Abstract
The Snail gene family includes Snai1, Snai2, and Snai3 that encode zinc finger-containing transcriptional repressors in mammals. The expression and localization of SNAI1 and SNAI2 have been studied extensively during folliculogenesis, ovulation, luteinization, and embryogenesis in mice. However, the role of SNAI3 is unknown. In this study, we investigated the expression of SNAI3 during these processes. Our immunohistochemistry data showed that SNAI3 first appeared in oocytes by postnatal day (PD) 9. Following this, SNAI3 was found to be expressed consistently in theca and interstitial cells, along with oocytes. In gonadotropin-treated immature mice, the expression of SNAI3 did not change significantly during follicular development. The expression of SNAI3 was reduced during ovulation, after which it increased gradually during luteinization. Similar results were obtained from western blot analyses. Furthermore, real-time polymerase chain reaction (RT-PCR) analyses revealed varying mRNA levels of different Snail factors at a given time in gonadotropin-induced ovaries. During early embryo cleavage, SNAI3 was localized to the nucleus, except the nucleolus at the germinal vesicle and one-cell stages. From two- to eight-cell stages, SNAI3 was localized only to the nucleolus. Thereafter, SNAI3 was detected only in the cytoplasm, except during the blastocyst stage when it was localized to the nucleus of the trophectoderm and the inner cell mass. RT-PCR results showed that the expression of Snail superfamily genes was decreased during the blastocyst stage. From the eight-cell to morula stage, when compaction occurs that is a prerequisite for blastocyst formation, Snai3 mRNA was expressed at very low levels and was opposite to the highest expression level of the compaction-related gene, E-cadherin, at the eight-cell stage. Taken together, our results suggest that SNAI3 likely plays some roles during folliculogenesis, luteinization, and early embryonic development.
Keywords: E-cadherin-catenin complex, Mouse, Ovaries, Pre-implantation embryos, SNAI3
In mammals, the oocyte pool that is generated early in life represents the entirety of the female reproductive potential over its life span [1, 2]. In mice, oocytes progress through the leptotene, zygotene, pachytene, and diplotene stages of meiotic prophase I, and are arrested at the dictyate stage in the neonatal ovary [3, 4]. To maintain an immature oocyte in the dormant state, it is enclosed by several flattened pre-granulosa cells to establish a functional unit called the primordial follicle [5]. Regulated oogenesis and folliculogenesis in the perinatal ovary are essential for fertility. Mature follicles develop gradually from primordial follicles, through primary, preantral, antral, to preovulatory stages, containing a fully grown oocyte and multiple layers of granulosa cells (GCs) [6, 7]. Subsequently, in response to the pre-ovulatory gonadotropin surge, some antral follicles ovulate for fertilization, while the remaining granulosa and theca cells of ovulated follicles undergo transformation to become corpora lutea (CL) through a process known as luteinization that sustains pregnancy in a fertile cycle [8].
Upon fertilization, mouse embryos develop from the one-cell to the blastocyst stage within 3.5 days [9]. Embryos develop in oviducts from one-cell to morulae, while blastocysts develop in the uterus. Before implantation in the uterus, blastocysts undergo a series of transformations, including cell division, apoptosis, and differentiation [10]. Mammalian oocytes and early embryos contain special nucleoli, known as nucleolus precursor bodies (NPBs). Unlike regular nucleoli that are composed of fibrillar centers, and granular and dense fibrillar components, NPBs are composed exclusively of dense fibrillar material of unknown composition [11, 12]. These atypical organelles originate from formerly transcriptionally active nucleoli of growing oocytes. Initially, oocytes increase enormously in size and synthesize large amounts of material (including rRNA), and the structure of the oocyte nucleolus resembles that of the somatic cell-type nucleolus. When oocytes reach full size, transcription ceases and the somatic cell-type nucleolus is transformed inside the NPB [13].
The Snail family of proteins are zinc-finger transcription factors with conserved structures across many species, including Drosophila and numerous vertebrates. These proteins are thought to be involved in various processes during cell differentiation [14, 15]. Three members of the Snail family have been described in mammals: Snai1 (also called Snail), Snai2 (Slug), and Snai3 (Smuc). These genes encode DNA binding zinc finger proteins. The C2H2 zinc fingers of the Snail proteins are similar and conserved with almost identical amino acid sequences. Members of the Snail family bind to E-box consensus sites (CAGGTG or CANNTG) to repress transcription [16]. This repressor activity not only depends on the zinc-finger region, but also on a SNAG (Snai/Gfi) domain [17].
It is interesting that studies have shown inverse correlations exist between E-cadherin, SNAI1, and SNAI2 in many different cell systems [8]. E-Cadherin is the main cell-cell adhesion molecule in maintaining the integrity of epithelial tissues, and mediates cell-cell adhesion during preimplantation development [18]. Redistribution of E-cadherin, and initiation of E-cadherin-mediated adhesion, are regulated by the catenins. β-Catenin binds α-catenin and the cytoplasmic domain of E-cadherin, which anchors the adhesion complex to the actin-based cytoskeleton to implement functions together [19, 20]. SNAI1 and SNAI2 are associated with follicular maturation, ovulation, luteinization, embryogenesis, and the epithelial-mesenchymal transition (EMT) in mice [21]. SNAI3 also actively represses transcription [16] and is expressed in skeletal muscle, thymus, and myeloid cells [16, 21]. Human SNAI3 has been identified via in silico analysis, contains the same SNAG and zinc finger domains as the mouse protein [22], and its distribution has been examined [23]. However, Snai3 knockout mice are viable and fertile [23]. Thus, the roles of SNAI3 in female reproduction, especially during embryogenesis, require further study.
To elucidate the location of SNAI3 during the development of follicles and pre-implantation embryos, and to explore whether there are redundant roles among SNAI1, SNAI2, and SNAI3, we examined the temporal and spatial expression of SNAI3 during follicular development, ovulation, luteinization, and embryogenesis. In addition, we examined the transcription of Snai1 and Snai2 during these processes. Our results indicate that SNAI3 might play important roles during follicular development, luteinization, and early embryonic development, and there may exist redundant roles among SNAI1, SNAI2, and SNAI3.
Materials and Methods
Animals
Mice (8 weeks old) were obtained from the Laboratory Animal Center of Shandong University (Ji’nan, Shandong, China). All mice were housed at Shandong Normal University at 23 ± 2˚C under a 12/12 h light/dark cycle with access to chow and water ad libitum. All experiments were performed with strict adherence to the Guidelines of Shandong Normal University for the Care and Use of Laboratory Animals.
Ovary, oocyte, and embryo collection
The day of delivery was designated as postnatal day (PD) 0. Ovaries from neonatal and prepubertal mice were collected on PD 1, 4, 9, 18, and 23. To stimulate follicular and luteal development, immature 21-day-old female mice were treated intraperitoneally with 5 IU of pregnant mare serum gonadotropin (PMSG) followed 48 h later by 5 IU human chorionic gonadotropin (hCG) (both from Ningbo Sansheng Pharmaceutical, Zhejiang, China). The ovaries of hormone-treated mice were collected at 24 and 48 h following PMSG injection, and at 16, 24, and 48 h following hCG administration.
For oocyte collection, adult female mice were treated with 10 IU PMSG and sacrificed 46 h later to isolate antral follicles. Fully grown germinal vesicle (GV) oocytes were then selected and collected from the antral follicles using a mouth pipette. For embryo collection, adult females were treated with 10 IU PMSG followed by hCG administration (10 IU) 48 h later, and mated with adult males. Zygotes, two- and four-cell, were retrieved from oviducts at 22, 42, and 55 h post-hCG. Because embryo compaction starts from the eight-cell stage and lasts about 10 h in mice, to study the association between SNAI3 and E-cadherin expression, and compaction, eight-cell embryos and morulae were recovered from oviducts at 66 and 88 h after hCG administration, respectively. Blastocysts were collected from uteri at 90–96 h post-hCG as described earlier [24].
Immunohistochemistry
Immunohistochemistry was carried out as described previously [25, 26]. Briefly, the collected tissues were immediately snap frozen in liquid nitrogen and stored at –80˚C. Ovaries were cryosectioned at 7 mm, fixed in acetone at –20˚C for 10 min, and incubated in 0.3% (v/v) Triton X-100 in phosphate-buffered saline (PBS), pH 7.2, for 20 min. Endogenous peroxidase activity was quenched by incubating the samples in 3% (v/v) H2O2 for 20 min, followed by washing with PBS. Sections were blocked with immunoglobulin G from the same animal species as the primary antibody for 1 h at room temperature (22–26ºC), and then incubated at 4˚C overnight with the primary antibody to SNAI3 (1:20 dilution, sc-10439; Santa Cruz Biotechnology, Santa Cruz, CA USA). Thereafter, the samples were incubated with a horseradish peroxidase-conjugated secondary antibody for 60 min at room temperature before being developed with a diaminobenzidine peroxidase substrate kit (ZLI-9033; ZSGB-BIO, Beijing, China). Negative controls were incubated with pre-immune serum instead of the primary antibody. Finally, the sections were counterstained with hematoxylin, dehydrated, mounted, and digitally photographed using a microscope (Olympus, Tokyo, Japan).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from oocytes and embryos with the E.Z.N.A. MicroElute Total RNA Kit (Omega Bio-tek, Guangzhou, China) or from ovaries using TRIzol Reagent (Tiangen Biotech, Beijing, China) as per the manufacturers’ instructions. At each time point, the numbers of CLs were roughly equal. Total RNA was treated with RNase-free DNase (Tiangen Biotech). Reverse transcription was performed with the Fast Quant RT Kit (Tiangen Biotech).
qRT-PCR was carried out using SYBR Green Master Mix on the LightCycler 96 System (Roche Diagnostics, Basel, Switzerland). The primers used are listed in Table 1. PCR was performed under the following conditions: 3 min at 95˚C, then 40 cycles of 15 sec at 95˚C, 30 sec at 59˚C, and 30 sec at 72˚C. This was followed by 5 min at 72˚C. Melting-curve analyses were performed to confirm product identity. PCRs were performed in triplicate, and values obtained are expressed relative to the abundance of endogenous β-actin in the same sample. The efficiency of PCR amplification was analyzed using LightCycler 96 software and data analysis was performed based on a calibrator sample using the ΔΔCt method [27].
Table 1. Target transcripts and primer sequences used for the quantification of mRNA levels in the ovaries and embryos.
| Primer | Sequence | Size (bp) |
|---|---|---|
| β-actin | 5’-TGTTACCAACTGGGACGACA-3’ | 165 |
| 5’-GGGGTGTTGAAGGTCTCAAA-3’ | ||
| α-catenin | 5’-ACTTGATGAATGCTGTTGTG-3’ | 177 |
| 5’-TAATCTTGGTCTGCGTCTC-3’ | ||
| β-catenin | 5’-CCGTTCGCCTTCATTATGG-3’ | 190 |
| 5’-GTGTCCTGATGTGCTCGTA-3’ | ||
| E-cadherin | 5’-GAGAGTGGAGAACGAGGAA-3’ | 171 |
| 5’-CGAGCGGTATAAGATGTGATT-3’ | ||
| SNAIL1 | 5’-CCATTCTCCTGCTCCCACTG-3’ | 100 |
| 5’-CCTGGCACTGGTATCTCTTCA-3’ | ||
| SNAIL2 | 5’-GCTCCACTCCACTCTCCTT-3’ | 122 |
| 5’-TCTGAACCACTGTGATCCTTG-3’ | ||
| SNAIL3 | 5’-GCAAGGAGTTGGCAGGAT-3’ | 129 |
| 5’-TGTGATTCGGCAGAAGAGG-3’ | ||
| Cyclin D2 | 5’-CAGGATGATGAAGTGAACACA-3’ | 159 |
| 5’-GGCGTTATGCTGCTCTTG-3’ | ||
| P21cip1 | 5’-AAGTGTGCCGTTGTCTCT-3’ | 110 |
| 5’-GTCAAAGTTCCACCGTTCT-3’ |
Western blot
Total protein was extracted from follicles and roughly equal numbers of CLs with radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate) containing protease inhibitors (Sigma-Aldrich, St. Louis, MO, USA). Proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrophoretically transferred onto a polyvinylidene fluoride membrane (Hybond, Amersham Pharmacia Biotech, Piscataway, NJ, USA). Following transfer, the membranes were incubated for 1.5 h in 5% nonfat dry milk at room temperature, then overnight at 4˚C with anti-SNAI3 polyclonal (1:500, sc-10439; Santa Cruz Biotechnology) and β-actin (1:5000, SC-47778; Santa Cruz Biotechnology) antibodies. After washing thrice in Tris-buffered saline Tween 20, the membranes were incubated for 1 h at room temperature with the appropriate secondary antibody in the same buffer. Finally, the proteins were detected using Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica, USA) with an enhanced chemiluminescence western blotting analysis system (Tanon 5500 Multi, Shanghai, China). β-Actin levels were measured as the internal loading control. The relative intensities of the bands were analyzed by Quantiscan software (Biosoft, Great Shelford, Cambridge, UK) and normalized to β-actin from the same blot.
Immunofluorescence
For immunofluorescence staining, embryos were fixed in 4% paraformaldehyde for 30 min and permeabilized in incubation medium (0.5% Triton X-100 in 3 mM MgCl2, 20 mM HEPES, 50 mM NaCl, 300 mM sucrose, and 0.02% NaN3, pH 7.4) for 30 min (blastocysts for 40 min). This was followed by incubating with 1% bovine serum albumin in PBS for 30 min at room temperature. The embryos were incubated with a goat anti-SNAI3 polyclonal antibody (1:50, sc-10439; Santa Cruz Biotechnology) overnight at 4˚C. After washing thrice in PBS for 5 min each, the embryos were incubated with fluorescein-isothiocyanate-conjugated rabbit anti-goat IgG (1:100, sc-2777; Santa Cruz Biotechnology) for 60 min at room temperature. The embryos were then washed thrice in PBS for 5 min each, stained with 10 μg/ml 4′,6-diamidino-2-phenylindole (Sigma-Aldrich), washed, and finally mounted on 1,4-diazabicyclo (2.2.2) octane hydrochloride-containing (Sigma-Aldrich) glass slides. The mounted embryos were observed under a confocal laser scanning microscope (TCS SPE, Leica, Wetzlar, Germany).
Statistical analyses
All statistical analyses were performed using GraphPad Prism (GraphPad Software, La Jolla, CA, USA). For the experiments described in Figs. 3, 4 and 6, one-way ANOVA followed by Dunnett’s test was performed to evaluate statistical significance. Data were considered statistically significant at P < 0.05. Graphs are presented as means ± standard error of the mean. All experiments were repeated independently at least three times.
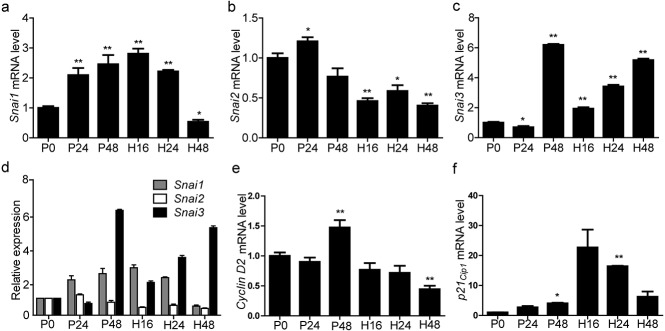
Fig. 3.
Quantitative real-time polymerase chain reaction analyses of Snai1, Snai2, and Snai3 mRNA expression in ovaries from mice treated with pregnant mare serum gonadotropin (P) for 0 h (P0), 24 h (P24), or 48 h (P48) followed by treatment with human chorionic gonadotropin (H) for 16 h (H16), 24 h (H24), or 48 h (H48). Relative expression of (a) Snai1, (b) Snai2, and (c) Snai3 mRNA to β-actin. (d) Comparison of Snai1, Snai2, and Snai3 expression. Relative expression of (e) cyclin D2 and (f) p21 mRNA to β-actin. Data are expressed as means ± SEM. n = 3. * P < 0.05, ** P < 0.01.
Fig. 4.
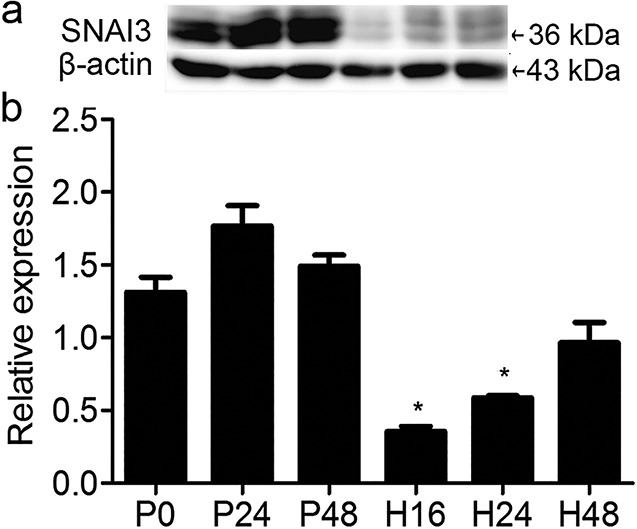
SNAI3 protein content in ovaries of immature mice at various stages following treatment with pregnant mare serum gonadotropin (P) for 0 h (P0), 24 h (P24), or 48 h (P48) followed by administration of human chorionic gonadotropin (H) for 16 h (H16), 24 h (H24), or 48 h (H48). (a) Western blot analysis of SNAI3 (36 kDa); β-actin was used as the loading control. (b) Relative expression of SNAI3 normalized to β-actin and compared with that at 0 h after P treatment (P0). Data are expressed as means ± SEM. n = 3. * P < 0.05, ** P < 0.01.
Fig. 6.
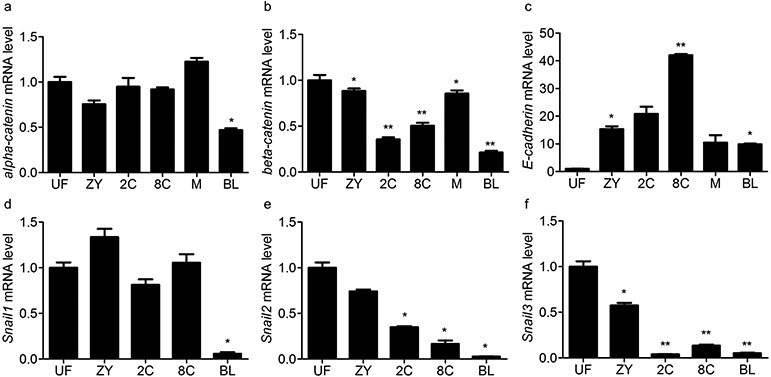
mRNA levels of α-catenin, β-catenin, E-cadherin, Snai1, Snai2, and Snai3 determined by the quantitative real-time polymerase chain reaction in eggs and pre-implantation embryos. UF, unfertilized eggs. ZY, zygote. 2C, two-cell embryos. 8C, eight-cell embryos. M, morulae. BL, blastocysts. At least 40 oocytes/embryos were evaluated in each group. Three independent assays were carried out for each gene. Data are presented as means ± SEM of triplicate simples. * P < 0.05, ** P < 0.01.
Results
Localization of SNAI3 during postnatal ovarian follicular development
The distribution of SNAI3 during ovarian development was detected by immunohistochemistry. The expression patterns of SNAI3 are shown in Figure 1. SNAI3 was not detected in the ovary at PDs 1 and 4 (Fig. 1a, b). It first appeared in oocytes on PD 9. At this time, SNAI3 was also found to be expressed in theca and interstitial cells. Following this, SNAI3 expression was detected consistently in these sites at all stages studied (PDs 18 and 23) (Fig. 1c–e).
Fig. 1.
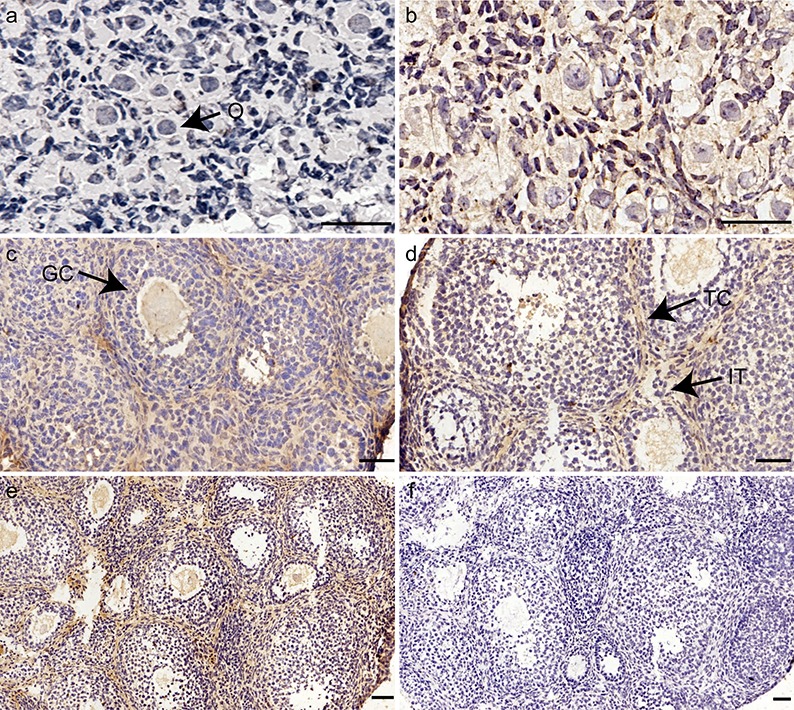
Immunohistochemical localization of SNAI3 during postnatal ovarian follicular development. Ovaries were collected from mice at (a) postnatal day (PD) 1, (b) PD 4, (c) PD 9, (d) PD 18, and (e) PD 23. SNAI3 was visualized with diaminobenzidine staining (brown). (a, b) SNAI3 immunoreactivity is not evident in the ovary (O, oocytes). (c, d) Positive staining for SNAI3 is evident in O, theca cells (TC), and interstitial cells (IT). (GC, granulosa cells.) (e) During ovarian development, immunostaining of SNAI3 is localized to IT, TC, and O. (f) Negative control. Bars = 50 μm.
Localization of SNAI3 during follicular maturation, ovulation, and luteinization
To stimulate synchronized follicular development, ovulation, and luteinization, immature 21-day-old female mice were injected with gonadotropins. The results demonstrated that in pre-antral, antral, and pre-ovulatory follicles, immunoreactivity of SNAI3 was evident in interstitial cells, theca cells, ovarian surface epithelial cells, and oocytes, which was similar to SNAI1 and SNAI2 (Fig. 2a–c). Following ovulation, GCs initiate a program of terminal differentiation and transform into luteal cells within a few hours. During this time, SNAI3 was evident in CL at 16 and 48 h following hCG treatment (Fig. 2d, e). As the CL regressed, the expression level of SNAI3 increased (Fig. 2e).
Fig. 2.
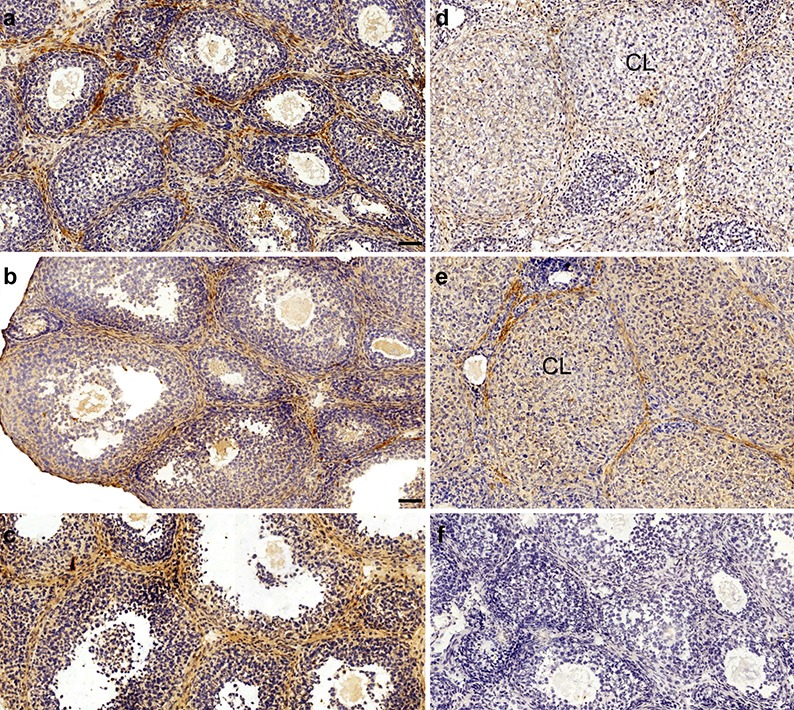
Ovarian localization of SNAI3 during gonadotropin-induced follicular maturation, ovulation, and luteinization in immature mice. (a) Postnatal day (PD) 21. (b) Treatment with pregnant mare serum gonadotropin (PMSG) for 24 h. (c) Treatment with PMSG for 48 h, followed by treatment with human chorionic gonadotropin (hCG) for (d) 16 h and (e) 48 h. (f) Negative control treated with nonimmune serum instead of the primary antibody. CL, corpus luteum. Bars = 50 μm.
Next, we examined the transcription of Snai1, Snai2,Snai3,cyclin D2, and p21 in ovaries from gonadotropin-treated mice and compared the mRNA levels by qRT-PCR. The results demonstrated that PMSG increased the transcription of Snai1 dramatically during follicular development and ovulation when compared with 0 h (P 0). However, following luteinization, the transcription of Snai1 decreased 48 h after hCG treatment (Fig. 3a). On the contrary, the expression of Snai2 did not change significantly during follicular development and decreased significantly 16 h after hCG treatment (Fig. 3b). The expression of Snai3 increased dramatically at 48 h following PMSG treatment and then increased steadily during ovulation and luteinization (Fig. 3c). When comparing the mRNA levels of Snai1, Snai2, and Snai3, we found that the expression of Snail superfamily members were different at a given time (Fig. 3d). We also determined the transcription of cyclin D2 and p21 and discovered that their expression was opposite to the levels of Snai3 after hCG treatment (Fig. 3e, f).
SNAI1 and SNAI2 are associated with follicular maturation, ovulation, and luteinization [24]. Therefore, we measured SNAI3 levels during these processes. We collected ovaries at 24 and 48 h post PMSG, and at 16, 24, and 48 h following hCG administration. The results showed that, in our model, the expression of SNAI3 did not change significantly during follicular development. After ovulation, SNAI3 expression decreased markedly at 16 h, after which the levels increased steadily at 24 and 48 h post hCG treatment (Fig. 4).
Subcellular localization of SNAI3 in oocytes and during early embryonic cleavage
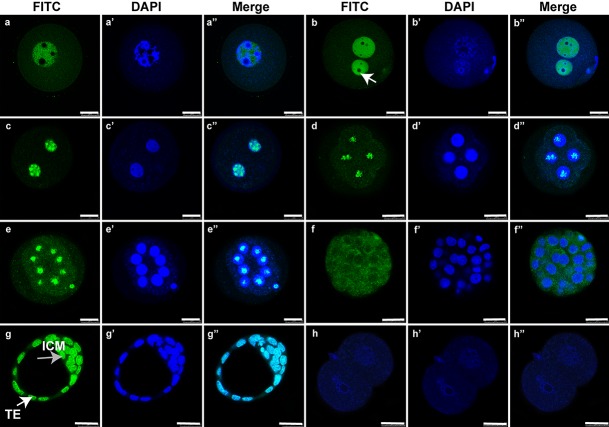
Our results indicated that SNAI3 was localized to the nucleus, except the nucleolus at the GV and one-cell stages. At these stages, GV oocytes and zygotes contained many NPBs (Fig. 5a, b”). SNAI3 was localized to the nucleolus from the two-cell stage to eight-cell embryos (Fig. 5c–e’’), after which SNAI3 was detected only in the cytoplasm (Fig. 5f–f’’). During the blastocyst stage, it was localized to the nucleus of the trophectoderm and in the inner cell mass (Fig. 5g–g’’).
Fig. 5.
Immunofluorescence localization of SNAI3 during the germinal vesicle (GV) stage and early embryonic cleavages. Immunostaining with an anti-SNAI3 antibody (green) and DNA staining with 4′,6-diamidino-2-phenylindole dihydrochloride (blue). (a–a’’) GV. (b–b’’) One-cell stage (white arrow indicates the nucleolus). (c–c’’) Two-cell stage. (d–d’’) Four-cell stage. (e–e’’) Eight-cell stage. (f–f’’) Morula. (g–g’’) Blastocyst (ICM, inner cell mass, TE/white arrow, trophectoderm). (h–h’’) Negative control for SNAI3. Bars = 25 μm.
qRT-PCR was performed to investigate the mRNA levels of α-catenin, β-catenin, E-cadherin,Snai1, Snai2, and Snai3 (Fig. 6). α-Catenin mRNA levels fluctuated during embryo development, and the expression of β-catenin was unstable. Their expression levels were lowest at the blastocyst stage. From eight-cell to morula stages, compaction occurs, which is a prerequisite for blastocyst formation. During this time, E-cadherin was expressed at its highest level at the eight-cell stage. Thereafter, it decreased and was maintained at low levels. The level of Snai1 mRNA changed slightly and reached its lowest level at the blastocyst stage, while Snai2 and Snai3 were expressed at very low levels from two-cell to the blastocyst stage.
Discussion
In this study, we determined the expression of SNAI3 in the ovaries of neonatal and gonadotropin-treated immature mice, as well as in GV oocytes and pre-implantation embryos. Our data showed that SNAI3 appeared in oocytes by PD 9, when it was also expressed in theca and interstitial cells, where it remained at later stages. In gonadotropin-induced immature mice, SNAI3 was expressed in CLs. Western blot analysis showed that SNAI3 did not change significantly during follicular development. SNAI3 was localized to the nucleus but not the nucleolus during the GV and one-cell stages. From two- to eight-cell stages, it was localized to the nucleolus, following which it was found in the cytoplasm. During the blastocyst stage, SNAI3 was located in the nucleus of the trophectoderm and inner cell mass. qRT-PCR results showed that the expression of Snail superfamily genes was decreased during the blastocyst stage. At the same time, the expression of compaction related genes (i.e., E-cadherin and catenins) changed greatly during early embryo development. Taken together, these results suggest that SNAI3 may play roles in the regulation of follicle development and early embryo cleavage.
SNAI3 in follicular development
Our results indicated that the localization of SNAI3 was distinct from the positive staining of oocytes for SNAI2 within primordial follicles, and was similar to SNAI1, as we have shown previously [24]. SNAI3 could not be detected in oocytes until PD 9 when oocyte growth begins [28]. Therefore, we speculated that SNAI3 might regulate primary oocyte development, but does not take part in establishment of the primordial follicle pool.
The pool of primordial follicles at birth represents the total population of germ cells available to female mammals during their entire reproductive life [1]. The zinc finger domains of SNAI3 bind to CAGGTG (CANNTG) E-box motifs and compete with the basic helix-loop-helix transcription factors [16], such as SOHLH2 and FIGα. In females, Sohlh2 transcripts are confined to oocytes of small follicles in the immature ovary. In adult ovaries, SOHLH2 protein is present in primordial follicles but is not detected in growing oocytes [29]. FIGα plays a key regulatory role in the expression of multiple oocyte-specific genes, including those that initiate folliculogenesis and those that encode the zona pellucida [30]. The persistence of SNAI3 in growing oocytes in our work indicates that SNAI3 may compete with SOHLH2 and FIGα to control follicular development. Because SNAI3 belongs to the Snail superfamily and its expression patterns are similar to SNAI1 and SNAI2 in primary, secondary, and mature follicles [24], we speculate that it has similar roles in promoting follicular development at these stages.
SNAI3 during formation of CL
When preantral follicles develop into Graafian follicles, GCs are sensitive to luteinizing hormone. Luteinizing hormone stimulates ovulation and acts as a signal for GCs to differentiate into CLs. The reprogramming of GCs into luteal cells demands they exit from the cell cycle and arrest predominantly in the G0/G1 phase [31]. Termination of cell proliferation during luteinization correlates with the loss of many cell cycle regulators, such as cyclin D2, that promote progression to the G1 phase by activating Cdk4 [31, 32]. The highly conserved promoter region of cyclin D2 contains two consensus E-box sequences that can compete for binding with members of the Snail family [33, 34]. Thus, Snail family proteins directly repress transcription of cyclin D2 to arrest the cell cycle.
A negative correlation was observed between SNAI3 and cyclin D2 in CLs, which led us to propose that SNAI3 participates in luteinization by repressing the promoter activity of cyclin D2 during transformation from GCs to luteal cells. In addition, members of the Snail family might function by cooperating with other E-box containing proteins, such as p21, or by binding to the E-box of proteins such as to c-Myc and arrest luteal cells in the G0/G1 phase.
P21 has been long known for inhibiting cyclin/Cdk [35, 36]. However, more recent data have shown that it has positive effects on cyclin/Cdk activation [37] through its “assembly factor” function. Because an inverse correlation was observed between SNAI3 and P21 in CLs, we speculated that SNAI3 had a potential influence on CL formation. The oncogene protein, c-Myc, binds specifically to 5′-CACGTG-3′ E-box sequences to activate transcription. c-Myc is expressed constitutively in human CLs and also has an effect on CL formation [38]. In summary, SNAI3 might block the cell cycle and promote CL formation, although this idea warrants further investigation.
SNAI3 expression during early embryonic cleavage
In GV oocytes, there are many NPBs that disappear with the onset of GV breakdown, but appear again after fertilization in the pro-nuclei of zygotes. NPBs in zygotes are of maternal origin and are essential for early development. Embryos originating from enucleolated oocytes form anucleolate nuclei and are arrested at the two-cell stage. When oocytes are enucleolated and the NPB is re-transplanted, healthy offspring can be obtained [39]. The reason for the developmental arrest of embryos lacking nucleoli is unclear, although it has been speculated that the problem might lie in the lack of, or aberrant, transcription.
SNAI3 is localized to nucleoli from the two-cell stage to eight-cell embryos. The only functions previously assigned to the nucleolus were rRNA synthesis and ribosome assembly [40,41,42]. But recently, a striking subnuclear distribution of c-Myc RNA transcripts was found. A major fraction of the sense-strand nuclear c-Myc transcript is localized to the nucleoli [43]. C-Myc is an oncogenic transcription factor that integrates the cell cycle machinery with cell adhesion, cellular metabolism, and apoptotic pathways [44]. Furthermore, the nucleolus is a key regulator of p53 stability. P53 is stable in a cell unless the nucleolus promotes its degradation through the ubiquitin pathway, with several knock-on effects through pathways controlled by the p53 status [45]. Relatively little is known about the spatial distribution and compartmentalization of RNA within nucleoli, or about the possible significance of subnuclear localization with respect to RNA processing, intranuclear turnover, or transport to the cytoplasm. The function of SNAI3 in the nucleoli is also unclear and further work is needed to delineate its role.
During embryo compaction, blastomeres acquire polarity and become flat [46, 47]. This compaction is mediated by the cell adhesion molecule, E-cadherin. SNAI1 represses the transcription of E-cadherin, both in vitro and in vivo, by binding to the E-box sequence of its promoter. Clustering of E-cadherin is thought to depend on linkage through cytoplasmic catenin to the actin cytoskeleton. Specifically, β-catenin binds directly to both the E-cadherin cytoplasmic domain [20] and to the actin-binding protein α-catenin [48]. In the current work, SNAI3 was localized to the cytoplasm during the morulae stage, when it may be unable to efficiently repress this activity, thus allowing compaction to proceed.
The EMT, in which polarized epithelial cells are converted into motile cells, is crucial for multicellular organisms to get past the blastula stage of embryonic development (embryogenesis), and is a defining structural change during organ development as well as carcinogenesis [49]. E-Cadherin at the adherens junction is a key molecular target of the EMT [49], and down-regulation of E-cadherin is thought to play a fundamental role during the EMT. SNAI1 is a direct repressor of E-cadherin [49], while SNAI2 represses E-cadherin to promote the EMT in various cancers. For example, it represses E-cadherin via p19Arf in prostate tumorigenesis [49].
Combined with our previous research [24], we found that SNAI3 shows similar tendencies as SNAI2 during early embryo development. Thus, because Snai3 mutant mice are viable and fertile [23], like Slug knockout mice [50], the SNAI family might play a similar role during embryogenesis, and there might be redundant roles among SNAI1, SNAI2, and SNAI3. However, whether SNAI3 and other family members repress the same or different sets of target genes, and how such regulation leads to the correct decision in various developmental and physiological processes, remains to be determined.
We noted an inconsistency between Snai3 mRNA levels and the amount of its protein during folliculogenesis and embryogenesis. Because gene expression is a complex process that is controlled at multiple steps, such as transcriptional regulation, post-transcriptional regulation (RNA processing, RNA stability), ribosome occupancy and density, tRNA availability, translational regulation, post-translational regulation, and protein stability, RNA and protein levels are not always well-correlated [51,52,53]. Moreover, the protein-per-mRNA ratio is different for different genes, and may even change for a given gene under different cellular conditions [54]. Therefore, the basis for this discrepancy needs considerable further investigation.
Conflict of interest
No financial conflict of interest exists. The authors do not have any potential conflict of interest on the discussed topic.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (NSFC: 31471399 and 31671199), National Key R&D Program of China (2017YFC1001403), and Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (20152515) to CZ. Support was also received by a grant from the Major Program of the National Natural Science Foundation of China (81490743) to Z-JC, and by the Shanghai Commission of Science and Technology (funding number: 17DZ2271100).
References
- 1.Kezele P, Nilsson E, Skinner MK. Cell-cell interactions in primordial follicle assembly and development. Front Biosci 2002; 7: d1990–d1996. [DOI] [PubMed] [Google Scholar]
- 2.Faddy MJ, Gosden RG, Gougeon A, Richardson SJ, Nelson JF. Accelerated disappearance of ovarian follicles in mid-life: implications for forecasting menopause. Hum Reprod 1992; 7: 1342–1346. [DOI] [PubMed] [Google Scholar]
- 3.Slizynski BM. Meiotic prophase in female mice. Nature 1957; 179: 638. [DOI] [PubMed] [Google Scholar]
- 4.Borum K. Oogenesis in the mouse. A study of the meiotic prophase. Exp Cell Res 1961; 24: 495–507. [DOI] [PubMed] [Google Scholar]
- 5.Pepling ME. Follicular assembly: mechanisms of action. Reproduction 2012; 143: 139–149. [DOI] [PubMed] [Google Scholar]
- 6.McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev 2000; 21: 200–214. [DOI] [PubMed] [Google Scholar]
- 7.Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev 2009; 30: 624–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fauser BC, Van Heusden AM. Manipulation of human ovarian function: physiological concepts and clinical consequences. Endocr Rev 1997; 18: 71–106. [DOI] [PubMed] [Google Scholar]
- 9.Bowman P, McLaren A. Cleavage rate of mouse embryos in vivo and in vitro. J Embryol Exp Morphol 1970; 24: 203–207. [PubMed] [Google Scholar]
- 10.Hardy K, Spanos S. Growth factor expression and function in the human and mouse preimplantation embryo. J Endocrinol 2002; 172: 221–236. [DOI] [PubMed] [Google Scholar]
- 11.Pochukalina G, Parfenov V. Nucleolus transformation in mouse antral follicles: distribution of coilin and components of RNA-polymerase I complex. Cell Tissue Biol 2008; 2: 522–530. [PubMed] [Google Scholar]
- 12.Biggiogera M, Martin TE, Gordon J, Amalric F, Fakan S. Physiologically inactive nucleoli contain nucleoplasmic ribonucleoproteins: immunoelectron microscopy of mouse spermatids and early embryos. Exp Cell Res 1994; 213: 55–63. [DOI] [PubMed] [Google Scholar]
- 13.Chouinard LA. A light- and electron-microscope study of the nucleolus during growth of the oocyte in the prepubertal mouse. J Cell Sci 1971; 9: 637–663. [DOI] [PubMed] [Google Scholar]
- 14.Sefton M, Sánchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development 1998; 125: 3111–3121. [DOI] [PubMed] [Google Scholar]
- 15.Boulay JL, Dennefeld C, Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature 1987; 330: 395–398. [DOI] [PubMed] [Google Scholar]
- 16.Kataoka H, Murayama T, Yokode M, Mori S, Sano H, Ozaki H, Yokota Y, Nishikawa S, Kita T. A novel snail-related transcription factor Smuc regulates basic helix-loop-helix transcription factor activities via specific E-box motifs. Nucleic Acids Res 2000; 28: 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2000; 2: 84–89. [DOI] [PubMed] [Google Scholar]
- 18.Fleming TP, Sheth B, Fesenko I. Cell adhesion in the preimplantation mammalian embryo and its role in trophectoderm differentiation and blastocyst morphogenesis. Front Biosci 2001; 6: D1000–D1007. [DOI] [PubMed] [Google Scholar]
- 19.Hirano S, Nose A, Hatta K, Kawakami A, Takeichi M. Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J Cell Biol 1987; 105: 2501–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber AH, Weis WI. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 2001; 105: 391–402. [DOI] [PubMed] [Google Scholar]
- 21.Newkirk KM, MacKenzie DA, Bakaletz AP, Hudson LG, Kusewitt DF. Microarray analysis demonstrates a role for Slug in epidermal homeostasis. J Invest Dermatol 2008; 128: 361–369. [DOI] [PubMed] [Google Scholar]
- 22.Katoh M, Katoh M. Identification and characterization of human SNAIL3 (SNAI3) gene in silico. Int J Mol Med 2003; 11: 383–388. [PubMed] [Google Scholar]
- 23.Bradley CK, Norton CR, Chen Y, Han X, Booth CJ, Yoon JK, Krebs LT, Gridley T. The snail family gene snai3 is not essential for embryogenesis in mice. PLoS ONE 2013; 8: e65344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo C, Meng X, Bai J, Chen C, Liu T, Liu S, Zhang C, Li W-P. Expression and localization of transcription factors SNAIL and SLUG in mouse ovaries and pre-implantation embryos. Cell Tissue Res 2014; 358: 585–595. [DOI] [PubMed] [Google Scholar]
- 25.Cui LL, Yang G, Pan J, Zhang C. Tumor necrosis factor α knockout increases fertility of mice. Theriogenology 2011; 75: 867–876. [DOI] [PubMed] [Google Scholar]
- 26.Guo T, Zhang L, Cheng D, Liu T, An L, Li WP, Zhang C. Low-density lipoprotein receptor affects the fertility of female mice. Reprod Fertil Dev 2015; 27: 1222–1232. [DOI] [PubMed] [Google Scholar]
- 27.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters H. The development of the mouse ovary from birth to maturity. Acta Endocrinol (Copenh) 1969; 62: 98–116. [DOI] [PubMed] [Google Scholar]
- 29.Ballow DJ, Xin Y, Choi Y, Pangas SA, Rajkovic A. Sohlh2 is a germ cell-specific bHLH transcription factor. Gene Expr Patterns 2006; 6: 1014–1018. [DOI] [PubMed] [Google Scholar]
- 30.Liang L, Soyal SM, Dean J. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 1997; 124: 4939–4947. [DOI] [PubMed] [Google Scholar]
- 31.Stocco C, Telleria C, Gibori G. The molecular control of corpus luteum formation, function, and regression. Endocr Rev 2007; 28: 117–149. [DOI] [PubMed] [Google Scholar]
- 32.Richards JS, Russell DL, Robker RL, Dajee M, Alliston TN. Molecular mechanisms of ovulation and luteinization. Mol Cell Endocrinol 1998; 145: 47–54. [DOI] [PubMed] [Google Scholar]
- 33.Mauhin V, Lutz Y, Dennefeld C, Alberga A. Definition of the DNA-binding site repertoire for the Drosophila transcription factor SNAIL. Nucleic Acids Res 1993; 21: 3951–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J 1999; 18: 5321–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Ried 1991; 49: 370. [DOI] [PubMed] [Google Scholar]
- 36.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993; 75: 805–816. [DOI] [PubMed] [Google Scholar]
- 37.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev 1997; 11: 847–862. [DOI] [PubMed] [Google Scholar]
- 38.Putowski LT, Skrzypczak M, Zielewicz J, Kaminski K, Jakowicki JA. The relevance of c-myc to the physiology of the human ovary. Gynecol Endocrinol 1997; 11: 5–10. [DOI] [PubMed] [Google Scholar]
- 39.Ogushi S, Palmieri C, Fulka H, Saitou M, Miyano T, Fulka J., Jr. The maternal nucleolus is essential for early embryonic development in mammals. Science 2008; 319: 613–616. [DOI] [PubMed] [Google Scholar]
- 40.Hadjiolov AA. Ribosome biogenesis in the life cycle of normal and cancer cells. The nucleolus and ribosome biogenesis. Springer; 1985: 165-196. [Google Scholar]
- 41.Hadjiolov AA, Nikolaev N. Maturation of ribosomal ribonucleic acids and the biogenesis of ribosomes. Prog Biophys Mol Biol 1976; 31: 95–144. [DOI] [PubMed] [Google Scholar]
- 42.Busch H, Smetana K. The Nucleolus. New York: Academic Press. 1970. [Google Scholar]
- 43.Bond VC, Wold B. Nucleolar localization of myc transcripts. Mol Cell Biol 1993; 13: 3221–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Exp Cell Res 1999; 253: 63–77. [DOI] [PubMed] [Google Scholar]
- 45.Matthews DA, Olson MO. What is new in the nucleolus?: workshop on the nucleolus: new perspectives. EMBO Rep 2006; 7: 870–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riethmacher D, Brinkmann V, Birchmeier C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc Natl Acad Sci USA 1995; 92: 855–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodall H. Manipulation of gap junctional communication during compaction of the mouse early embryo. J Embryol Exp Morphol 1986; 91: 283–296. [PubMed] [Google Scholar]
- 48.Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci 1994; 107: 3655–3663. [DOI] [PubMed] [Google Scholar]
- 49.Katoh M. Epithelial-mesenchymal transition in gastric cancer (Review). Int J Oncol 2005; 27: 1677–1683. [PubMed] [Google Scholar]
- 50.Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol 1998; 198: 277–285. [PubMed] [Google Scholar]
- 51.Gupta S, Verma S, Mantri S, Berman NE, Sandhir R. Targeting microRNAs in prevention and treatment of neurodegenerative disorders. Drug Dev Res 2015; 76: 397–418. [DOI] [PubMed] [Google Scholar]
- 52.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst 2009; 5: 1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 2003; 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol 2004; 2: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]