang S, Lin HK. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Fukushima H, Inuzuka H, Wan L, Liu P, Gao D, Sarkar FH, Wei W. Skp2 is a promising therapeutic target in breast cancer. Front Oncol. 2012;1 doi: 10.3389/fonc.2011.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Wang H, Ma J, Xu J, Sheng C, Yang S, Sun L, Ni Q. The expression and prognosis of Emi1 and Skp2 in breast carcinoma: associated with PI3K/Akt pathway and cell proliferation. Med Oncol. 2013;30:735. doi: 10.1007/s12032-013-0735-0. [DOI] [PubMed] [Google Scholar]
- 40.Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nature cell biology. 2009;11:397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichert M, Saur D, Hamacher R, Schmid RM, Schneider G. Phosphoinositide-3-kinase signaling controls S-phase kinase-associated protein 2 transcription via E2F1 in pancreatic ductal adenocarcinoma cells. Cancer Res. 2007;67:4149–4156. doi: 10.1158/0008-5472.CAN-06-4484. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, Lee SW, Zhang X, Han F, Kwan SY, Yuan X, Yang WL, Jeong YS, Rezaeian AH, Gao Y, Zeng YX, Lin HK. Foxo3a transcription factor is a negative regulator of Skp2 and Skp2 SCF complex. Oncogene. 2013;32:78–85. doi: 10.1038/onc.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogueira V, Sundararajan D, Kwan JM, Peng XD, Sarvepalli N, Sonenberg N, Hay N. Akt-dependent Skp2 mRNA translation is required for exiting contact inhibition, oncogenesis, and adipogenesis. EMBO J. 2012;31:1134–1146. doi: 10.1038/emboj.2011.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, Yang WL, Erdjument-Bromage H, Nakayama KI, Nimer S, Tempst P, Pandolfi PP. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–432. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez CG, Ma CX, Crowder RJ, Guintoli T, Phommaly C, Gao F, Lin L, Ellis MJ. Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 2011;13:R21. doi: 10.1186/bcr2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, Dunn IF, Schinzel AC, Sandy P, Hoersch S, Sheng Q, Gupta PB, Boehm JS, Reiling JH, Silver S, Lu Y, Stemke-Hale K, Dutta B, Joy C, Sahin AA, Gonzalez-Angulo AM, Lluch A, Rameh LE, Jacks T, Root DE, Lander ES, Mills GB, Hahn WC, Sellers WR, Garraway LA. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasser JA, Inuzuka H, Lau AW, Wei W, Beroukhim R, Toker A. SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Molecular cell. 2014;56:595–607. doi: 10.1016/j.molcel.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H, Craig-Mueller N, Colinge J, Duernberger G, Nijman SM. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol. 2011;7:787–793. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarmento LM, Huang H, Limon A, Gordon W, Fernandes J, Tavares MJ, Miele L, Cardoso AA, Classon M, Carlesso N. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J Exp Med. 2005;202:157–168. doi: 10.1084/jem.20050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pavlides SC, Huang KT, Reid DA, Wu L, Blank SV, Mittal K, Guo L, Rothenberg E, Rueda B, Cardozo T, Gold LI. Inhibitors of SCF-Skp2/Cks1 E3 ligase block estrogen-induced growth stimulation and degradation of nuclear p27kip1: therapeutic potential for endometrial cancer. Endocrinology. 2013;154:4030–4045. doi: 10.1210/en.2013-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaussade C, Rewcastle GW, Kendall JD, Denny WA, Cho K, Gronning LM, Chong ML, Anagnostou SH, Jackson SP, Daniele N, Shepherd PR. Evidence for functional redundancy of class IA PI3K isoforms in insulin signalling. Biochem J. 2007;404:449–458. doi: 10.1042/BJ20070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapeller R, Toker A, Cantley LC, Carpenter CL. Phosphoinositide 3-kinase binds constitutively to alpha/beta-tubulin and binds to gamma-tubulin in response to insulin. The Journal of biological chemistry. 1995;270:25985–25991. doi: 10.1074/jbc.270.43.25985. [DOI] [PubMed] [Google Scholar]
- 56.Debnath J, Walker SJ, Brugge JS. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J Cell Biol. 2003;163:315–326. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Reactivation of AKT in response to PIK3CA depletion and inhibition.