Abstract
Anti-diabetes activity of Catharsius molossus (Ca, a type of dung beetle) glycosaminoglycan (G) was evaluated to reduce glucose, creatinine kinase, triglyceride and free fatty acid levels in db mice. Diabetic mice in six groups were administrated intraperitoneally: Db heterozygous (Normal), Db homozygous (CON), Heuchys sanguinea glycosaminoglycan (HEG, 5 mg/kg), dung beetle glycosaminoglycan (CaG, 5 mg/kg), bumblebee (Bombus ignitus) queen glycosaminoglycan (IQG, 5 mg/kg) and metformin (10 mg/kg), for 1 month. Biochemical analyses in the serum were evaluated to determine their anti-diabetic and anti-inflammatory actions in db mice after 1 month treatment with HEG, CaG or IQG treatments. Blood glucose level was decreased by treatment with CaG. CaG produced significant anti-diabetic actions by inhiting creatinine kinase and alkaline phosphatase levels. As diabetic parameters, serum glucose level, total cholesterol and triglyceride were significantly decreased in CaG5-treated group compared to the controls. Dung beetle glycosaminoglycan, compared to the control, could be a potential therapeutic agent with anti-diabetic activity in diabetic mice. CaG5-treated group, compared to the control, showed the up-regulation of 48 genes including mitochondrial yen coded tRNA lysine (mt-TK), cytochrome P450, family 8/2, subfamily b, polypeptide 1 (Cyp8b1), and down-regulation of 79 genes including S100 calcium binding protein A9 (S100a9) and immunoglobulin kappa chain complex (Igk), and 3-hydroxy-3-methylglutaryl-CoenzymeAsynthase1 (Hmgcs1). Moreover, mitochondrial thymidine kinase (mt-TK), was up-regulated, and calgranulin A (S100a9) were down-regulated by CaG5 treatment, indicating a potential therapeutic use for anti-diabetic agent.
Keywords: Anti-diabetic effect, Queen of B. ignitus, Catharsius molossus, Glycosaminoglycan, Microarray
INTRODUCTION
Diabetes mellitus is a metabolic disease associated with systemic damages to microvessel of kidney and other organs (1). With decreasing morbidity, many drugs from natural product and synthetic chemicals have been developed as anti-diabetic agents. Early stages of diabetic nephropathy are characterized by low urinary excretion of albumin, with glomerular basement membrane functioning by charge-dependent permeability due to the presence of anionic constituents, especially heparin sulfate [HS, a type of glycosaminoglycan (GAG)] proteoglycans (2). Among inherited metabolic disorders, particularly mucopolysaccharidoses, GAG degradation pathways are disrupted due to enzyme deficiency (3). Meanwhile, cellular oxidative damage could be repaired by expression of antioxidative enzymes (including superoxide dismutase and glutathione peroxidase) so that peroxided glycosylated lipids, proteins and nucleic acids could not be produced anymore (1). Sulodexide, a highly purified mixture of glycosaminoglycans composed of low molecular weight heparin and dermatan sulfate, has been recently developed for treatment of diabetic nephropathy. Clinical studies have suggested that Sulodexide therapy offers renal protection, capable of reducing urinary albumin excretion (4). According to a recent report, similar to Sulodexide (commercial glycosaminoglycan), some insect GAGs can also increase anti-oxidant activity and diminish cellular oxidative damage. After treatment with bumblebee queen glycosamino-glycan (IQG), activities of anti-oxidative enzyme, SOD and catalase are increased whereas oxidative stress parameter, carbonyl content (protein oxidative damage) and malondialdehyde (MDA, lipid denaturant) in high-fat diet rat hepatocyte were diminished by (5). Therefore, we designed this study to determine whether insect glycosaminoglycans might have a role in treating diabetes or increasing antioxidant enzyme activity under diabetes condition using, Homo diabetic (db) mice. An insect glycosaminoglycan was been prepared and purified from black and scarlet cicada (Huechys sanguinea) (6). Another insect glycosaminoglycan isolated from dung beetle (Catharsius molossus, Ca) also possesses anti-aging activities. It can reduce serum level of creatinine kinase with aortic vasorelaxant activities. It can also maintain normal glucose level in aged rat (7). As an apicultural product, bumblebee (Bombus ignitus) queen (BIQ) glycosaminoglycan is a potential agent for treating obesity in high-fat diet rats (5). Anti-diabetic gene targets of glycosaminoglycan in db mice have not been fully elucidated. In this study, we found that glycosaminoglycans such as HEG, CaG and IQG displayed anti-oxidant and anti-diabetic properties. They also changed gene expression profiling in db mice. The glycosaminoglycans might hold great promise as antidiabetic agents by reducing cellular oxidative damages.
MATERIALS AND METHODS
Materials
Preparation of insect glycosaminoglycans
Dried C. molossus and H. sanguinea, were purchased at a local market in China. These crude drugs were obtained from insects identified by insect classification experts in the Department of Agricultural Biology, National Academy of Agricultural Science, South Korea. Bumble bee (B. ignitus) queen was reared and freeze-dried, in the same institute. Metformin was purchased from CJ Healthcare Co (Seoul, Korea).
Dried insect (1 kg each) was soaked and extracted three times with ethanol by ultrasonification for 30 min. Residues were separated from alcohol extracts and were defatted twice with two volumes of acetone. Approximately 200 g of dried, defatted and pulverized powder was suspended in 2 L of 0.05 M sodium carbonate buffer (pH 9). The suspension was incubated at 60°C for 48 hr after adding 28 mL (1.4%) of Alcalase (Sigma Aldrich, St. Louis, MO, USA) for protein enzymatic hydrolysis. The digestion mixture was cooled to 4°C. Trichloroacetic acid was added to a final concentration of 5%. The sample was allowed to stand for 1 hr followed by centrifugation at 8000 ×g for 30 min. Three volumes of 5% potassium acetate in ethanol were added to one volume of supernatant, stored overnight at 4°C, and then centrifuged at 8,000 g for 30 min. The precipitate (20 g) was dissolved in 40 mL of 0.2 M NaCl and centrifuged at 8,000 g for 30 min. Cetylpyridinium chloride (5%) was added to 0.2 volumes of the supernatant followed by centrifugation (8,000 g for 30 min). The precipitate was dissolved in 20 mL of 2.5 M NaCl. Five volumes of ethanol were added followed by centrifugation at 8,000 g for 30 min. The precipitate was then dissolved in water and dialyzed against 100 volumes of water (8). The dialyzed crude GAG was freeze-dried to obtain a concentration of up to 0.9%. Crude GAG was loaded onto a DEAE Sephadex A-25 gel chromatography column (40 × 1.2 cm) equilibrated with 50 mM phosphate buffer (pH 7.4). Fractions were eluted using a linear sodium chloride gradient from 0 to 2.5 M NaCl in phosphate buffer at a flow rate of 20 mL/hr. Dialyzed glycan was freeze-dried to obtain pure GAG.
Animals
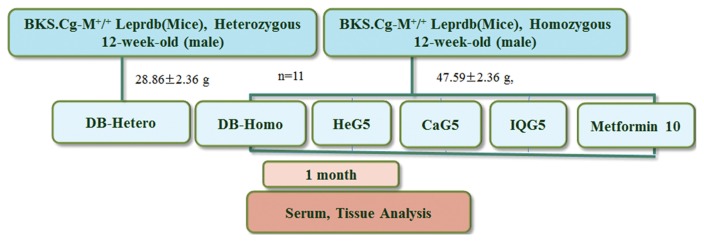
BKS.Cg-m+/+Leprdb (db/db), heterozygous and homozygous male db mice at 12-weeks of age, were purchased from Samtako Co. Ltd (Osan, Korea). All procedures were in accordance with NIH Guidelines for Care and Use of Laboratory Animals. All experiments were approved by Laboratory Animals’ Ethical Committee of the National Academy of Agricultural Science (NIAS201605), Rural Development Administration, South Korea. All procedures followed national guidelines for the care and use of animals (individual housing). Mice were acclimated for 6 months under normal husbandry conditions (temperature, 23 ± 2°C; humidity, 55 ± 10%; and light/dark cycle, 12 hr/12 hr). They were provided free access to normal diet (D10001, AIN-76A rodent diet, Research Diet Inc., New Brunswick, NJ, USA) and water free access. These mice were allocated into two control groups and three treatment groups (11 mice per group). They were distributed according to similarity in weight (28.86 ± 2.36 g). Treatments were given in PBS daily. Each treatment was administrated intraperitoneally. The following treatment groups were used: 1) Normal (DB-Hetero), 2) Control (DB-Homo), 3) 5 mg/kg HeG (HeG5), 4) 5 mg/kg CaG (CaG5), 5) 5 mg/kg IQG (IQG5), and 6) 10 mg/kg Metformin (Metformin10). Mice in each group were maintained with normal diet (AIN-76A rodent diet, Research Diet). Animal experimental design is shown in Fig. 1.
Fig. 1.
Animal experimental design. HeG5; H. sanguinea (Hongrangja, Korean name) glycosaminoglycan 5mg/kg; C. molossus (dung beetle) glycosaminoglycan 5mg/kg; IQG5: B. ignitus (a type of bumblebee) queen glycosaminoglycan 5mg/kg, and Metformin10: Metformin 10mg/kg.
Body weight and blood glucose detection
Body weights were measured every week. Glucose levels were recorded weekly with glucose stick and using a blood glucose Nocodingone detector (Theragen Etex Co. Ltd., Sungnam, Korea). Serum glucose level at the last day of treated schedule, after one-month of treatment was recorded using an auto-analyzer. As a positive control, metformin, a hypoglycemic reagent for type 2 diabetes, was used in this study.
Organ and adipose tissue weights
Absolute and relative weights (organ-to-body ratio) were measured for adrenal glands, kidneys, heart, liver, lung, spleen, stomach and pancreas. Abdominal fat to-body weight ratio was also determined. These measurements were made after sacrifice at the end of the one-month treatment period.
Blood sampling and serum assay
After treatment with HeG5, CaG5, IQG5, or metformin10 treatment, for 1-month, all non-diabetic heterozygous control mice, db control mice, and db treated mice were sacrificed for serum assay and DNA microarray. Approximately 1 mL of blood was collected from the posterior vena cava under light CO2 inhalation and used for serum chemistry measurements The following parameters were examined: albumin, hyaluronic acid (HA), free fatty acid (FFA), alkaline phosphatase (ALP), glutamic oxaloacetic transaminase (AST), glutamic pyruvic transaminase (ALT), lactic dehydrogenase (LDH), creatinine phosphokinase (CK), glucose, total cholesterol triglyceride, high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, creatinine, blood urea nitrogen (BUN), total protein, sodium (Na), chloride (Cl), c-reactive protein (CRP), calcium (ca), potassium (K), total IgE, C-peptide and insulin. These parameters were evaluated using an autoanalyzer (7060 Automatic Clinical Analyzer, Hitachi, Tokyo, Japan).
Adipocyte density
Excised organs and adipose tissue were fixed with 10% neutral formalin. After paraffin embedding and sectioning, sections were stained with hematoxylin and eosin (H&E), and Toluidine blue O, examined by light microscopy (CRT6000, Leica, Hesse, Germany), and photographed. Adipocyte densities (cells/mm2) were determined for treated and control tissue by toluidine blue O stain (original magnification, ×400).
Oxidative protein damage
Liver homogenate and blood were centrifuged (8,000 g for 30 min). Supernatants were used for determination of carbonyl content. Protein oxidative stress was evaluated by measuring protein carbonyl content in the blood. Carbonyl content was determined with an enzyme-linked immunosorbent assay (ELISA) using OxiSelectTM protein carbonyl ELISA kit (Cell Biolabs, Inc., San Diego, CA, USA) according to the manufacturer’s protocol. CAT activity (U/mg protein) was measured based on CAT-mediated decomposition of H2O2 (9).
Liver homogenate preparation for oxidative enzyme detection
Six groups (DB-Hetero, DB-Homo, HeG5, CaG5, IQG5, Metformin10) of liver tissues were homogenized on ice in a 10-fold volume lysis buffer PRO-PREPTM protein extraction solution (iNtRON, Busan, Korea). The supernatant of each liver homogenate after centrifugation (800 g, 10 min) was assayed for catalase, glutathione peroxidase, glutathione s-transferase and superoxide dismutase activities using OxiSelectTM ELISA kit (Cell Biolabs, Inc.) according to the assay manual.
RNA preparation and quantitative real-time PCR analysis
Total RNA was isolated from liver tissue using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA concentration and purity were measured using a UV/Vis spectrophotometer (Beckman Coulter Co., Miami, FL, USA). Complementary DNA (cDNA) was synthesized from 1 mg of total RNA using high capacity cDNA Reverse Transcription Kit (Amersham Biosciences Co., Piscataway, NJ, USA). Real-time polymerase chain reaction (PCR) amplification was performed using Power SYBR Green Master Mix on a 7500 Real-Time PCR System (Applied Biosystems, Fostercity, CA, USA), according to the manufacturer’s instructions. For detection of target gene transcripts, we designed specific forward and reverse oligonucleotide primers using Beacon Designer software (PREMIER Biosoft, Palo Alto, CA, USA). The primer sequences are listed in Table 1. Target mRNA levels were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control to qualify the relative expression of target mRNA according to cycling threshold method. All samples were analyzed in triplicate. Primer sequences for amplification of genes involved in cell repair mechanism and GAPDH internal standard (Table 1).
Table 1.
Primer sequences for amplication of genes involved in glycogen metabolism and GAPDH internal standard
| Gene symbol | Gene name | Primer sequence |
|---|---|---|
| Gpx1 | Glutathione peroxidase 1 | 5′-CCAACACCCAGTGACGACC-3′ 5′-CTCAAAGTTCCAGGCAATGTC-3′ |
| Heparanase | Heparanase | 5′-ACTTGAAGGTACCGCCTCCG-3′ 5′-GAAGCTCTGGAACTCGGCAA-3′ |
| GSK3b | Glycogen synthase kinase 3 beta | 5′-tccgaggagagcccaatgtt-3′ 5′-acaccactgtccccaggaaa-3′ |
| SAA4 | Serum amyloid A4 | 5′-gaaggcacggatgaagacgg-3′ 5′-acaggctccatccatcctcc-3′ |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 5′-GTGGAGATTGTTGCCATCAACGA-3′ 5′-CCCATTCTCGGCCTTGACTGT-3′ |
DNA microarray procedure
Microarray hybridization was performed for mouse liver samples. Total RNA was isolated from mice liver tissue using a Qiagen RNeasy Midi Kit (Qiagen, Valencia, CA, USA). A FairPlayTM microarray labeling kit (Stratagene, La Jolla, CA, USA) was used for DNA labeling according to the manufacturer’s instructions. Labeled DNA was loaded onto a microarray chip. A hybridization chamber was assembled with the microarray chip of Mouse Gene 2.0 ST Arrays (Affymetrix Inc., Santa Clara, CA, USA), and submerged in a water bath at 60°C overnight. The microarray chip was washed with wash buffers I, II, and III. The slide was then dried by centrifuging and scanned with a BMS Array Scanner (Applied Precision Array WoRx eBiochip Reader (BioRad, Dallas, TX, USA) (10).
Statistical analysis
Means and standard errors of parameters were determined for each group using analysis of variance (ANOVA). Student’s t-test was used to determine significant differences between control and treated groups. A p value of less than 0.05 was considered statistically significant.
RESULTS
Yield of insect glycosaminoglycan
From 1 kg of each dried insect, the yield of freeze-dried GAG powder was about 8.8 g (0.88%) for HEG, 1.52 g (0.15%) for CaG, and 2.8 g (0.28%) for IQG. Insect shells left after preparation of alcoholic extract from these three species of insects were dried. About 1 g was obtained after drying. Proteins were removed with proteases. After removing impurities by precipitation, supernatant was dialyzed with distilled water. For treating db mice for diabetic study, protein was prepared by collecting sugar acid fraction followed by purification with salt gradient (0 to 2.5 M NaCl in phosphate buffer) via strong anion exchange (SAX) and gel filtration chromatography using DEAE Sephadex A-25. Fraction containing uronic acids were collected. To determine GAG purity, digested GAG, oligosaccharides mixture by GAG enzymes (heparinase I, II, III, etc) was fractionated with SAX high performance liquid chromatography, affording oligosaccharides that were sufficiently pure for structural characterization by LC-MS.
Body weight change
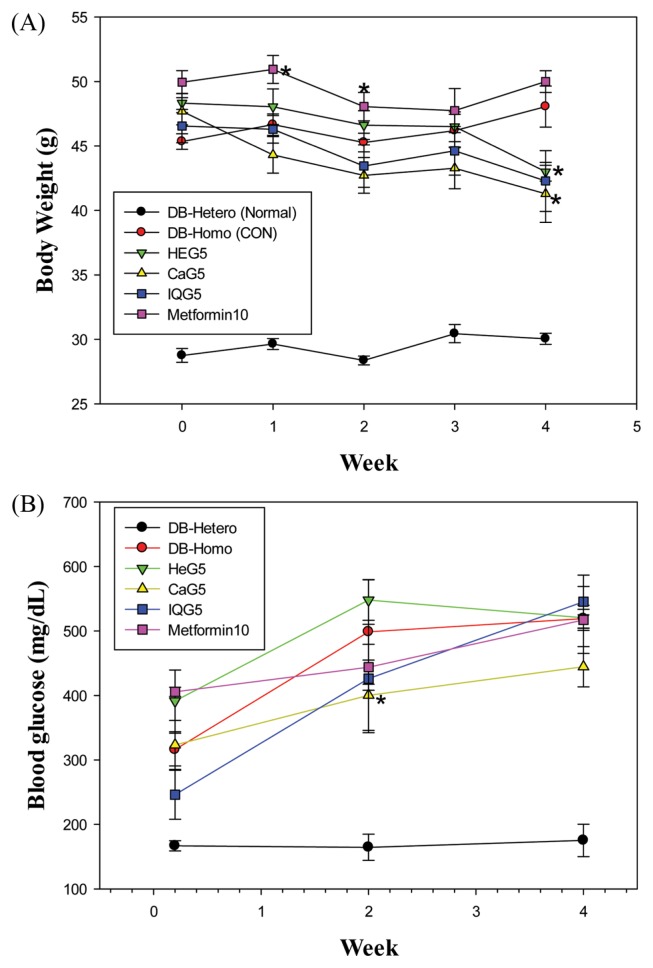
There were no significantly differences in total mean body weight between control and treatment groups at the beginning of treatment (Fig. 2A): DB-Hetero, 28.8 ± 0.5 g; DB-Homo, 45.4 ± 0.6 g; HEG5, 48.3 ± 0.8 g; CaG5, 47.7 ± 1.1 g; IQG5, 46.6 ± 1.3 g; and Metformin 10, 50.0 ± 0.9 g. However, body weights of mice at the end of 4-week treatment were lower than those of control mice in Db-Homo groups: DB-Hetero, 30.1 ± 0.4 g; DB-Homo, 48.1 ± 1.6 g (100%); HEG5, 43.0 ± 0.7 g (89.4%, HeG5 vs. DB-Homo, p < 0.05); CaG5, 41.3 ± 2.2 g (86.0%, CaG5 vs. DB-Homo, p < 0.05); IQG5, 42.3 ± 2.4 g, 87.3%); and Metformin 10, 50.0 ± 0.8.
Fig. 2.
Body weight and blood glucose level of db mice treated with dung beetle glycosaminoglycan for a one month. [sample group vs. CON (DB-Homo), *p<0.05].
Effect of CaG5 on blood glucose level
First, we examined blood glucose levels after a single administration of CaG in db/db mice and observed a reduction in blood glucose levels as shown previously (11). Glucose levels of homozygous db mice were increased with aging, from 316 mg/dL at age of 12-week to 519 mg/dL at age of 17-week. Mean glucose levels in treatment groups were as follows: DB-Hetero (normal), 168.85 ± 5.66 mg/dL; DB-homo, 508.9 ± 14.5 mg/dL; HEG5, 486.7 ± 83.5 mg/dL; CaG5, 389.4 mg/dL; IQG5, 414.6 ± 137.1 mg/dL; and Metformin 10, 455.6 ± 56.7 mg/dL (Fig. 2B). Although metformin did not present better efficacy in lowering blood glucose level than other GAGs because metformin was used at a low dose, blood glucose levels in GaG5 treated groups were significantly lower than those in the control group.
Abdominal fat weight and adipocyte density
Abdominal fat weight (g) of each treated GAG group was significantly differences from DB-Hetero (normal db mice) group but was no significantly difference from DB-Homo group: DB-Hetero, 0.55 ± 0.26; DB-Homo, 3.34 ± 0.50; HEG5, 3.35 ± 0.22; CaG5, 2.99 ± 0.55; IQG5, 2.80 ± 1.39; Metformin 10, 3.95 ± 0.53.
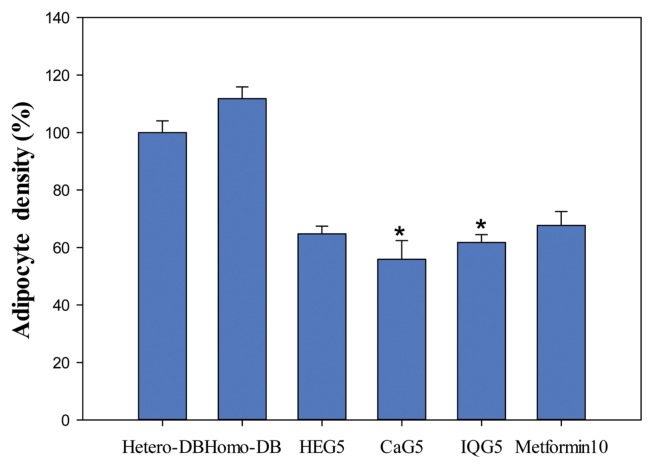
Adipocyte density tissues from treated db mice were significantly reduced by DB-Homo, 38 ± 1.40 (100%); HEG5, 22 ± 0.92 (57.9%); CaG5, 19 ± 2.22 (50.0%, CaG5 vs. DB-Homo, p < 0.05); IQG5, 21 ± 0.92 (55.3%, IQG5 vs. DB-Homo, p < 0.05) and Metformin 10, 23 ± 0.35 (60.5%) (Fig. 3).
Fig. 3.
Adipose tissue ratio (%) of db mice treated with C. molossus glycosaminoglycan. The adipocyte cell density was counted from db mouse liver tissue toluidine blue O stained depots (*p<0.05).
Sero-biochemical finding of db mice after treatment with insect GAG
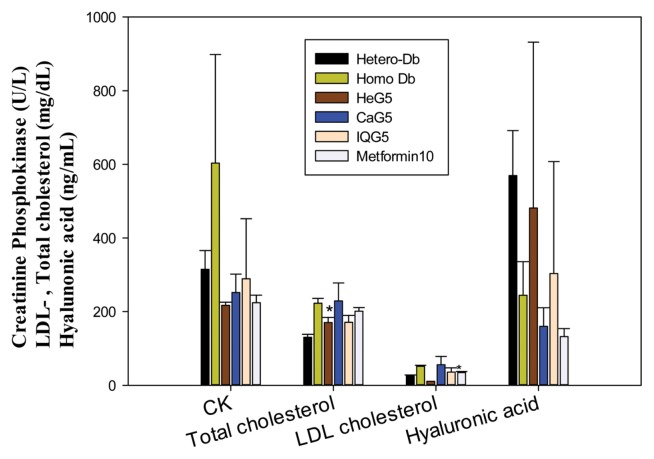
Sera parameters from the CaG5- and IQG5-treated groups (Table 2) were also significantly lower than those in the control group at one month after treatment. Serum albumin levels (g/dL) in GAG treated groups were significantly lower than those in the control: DB-Homo (CON), 3.8 ± 0.3; GaG5, 3.23 ± 0.15 (CaG5 vs. CON, p < 0.05); IQG5, 2.83 ± 0.17 (IQG5 vs. CON, p < 0.01). Free fatty acid levels in IQG-treatment db mice (μEq/L) were also significantly lower than those in the control: CON, 1310.3 ± 125.6; IQG5, 765.8 ± 121.4 (IQG5 vs. CON, p < 0.001). Creatinine phosphokinase levels (U/L) were also decreased: DB-Hetero, 314.75 ± 51.3; DB-Homo, 603.3 ± 294.8; HEG5, 217 ± 8.4; CaG5, 251.7 ± 49.9; IQG5, 288.75 ± 163.5; and Metformin 10, 224 ± 20.52. Alkaline phosphatase (ALP) levels (U/L) in IQG5- and CaG5-treated groups were also significantly lower than those in the control groups: CON, 109.33 ± 12.34; CaG5, 73.3 ± 7.0 (CaG5 vs. CON, p < 0.05); IQG5, 70.0 ± 12.6 (IQG5 vs. CON, p < 0.001). Mean total cholesterol levels (mg/dL) in HeG- or IQG-treated db mice were also significantly lower than those in the control group: CON, 222.7 ± 13.2; HeG5, 170 ± 14.1 (HeG5 vs. CON, p < 0.05); IQG5, 171 ± 18.7 (IQG5 vs. CON, p < 0.05) (Table 2, Fig. 4). These serological results showed that these GAGs could play a role in the cure against increased blood glucose, hyperglycemia, and complicated diseases.
Table 2.
Serological findings of db mice treated intraperitoneally with Heuchys, C. molossus or queen of B. ignitus, glycosaminoglycan over an one month
| Function | Parameter | Unit | DB-Hetero | DB-Homo (CON) | HeG5 | CaG5 | IQG5 | Metformin10 |
|---|---|---|---|---|---|---|---|---|
| Tonic | Albumin | g/dL | 2.88 ± 0.05 | 3.8 ± 0.3 | 3.8 ± 0 | 3.23 ± 0.15* | 2.83 ± 0.17** | 3.73 ± 0.15 |
| Fatty liver | Free fatty acid | μEq/L | 712.75 ± 53.34 | 1310.3 ± 125.6 | 1749.5 ± 61.5 | 1093.7 ± 164.4 | 765.8 ± 121.4** | 1016 ± 30.51 |
| Hepatitis | ALP | U/L | 60 ± 16.73 | 109.33 ± 12.34 | 114 ± 17.0 | 73.3 ± 7.0* | 70 ± 12.6** | 127 ± 13.45 |
| AST (SGOT) | U/L | 92 ± 16.47 | 110.5 ± 14.85 | 123 ± 50.9 | 109.3 ± 2.5 | 129.5 ± 65.6 | 120 ± 20.07 | |
| ALT (SGPT) | U/L | 39.75 ± 3.4 | 77 ± 11.31 | 74.5 ± 17.7 | 68.7 ± 6.4 | 92.5 ± 66.7 | 91.33 ± 20.98 | |
| Heart function | CK (SE) | U/L | 314.75 ± 51.3 | 603.3 ± 294.8 | 217 ± 8.4 | 251.7 ± 49.9 | 288.75 ± 163.5 | 224 ± 20.52 |
| Diabetes | Glucose (S) | mg/dL | 195 ± 24.45** | 660.3 ± 78.8 | 732 ± 25.5 | 543 ± 139.4 | 663.3 ± 73.0 | 750 ± 0 |
| Lipidemia | Cholesterol, total | mg/dL | 130.25 ± 7.85 | 222.7 ± 13.2 | 170 ± 14.1* | 228.7 ± 48.9 | 171 ± 18.7* | 201 ± 9.85 |
| Triglyceride | mg/dL | 126.5 ± 12.12 | 253.5 ± 43.4 | 274 ± 8.49 | 229 ± 28.8 | 240.3 ± 22.8 | 205 ± 15.72 | |
| LDL cholesterol | mg/dL | 25 ± 2.83 | 51.33 ± 3.06 | 11 ± 0 | 55.7 ± 22.3 | 35.8 ± 11.6 | 34.33 ± 3.06* | |
| HDL cholesterolL | mg/d | 90 ± 10.55 | 120 ± 0 | 120 ± 0 | 120 ± 0 | 108 ± 8.9 | 120 ± 0 | |
| Nepritis | Creatinine | mg/dL | 0.2 ± 0.003 | 0.32 ± 0.03 | 0.34 ± 0.01 | 0.29 ± 0.06 | 0.28 ± 0.03 | 0.36 ± 0.02 |
| BUN | mg/dL | 23.63 ± 0.74 | 23.1 ± 4.01 | 24.2 ± 2.4 | 24.2 ± 2.6 | 19.2 ± 3.7 | 22.3 ± 3.12 | |
| Hypertension | Na (Sodium) | mmol/L | 148 ± 2.16 | 147 ± 1.73 | 153.5 ± 6.4 | 150.7 ± 4.7 | 148 ± 3.5 | 151.33 ± 2.52 |
| Calcium | mg/dL | 11.8 ± 0.37** | 13.83 ± 0.23 | 13.8 ± 0.28 | 14.5 ± 0.7 | 14.05 ± 0.7 | 14.2 ± 0.56 | |
| Cl (Chloride) | mmol/L | 103 ± 3.46* | 94.33 ± 2.89 | 99.0 ± 5.66 | 99.0 ± 6.9 | 96.3 ± 3.9 | 102.67 ± 2.08* | |
| Edema | Protein, total (S) | mmol/L | 5.68 ± 0.19** | 7.87 ± 0.31 | 7.7 ± 0.14 | 7.53 ± 0.67 | 7.05 ± 0.56 | 7.57 ± 0.21 |
| Rheumatis | CRP (HS) | mg/L | 0.15 ± 0.06 | 0.3 ± 0.1 | 0.85 ± 0.21 | 0.37 ± 0.12 | 0.65 ± 0.25 | 0.67 ± 0.06* |
Each value represents mean ± SD statistically significant from control (*p<0.05, **p < 0.01).
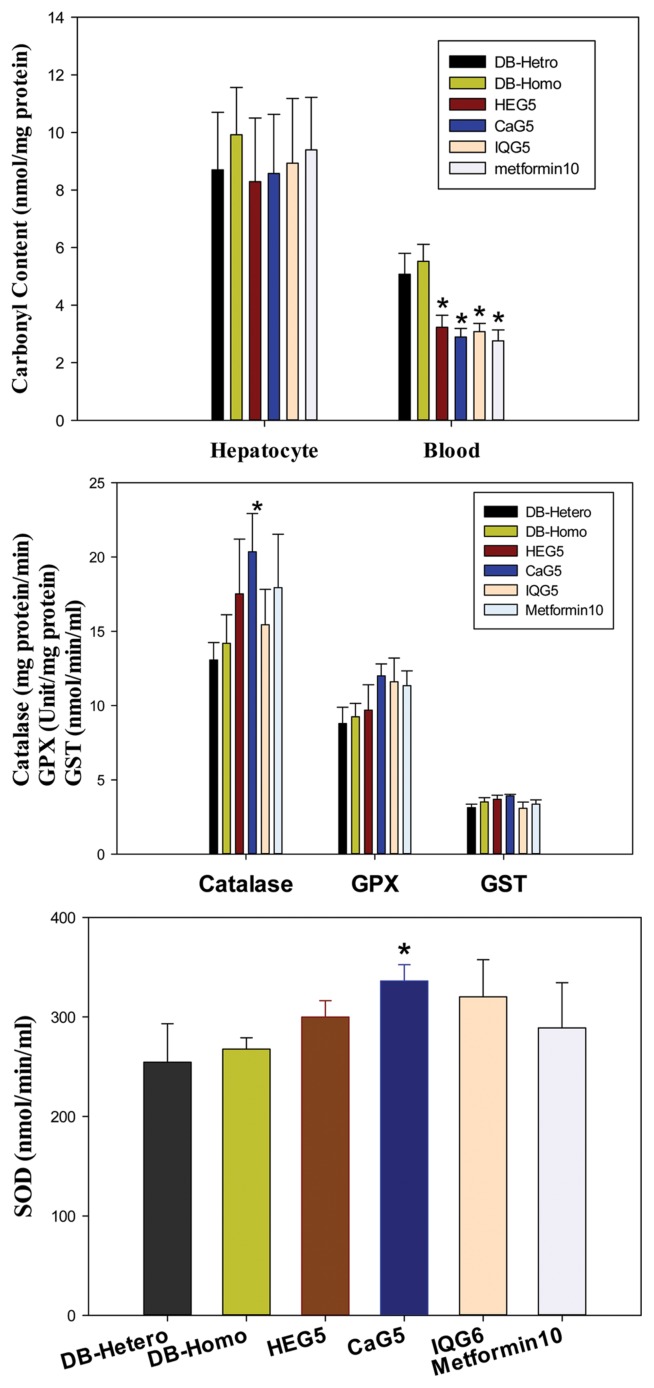
Fig. 4.
Anti-oxidative effect of dung beetle glycosaminoglycan on proteins carbonyl, catalase, Glutathione peroxidase (GPX) or glutathione s-transferase (GTS) content, Super oxide dismutase (SOD) (*p<0.05).
Decrease of oxidative damage
Protein oxidative cellular stress was evaluated by measuring carbonyl content in blood neutrophil. Protein carbonyl concentrations in blood were decreased after treatment with insect glycosaminoglycans. Decreased ratios were 58.51%, 52.36%, 55.80% and 50% for HeG5 (HeG5 vs. CON, p < 0.05), CaG5 (CaG5 vs. CON, p < 0.05), IQG5 (IQG5 vs. CON, p < 0.05), and Metformin10 (Metformin10 vs. CON, p < 0.05) groups, respectively. However, there were no statistical differences in hepatocyte carbonyl contents between control and treatment groups of db mice (Table 3, Fig. 5). Therefore, protein oxidative damages could be reduced by theses GAGs.
Table 3.
Antioxidant enzyme activities of dung beetle glycosaminoglycan in hepatocyte of db (+leptin) mice
| Anti-Oxydative enzyme | Unit | DB-Hetero | DB-Homo | HEG5 | CaG5 | IQG5 | Metformin10 | |
|---|---|---|---|---|---|---|---|---|
| Catalase | mg protein/min | 13.07 ± 1.17 | 14.19 ± 1.92 | 17.52 ± 3.69 | 20.35 ± 2.58* | 15.44 ± 2.38 | 17.93 ± 3.6 | |
| Glutathione peroxidase | unit/mg protein | 8.79 ± 1.1 | 9.25 ± 0.9 | 9.7 ± 1.7 | 12.0 ± 0.8 | 11.6 ± 1.6 | 11.34 ± 1.0 | |
| Glutathione-s-transeferasex | nmol/min/ml | 3.13 ± 0.24 | 3.51 ± 0.29 | 3.69 ± 0.28 | 3.93 ± 0.09 | 3.09 ± 0.41 | 3.37 ± 0.28 | |
| Superoxide dismutase | nmol/min/ml | 254.58 ± 38.57 | 267.57 ± 11.49 | 299.87 ± 16.44 | 336.11 ± 16.37* | 320.28 ± 37.38 | 289.01 ± 45.47 | |
| Carbonyl content | Hepatocyte | nmol/mg protein | 8.7 ± 2.0 | 9.92 ± 1.64 | 8.29 ± 2.21 | 8.57 ± 2.06 | 8.93 ± 2.25 | 9.39 ± 1.83 |
| Blood | nmol/mg protein | 5.07 ± 0.73 | 5.52 ± 0.59 | 3.23 ± 0.42* | 2.89 ± 0.3* | 3.08 ± 0.29* | 2.76 ± 0.38* | |
Each value represents mean ± SE statistically significant from control (*p<0.05).
Fig. 5.
Sero-biochemical detection of creatinine phosphokinase, LDL−/total cholesterol, hyaluronic acid in CaG-treated db mice (*p < 0.05).
Oxidative enzyme (catalase, GPx, GST, SOD) quantitation
Oxidative enzyme is a free radical scavenger enzyme. Level of enzymes such as catalase, GPx, GST, and SOD enzymes were increased by GAG at one month after treatment.
Catalase activites (mg protein/min) in db mice hepatocytes after one month of GAG treatment were as follows: db mice (CON), 14.19 ± 1.92; HeG5, 17.52 ± 3.69 (HeG5/CON: 123%); CaG5, 20.35 ± 2.58 (143%, CaG5 vs. CON, p < 0.05); IQG5, 15.44 ± 2.38; and Metformin10, 17.93 ± 3.6 (Met/CON: 126%). Catalase activities in all GAG-treated hepatocyte groups were increased compared to those in the control group. Glutathione peroxidase activities (Unit/mg protein) in all GAG treated groups were also increased: db mice (CON), 9.25 ± 0.9; HeG5, 9.7 ± 1.7; CaG5, 12.0 ± 0.8 (CaG5/CON: 129.7%); IQG5, 11.6 ± 1.6 (IQG5/CON: 125.4%); Metformin10, 11.34 ± 1.0 (Met/CON: 122.6%). Glutathione-s-transferase activities (nmol/min/ml) were as follows: CON, 3.51 ± 0.29; HeG5, 3.69 ± 0.28; CaG5, 3.93 ± 0.09 (CaG5/CON: 112.0%); IQG5, 3.09 ± 0.41; and Metformin10, 3.37 ± 0.28. Superoxide dismutase activities (nmol/min/ml) were significantly increased in treatment groups: DB-Hetero (normal mice), 254.58 ± 38.57; db mice (CON), 267.57 ± 11.49; HeG5, 299.87 ± 16.44; CaG5, 336.11 ± 16.37 (125.6%, CaG5 vs. CON, p < 0.05); IQG5, 320.28 ± 37.38; and Metformin10, 289.01 ± 45.47 (Table 3, Fig. 4).
Gene expression by quantitative real-time PCR analysis
Some genes were elucidated by real-time PCR. Data are presented as PCR cycle number (Ct value) when concentration of PCR amplicon is equilibrated by sample amplicon. Median CT values of five samples (DB-Homo, HeG5, CaG5, IQG5, and Metformin10) are presented. Mean Ct value of treatment groups were as follows: heparanase: CON, 35.4; HeG5, 35.3; CaG5, 35.6: IQG5, 35.2; Metformin10, 35.4; Glutathione peroxidase 1 (Gpx1): CON, 26.9; HeG5, 27.0; CaG5, 26.8: IQG5, 26.8; Metformin10, 26.8; glycogen synthase kinase 3 beta (GSK3b): CON, 26.3; HeG5, 26.5; CaG5, 26.4: IQG5, 26.2; Metformin10, 26.8. There were no significant differences in gene expression between treatment groups and the control group (Ct value > 2.0).
Gene expression profiling by DNA microarray
Microarray analysis using a Mouse 28K cDNA clone array was performed in order to obtain gene-expression profiles for HeG, CaG, IQG, and metformin-treated db mice livers. This might provide information for potential anti-diabetic markers (Table 4, 5). Compared to the control group, 49, 48, 279 and 225 genes were upregulated (> 2 fold increase, Table 3) 83, 79, 679 and 110 genes were downregulated (> 2-fold decrease, Table 5) in HeG, CaG, IQG, and metformin-treated groups. Therefore, IQG-treatment showed the most effect on gene expression in the liver of db mice. An overview of these genes revealed 19 functional categories, including genes, 49 homeostatic process related genes that showed more changed in expression. Furthermore IQG- and metformin-treated group genes showed more changes in expression, especially in case of IQG-treated group. A total of 71 genes related to lipid metabolism were changed in expression in IQG-, and metformin-treated groups, especially in IQG-treated group. Only three genes showed changes in expression compared to control group. They were associated with cell growth (Table 4, 5).
Table 4.
Upregulated genes differentially expressed in liver tissue of melanoma induced mice treated with GAG over a 1-month period
| Index | Gene description | Genesymbol | HeG5 | CaG5 | IQG5 | Met10 |
|---|---|---|---|---|---|---|
| 1 | Mitochondriallyen coded tRNAlysine | mt-Tk | 2.19 | 9.64 | 1.76 | 1.28 |
| 2 | Cytochrome P450, family8, subfamilyb, polypeptide1 | Cyp8b1 | 3.35 | 8.34 | 1.25 | 1.48 |
| 3 | 5′Nucleotidase, ecto | Nt5e | 2.01 | 6.34 | 1.21 | 1.61 |
| 4 | Myo-inositol1-phosphatesynthaseA1 | Isyna1 | 2.23 | 6.28 | 1.43 | 1.61 |
| 5 | CytochromeP450, family2, subfamilyb, polypeptide10 | Cyp2b10 | 3.17 | 4.86 | 1.32 | 1.32 |
| 6 | Immunoglobulin kappa variable 4–55 | Igkv4-55 | 3.23 | 4.77 | 1.03 | 1.79 |
| 7 | Immunoglobulin kappa chainvariable 4–72 | Igkv4-72 | 2.20 | 4.50 | 1.23 | 1.08 |
| 8 | Gprotein-coupled receptor, familyC, group5, memberA | Gprc5a | 2.20 | 4.50 | 1.12 | 1.36 |
| 9 | Murinoglobulin, pseudogene1 | Mug-ps1 | 2.04 | 4.46 | 1.70 | 1.12 |
| 10 | Immunoglobulin kappa chain complex | Igk | 2.10 | 4.44 | 1.16 | 1.64 |
| 11 | PTC7 protein phosphatase homolog (S.cerevisiae) | Pptc7 | 2.12 | 4.18 | 1.35 | 1.71 |
| 12 | Thioredoxinin teracting protein | Txnip | 3.03 | 3.97 | 1.00 | 1.46 |
| 13 | CD59a antigen | Cd59a | 2.12 | 3.90 | 1.87 | 1.80 |
| 14 | Odorant binding protein2A | Obp2a | 2.01 | 3.80 | 1.15 | 1.91 |
| 15 | CytochromeP450, family2, subfamilyc, polypeptide39 | Cyp2c39 | 2.57 | 3.75 | 1.07 | 1.67 |
| 16 | Angiopoietin-like4 | Angptl4 | 2.05 | 3.75 | 1.07 | 1.35 |
| 17 | CytochromeP450, family39, subfamilya, polypeptide1 | Cyp39a1 | 2.00 | 3.74 | 1.09 | 1.55 |
| 18 | MicroRNA5125 | Mir5125 | 2.06 | 3.71 | 1.12 | 1.14 |
| 19 | Serine/arginine repetitive matrix2 | Srrm2 | 2.38 | 3.62 | 1.05 | 1.12 |
| 20 | StefinA2 like1 | Stfa2l1 | 2.03 | 3.62 | 1.46 | 1.32 |
| 21 | Immunoglobulin heavy joining2| immunoglobulin heavy variable14-2 | Ighj2|Ighv14-2|Ighm|Ighj1 | 2.28 | 3.42 | 1.99 | 1.31 |
| 22 | Solute carrier family13 (sodium-dependent citrate transporter), member5 | Slc13a5 | 2.02 | 3.40 | 1.01 | 1.34 |
| 23 | Vanin1 | Vnn1 | 2.12 | 3.33 | 1.27 | 1.81 |
| 24 | Chitinase3-like1 | Chi3l1 | 2.82 | 3.28 | 1.28 | 1.89 |
| 25 | Amino carboxy muconatesemialdehyde decarboxylase | Acmsd | 2.43 | 3.23 | 1.09 | 1.84 |
| 26 | Predictedgene5068 | Gm5068 | 1.52 | 3.22 | 1.97 | 1.01 |
| 27 | Retinol binding protein1, cellular | Rbp1 | 1.81 | 3.21 | 1.16 | 1.04 |
| 28 | Ubiquitin specific peptidase2 | Usp2 | 1.95 | 3.14 | 1.45 | 1.95 |
| 29 | Homocysteine-inducible, endoplasmicreticulumstress-inducible, ubiquitin-likedomainmember1 | Herpud1 | 1.64 | 3.14 | 1.56 | 1.52 |
| 30 | Predictedgene15889 | Gm15889 | 1.33 | 3.14 | 1.48 | 1.33 |
| 31 | Tumor necrosis factor receptor superfamily,member23 | Tnfrsf23 | 1.34 | 3.13 | 1.42 | 1.04 |
| 32 | RIKENcDNA1810008I18 gene | 1810008I18Rik | 1.77 | 3.06 | 1.29 | 1.31 |
| 33 | Immunoglobulin kappa chain complex| immunoglobu-linkappavariable8-19| immunoglobulinkappajoining5 | Igk|Igkv8-19|Igkj5 | 1.99 | 3.05 | 1.72 | 1.62 |
| 34 | Cytochrome P450, family51 | Cyp51 | 1.50 | 2.94 | 1.79 | 1.49 |
| 35 | Secreted phosphoprotein1 | Spp1 | 1.91 | 2.88 | 1.75 | 1.23 |
| 36 | Phosphogluconated ehydrogenase | Pgd | 1.31 | 2.85 | 1.21 | 1.16 |
| 37 | GlutathioneS-transferase, mu3 | Gstm3 | 1.40 | 2.84 | 1.54 | 1.42 |
| 38 | Farnesyl diphosphate synthetase| predictedgene3571 | Fdps|Gm3571 | 1.84 | 2.76 | 1.01 | 1.36 |
| 39 | Carbonic anhydrase3 | Car3 | 1.66 | 2.76 | 1.80 | 1.86 |
| 40 | Olfactory receptor205 | Olfr205 | 1.99 | 2.73 | 1.34 | 1.42 |
Table 5.
Downregulated genes differentially expressed in liver tissue of melanoma induced mice treated with GAG over a 1-month period
| Index | Gene description | Genesymbol | HeG5 | CaG5 | IQG5 | Met10 |
|---|---|---|---|---|---|---|
| 1 | S100 calcium binding proteinA8 (calgranulinA) | S100a8 | −5.17 | −4.86 | −2.33 | −2.99 |
| 2 | Immunoglobulin kappachaincomplex | immunoglobulinkappavariable6-23 | Igk|Igkv6-23|Igk-V28|Igkc | −6.61 | −4.50 | 1.02 | −2.30 |
| 3 | Interferon-inducibleGTPase1-like|cDNAsequence-BC023105 | LOC630751|BC023105 | −3.68 | −4.50 | −6.16 | −3.24 |
| 4 | Fatty acid synthase | Fasn | −2.82 | −4.46 | −1.35 | −1.06 |
| 5 | Immunoglobulinheavyconstantmu|immunoglobulinheavychain (J558family)| immunoglobulinheavyconstantgamma1 (G1mmarker) | Ighm|Igh-VJ558|Ighg1 | −5.44 | −4.18 | −1.88 | −4.86 |
| 6 | CyclinD1 | Ccnd1 | −3.27 | −3.80 | −1.87 | −6.24 |
| 7 | Neutrophilic granule protein | Ngp | −3.77 | −3.75 | −3.46 | −3.10 |
| 8 | Immunoglobulin heavychain (X24family)| immunoglobulin heavychain (J558family) | Igh-VX24|Igh-VJ558|Igha|Ighm|Ighg|Ighg3|Ighj4|Ighv14-2 | −4.52 | −3.74 | −1.07 | −2.50 |
| 9 | S100 calcium binding proteinA9 (calgranulinB) | S100a9 | −4.24 | −3.71 | −4.22 | −3.09 |
| 10 | Lymphocyte antigen6 complex, locusD | Ly6d | −1.51 | −3.62 | −2.25 | −2.37 |
| 11 | Tubulin, beta2A classIIA | Tubb2a | −1.94 | −3.56 | −1.95 | −1.28 |
| 12 | 3-Hydroxy-3-methylglutaryl-CoenzymeA synthase1 | Hmgcs1 | −1.31 | −3.42 | −2.02 | −1.10 |
| 13 | Lactotransferrin | Ltf | −3.18 | −3.33 | −2.70 | −2.94 |
| 14 | RIKENcDNA9030619P08gene | 9030619P08Rik | −2.41 | −3.28 | −3.34 | −1.86 |
| 15 | InterferoninducibleGTPase1 | cDNAsequenceBC023105 | Iigp1|BC023105 | −2.77 | −3.23 | −3.63 | −2.43 |
| 16 | Lymphocyte antigen 6complex, locusC2 | Ly6c2 | −2.63 | −3.22 | −2.41 | −2.08 |
| 17 | Murinoglobulin2 | Mug2 | −4.48 | −3.21 | −15.04 | −1.00 |
| 18 | Squalene epoxidase | Sqle | −1.62 | −3.14 | −1.32 | −1.62 |
| 19 | Immunoglobulin kappachaincomplex | V(kappa)gene-product|immunoglobulinkappachainvariable28 (V28) |immunoglobulinkappajoining1 | Igk|LOC672450|Igk-V28|Igkj1 | −3.22 | −3.14 | −3.47 | −2.20 |
| 20 | Monooxygenase, DBH-like1 | Moxd1 | −2.94 | −3.14 | −1.91 | −2.44 |
| 21 | Glycoprotein49A | Gp49a | −3.06 | −3.06 | −2.73 | −2.07 |
| 22 | Farnesyl diphosphate synthetase | predictedgene3571 | Fdps|Gm3571 | −1.84 | −3.05 | −1.01 | −1.36 |
| 23 | S100 calciumbinding proteinA11 (calgizzarin) | predictedgene12854| predictedgene5068 | S100a11|Gm12854|Gm5068 | −1.69 | −2.85 | −2.16 | −1.33 |
| 24 | Heatshockprotein8 | Hspa8 | −2.42 | −2.84 | −1.95 | −2.15 |
| 25 | Immunoglobulin heavychain (gammapolypeptide) |immunoglobulin heavyconstantmu|immunoglobulinheavyconstantgamma2B | Ighg|Ighm|Ighg2b | −2.73 | −2.73 | −1.26 | −1.75 |
| 26 | SerumamyloidP-component | Apcs | −3.06 | −2.67 | −4.56 | −2.12 |
| 27 | ATPcitratelyase | Acly | −1.86 | −2.64 | −1.20 | −1.03 |
| 28 | U1b2 small nuclear RNA| U1b6 small nuclear RNA|U1b1smallnuclearRNA | Rnu1b2|Rnu1b6|Rnu1b1 | −2.95 | −2.61 | −3.28 | −2.55 |
| 29 | CytochromeP450, family51 | Cyp51 | −1.50 | −2.60 | −1.79 | −1.49 |
| 30 | Small nucleolar RNA, H/ACA box62 | Snora62 | −2.17 | −2.58 | −2.67 | −3.66 |
| 31 | Interleukin1 beta | Il1b | −2.34 | −2.53 | −1.89 | −1.51 |
| 32 | Immunoglobulin kappa chain complex | Igk | −2.15 | −2.51 | −1.41 | −1.93 |
| 33 | Immunoglobulin heavy chain (J558family) | Igh-VJ558 | −2.74 | −2.50 | −1.57 | −2.28 |
| 34 | Leukocyte immunoglobulin-like receptor, subfamilyB, member4 | Lilrb4 | −2.46 | −2.49 | −4.86 | −1.44 |
| 35 | Olfactoryreceptor205 | Olfr205 | −1.98 | −2.47 | −1.34 | −1.42 |
| 36 | Oredictedgene10106| predictedgene10717| predictedgene10721|predictedgene10716 | Gm10106|Gm10717|Gm10721|Gm10716 | −1.89 | −2.44 | −11.26 | −3.45 |
| 37 | Predictedgene15998 | Gm15998 | −2.00 | −2.41 | −2.45 | −1.24 |
| 38 | Elastase, neutrophil expressed | Elane | −1.93 | −2.39 | −1.04 | −1.76 |
| 39 | Resistinlike gamma | Retnlg | −2.37 | −2.37 | −1.36 | −2.37 |
| 40 | Serine (orcysteine) peptidase inhibitor, cladeA (alpha-1antiproteinase, antitrypsin), member7 | Serpina7 | −1.91 | −2.33 | −3.55 | −1.71 |
Compared to the control group, mitochondriallyen coded tRNA lysine (mt-TK) gene, encoding a mitochondrial thymidine kinase, was increased about 10-fold in CaG5-treated group, 1.8-fold in the IQ5 group, and 2.2-fold in the HeG5 group. In liver tissues of CaG5-treated mice, cytochrome P450, family 8, subfamily b, polypeptide1 (Cyp8b1) was upregulated about 8.3-fold compared to that in control group. CaG5 treated mice group, compared to the control, 48 genes were up-regulated, including 5’ nucleotidase ecto, myo-inositol1-phosphatate synthase A1, and cytochrome p450, family2, subfamily b, polypeptide10 (Table 4), While, 79 genes were down-regulated, including S100 calcium binding protein A8 (calgranulin A), immunoglobulin kappa chain complex (Igk, IGkv6-23), interferon-inducible GTPase1-like, fatty acid synthase, immunoglobulin heavy constant mu, and cyclin D1 (Table 5). These data indicate that mitochondrial thymidine kinase and cytochrome p450 family 8/2 as upregulated genes and calgranulin A and immunoglobulin kappa chain complex as downregulated genes might be markers of potential therapeutics that work as anti-diabetic agents.
DISCUSSION
Diabetes is a chronic disease that cannot be easily cured by therapeutic agents. Glycosaminoglycan could play a role in the cure against increased blood glucose, hyperglycemia and complicated diseases. Glomerular basement membrane is mainly consisted of type 4 collagen, laminin and heparan sulfate proteoglycan. Heparanase plays a role in beta-cell failure, in addition to its ability to increase glomerular basement membrane permeability due to loss of negatively charged heparin sulfate, leading to urinary protein excretion (11,12). Results of the present study revealed that, after, treatment with each glycosaminoglycan, sera total protein levels were decreased (HEG5, 97.8%; CaG5, 95.7%; IQG5, 89.6% and Metformin10, 96.1%) compared to those in DB-Homo group. Furthermore, levels of total cholesterol, LDL cholesterol, and alkaline phosphatase were also decreased by these GAGs, demonstrating their anti-lipedema effects. It has been reported that only high-dose (400 mg/kg) metformin can significantly decrease blood glucose levels compared to vehicle alone. In contrast, little reduction, if any, was seen in mice treated with low-dose (50 mg/kg) of metformin (13). In the present study, db mice (12~16 weeks age) showed such a very high glucose state in severe diabetes that metformin did not recover blood glucose to normal level. Accordingly, the initial treatment dose at 10 mg/kg in db mice, equivalent of 600 mg/60 kg (commercially sold DiabexTM at a dose of 500 mg/kg/day or 1,000 mg/kg/day) (14) in men, might not be sufficient enough to lower the blood glucose level. Metformin is widely used as a hypoglycemic reagent for type 2 diabetes. Nowadays, statin-metformin (antilipidemic-antidiabetic) combined complex drug preparation is used more in diabetes complex diseases than metformin.
Levels of anti-oxidative enzymes and activities of catalase, glutathione peroxidase, and glutathione-s-transferase were increased by these GaGs. Therefore, cellular oxidative stress by free radical damage could be scavenged with the help of these antioxidant enzymes. These cellular oxidative damage repairs could contribute to diabetic disease control not only by repairing hepatocytes, but also by repairing other organ cells including pancreatic and kidney cells. Recently, antioxidant and antithrombotic treatment has become a trend therapy for diabetic kidney disease besides the use classic agents, such as aspirin and novel drugs such as sulodexide (a type of glycosaminoglycan) and Chinese medicine such as lumbrikinase that shows beneficial effects on diabetic patients (1). In our db mice experiment, CaG5 appeared to have anti-oxidant activity, increasing activity of catalase by 143%, GPX by 129.7%, GST by 112.0% and SOD by 125.6%. As a cellular oxidative damage, protein oxidative damage was also reduced (HEG5, 58.5%; CaG5, 52.4%; IQG5, 55.8% and Metformin10, 50.0% ) by these GaGs based on blood neutrophil carbonyl content.
The most upregulated gene found in this study was a mitochondriallyen coded tRNA lysine (mt-TK) gene, an electron transporter related gene. It encodes a mitochondrial thymidine kinase that plays a role in aminoglycosideinduced nonsyndromic disorder. It affects the translation of all mitochondrial DNA encoded proteins and impairs the assembly of the electron transport chain complexes, leading to decreased mitochondrial respiratory function (15). The 2nd most upregulated gene in this study was found to be cytochrome P450, family 8, subfamily b, polypeptide 1, sterol 12 alpha-hydroxylase (cyp8b) is a key enzyme for regulating cholic acid/chendeoxycholic acid ratio in bile biosynthesis (16). It is related to antilipedema and lipid digestion. Whereas, loss of Cyp8b1 improves glucose homeostasis by increasing GLP-1and Cyp8b1 results in improved glucose tolerance, insulin sensitivity, and beta-cell function, mediated by absence of cholic acid in Cyp8b12/2 mice (17).
Overviewing of acquired microarray data of db mice after GAG treatment, upregulated genes such as mitochondrial thymidine kinase (mt-TK), cytochrome P450, family 8/2- (Cyp8b1/2b10), and immunoglobulin kappa variable 4–55/72 (Igkv4–55/4–72) are related to decrease of cellular oxidative stress and increase of anti-oxidant enzyme capacity and quantity. In fact, a mitochondrial disease by a point mutation of mitochondrial DNA in mt-TK genes can be partially restored by coenzyme Q10. This indicates that glycosaminoglycan with antioxidant activities might be useful as therapeutic agents.
The most downregulated gene based on DNA microarray in this study was found to be S 100 calcium binding protein A8 (S100A8, calgranulin A), an inflammatory molecules. A heterodimer of calprotectin with S100A9 has been reported (18). Decrease of neutrophil-derived S100 calcium - binding proteins A8/A9 could depress reticulated thrombocytosis and atherogenesis in diabetes suggesting suppress inflammatory molecule S100A9/A8 might have anti-diabetic effect (19). The 2nd most downregulated gene found in this study was immunoglobulin kappa chain complex (Igk, IGkv6–23), an immunoglobulin kappa variant 6–23 in house mouse (Mus musculus) (20).
In sera data of this study, creatinine kinase level as a cardiac inflammation marker was reduced by GAG treatment. GPRC5 is a potential tumor suppressor and oncogene with emerging roles in diseases (21). GPRC5 in this db mice study was upregulated with tumor suppression potential as a similar cure function. Cyclin D1 plays an important role in the regulation of G1 progression of somatic cell cycle by functioning as a regulatory submit of cdk4 and cdk6 (22). Increased expression of cyclinD (CcnD10) and Myc (MYC) genes are known to be involved in liver cancer. Results of this study showed their downregulation, indicating that these GAG could prevent liver cancer in db mice as indicated in a previous study (23).
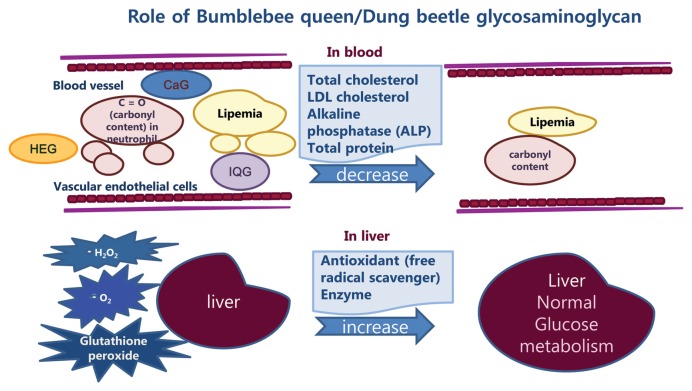
These results from sero-biochemical, hepatocellular antioxidant assay, DNA gene expression profiling and db mice clinical data suggest CaG, IQG and HeG could be used as natural anti-diabetic agents and functional food, like Sulodexide (24). Proposed mechanisms involved in the decrease of cellular oxidative stress in blood and liver tissues with increase of anti-oxidant enzyme in db mice after treatment with GAG are summarized in Fig. 6.
Fig. 6.
Summary of GAG’s mechanism of action in db mice.
ACKNOWLEDGMENTS
The authors acknowledge National Academy of Agricultural Science for financial (RDA, PJ011853) and technical supports.
REFERENCES
- 1.Yan W, Zhou B, Shen Y, Xu G. Antioxidant and antithrombotic therapies for diabetic kidney disease. Iran J Kidney Dis. 2015;9:413–420. [PubMed] [Google Scholar]
- 2.Lepedda AJ, Muro PD, Capobianco G, Formato M. Significance of urinary glycosaminoglycans/proteoglycans in the evaluation of type 1 and type 2 diabetes complications. J Diabetes Complicat. 2017;31:149–155. doi: 10.1016/j.jdiacomp.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 3.Kubaski F, Osago H, Mason RW, Yamaguchi S, Kobayashi H, Tsuchiya M, Orii T, Tomatsu S. Glycosaminoglycans detection methods: application of mass spectrometry. Mol Genet Metab. 2017;120:67–77. doi: 10.1016/j.ymgme.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li R, Xing J, Mu X, Wang H, Zhang L, Zhao Y, Zhang Y. Sulodexide therapy for the treatment of diabetic nephropathy, a meta-analysis and literature review. Drug Des Devel Ther. 2015;9:6275–6283. doi: 10.2147/DDDT.S87973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn MY, Kim BJ, Kim HJ, Yoon HJ, Jee SD, Hwang JS, Park KK. Anti-obesity effect of Bombus ignitus queen glycosaminoglycans in rats on a high-fat diet. Int J Mol Sci. 2017;18:E681. doi: 10.3390/ijms18030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn MY, Kim BJ, Kim HJ, Hwang JS, Jung YS, Park KK. Anti-aging effect and gene expression profiling of dung beetle glycosaminoglycan in aged rats. Biomater Res. 2017;21:5. doi: 10.1186/s40824-017-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia L, Bai L, Yi L, Liu B, Chu C, Liang Z, Li P, Jiang Z, Zhao Z. Authentication of the 31 species of toxic and potent chinese materia medica (T/PCMM) by microscopic technique, part 1: three kinds of toxic and potent animal CMM. Microsc Res Tech. 2007;70:960–968. doi: 10.1002/jemt.20500. [DOI] [PubMed] [Google Scholar]
- 8.Kim YS, Jo YY, Chang IM, Toida T, Park Y, Linhardt RJ. A new glycosaminoglycan from the giant african snail Achatina fulica. J Biol Chem. 1996;271:11750–11755. doi: 10.1074/jbc.271.20.11750. [DOI] [PubMed] [Google Scholar]
- 9.Fratz EJ, Hunter GA, Ferreira GC. Expression of murine 5-aminolevulinate synthase variants causes protoporphyrin IX accumulation and light-induced mammalian cell death. PLoS ONE. 2014;9:e93078. doi: 10.1371/journal.pone.0093078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song J, Liu H, Ressom HW, Tiwari S, Ecelbarger CM. Chronic Rosigiltazone therapy normalizes expression of ACE1, SCD1 and other genes in the kidney of obese Zucker rats as determined by microarray by microarray analysis. Exp Clin Endocrinol Diabetes. 2008;116:315–325. doi: 10.1055/s-2008-1042429. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Wan A, Rodrigues B. The function of heparanase in diabetes and its complications. Can J Diabetes. 2013;37:332–338. doi: 10.1016/j.jcjd.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Ahn MY, Hwang JS, Yoon HJ, Yun EY. Korean patient 10-120419. 2012. Anti-diabetic composition comprising glycosaminoglycans derived insect. [Google Scholar]
- 13.Heishi M, Ichihara J, Teramoto R, Itakura Y, Hayashi K, Ishikawa H, Gomi H, Sakai J, Kanaoka M, Taiji M, Kimura T. Global gene expression analysis in liver of obese diabetic db/db mice treated with metformin. Diabetologia. 2006;49:1647–1655. doi: 10.1007/s00125-006-0271-y. [DOI] [PubMed] [Google Scholar]
- 14.Dou J, Ma J, Liu J, Wang C, Johnsson E, Yao H, Zhao J, Pan C. Efficacy and safety of saxagliptin in combination with metformin as initial therapy in Chinese patients with type 2 diabetes: results from the START study, a multicenter, randomized, double-blind, active-controlled, phase 3 trial. Diabetes Obes Metab. 2018;20:590–598. doi: 10.1111/dom.13117. [DOI] [PubMed] [Google Scholar]
- 15.Mata MDI, Garrido-Maraver J, Cotán D, Cordero MD, Oropesa-Ávila M, Izquierdo LG, Miguel MD, Lorite JB, Infante ER, Ybot P, Jackson S, Sanchez-Alcázar JA. Recovery of MERRF fibroblasts and cybrids pathophysiology by Coenzyme Q10. Neurotherapeutics. 2012;9:446–463. doi: 10.1007/s13311-012-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida H, Yamashita C, Kuruta Y, Yoshida Y, Noshiro M. Insulin is a dominant suppressor of sterol 12a-hydroxylase P450 (CYP8B) expression in rat liver possible role of insulin in Circadian rhythm of CYP8B. J Biochem. 2000;127:57–64. doi: 10.1093/oxfordjournals.jbchem.a022584. [DOI] [PubMed] [Google Scholar]
- 17.Kaur A, Patankar JV, de Haan W, Ruddle P, Wijesekara N, Groen AK, Verchere CB, Singaraja RR, Hayden MR. Loss of cyp8b1 improves glucose homeostasis by increasing GLP-1. Diabetes. 2015;64:1168–1179. doi: 10.2337/db14-0716. [DOI] [PubMed] [Google Scholar]
- 18.Mortensen OH, Nielsen AR, Erikstrup C, Plomgaard P, Fischer CP, Krogh-Madsen R, Lindegaard B, Petersen AM, Taudorf S, Pedersen BK. Calprotectin - a novel marker of obesity. PLoS ONE. 2009;4:e7419. doi: 10.1371/journal.pone.0007419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraakman MJ, Lee MKS, Al-Sharea A, Dragoljevic D, Barrett TJ, Montenont E, Basu D, Heywood S, Kammoun HL, Flynn M, Whillas A, Hanssen NMJ, Febbraio MA, Westein E, Fisher EA, Chin-Dusting J, Cooper ME, Berger JS, Goldberg IJ, Nagareddy PR, Murphy AJ. Neutrophil-derived S100 calcium-binding proteins A8/A9 promote reticulated thrombocytosis and atherogenesis in diabetes. J Clin Invest. 2017;127:2133–2147. doi: 10.1172/JCI92450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giudicelli V, Chaume D, Lefranc M. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Rigoutsos I. The emerging roles of GPRC5A in diseases. Oncoscience. 2014;1:765–776. doi: 10.18632/oncoscience.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, Kubota M, Shinohara K, Yasuda K, Kato J. In vivo analysis of the cyclin D1 promotor during early embryogenesis in Xenopus. Cell Struct Funct. 2003;28:165–177. doi: 10.1247/csf.28.165. [DOI] [PubMed] [Google Scholar]
- 23.Chowdhury KH, Montgomery MK, Morris MJ, Cognard E, Shepherd RR, Smith GC. Glucagon phosphorylate serine 552 of beta-catenin leading to increased expression of cyclinD1 and C-Myc in isolated rat liver. Arch Physiol Biochem. 2015;121:88–96. doi: 10.3109/13813455.2015.1048693. [DOI] [PubMed] [Google Scholar]
- 24.Lauver DA, Lucchesi BR. Sulodexide: a renewed interest in this glycosaminoglycan. Cardiovasc Drug Rev. 2006;24:214–226. doi: 10.1111/j.1527-3466.2006.00214.x. [DOI] [PubMed] [Google Scholar]