Abstract
Reproduction is an important trait in sheep breeding as well as in other livestock. However, despite its importance the genetic mechanisms of litter size in domestic sheep (Ovis aries) are still poorly understood. To explore genetic mechanisms underlying the variation in litter size, we conducted multiple independent genome-wide association studies in five sheep breeds of high prolificacy (Wadi, Hu, Icelandic, Finnsheep, and Romanov) and one low prolificacy (Texel) using the Ovine Infinium HD BeadChip, respectively. We identified different sets of candidate genes associated with litter size in different breeds: BMPR1B, FBN1, and MMP2 in Wadi; GRIA2, SMAD1, and CTNNB1 in Hu; NCOA1 in Icelandic; INHBB, NF1, FLT1, PTGS2, and PLCB3 in Finnsheep; ESR2 in Romanov and ESR1, GHR, ETS1, MMP15, FLI1, and SPP1 in Texel. Further annotation of genes and bioinformatics analyses revealed that different biological pathways could be involved in the variation in litter size of females: hormone secretion (FSH and LH) in Wadi and Hu, placenta and embryonic lethality in Icelandic, folliculogenesis and LH signaling in Finnsheep, ovulation and preovulatory follicle maturation in Romanov, and estrogen and follicular growth in Texel. Taken together, our results provide new insights into the genetic mechanisms underlying the prolificacy trait in sheep and other mammals, suggesting targets for selection where the aim is to increase prolificacy in breeding projects.
Keywords: sheep, prolificacy, genome-wide association study, biological pathways, regulation
Introduction
Reproduction is one of the most important traits in livestock production particularly for females. Selection for higher prolificacy in domestic sheep (Ovis aries) has led to variable litter size (LS) within and among breeds. For example, individual litter size of 1 to 8 has been recorded in the Hu sheep and Finnsheep (Yue, 1996; Davis et al., 2006a).
Previous studies reported that the exceptional prolificacy of the Booroola Merino was attributed to a single major gene, while a number of mutations of a major effect on litter size have been identified in other sheep breeds (Table 1; see also Xu and Li, 2017). Vage et al. (2013) detected a mutation FecGF in gene GDF9 strongly associated with litter size in Norwegian White Sheep and Finnish Landrace (Finnsheep) using a genome-wide association analysis. Demars et al. (2013) reported the mutations FecXGr in Grivette sheep and FecXO in Olkuska sheep associated with the highly prolific phenotype by a genome-wide association analysis. Cao et al. (2016) found that nine candidate genes including the well-known FecB mutation played important roles in the variable litter size in Hu and Small-tailed Han sheep through methylated DNA-immunoprecipitation sequencing data. Miao et al. (2016) identified a set of differentially expressed genes (e.g., FecB) between low- and high-prolificacy breeds (Dorset vs. Small-tailed Han sheep) through implementing integrated analysis of miRNAs and lncRNAs. Lassoued et al. (2017) found the mutation FecXBar associated with the prolificacy in Tunisian Barbarine. Despite its great importance the genetic mechanisms of the high prolificacy trait in domestic sheep are still poorly understood, partly due to shortage of studies conducted across multiple prolific sheep breeds. To date, numerous fecundity-associated mutations have been identified in different sheep breeds, but very few mutations have been consistently detected across the breeds. Despite the reproduction of ewes can be affected by the complex interactions of environmental conditions (i.e., climate, density, and food abundance) (Wilson et al., 2009), previous studies suggested that genetic factor could play important roles in the variable litter size of ewes.
Table 1.
Genetics variants associated with the fecundity in sheep.
| Gene | Mutation | Name, allele symbol | Founder breeds | Reference |
|---|---|---|---|---|
| BMP15 | V299D | Inverdale, FecXI | Romney, Inverdale | Galloway et al., 2000 |
| Q291Ter | Hanna, FecXH | Romney | Galloway et al., 2000 | |
| S367I | Belclare, FecXB | Belclare | Hanrahan et al., 2004 | |
| Q239R | Galway, FecXG | Belclare, Cambridge, Small-tailed Han | Hanrahan et al., 2004 | |
| C321Y | Lacaune, FecXL | Lacaune | Bodin et al., 2007 | |
| ΔP154S159 | Rasa Aragonesa, FecXR | Rasa Aragonesa | Martinez-Royo et al., 2008; Monteagudo et al., 2009 | |
| T317I | Grivette, FecXGr | Grivette (France) | Demars et al., 2013 | |
| N337H | Olkuska, FecXO | Olkuska (Poland) | Demars et al., 2013 | |
| c.301G > T, c.310insC, c.302_304delCTA | Barbarine, FecXBar | Tunisian Barbarine | Lassoued et al., 2017 | |
| Unknown | Woodlands, FecXW | Woodlands | Feary et al., 2007 | |
| BMPR1B | Q249R | Booroola, FecBB | Booroola Merino, Garole, Javanese, Small-tailed Han, Wadi, Hu | Mulsant et al., 2001; Souza et al., 2001; Wilson et al., 2001; Chu et al., 2011; Zhang et al., 2011; Cao et al., 2016 |
| GDF9 | S395F | High Fertility, FecGH | Belclare, Cambridge | Hanrahan et al., 2004 |
| S427R | Thoka, FecGT | Icelandic | Nicol et al., 2009 | |
| F345C | Embrapa, FecGE | Santa Ines | Silva et al., 2011 | |
| V371M | FecGF | Norwegian White Sheep, Finnsheep Landrace, Belclare | Vage et al., 2013; Mullen and Hanrahan, 2014 | |
| R315C | Vacaria, FecGV | Brazilian sheep | de Souza et al., 2012 | |
| R87H | FecGI | Baluchi | Moradband et al., 2011 | |
| B4GALNT2 | Lacaune, FecLL | Lacaune | Drouilhet et al., 2013 | |
| Woodlands | Wood-land, FecX2W | Coopworth | Davis, 2005 | |
| OLKUSKA | Olkuska | Davis, 2004 | ||
| BELLE-ILE | Belle-Ile | Davis, 2005 | ||
| Unknown | FecW | Davis et al., 2006b |
In this study, we conducted multiple independent genome-wide association studies (GWAS) on litter size in the sheep breeds of high (Wadi, Hu, Icelandic, Finnsheep, and Romanov) and low (Texel) prolificacy with a litter size ranging from 1 to 6 from different geographic regions (Figure 1A) and genetic origins (Figure 1B) of the world, respectively. Wadi sheep is a high-prolificacy native breed from the Shandong Province of China (Peng et al., 2017). Hu sheep is famous for early sexual maturity and high fecundity, and are distributed in the Taihu Lake area of Eastern China (Yue, 1996). Icelandic and Finnsheep (Finnish Landrace) sheep are northern European high-fecundity breeds (Mullen and Hanrahan, 2014; Eiriksson and Sigurdsson, 2017). Romanov sheep from the Volga Valley shows outstanding reproduction qualities: early sexual maturity, out-of-season breeding and extraordinary prolificacy (Deniskova et al., 2017). The Texel sheep is a relatively low-prolificacy breed originally from the island of Texel in the Netherlands and excels in muscle growth and lean carcasses (Casas et al., 2004). Our results will be important for further genetic improvement of the trait and for better understanding the molecular basis of reproduction in sheep as well as other mammals.
FIGURE 1.
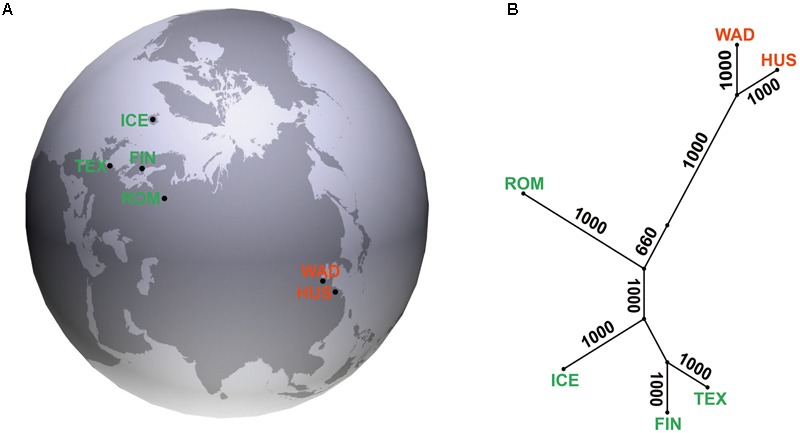
(A) Geographic locations for five sheep breeds of high (WAD, Wadi sheep; HUS, Hu sheep; ICE, Icelandic sheep; FIN, Finnsheep; and ROM, Romanov sheep) and one low (TEX, Texel sheep) prolificacy. (B) Neighbor-joining tree of the six sheep breeds with 1000 bootstrap replicates.
Materials and Methods
Sample Collection and Phenotyping
A total of 522 ewes from five sheep breeds of high (Wadi, n = 160; Hu, n = 117; Icelandic, n = 54; Finnsheep, n = 54; and Romanov, n = 78) and one low (Texel, n = 59) prolificacy were collected from farms in China, Iceland, Finland, and Russia (Figure 1A). Animals included were as unrelated as possible based on analysis of pedigree records and farmers’ knowledge. Data for the phenotype of litter size and the total number of litters collected from farm records are shown in Figure 2. The litter size ranged from 1 to 6 based on parity from 1 to 11 in six sheep breeds. Genomic DNA was extracted from the ear marginal tissues following a standard phenol/chloroform method and was diluted to 50 ng/μl for the SNP BeadChip genotyping (Köchl et al., 2005), except for the Icelandic samples which were isolated from whole-blood using MasterPureTM Complete DNA Purification Kit (Epicentre Biotech) following the manufacturers protocol.
FIGURE 2.
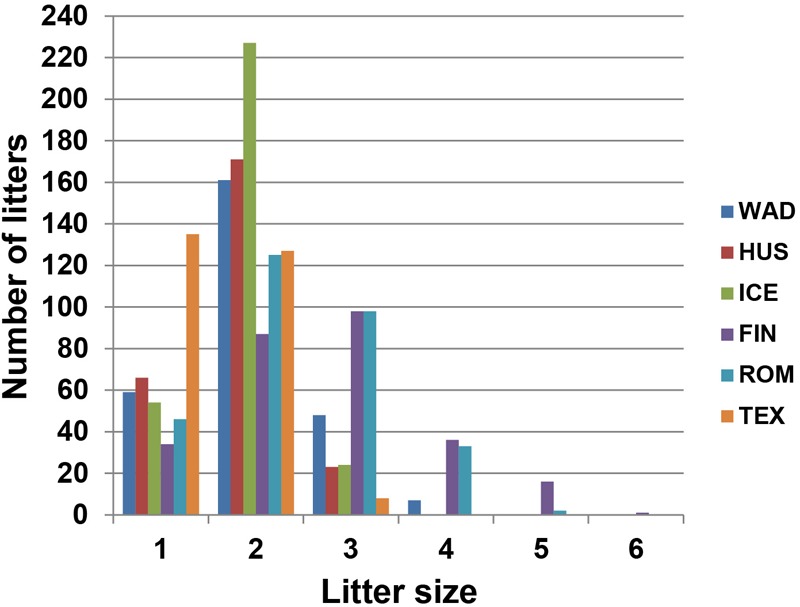
Phenotypic distribution of litter size in the six sheep breeds (WAD, Wadi sheep; HUS, Hu sheep; ICE, Icelandic sheep; FIN, Finnsheep; ROM, Romanov sheep; and TEX, Texel sheep).
Genotyping and Quality Control
All the samples were genotyped using the Ovine Infinium HD BeadChip according to the manufacturer’s protocol. Genotypes of a total of 606,006 SNPs were obtained (genotype and phenotype datasets1). We implemented quality control of these SNPs using PLINK v1.07 software (Purcell et al., 2007). The SNPs or individuals were excluded if they met any of the criteria: (1) no chromosomal or physical location, (2) call rate < 0.95, (3) missing genotype frequency > 0.05, and/or (4) minor allele frequency (MAF) < 0.05. SNPs were excluded from the analysis if a p-value of Fisher’s exact test for Hardy–Weinberg equilibrium less than 0.001.
Genetic Relationships and Population Structure
To investigate the genetic relationships and population structure among the six domestic sheep, we performed global FST, neighbor-joining (NJ) tree and principle component analysis (PCA). The global FST value was calculated using GENEPOP v4.2 (Raymond and Rousset, 1995). The genetic distances between populations were calculated using an identity by state (IBS) similarity matrix (Kang et al., 2010). Then, the distances were used to construct a NJ tree with 1000 bootstraps using the package PHYLIP v.3.695 (Felsenstein, 1989). In addition, PCA was conducted using the SmartPCA program from the EIGENSOFT package version 4.2 (Patterson et al., 2006) based on the genotypes data.
Genome-Wide Association Analysis
To explore genetic structure within the breeds, multidimensional scaling (MDS) analysis was performed based on the independent SNPs using PLINK v1.07. Firstly, we implemented the option of ‘indep-pairwise 50 5 0.05’ in PLINK v1.07, which calculated pairwise linkage disequilibrium (LD) in a 50-SNP-window shifted at a pace of five SNPs. If the LD estimate was r2 > 0.05, one of the pairs of SNPs was removed (Purcell et al., 2007). The independent SNPs retained by the LD criteria were then used in the MDS analysis, and the results were plotted using the GenABEL package in R v3.2.2 (Aulchenko et al., 2007).
We performed genome-wide association studies within five sheep breeds of high prolificacy (Wadi, Hu, Icelandic, Finnsheep, and Romanov) and one low prolificacy (Texel) using the case/control design. We ranked all individuals within the breeds according to their litter size from the highest to lowest. Then, we selected individuals from two tails for each breed as ‘case’ and ‘control,’ respectively. Based on the distribution of phenotypes, 114 samples (LS ≥ 2) in Wadi, 66 samples (LS ≥ 2) in Hu, 20 samples (LS > 2) in Icelandic, 37 samples (LS ≥ 2.5) in Finnsheep, 40 samples (LS ≥ 2.5) in Romanov and 28 samples (LS ≥ 1.6) in Texel sheep were selected as ‘cases,’ while 28 samples (LS = 1) in Wadi, 15 samples (LS = 1) in Hu, 15 samples (LS ≤ 1.75) in Icelandic, 9 samples (LS ≤ 2) in Finnsheep, 26 samples (LS ≤ 2) in Romanov and 14 samples (LS ≤ 1.33) in Texel sheep were selected as ‘controls.’ In the GWAS, we used the function of “qtscore” in the GenABEL package. Associated SNPs were identified at both the genome-wide and chromosome-wise significance levels (p < 0.05) after the Bonferroni correction (Bonferroni, 1936). To account for systematic biases caused by within-population substructure, the first and second dimensions from the MDS analyses were used as the covariates (Price et al., 2006). The correlation analysis between litter size and parity within breeds showed that there were significant effects between litter size and parity in four breeds (Wadi, Hu, Icelandic, and Texel), and the effect of parity 1 on litter size was less than that of parities 2 through 10 (Supplementary Table S1 and Supplementary Figure S1). However, the parity of individuals within breeds was different, and we mainly focused on the mean of litter size of individual (total litter size/parity) in per breed. Therefore, we excluded the effect of parity from the model. The Quantile–Quantile (Q–Q) plots were visualized by plotting the distribution of obtained vs. expected genome-wide p-values. For genotype effect of potential SNPs on litter size in each breed, differences between means were analyzed by the Student’s t-test. The p < 0.05 was considered statistically significant. All the results were presented as mean ± standard error (SE). We implemented pairwise tests of linkage disequilibrium (LD) between the most significant SNPs and their flanking SNPs within approximately 1 Mb upstream and downstream using PLINK v1.07. Regional association plots were generated using the R package v3.2.2.
Bioinformatics Analysis
We annotated the genes associated with litter size in each breed using the O. aries assembly Oar_v.4.02. Further, we submitted the genes to the DAVID (database for annotation, visualization and integrated discovery) database3 for gene ontology (GO) enrichment and pathways analyses (Huang et al., 2009a,b). The p-value of 0.1 and at least two genes from the input gene list in the enriched category were considered for the enriched GO terms. Also, we investigated the protein–protein interaction network for the candidate genes using the STRING database version 10.5 (Szklarczyk et al., 2017). In addition, differential expressions of the candidate genes in various tissues were examined using the EMBL-EBI Expression Atlas database4 (Petryszak et al., 2016).
Results
Population Relationship and Differentiation
Pairwise FST value varied from 0.023 to 0.104 among the populations with the least genetic differentiation observed between Wadi and Hu sheep breeds (Supplementary Table S2). The NJ tree showed that these breeds were clustered into two major groups according to their Chinese and European origins (Figure 1B). A similar geographic pattern was seen in the PCA analyses with the grouping of Wadi and Hu sheep separated from the other four European breeds (Supplementary Figure S2).
Genome-Wide Association Analysis
After the quality control, 508,444 SNPs and 114 individuals (91 cases vs. 23 controls) in Wadi, 506,031 SNPs and 80 individuals (66 cases vs. 14 controls) in Hu, 443,125 SNPs and 23 individuals (8 cases vs. 15 controls) in Icelandic, 492,165 SNPs and 37 individuals (28 cases vs. 9 controls) in Finnsheep, 465,794 SNPs and 38 individuals (29 cases vs. 9 controls) in Romanov, 475,955 SNPs and 39 individuals (28 cases vs. 11 controls) in Texel sheep were retained in the working dataset for the GWAS. We did find several animals outlying the clusters of cases, which might cause biases in the association analyses (Supplementary Figure S3). We have repeated the association analyses without these animals, and found the results are very similar. Thus, we did not exclude these animals in the association analyses due to the small sample size for the breeds. The resulting genomic inflation factors were equal to 1.07 in Wadi, 1.14 in Hu, 1.12 in Icelandic, 1.14 in Finnsheep, 1.10 in Romanov, and 1.05 in Texel sheep, suggesting well-controlled population stratifications (Supplementary Figure S4).
In Wadi sheep, we detected 59 and 8 SNPs at the chromosome-wise and genome-wide (p < 1.92 × 10-6) 5% significance after the Bonferroni correction, respectively (Figure 3A and Supplementary Tables S3, S4). We observed a high level of LD between the top significant SNP rs416717560 and rs421635584 located in gene BMPR1B (Figure 4A). For the SNP rs416717560, average litter size of individuals with the G/G genotype (n = 115, LS = 2.05 ± 0.06) was significantly (p < 0.01) higher than that of the ewes with the A/G (n = 15, LS = 1.47 ± 0.16) genotype (Figure 5A). Also, we found three additional significant SNPs (rs429416173, rs402803857, and rs160917020) neighboring genes BMPR1B, FBN1, and MMP2 (Table 2 and Supplementary Table S3).
FIGURE 3.
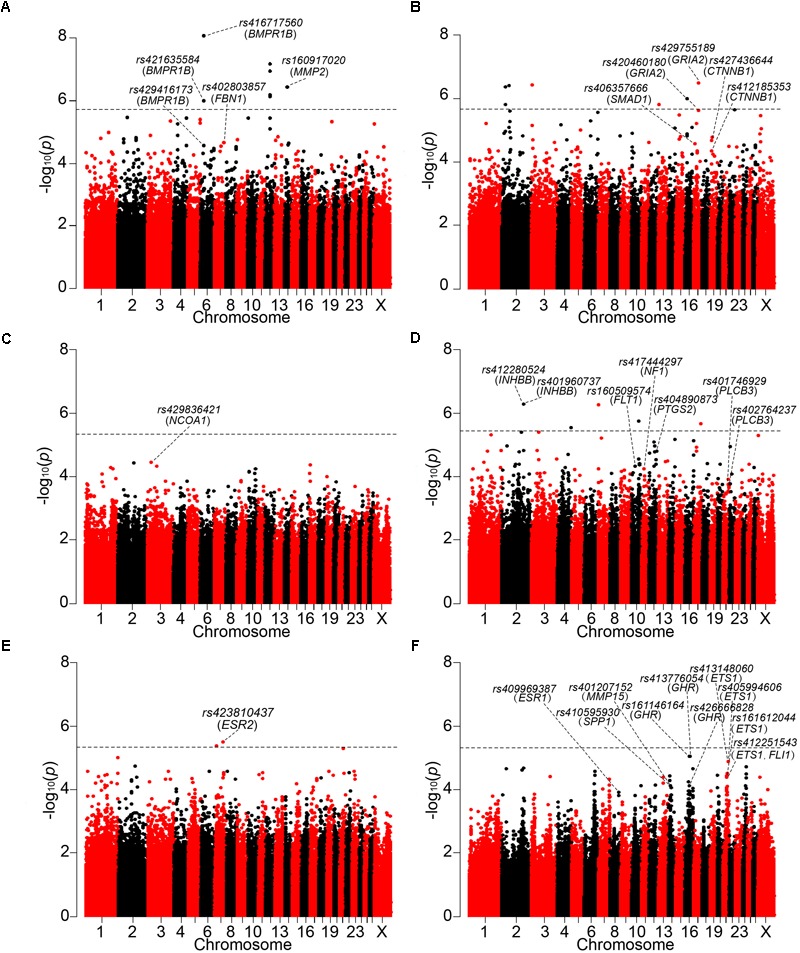
Manhattan plots of GWAS are shown on (A) Wadi, (B) Hu, (C) Icelandic, (D) Finnsheep, (E) Romanov and (F) Texel sheep. The 5% genome-wide significant threshold value is indicated by a dotted line. The significant SNPs surrounding the genes previously reported to be associated with reproduction are annotated at the chromosome-wise and genome-wide 5% significance after the Bonferroni correction.
FIGURE 4.
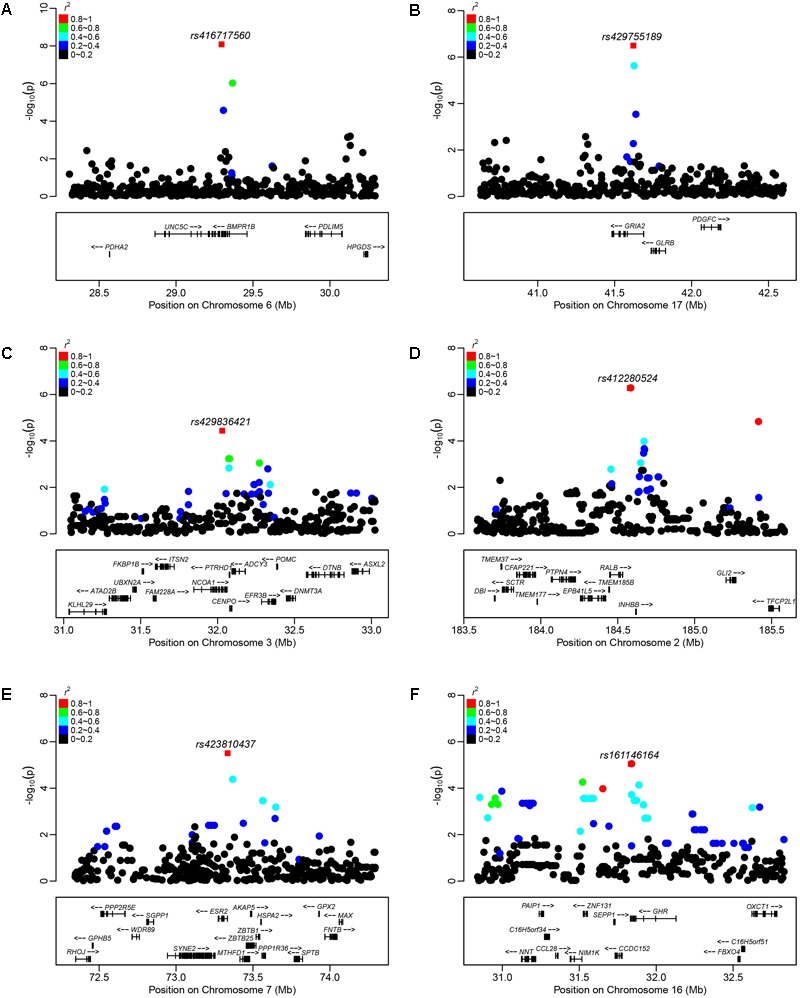
Plots of regional association results for the top significant SNP (red square) and their near SNPs in (A) Wadi, (B) Hu, (C) Icelandic, (D) Finnsheep, (E) Romanov, and (F) Texel sheep. Different colors represent the r2 values of pair-wise LD estimates.
FIGURE 5.
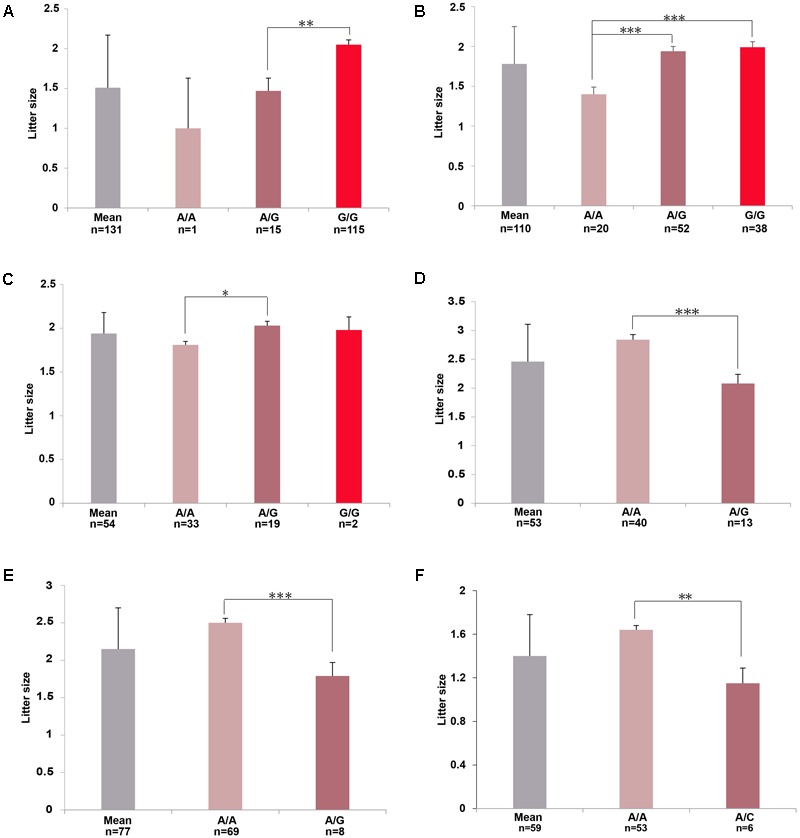
Genotypic distributions of the top significant SNPs for the litter size (LS) phenotype in (A) Wadi, (B) Hu, (C) Icelandic, (D) Finnsheep, (E) Romanov, and (F) Texel sheep, respectively. The means LS were calculated for various breeds. Number of ewes per group of genotype is mentioned. Pairwise statistical comparisons between means of genotype’s clades were performed using Student’s t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Table 2.
Genome-wide and chromosome-wise significant SNPs and associated genes.
| Population | SNP | Chr | Position (bp) | MAF | p-unadjusted | p-adjusted | Genes | Location |
|---|---|---|---|---|---|---|---|---|
| Wadi | rs416717560∗ | 6 | 29295803 | 0.07 | 3.65E-08 | 8.19E-09 | BMPR1B1 | 3′UTR |
| rs421635584∗ | 6 | 29361782 | 0.05 | 4.36E-06 | 9.78E-07 | BMPR1B1 | Intron | |
| rs429416173 | 6 | 29302788 | 0.2 | 7.55E-05 | 2.75E-05 | BMPR1B1 | CDS | |
| rs402803857 | 7 | 58598895 | 0.1 | 4.96E-05 | 2.93E-05 | FBN11 | Intron | |
| rs160917020∗ | 14 | 23133427 | 0.19 | 1.10E-06 | 3.71E-07 | MMP2 | Downstream | |
| Hu | rs429755189∗ | 17 | 41621298 | 0.43 | 1.94E-06 | 3.21E-07 | GRIA21 | Intron |
| rs420460180 | 17 | 41621269 | 0.29 | 8.50E-06 | 2.43E-06 | GRIA21 | Intron | |
| rs406357666 | 17 | 12487861 | 0.19 | 1.40E-05 | 2.66E-05 | SMAD11 | Intron | |
| rs427436644 | 19 | 13639996 | 0.32 | 7.69E-05 | 2.14E-05 | CTNNB1 | Downstream | |
| rs412185353 | 19 | 13641870 | 0.33 | 1.51E-04 | 4.49E-05 | CTNNB1 | Downstream | |
| Icelandic | rs429836421 | 3 | 32030054 | 0.16 | 4.55E-05 | 3.63E-05 | NCOA11 | Intron |
| Finnsheep | rs412280524∗ | 2 | 184578329 | 0.09 | 2.62E-05 | 5.32E-07 | INHBB | Downstream |
| rs401960737∗ | 2 | 184579671 | 0.09 | 2.62E-05 | 5.32E-07 | INHBB | Downstream | |
| rs160509574 | 10 | 31933001 | 0.27 | 1.50E-05 | 4.71E-05 | FLT11 | Intron | |
| rs417444297 | 11 | 18552961 | 0.11 | 4.20E-05 | 5.65E-05 | NF1 | Downstream | |
| rs404890873 | 12 | 65662842 | 0.05 | 1.87E-04 | 1.59E-05 | PTGS2 | Upstream | |
| rs401746929 | 21 | 41915064 | 0.08 | 1.85E-03 | 1.75E-04 | PLCB3 | Upstream | |
| rs402764237 | 21 | 41919836 | 0.08 | 1.85E-03 | 1.75E-04 | PLCB3 | Upstream | |
| Romanov | rs423810437∗ | 7 | 73335157 | 0.07 | 1.65E-05 | 3.12E-06 | ESR21 | 5′ flanking region |
| Texel | rs409969387 | 8 | 75353388 | 0.08 | 1.11E-03 | 1.21E-04 | ESR1 | Intron |
| rs410595930 | 14 | 23645021 | 0.06 | 1.33E-04 | 1.46E-04 | SPP11 | Intron | |
| rs401207152 | 14 | 25147418 | 0.06 | 1.33E-04 | 1.46E-04 | MMP15 | Downstream | |
| rs161146164 | 16 | 31834495 | 0.06 | 1.33E-04 | 9.11E-06 | GHR1 | CDS | |
| rs413776054 | 16 | 31834942 | 0.06 | 1.33E-04 | 9.11E-06 | GHR | CDS | |
| rs426666828 | 16 | 31882869 | 0.18 | 1.88E-04 | 7.54E-05 | GHR1 | Intron | |
| rs413148060 | 21 | 30950537 | 0.15 | 1.02E-04 | 4.17E-05 | ETS1 | Upstream | |
| rs405994606 | 21 | 31001548 | 0.15 | 1.02E-04 | 4.17E-05 | ETS11 | Intron | |
| rs161612044 | 21 | 31009743 | 0.14 | 5.41E-04 | 1.01E-04 | ETS11 | Intron | |
| rs412251543 | 21 | 31178275 | 0.1 | 4.01E-03 | 1.46E-04 | ETS1/FLI1 | Upstream/Downstream |
For genes the best SNP of which is located outside of upstream/downstream 150 kb region. Chr., chromosome; MAF, Minor Allele Frequency. The p-unadjusted corresponds to exact p for the Fisher’s test. The p-adjusted corresponds to the corrected significance of GWAS after principle component adjustment. The SNPs with symbol (∗) denote that bonferroni-corrected genome-wide significant SNPs. The genes with symbol (1) denote that the SNPs are intragenic, otherwise they are the nearest genes upstream and downstream of the tested SNPs.
In Hu sheep, we identified 98 and 9 SNPs at the chromosome-wise and genome-wide (p < 2.18 × 10-6) 5% significance after Bonferroni correction (Figure 3B and Supplementary Tables S3, S4). The regional plot showed that the top significant SNPs rs429755189 and rs420460180 on chromosome 17 were in an LD block that contained gene GRIA2 (Figure 4B). For the rs429755189, average litter size of individuals with the genotypes G/G (n = 38, LS = 1.99 ± 0.07) and A/G (n = 52, LS = 1.94 ± 0.06) were significantly (p < 0.001) higher than that of ewes with the genotype A/A (n = 20, LS = 1.40 ± 0.09) in the present population (Figure 5B). Among these significant SNPs, 3 (rs406357666, rs427436644 and rs412185353) are located within the genes SMAD1 and CTNNB1 (Table 2 and Supplementary Table S3).
In Icelandic sheep, we found 22 SNPs at the chromosome-wise 5% significance after the Bonferroni correction (Figure 3C and Supplementary Tables S3, S4). The top significant SNP rs429836421 on chromosome 3 was located within gene NCOA1 (Figure 4C). For rs429836421, average litter size of individuals with the A/G genotype (n = 19, LS = 2.03 ± 0.05) is significantly (p < 0.05) higher than that of the ewes with the genotype A/A (n = 33, LS = 1.81 ± 0.04) (Figure 5C).
In Finnsheep, we detected 102 and 6 SNPs at the chromosome-wise and genome-wide (p < 3.64 × 10-6) 5% significance after the Bonferroni correction, respectively (Figure 3D and Supplementary Tables S3, S4). The regional plot revealed strong LD between the top significant SNP rs412280524 and its neighboring SNPs rs401960737 and rs407751830 harbored gene INHBB (Figure 4D). For the SNP rs412280524, litter size of ewes with the genotype A/A (n = 40, LS = 2.84 ± 0.09) is significantly (p < 0.001) higher than that of the ewes with the genotype A/G (n = 13, LS = 2.08 ± 0.16) (Figure 5D). Also, five additional significant SNPs (rs160509574, rs417444297, rs404890873, rs401746929, and rs402764237) were found to be located near to genes FLT1, NF1, PTGS2, and PLCB3 (Table 2 and Supplementary Table S3).
In Romanov sheep, we identified 77 and 2 SNPs at the chromosome-wise and genome-wide (p < 4.56 × 10-6) 5% significance after the Bonferroni correction (Figure 3E and Supplementary Tables S3, S4). The top significant SNP rs423810437 on chromosome 7 was in the gene ESR2 (Figure 4E). For rs423810437, litter size of ewes with the genotype A/A (n = 69, LS = 2.50 ± 0.06) is significantly (p < 0.001) higher than that of the ewes with the genotype A/G (n = 8, LS = 1.79 ± 0.18) (Figure 5E).
In Texel sheep, we observed 133 SNPs at the chromosome-wise 5% significance after the Bonferroni correction (Figure 3F and Supplementary Tables S3, S4). The regional plot showed that the top significant SNPs rs161146164 and rs413776054 on chromosome 16 were in a strong LD region containing one functional gene GHR (Figure 4F). For rs161146164, litter size of ewes with the genotype A/A (n = 53, LS = 1.64 ± 0.05) is significantly (p < 0.01) higher than that of the ewes with the genotype A/C (n = 6, LS = 1.15 ± 0.14) (Figure 5F). The two mutations (rs161146164, Asn > His; rs413776054, Pro > Ser) cause the amino acid change in coding region of the GHR gene. In addition, we found eight additional significant SNPs (rs426666828, rs409969387, rs410595930, rs401207152, rs413148060, rs405994606, rs161612044, and rs412251543) surrounding genes ESR1, ETS1, FLI1, SPP1, and MMP15 (Table 2 and Supplementary Table S3).
In addition to the source breed where the target SNPs have been detected, we further assessed genotype effect of the most significant SNPs on litter size in the other five sheep breeds. In general, genotypes of the target SNPs did not show significant association with increased litter size in the breeds other than the source breed (Supplementary Table S7). Nevertheless, we observed some exceptions. For example, the genotype A/G of rs429836421, which was identified in Icelandic sheep, showed significant associations with increased litter size in both Icelandic and Hu sheep breeds. However, a lack of homozygotes for the SNPs such as the genotype G/G for rs412280524 in Finnsheep, G/G for rs423810437 in Romanov and C/C for rs161146164 in Texel sheep could be because of low frequency of the mutations and small sample size.
Bioinformatics Analysis
We found significantly (p < 0.1) enriched GO terms associated with reproduction for the candidate genes. The GO clusters were primarily enriched in the categories of ovarian and oocyte development (PTGS2, BMPR1B, INHBB, CTNNB1, MMP2, MMP15, FBN1, GHR, and SPP1), phospholipase C activity (FLT1 and ESR1), SMAD protein (INHBB and SMAD1) and BMP signaling (SMAD1 and BMPR1B) and positive regulation of transcription (NCOA1, FLI1, ESR1, ESR2, CTNNB1, ETS1, and BMPR1B), all of which are involved in the folliculogenesis, follicle growth and granulosa cell proliferation (Figure 6 and Supplementary Table S5). Another relevant GO category was hindbrain development (SMAD1 and CTNNB1), which participated in regulating ovulation (Baird et al., 2006). In addition, we detected 11 genes (i.e., PLCB3, ESR1, ESR2, MMP2, NCOA1, CTNNB1, INHBB, SMAD1, BMPR1B, PTGS2, and GRIA2) involved in estrogen, thyroid hormone, TGF-beta, retrograde endocannabinoid and hippo signaling pathways, and these pathways played important roles in regulating follicle growth and ovulation in livestock (Supplementary Table S5). However, we observed different GO terms for the candidate genes in different sheep breeds. For example, I-SMAD binding were enriched in Hu sheep, and chromatin binding were enriched in Texel sheep (Supplementary Table S6). In the gene network analysis, we observed that 16 genes (i.e., BMPR1B, FBN1, MMP2, SMAD1, CTNNB1, GRIA2, NCOA1, FLT1, NF1, PTGS2, PLCB3, ESR2, ESR1, ETS1, SPP1, and GHR) showed protein–protein interactions in the network (Figure 7). Expression data further showed that the genes BMPR1B, FBN1, MMP2, GRIA2, SMAD1, CTNNB1, NCOA1, NF1, FLT1, PTGS2, PLCB3, ESR2, ESR1, GHR, ETS1, MMP15, FLI1, and SPP1 were either highly or moderately expressed in reproduction-related tissues such as ovary, uterine cervix, placenta, corpus luteum, cerebellum, pituitary gland or uterus in sheep (Figure 8). Also, gene INHBB showed a high expression in ovary and uterus of Mus musculus5.
FIGURE 6.
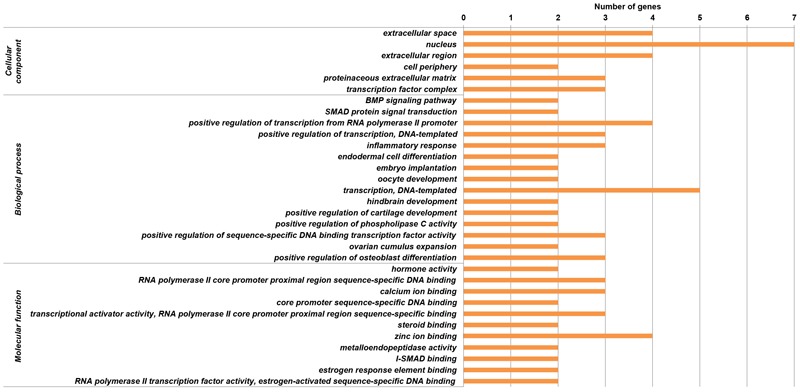
Gene ontology (GO) enrichments based on the functional genes surrounding the significant SNPs at the chromosome-wise 5% level.
FIGURE 7.
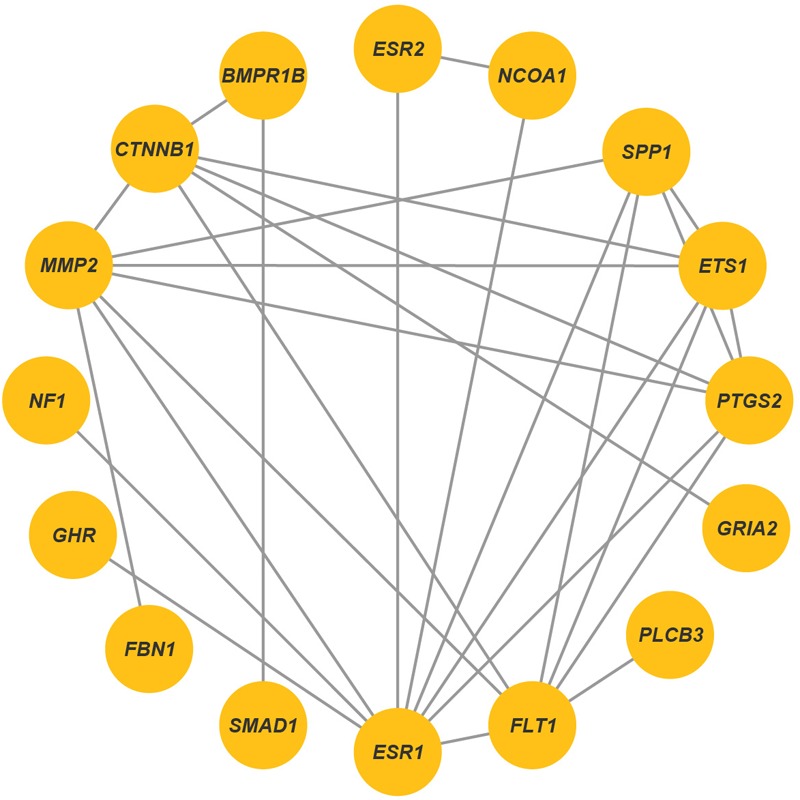
Protein–protein interaction networks identified by using STRING database. Each line indicated known signaling pathways and protein complexes.
FIGURE 8.
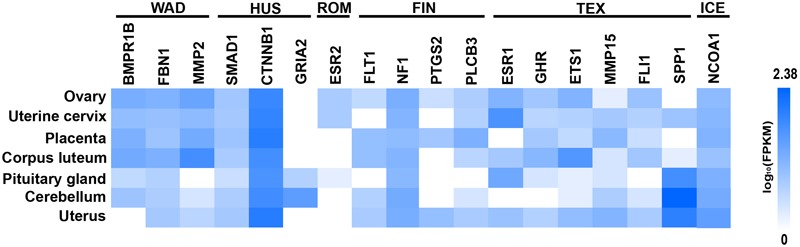
Heatmap of the candidate genes identified from six sheep breeds (WAD, Wadi sheep; HUS, Hu sheep; ICE, Icelandic sheep, FIN, Finnsheep, ROM, Romanov sheep, and TEX, Texel sheep) enriched for expression in different ewes tissues deposited in the EBI Gene Expression Atlas database. The FPKM (fragments per kilobase of transcript per million mapped reads) value is used to measure the expression level.
Discussion
In this study, we conducted multiple independent GWAS in different sheep breeds to investigate the genetic mechanisms underlying the litter size in sheep. Coupled with population relationship and bioinformatics analyses, the GWAS identified different genes associated with the litter size in different breeds and revealed their differentially genetic regulation mechanisms associated with follicle growth and ovulation in the reproduction of ewes.
The diverse biological pathways identified from the novel genes annotation play an important role in follicle growth and ovulation of females in different sheep breeds (Figure 9). The three genes identified in Wadi sheep, BMPR1B, FBN1, and MMP2, all play a crucial role in regulating hormone secretion (Mulsant et al., 2001; Basini et al., 2011; Zhang et al., 2011; Zhai et al., 2013). For example, BMPR1B gene can lead to an increased density of the follicle-stimulating hormone (FSH) and luteinizing hormone (LH) receptors with a concurrent reduction in apoptosis to increase the ovulation rate of ewes (Regan et al., 2015; Hu et al., 2016). As the main component of microfibrils in the extracellular matrix, the gene FBN1 regulates cumulus cell apoptosis by reducing the expression level of BMP15 involved in estrogen signaling in porcine ovaries (Zhai et al., 2013). The MMP2 gene plays a key role in ovulation and follicle atresia by regulating FSH and insulin like growth factor 1 (IGF1) (Knapp and Sun, 2017). In Hu sheep, the three genes GRIA2, SMAD1, and CTNNB1 are related to estrogen response element (Chang et al., 2013; Kumar et al., 2016; Vastagh et al., 2016). For example, the gene GRIA2 has been shown to participate in the glutamatergic pathway that regulates gonadotropin-releasing hormone (GnRH), a known prerequisite of the subsequent hormonal cascade inducing the ovulation in mice (Vastagh et al., 2016). The gene SMAD1 encodes an intracellular BMP signaling molecule, which is involved in mediating ovulation rate of ewes (Xu et al., 2010). The CTNNB1 gene enhances FSH and LH actions in follicles by stimulating WNT/CTNNB1 pathway and G protein-coupled gonadotropin receptors in female (Fan et al., 2010). In Icelandic sheep, the gene NCOA1 can alter the expression of multiple key genes PBP, AIB3, and FGFR2, which are important for aberrant labyrinth morphogenesis of the placenta and embryonic lethality (Chen et al., 2010; Huang et al., 2011). In Finnsheep, the five candidate genes INHBB, NF1, FLT1, PTGS2, and PLCB3 played important roles in the development of folliculogenesis and LH signaling (Ding et al., 2006; Tal et al., 2014; De Cesaro et al., 2015; Ben Sassi et al., 2016; Cadoret et al., 2017). For example, the INHBB gene encodes an inhibitor of apoptosis, which regulates porcine ovarian follicular atresia (Terenina et al., 2017). The coding region of gene NF1 presents non-CpG methylation in the murine oocyte, which plays a critical role in mammalian development (Haines et al., 2001). The FLT1 gene has an important role in the activity of vascular endothelial growth factor that linked to folliculogenesis (Celik-Ozenci et al., 2003). The PTGS2 gene plays a critical role in the ovulation by stimulating LH signaling in zebrafish (Tang et al., 2017). The PLCB3 gene is highly expressed in bovine cells of the ovulatory-sized follicles, with the role of activating LH/LHR signaling (Castilho et al., 2014). In Romanov sheep, the gene ESR2 activates ovulation and regulates preovulatory follicle maturation through regulating estrogen response element (Laliotis et al., 2017; Rumi et al., 2017). In Texel sheep, the six candidate genes ESR1, GHR, ETS1, MMP15, FLI1, and SPP1 are relevant to estrogen and follicular growth (Putnova et al., 2001; Bachelot et al., 2002; Munoz et al., 2007; Xiao et al., 2009; Hatzirodos et al., 2015; Ogiwara and Takahashi, 2017). As a key gene affecting estrogen biosynthesis, ESR1 gene functions similarly to ESR2, and is critical for follicular growth and successful ovulation in ewes (Foroughinia et al., 2017). The GHR gene plays a role in follicular growth through stimulating IGF1 in mice (Bachelot et al., 2002). The ETS1 gene was linked to the regulator of protein signaling protein-2 (RGS2) involved in the ovulation in bovine (Sayasith et al., 2014). As a proteolytic enzyme gene, the MMP15 gene has been shown to mediate LH and its receptor in the preovulatory follicles of teleost medaka (Ogiwara and Takahashi, 2017). The FLI1 gene encodes a critical transcription factor, which regulates gene ETS1 (Vo et al., 2017). The SPP1 gene accounts for establishing and maintaining cellular interactions between steroidogenic and non-steroidogenic cells during the development of corpus luteum (Poole et al., 2013). In addition, the GO categories as well as protein–protein network and expression analysis showed that these genes played an essential role in follicle growth and ovulation of ewes. However, further expression analyses of these genes in each breed are necessary in future study. Taken together, the apparent difference for the litter size among the breeds might be explained by diverse regulation mechanisms.
FIGURE 9.
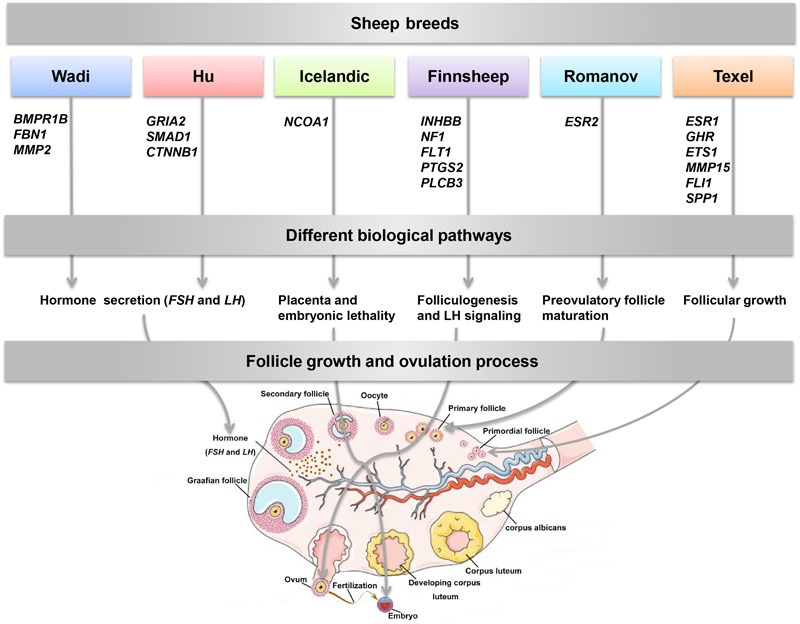
Follicle growth and ovulation process for the role of the candidate genes identified from six sheep breeds in litter size.
Also, we calculated genetic differentiation among populations using the global FST, PCA, and NJ tree methods to obtain a refined picture of population genetic relationships. The result showed that the genetic groups were consistent with the geographic origins of the breeds. The different genetic mechanisms associated with physiological processes for the litter size among sheep breeds could be related to the various environments in different geographic regions.
We noticed that previous studies had identified several genes of major effect such as BMPR1B, BMP15, and GDF9 for the prolificacy in ewes (Table 1). Different from early investigations, we detected a set of novel genes for the litter size in ewes. The main reason could be that most of early studies are based on genome-wide selection tests between prolific and non-prolific breeds using a lower density of SNPs. Instead, here we implemented GWAS within specific sheep breeds of high or low prolificacy using a high density SNP BeadChip array, which should lead to more reliable associations. In addition, the difference in threshold value used to define the ‘case’ and ‘control’ groups for each breed was also another potentially influential factor. When we implemented the GWAS using a two-step approach via the general linear model and genome-wide efficient mixed-model analysis (GEMMA), we did not find interesting candidate genes associated with reproduction across the six breeds (see Supplementary Material for further details). The fact that no candidate genes associated with reproduction were detected could be due to that the power to detect such associations will be weak when treating the trait of interest as quantitative given the small sample size. Also, these populations could have been subjected to selection on litter size through environmental variables such as climate and diet. However, we did not obtain data for local environmental variables in our data. Thus, environmental variables as well as the age of reproduction for the ewes were not taken into account in the model of the GWAS, which would be essential for future study.
Conclusion
We revealed a set of novel functional genes for the litter size in different sheep breeds across the world. Our results suggested differentially genetic regulation mechanisms for the functional genes in the reproduction of sheep. The significant SNPs and genes identified here are useful for future molecular-based breeding for a higher fertility. Also, our results provide important insights into the regulation of reproduction in sheep and other mammals.
Author Contributions
M-HL conceived and designed the project. FW, Z-QS, Y-LR, MS, EE, JH, JK, and TK collected the samples. X-LX extracted the DNA. JK provided help in Beadchip genotyping. S-SX and LG analyzed the data. S-SX wrote the paper with contributions from M-HL. All authors reviewed and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 91731309 and 31661143014), the Taishan Scholars Program of Shandong Province (No. ts201511085), the National Transgenic Breeding Project of China (2014ZX0800952B), the Academy of Finland (Grant No. 250633), and the Climate Genomics for Farm Animal Adaptation (ClimGen) Project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00118/full#supplementary-material
Parity effect for litter size in the six breeds. X-axis is labeled as the number of parity and Y-axis represents litter size. Pairwise statistical comparisons between means of litter size in parity’s clades were performed using Student’s t-test. ∗p < 0.05; ∗∗p < 0.01, and ∗∗∗p < 0.001.
Principle component plots for 522 ewes from the six sheep breeds (WAD: Wadi sheep, HUS: Hu sheep, ICE: Icelandic sheep, FIN: Finnish sheep, ROM: Romanov sheep, and TEX: Texel sheep), respectively.
Multidimensional scaling (MDS) plots in (a) Wadi, (b) Hu, (c) Icelandic, (d) Finnish, (e) Romanov, and (f) Texel sheep. The red squares indicate animals from the case group (highly prolific ewes), and the purple dots represent animals in the control group (normally prolific ewes).
Q–Q (quantile–quantile) plots of GWAS in (a) Wadi, (b) Hu, (c) Icelandic, (d) Finnish, (e) Romanov, and (f) Texel sheep. Gray and black rings represent association statistics before and after correction for population stratification, respectively.
Parity effect for litter size and pairwise statistical comparisons between means of litter size in parity’s clades in the six breeds.
Pairwise FST value among six breeds.
Bonferroni-corrected 5% chromosome-wise significance threshold in the six sheep breeds, respectively.
Bonferroni-corrected genome-wide and chromosome-wise significant SNPs and their nearest gene based on the GWAS.
GO enrichment analysis of the genes associated with the target SNPs at the chromosome-wise level as identified by the GWAS.
GO enrichments of the novel genes identified by the GWAS at the chromosome-wise level for the six sheep breeds, respectively.
Genotype effects of the most significant SNPs on litter size in six sheep breeds, respectively.
References
- Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M. (2007). GenABEL: an R library for genome-wide association analysis. Bioinformatics 23 1294–1296. 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- Bachelot A., Monget P., Imbert-Bollore P., Coshigano K., Kopchick J. J., Kelly P. A., et al. (2002). Growth hormone is required for ovarian follicular growth. Endocrinology 143 4104–4112. 10.1210/en.2002-220087 [DOI] [PubMed] [Google Scholar]
- Baird D. T., Cnattingius S., Collins J., Evers J. L. H., Glasier A., Heitmann B. L., et al. (2006). Nutrition and reproduction in women. Hum. Reprod. Update 12 193–207. 10.1093/humupd/dmk003 [DOI] [PubMed] [Google Scholar]
- Basini G., Bussolati S., Baioni L., Grasselli F. (2011). Gelatinases (MMP2 and MMP9) in swine antral follicle. Biofactors 37 117–120. 10.1002/biof.153 [DOI] [PubMed] [Google Scholar]
- Ben Sassi N., Gonzalez-Recio O., de Paz-del Rio R., Rodriguez-Ramilo S. T., Fernandez A. I. (2016). Associated effects of copy number variants on economically important traits in Spanish Holstein dairy cattle. J. Dairy Sci. 99 6371–6380. 10.3168/jds.2015-10487 [DOI] [PubMed] [Google Scholar]
- Bodin L., Di Pasquale E., Fabre S., Bontoux M., Monget P., Persani L., et al. (2007). A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep. Endocrinology 148 393–400. 10.1210/en.2006-0764 [DOI] [PubMed] [Google Scholar]
- Bonferroni C. E. (1936). Teoria statistica delle classi e calcolo delle probabilita’. Pubbl. R Ist. Sup. Sci. Econ. Commer. Fir. 8 3–62. [Google Scholar]
- Cadoret V., Frapsauce C., Jarrier P., Maillard V., Bonnet A., Locatelli Y., et al. (2017). Molecular evidence that follicle development is accelerated in vitro compared to in vivo. Reproduction 153 493–508. 10.1530/Rep-16-0627 [DOI] [PubMed] [Google Scholar]
- Cao J. X., Wei C. H., Zhang S. Z., Capellini T. D., Zhang L., Zhao F. P., et al. (2016). Screening of reproduction-related single-nucleotide variations from MeDIP-seq data in sheep. Mol. Reprod. Dev. 83 958–967. 10.1002/mrd.22734 [DOI] [PubMed] [Google Scholar]
- Casas E., Freking B. A., Leymaster K. A. (2004). Evaluation of Dorset, Finnsheep, Romanov, Texel, and Montadale breeds of sheep: II. Reproduction of F1 ewes in fall mating seasons. J. Anim. Sci. 82 1280–1289. 10.2527/2004.8251280 [DOI] [PubMed] [Google Scholar]
- Castilho A. C., Nogueira M. F., Fontes P. K., Machado M. F., Satrapa R. A., Razza E. M., et al. (2014). Ovarian superstimulation using FSH combined with equine chorionic gonadotropin (eCG) upregulates mRNA-encoding proteins involved with LH receptor intracellular signaling in granulosa cells from Nelore cows. Theriogenology 82 1199–1205. 10.1016/j.theriogenology.2014.06.011 [DOI] [PubMed] [Google Scholar]
- Celik-Ozenci C., Akkoyunlu G., Kayisli U. A., Arici A., Demir R. (2003). Localization of vascular endothelial growth factor in the zona pellucida of developing ovarian follicles in the rat: a possible role in destiny of follicles. Histochem. Cell Biol. 120 383–390. 10.1007/s00418-003-0586-4 [DOI] [PubMed] [Google Scholar]
- Chang H. M., Cheng J. C., Klausen C., Leung P. C. K. (2013). BMP15 suppresses progesterone production by down-regulating StAR via ALK3 in human granulosa cells. Mol. Endocrinol. 27 2093–2104. 10.1210/me.2013-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu Z., Xu J. (2010). The cooperative function of nuclear receptor coactivator 1 (NCOA1) and NCOA3 in placental development and embryo survival. Mol. Endocrinol. 24 1917–1934. 10.1210/me.2010-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M. X., Jia L. H., Zhang Y. J., Jin M., Chen H. Q., Fang L., et al. (2011). Polymorphisms of coding region of BMPR-IB gene and their relationship with litter size in sheep. Mol. Biol. Rep. 38 4071–4076. 10.1007/s11033-010-0526-z [DOI] [PubMed] [Google Scholar]
- Davis G. H. (2004). Fecundity genes in sheep. Anim. Reprod. Sci. 82–83 247–253. 10.1016/j.anireprosci.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Davis G. H. (2005). Major genes affecting ovulation rate in sheep. Genet. Sel. Evol. 37 S11–S23. 10.1051/gse:2004026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis G. H., Balakrishnan L., Ross I. K., Wilson T., Galloway S. M., Lumsden B. M., et al. (2006a). Investigation of the Booroola (FecB) and Inverdale (FecXI) mutations in 21 prolific breeds strains of sheep sampled in 13 countries. Anim. Reprod. Sci. 92 87–96. 10.1016/j.anireprosci.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Davis G. H., Farquhar P. A., O’Connell A. R., Everett-Hincks J. M., Wishart P. J., Galloway S. M., et al. (2006b). A putative autosomal gene increasing ovulation rate in Romney sheep. Anim. Reprod. Sci. 92 65–73. [DOI] [PubMed] [Google Scholar]
- De Cesaro M. P., Macedo M. P., Santos J. T., Rosa P. R., Ludke C. A., Rissi V. B., et al. (2015). Natriuretic peptides stimulate oocyte meiotic resumption in bovine. Anim. Reprod. Sci. 159 52–59. 10.1016/j.anireprosci.2015.05.012 [DOI] [PubMed] [Google Scholar]
- de Souza C. J. H., Mcneilly A. S., Benavides M. V., Melo E. D. O., Moraes J. C. F. (2012). “Novel GDF9 polymorphism determining higher ovulation rate and litter size in sheep,” in Proceedings of the Society for Reproduction and Fertility Annual Conference Edinburgh. [Google Scholar]
- Demars J., Fabre S., Sarry J., Rossetti R., Gilbert H., Persani L., et al. (2013). Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep. PLoS Genet. 9:e1003482. 10.1371/journal.pgen.1003482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniskova T., Dotsev A. V., Selionova M., Wimmers K., Reyer H., Kharzinova V. R., et al. (2017). Whole-genome single nucleotide polymorphism study of Romanov sheep. J. Anim. Sci. 95 339–340. 10.2527/asasann.2017.696 [DOI] [Google Scholar]
- Ding N. S., Ren D. R., Guo Y. M., Ren J., Yan Y., Ma J. W., et al. (2006). Genetic variation of porcine prostaglandin-endoperoxide synthase 2 (PTGS2) gene and its association with reproductive traits in an Erhualian × Duroc F2 population. Yi Chuan Xue Bao 33 213–219. 10.1016/S0379-4172(06)60042-5 [DOI] [PubMed] [Google Scholar]
- Drouilhet L., Mansanet C., Sarry J., Tabet K., Bardou P., Woloszyn F., et al. (2013). The highly prolific phenotype of Lacaune sheep is associated with an ectopic expression of the B4GALNT2 gene within the ovary. PLoS Genet. 9:e1003809. 10.1371/journal.pgen.1003809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiriksson J. H., Sigurdsson A. (2017). Sources of bias, genetic trend and changes in genetic correlation in carcass and ultrasound traits in the Icelandic sheep population. Iceland. Agric. Sci. 30 3–12. 10.16886/Ias.2017.01 [DOI] [Google Scholar]
- Fan H. Y., O’Connor A., Shitanaka M., Shimada M., Liu Z., Richards J. S. (2010). Beta-catenin (CTNNB1) promotes preovulatory follicular development but represses LH-mediated ovulation and luteinization. Mol. Endocrinol. 24 1529–1542. 10.1210/me.2010-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feary E. S., Juengel J. L., Smith P., French M. C., O’Connell A. R., Lawrence S. B., et al. (2007). Patterns of expression of messenger RNAs encoding GDF9, BMP15, TGFBR1, BMPR1B, and BMPR2 during follicular development and characterization of ovarian follicular populations in ewes carrying the Woodlands FecX2W mutation. Biol. Reprod. 77 990–998. 10.1095/biolreprod.107.062752 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP-phylogeny inference package (version 3.2). Cladistics 5 164–166. 10.1111/j.1096-0031.1989.tb00562.x [DOI] [Google Scholar]
- Foroughinia G., Fazileh A., Eghbalsaied S. (2017). Expression of genes involved in BMP and estrogen signaling and AMPK production can be important factors affecting total number of antral follicles in ewes. Theriogenology 91 36–43. 10.1016/j.theriogenology.2016.12.023 [DOI] [PubMed] [Google Scholar]
- Galloway S. M., McNatty K. P., Cambridge L. M., Laitinen M. P., Juengel J. L., Jokiranta T. S., et al. (2000). Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat. Genet. 25 279–283. 10.1038/77033 [DOI] [PubMed] [Google Scholar]
- Haines T. R., Rodenhiser D. I., Ainsworth P. J. (2001). Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev. Biol. 240 585–598. 10.1006/dbio.2001.0504 [DOI] [PubMed] [Google Scholar]
- Hanrahan J. P., Gregan S. M., Mulsant P., Mullen M., Davis G. H., Powell R., et al. (2004). Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol. Reprod. 70 900–909. 10.1095/biolreprod.103.023093 [DOI] [PubMed] [Google Scholar]
- Hatzirodos N., Hummitzsch K., Irving-Rodgers H. F., Rodgers R. J. (2015). Transcriptome comparisons identify new cell markers for theca interna and granulosa cells from small and large antral ovarian follicles. PLoS One 10:e0119800. 10.1371/journal.pone.0119800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009b). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. Y., Li S. F., Zhao Z. H., Liang Z., Zhang J., Ding Y. R. (2011). Association of polymorphisms for nuclear receptor coactivator 1 gene with egg production traits in the maternal line of Shaobo hens. Br. Poult. Sci. 52 328–332. 10.1080/00071668.2011.577057 [DOI] [PubMed] [Google Scholar]
- Hu X. J., Pokharel K., Peippo J., Ghanem N., Zhaboyev I., Kantanen J., et al. (2016). Identification and characterization of miRNAs in the ovaries of a highly prolific sheep breed. Anim. Genet. 47 234–239. 10.1111/age.12385 [DOI] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S. Y., Freimer N. B., et al. (2010). Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42 348–354. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp E., Sun J. J. (2017). Steroid signaling in mature follicles is important for Drosophila ovulation. Proc. Natl. Acad. Sci. U.S.A. 114 699–704. 10.1073/pnas.1614383114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köchl S., Niederstätter H., Parson W. (2005). DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Forensic DNA Typing Protoc. 297 13–30. 10.1385/1-59259-867-6:013 [DOI] [PubMed] [Google Scholar]
- Kumar M., Camlin N. J., Holt J. E., Teixeira J. M., McLaughlin E. A., Tanwar P. S. (2016). Germ cell specific overactivation of WNT/beta catenin signalling has no effect on folliculogenesis but causes fertility defects due to abnormal foetal development. Sci. Rep. 6:27273. 10.1038/srep27273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliotis G. P., Marantidis A., Avdi M. (2017). Association of BF, RBP4, and ESR2 genotypes with litter size in an autochthonous pig population. Anim. Biotechnol. 28 138–143. 10.1080/10495398.2016.1242490 [DOI] [PubMed] [Google Scholar]
- Lassoued N., Benkhlil Z., Woloszyn F., Rejeb A., Aouina M., Rekik M., et al. (2017). FecX(Bar) a novel BMP15 mutation responsible for prolificacy and female sterility in Tunisian Barbarine sheep. BMC Genet. 18:43. 10.1186/s12863-017-0510-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Royo A., Jurado J. J., Smulders J. P., Marti J. I., Alabart J. L., Roche A., et al. (2008). A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep. Anim. Genet. 39 294–297. 10.1111/j.1365-2052.2008.01707.x [DOI] [PubMed] [Google Scholar]
- Miao X., Luo Q., Zhao H., Qin X. (2016). Ovarian transcriptomic study reveals the differential regulation of miRNAs and lncRNAs related to fecundity in different sheep. Sci. Rep. 6:35299. 10.1038/srep35299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteagudo L. V., Ponz R., Tejedor M. T., Lavina A., Sierra I. (2009). A 17 bp deletion in the Bone Morphogenetic Protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed. Anim. Reprod. Sci. 110 139–146. 10.1016/j.anireprosci.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Moradband F., Rahimi G., Gholizadeh M. (2011). Association of polymorphisms in fecundity genes of GDF9, BMP15 and BMP15-1B with litter size in Iranian Baluchi sheep. Asian Australas. J. Anim. Sci. 24 1179–1183. 10.5713/ajas.2011.10453 [DOI] [Google Scholar]
- Mullen M. P., Hanrahan J. P. (2014). Direct evidence on the contribution of a missense mutation in GDF9 to variation in ovulation rate of Finnish. PLoS One 9:e95251. 10.1371/journal.pone.0095251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulsant P., Lecerf F., Fabre S., Schibler L., Monget P., Lanneluc I., et al. (2001). Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc. Natl. Acad. Sci. U.S.A. 98 5104–5109. 10.1073/pnas.091577598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz G., Ovilo C., Estelle J., Silio L., Fernandez A., Rodriguez C. (2007). Association with litter size of new polymorphisms on ESR1 and ESR2 genes in a Chinese-European pig line. Genet. Sel. Evol. 39 195–206. 10.1186/1297-9686-39-2-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol L., Bishop S. C., Pong-Wong R., Bendixen C., Holm L. E., Rhind S. M., et al. (2009). Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep. Reproduction 138 921–933. 10.1530/REP-09-0193 [DOI] [PubMed] [Google Scholar]
- Ogiwara K., Takahashi T. (2017). Involvement of the nuclear progestin receptor in LH-induced expression of membrane type 2-matrix metalloproteinase required for follicle rupture during ovulation in the medaka, Oryzias latipes. Mol. Cell. Endocrinol. 450 54–63. 10.1016/j.mce.2017.04.016 [DOI] [PubMed] [Google Scholar]
- Patterson N., Price A. L., Reich D. (2006). Population structure and eigenanalysis. PLoS Genet. 2:e190. 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W. F., Xu S. S., Ren X., Lv F. H., Xie X. L., Zhao Y. X., et al. (2017). A genome-wide association study reveals candidate genes for the supernumerary nipple phenotype in sheep (Ovis aries). Anim. Genet. 48 570–579. 10.1111/age.12575 [DOI] [PubMed] [Google Scholar]
- Petryszak R., Keays M., Tang Y. A., Fonseca N. A., Barrera E., Burdett T., et al. (2016). Expression Atlas update-an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44 D746–D752. 10.1093/nar/gkv1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole D. H., Ndiaye K., Pate J. L. (2013). Expression and regulation of secreted phosphoprotein 1 in the bovine corpus luteum and effects on T lymphocyte chemotaxis. Reproduction 146 527–537. 10.1530/Rep-13-0190 [DOI] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnova L., Kolarikova O., Knoll A., Dvorák J. (2001). Association study of osteopontin (SPP1) and estrogen receptor (ESR) genes with reproduction traits in pigs. Acta Univ. Agric. Silvic. Mendel. Brun. 49 69–74. [Google Scholar]
- Raymond M., Rousset F. (1995). Genepop (version-1.2)-population-genetics software for exact tests and ecumenicism. J. Hered. 86 248–249. 10.1093/oxfordjournals.jhered.a111573 [DOI] [Google Scholar]
- Regan S. L., McFarlane J. R., O’Shea T., Andronicos N., Arfuso F., Dharmarajan A., et al. (2015). Flow cytometric analysis of FSHR, BMRR1B, LHR and apoptosis in granulosa cells and ovulation rate in merino sheep. Reproduction 150 151–163. 10.1530/Rep-14-0581 [DOI] [PubMed] [Google Scholar]
- Rumi M. A. K., Singh P., Roby K. F., Zhao X., Iqbal K., Ratri A., et al. (2017). Defining the role of estrogen receptor beta in the regulation of female fertility. Endocrinology 158 2330–2343. 10.1210/en.2016-1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayasith K., Sirois J., Lussier J. G. (2014). Expression and regulation of regulator of G-protein signaling protein-2 (RGS2) in equine and bovine follicles prior to ovulation: molecular characterization of RGS2 transactivation in bovine granulosa cells. Biol. Reprod. 91 75–86. 10.1095/biolreprod.114.121186 [DOI] [PubMed] [Google Scholar]
- Silva B. D. M., Castro E. A., Souza C. J. H., Paiva S. R., Sartori R., Franco M. M., et al. (2011). A new polymorphism in the growth and differentiation factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep. Anim. Genet. 42 89–92. 10.1111/j.1365-2052.2010.02078.x [DOI] [PubMed] [Google Scholar]
- Souza C. J. H., MacDougall C., Campbell B. K., McNeilly A. S., Baird D. T. (2001). The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. J. Endocrinol. 169 R1–R6. 10.1677/joe.0.169R001 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J. H., Cook H., Kuhn M., Wyder S., Simonovic M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45 D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal R., Seifer D. B., Grazi R. V., Malter H. E. (2014). Follicular fluid placental growth factor is increased in polycystic ovarian syndrome: correlation with ovarian stimulation. Reprod. Biol. Endocrinol. 12 1–7. 10.1186/1477-7827-12-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Liu Y., Li J., Li G., Chen Y., Yin Y., et al. (2017). LH signaling induced ptgs2a expression is required for ovulation in zebrafish. Mol. Cell. Endocrinol. 447 125–133. 10.1016/j.mce.2017.02.042 [DOI] [PubMed] [Google Scholar]
- Terenina E., Fabre S., Bonnet A., Monniaux D., Robert-Granie C., SanCristobal M., et al. (2017). Differentially expressed genes and gene networks involved in pig ovarian follicular atresia. Physiol. Genomics 49 67–80. 10.1152/physiolgenomics.00069.2016 [DOI] [PubMed] [Google Scholar]
- Vage D. I., Husdal M., Kent M. P., Klemetsdal G., Boman I. A. (2013). A missense mutation in growth differentiation factor 9 (GDF9) is strongly associated with litter size in sheep. BMC Genet. 14:1. 10.1186/1471-2156-14-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastagh C., Rodolosse A., Solymosi N., Liposits Z. (2016). Altered expression of genes encoding neurotransmitter receptors in GnRH neurons of proestrous mice. Front. Cell. Neurosci. 10:230. 10.3389/Fncel.2016.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo K. K., Jarocha D. J., Lyde R. B., Hayes V., Thom C. S., Sullivan S. K., et al. (2017). FLI1 level during megakaryopoiesis affects thrombopoiesis and platelet biology. Blood 129 3486–3494. 10.1182/blood-2017-02-770958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J., Pemberton J. M., Pilkington J. G., Clutton-Brock T. H., Kruuk L. E. (2009). Trading offspring size for number in a variable environment: selection on reproductive investment in female Soay sheep. J. Anim. Ecol. 78 354–364. 10.1111/j.1365-2656.2008.01489.x [DOI] [PubMed] [Google Scholar]
- Wilson T., Wu X. Y., Juengel J. L., Ross I. K., Lumsden J. M., Lord E. A., et al. (2001). Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol. Reprod. 64 1225–1235. 10.1095/biolreprod64.4.1225 [DOI] [PubMed] [Google Scholar]
- Xiao X. H., Liu A. Q., Wen H. X., Tian Y. J., Ni J., Liu G. Y. (2009). Expression and localization of transcription factor Ets-1 in the rat ovary during the estrous cycle and pregnancy. Fertil. Steril. 91 1998–2005. 10.1016/j.fertnstert.2008.02.166 [DOI] [PubMed] [Google Scholar]
- Xu S.-S., Li M.-H. (2017). Recent advances in understanding genetic variants associated with economically important traits in sheep (Ovis aries) revealed by high-throughput screening technologies. Front. Agr. Sci. Eng. 4 279–288. 10.15302/J-FASE-2017151 [DOI] [Google Scholar]
- Xu Y. F., Li E. L., Han Y. D., Chen L., Xie Z. A. (2010). Differential expression of mRNAs encoding BMP/Smad pathway molecules in antral follicles of high- and low-fecundity Hu sheep. Anim. Reprod. Sci. 120 47–55. 10.1016/j.anireprosci.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Yue G. H. (1996). Reproductive characteristics of Chinese Hu sheep. Anim. Reprod. Sci. 44 223–230. 10.1016/0378-4320(96)01562-X 24841789 [DOI] [Google Scholar]
- Zhai B., Liu H. Y., Li X., Dai L. S., Gao Y., Li C. H., et al. (2013). BMP15 prevents cumulus cell apoptosis through CCL2 and FBN1 in porcine ovaries. Cell. Physiol. Biochem. 32 264–278. 10.1159/000354435 [DOI] [PubMed] [Google Scholar]
- Zhang C. S., Geng L. Y., Du L. X., Liu Z. Z., Fu Z. X., Feng M. S., et al. (2011). Polymorphic study of FecX(G), FecG(H) and Fec(B) mutations in four domestic sheep breeds in the Lower Yellow River Valley of China. J. Anim. Vet. Adv. 10 2198–2201. 10.3923/javaa.2011.2198.2201 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Parity effect for litter size in the six breeds. X-axis is labeled as the number of parity and Y-axis represents litter size. Pairwise statistical comparisons between means of litter size in parity’s clades were performed using Student’s t-test. ∗p < 0.05; ∗∗p < 0.01, and ∗∗∗p < 0.001.
Principle component plots for 522 ewes from the six sheep breeds (WAD: Wadi sheep, HUS: Hu sheep, ICE: Icelandic sheep, FIN: Finnish sheep, ROM: Romanov sheep, and TEX: Texel sheep), respectively.
Multidimensional scaling (MDS) plots in (a) Wadi, (b) Hu, (c) Icelandic, (d) Finnish, (e) Romanov, and (f) Texel sheep. The red squares indicate animals from the case group (highly prolific ewes), and the purple dots represent animals in the control group (normally prolific ewes).
Q–Q (quantile–quantile) plots of GWAS in (a) Wadi, (b) Hu, (c) Icelandic, (d) Finnish, (e) Romanov, and (f) Texel sheep. Gray and black rings represent association statistics before and after correction for population stratification, respectively.
Parity effect for litter size and pairwise statistical comparisons between means of litter size in parity’s clades in the six breeds.
Pairwise FST value among six breeds.
Bonferroni-corrected 5% chromosome-wise significance threshold in the six sheep breeds, respectively.
Bonferroni-corrected genome-wide and chromosome-wise significant SNPs and their nearest gene based on the GWAS.
GO enrichment analysis of the genes associated with the target SNPs at the chromosome-wise level as identified by the GWAS.
GO enrichments of the novel genes identified by the GWAS at the chromosome-wise level for the six sheep breeds, respectively.
Genotype effects of the most significant SNPs on litter size in six sheep breeds, respectively.


