Abstract
Aim
This research aims to evaluate the predictive performance of a published allopurinol dosing tool.
Methods
Allopurinol dose predictions were compared to the actual dose required to achieve serum urate (SU) <0.36 mmol l−1 using mean prediction error. The influence of patient factors on dose predictions was explored using multilinear regression.
Results
Allopurinol doses were overpredicted by the dosing tool; however, this was minimal in patients without diuretic therapy (MPE 63 mg day−1, 95% CI 40–87) compared to those receiving diuretics (MPE 295 mg day−1, 95% CI 260–330, P < 0.0001). ABCG2 genotype (rs2231142, G>T) had an important impact on the dose predictions (MPE 201, 107, 15 mg day−1 for GG, GT and TT, respectively, P < 0.0001). Diuretic use and ABCG2 genotype explained 53% of the variability in prediction error (R 2 = 0.53, P = 0.0004).
Conclusions
The dosing tool produced acceptable maintenance dose predictions for patients not taking diuretics. Inclusion of ABCG2 genotype and a revised adjustment for diuretics would further improve the performance of the dosing tool.
Keywords: ABCG2, allopurinol, diuretics, genotype, gout, urate
What is Already Known about this Subject
The maintenance dose of allopurinol required to achieve target SU (<0.36 mmol l−1) is highly variable between individuals.
To date, there has been little research to identify the patient factors that can predict allopurinol dose requirements and could therefore aid dosing decisions in the clinic.
A recently published dosing tool for allopurinol suggested that renal function, weight and diuretic use would determine maintenance dose requirements, but this tool has not been evaluated against data from an independent cohort of people with gout.
What this Study Adds
The allopurinol dosing tool produced reasonably accurate and precise maintenance dose predictions for patients not taking diuretics.
Concomitant diuretic therapy resulted in a profound overprediction of allopurinol dose and, for this reason, we recommend that the non‐diuretic dose predictions from the published dosing tool be used for all patients, regardless of diuretic use.
Inclusion of ABCG2 genotype (rs2231142, G>T) and a revised adjustment for diuretics would further improve the performance of the dosing tool.
Introduction
Treat‐to‐target serum urate (SU) is a key strategy in the long‐term management of gout 1, 2. While http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6795 is a first line urate‐lowering therapy 2, many people fail to achieve target SU due to restricted dosing, particularly in those with kidney disease 3, 4, 5. There is increasing evidence to support the use of allopurinol at doses above those based on kidney function to achieve target SU 6, 7. The initiation of allopurinol therapy requires the use of low doses, with a gradual increase based on serum urate response and tolerability. However, the dose of allopurinol that will ultimately be required to achieve target SU (<0.36 mmol l−1) is highly variable between individuals 3, 4, 5, 6, 7, 8.
The patient factors that determine allopurinol dose requirements are not well understood. Recent studies have found that renal function, diuretic use, probenecid use, and body weight have an important impact on oxypurinol clearance (the active metabolite of allopurinol) 9, 10. Some cardiovascular drugs have been associated with lower serum urate concentrations and may enhance allopurinol response (e.g. losartan 11), while others are reported to increase hyperuricemia risk and may be anticipated to diminish allopurinol response (e.g. beta‐blockers 11).
Recent work on the genetic determinants of gout have found that variants of apical and basolateral urate transporters in the kidney and gut are associated with hyperuricemia and gout risk 12, 13. In the case of http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=792, which encodes the efflux transporter Breast Cancer Resistant Protein (BRCP), a reduced function allele rs2231142 (Q141K, G>T) has been associated with poor allopurinol response 14, hyperuricemia 15, and the presence of tophi 16. To date, there has been little research designed to quantify the impact of these factors on allopurinol maintenance dose requirements and which could therefore aid dosing decisions in the clinic.
We have previously developed a pharmacokinetic‐pharmacodynamic (PKPD) model for allopurinol in people with gout 8. We identified body weight, diuretic use and, to a lesser extent, kidney function estimated using creatinine clearance (CLcr, as defined by Cockcroft and Gault 17) as key factors influencing allopurinol urate‐lowering response. Model predictions of urate lowering under different values of CLcr, weight, and with and without concomitant diuretics, were used to produce a dosing tool to guide dosing decisions. The tool, in the form of a dosing table, is presented in the Supplementary Material (herein termed the ‘Otago dosing tool’). To date, the tool has not been evaluated against data from an independent cohort of people with gout. The aims of this study were: (1) to evaluate the predictive performance of the Otago dosing tool against an evaluation dataset, and (2) to determine the influence of patient factors (including genetic variants in urate transporters 12, 13) on predicted doses.
Methods
Evaluation dataset
Data from a published randomized controlled trial 6, 7 (RCT) comparing CLcr‐based dosing (based on Hande et al. 18) to a dose escalation protocol were analysed. In brief, 183 people with gout on allopurinol with SU ≥ 0.36 mmol l−1 were randomized to either continue their current dose of allopurinol for 12 months and then undergo dose escalation or to begin allopurinol dose escalation immediately. During dose‐escalation, allopurinol was increased monthly until SU was <0.36 mmol l−1. Blood samples for the measurement of SU, creatinine, and plasma oxypurinol were obtained at least three‐monthly. Co‐author LKS was the principle investigator of the RCT. The study was approved by the New Zealand Health and Disabilities Ethics Committee and prospectively registered with the Australian New Zealand Clinical Trials Registry (ANZCTR12611000845932). All participants gave written, informed consent.
Genotyping
Single nucleotide polymorphisms (SNPs) in urate transporter variants associated with hyperuricemia or gout 12, 13 were analysed using TaqMan® assays (Applied Biosystems, Foster City, CA, USA); rs11942223 (SLC2A9), rs2231142 (Q141K, ABCG2), rs10011796 (ABCG2), rs1183201 (NPT1/SLC17A1), rs17300741 (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1030) and rs3825018 (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1031). Details of the genotypes are presented in the Supplementary Material.
Definition of observed dose
The observed dose was defined as the dose required by the participant to achieve SU < 0.36 mmol l−1 on two consecutive visits. Participants were excluded if they failed to achieve the SU target on two consecutive visits while taking the same dose of allopurinol, were receiving dialysis, or had plasma oxypurinol <20 μmol l−1, below which nonadherence is considered likely 3, 4. Patient characteristics were recorded for each participant on the same clinic visit as the observed dose. For missing patient characteristics, the last observation was carried forward. In the case of missing genotype data, the patient was excluded from the analysis.
Definition of model‐predicted dose
Model‐predicted doses for each participant were determined by referring to the published Otago dosing tool (see Supplementary Material). Renal function, weight and diuretic use were recorded for each patient on the second of two clinic visits where the target urate was achieved (as above). In this way, the predicted dose and observed dose were aligned to the same clinic visit. The dose predictions are intended to signal the likely allopurinol dose required to achieve SU < 0.36 mmol l−1 given differences in kidney function, weight and diuretic use.
Data analysis
The observed and predicted maintenance doses were compared using mean prediction error (MPE) as a measure of bias 19, given by:
where pe i is the prediction error for the ith individual, and N is the number of participants. If the 95% confidence interval of MPE included zero, then no bias was concluded. The proportion of dose predictions within 100 mg from the observed dose was determined as a measure of precision.
Further analyses explored the influence of kidney function, weight, ethnicity, sex, concomitant drugs and urate transporter genotype on the predictive performance of the model‐based dosing tool. For categorical variables, the marginal influence on MPE was assessed using a t‐test and/or ANOVA. The marginal influence of continuous variables on prediction error was assessed using linear regression analysis in Prism (v7.03, GraphPad, La Jolla, CA, USA). A significant relationship was concluded if the 95% confidence interval of the slope of the regression line did not include zero. P‐values were considered significant if P < 0.05. A multilinear regression analysis to assess the joint influence of patient factors on the variability in prediction error was conducted in R (v3.2.2, The R Foundation).
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY 20, and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 21.
Results
Allopurinol doses and SU concentrations were available for 183 people with gout. Thirty‐four participants who did not achieve target SU, three receiving dialysis, and two with plasma oxypurinol <20 μmol l−1, were excluded from the analysis. Data for one participant with an implausible dose–response relationship was also excluded (assumed nonadherence). The final dataset included 143 participants. Demographics and clinical features are outlined in Table 1. Genotype information is summarized in the Supplemental Material.
Table 1.
Demographic and clinical details of the external dataset
| Total (n = 143) | |
|---|---|
| Males (n [%]) | 126 [88%] |
| Age (years) | 62 [23–86] |
| Weight (kg) | 104 [64–177] |
| European (n [%]) | 70 [49%] |
| Pacific Island (n [%]) | 39 [27%] |
| Māori (n [%]) | 27 [19%] |
| South Asian (n [%]) a | 4 [3%] |
| East Asian (n [%]) | 3 [2%] |
| CLcr (ml min −1 ) b | 61 [11–150] |
| Diuretics (n [%]) c | 53 [37%] |
| Beta‐blockers (n [%]) | 60 [42%] |
| Angiotensin converting enzyme inhibitors (n [%]) | 78 [55%] |
| Angiotensin II receptor blockers (n [%]) | 17 [12%] |
| Calcium channel blockers (n [%]) | 33 [23%] |
| HMG‐CoA reductase inhibitor (n [%]) | 75 [52%] |
| Uricosuric (n [%]) | 5 [3.5%] |
| Allopurinol dose at target SU (mg day −1 ) | 350 [100–700] |
| Oxypurinol at target SU (μmol l −1 ) d | 105 [30–512] |
| Pretreatment urate (mmol l −1 ) e | 0.61 [0.36–0.89] |
| Urate at target (mmol l −1 ) | 5.4 [3.5–6.0] |
All data expressed as median [range] unless otherwise stated. CLcr = creatinine clearance.
Subjects of Southern Asian ancestry were all Indian.
Creatinine clearance determined using the Cockcroft‐Gault equation 17.
Diuretics included n = 40 loop (39 × frusemide, 1 × bumetanide), n = 13 thiazides (10 × hydrochlorothiazide, 3 × bendrofluazide) and n = 1 spironolactone.
Oxypurinol plasma concentrations were measured for 110 individuals.
Pretreatment urate concentrations were available for 53 individuals.
A summary of the MPE, and imprecision results are presented in Table 2 and Figure 1. MPE was greater than zero indicating an overpredicted dose (MPE 150 mg day−1, 95% CI 125–175) (Table 2, Figure 1A). The overprediction was substantially lower in participants without concomitant diuretic therapy (MPE 63 mg day−1, 95% CI 40–87). A profound overprediction was observed in participants taking diuretics (MPE 295 mg day−1, 95% CI 260–330) (Table 2, Figure 1B). The predicted doses were within 100 mg of the observed allopurinol dose in 68% of participants not receiving diuretics compared to only 13% receiving diuretics. No differences were observed between loop and thiazide diuretics (Table 2). No other patient characteristics were found to influence the dose predictions (see Supplementary Material for plots of prediction error against CLcr and weight).
Table 2.
A summary of observed and predicted doses, mean prediction error, and precision
| Observed dose a (mg day −1 ) | Predicted dose a (mg day −1 ) |
MPE
(mg day −1 ) |
95% CI (MPE) |
RMSE
(mg day −1 ) |
± 100 mg of observed (%) | |
|---|---|---|---|---|---|---|
| All data (n = 143) | 385 | 535 | 150 | 125–175 | 222 | 47% |
| Diuretic therapy b, c | ||||||
| No diuretic ( n = 89) | 391 | 454 | 63 | 40–87 | 129 | 68% |
| Diuretics ( n = 54) | 373 | 668 | 295 | 260–330 | 317 | 13% |
| Loop ( n = 40) | 381 | 672 | 291 | 247–335 | – | |
| Thiazides ( n = 13) | 369 | 661 | 292 | 237–347 | – | |
| Transporter genotype d, e | ||||||
| ABCG2 GG ( n = 80) | 344 | 545 | 201 | 166–236 | 255 | 40% |
| No diuretic ( n = 44) | 351 | 448 | 97 | 68–126 | 135 | 68% |
| Diuretics ( n = 36) | 335 | 664 | 329 | 289–369 | 349 | 6% |
| ABCG2 GT ( n = 49) | 427 | 534 | 107 | 68–146 | 171 | 59% |
| No diuretic ( n = 35) | 419 | 474 | 55 | 22–89 | 111 | 71% |
| Diuretics ( n = 14) | 446 | 682 | 236 | 159–312 | 268 | 21% |
| ABCG2 TT ( n = 13) | 481 | 496 | 15 | −99–129 | 182 | 38% |
| No diuretic ( n = 9) | 489 | 422 | −67 | −187–53 | 162 | 44% |
| Diuretics ( n = 4) | 463 | 663 | 200 | 28–372 | 221 | 25% |
Observed and predicted doses expressed as the mean.
Diuretic therapy included loop (frusemide) and thiazide diuretics. One individual was taking spironolactone.
Differences in PE between those taking and not taking diuretics assessed by a t‐test (P < 0.0001).
Differences in PE between ABCG2 genotypes (GG, GT and TT) assessed using a one‐way ANOVA (P < 0.0001). Post‐hoc t‐tests assessed differences in prediction error between ABCG2 genotypes: GG vs. GT (P = 0.007), GG vs. TT (P = 0.0002), and GT vs. TT (P = 0.05). Differences in PE for those with and without concomitant diuretic therapy stratified by ABCG2 genotype was assessed by t‐test as follows: GG genotype no diuretic vs. diuretic (P < 0.0001), GT genotype no diuretic vs. diuretic (P < 0.001) and TT genotype no diuretic vs. diuretic (P = 0.0107).
One patient with missing ABCG2 genotype data was omitted from the analysis.
CI, confidence interval; MPE, mean prediction error.
Figure 1.
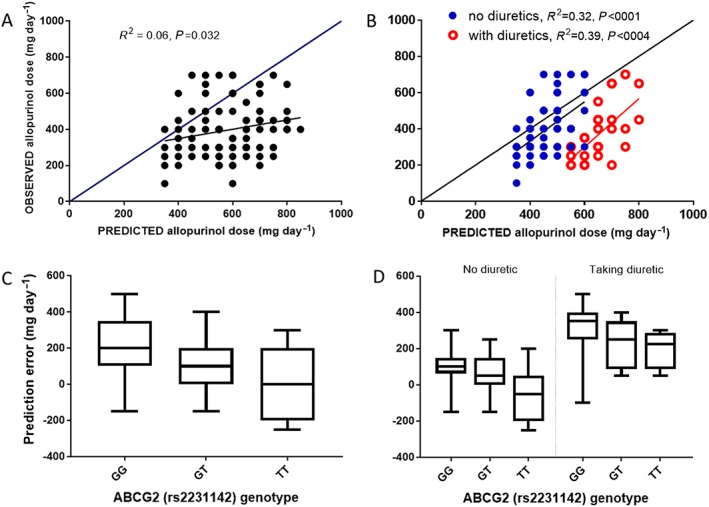
Data plots for observed doses, predicted doses and prediction error. A: Relationship between the observed and predicted allopurinol maintenance doses. B: Relationship between the observed and predicted allopurinol maintenance doses in those with and without concomitant diuretics. C: Box plots of raw prediction error for each genotype of ABCG2 (rs2231142). Differences in prediction error between ABCG2 genotypes were assessed by ANOVA (P < 0.0001) and by t‐test, i.e. GG vs. GT (P = 0.007), GG vs. TT (P = 0.0002) and GT vs. TT (P = 0.05). D: Box plots of raw prediction error for each genotype of ABCG2 (rs2231142) in those with and without concomitant diuretics. Differences in prediction error between ABCG2 genotypes for those not taking diuretics were assessed by ANOVA (P = 0.0002) and by t‐test, i.e. GG vs. GT (P = 0.0001), GG vs. TT (P = 0.065), and, GT vs. TT (P = 0.0052). Differences in prediction error between ABCG2 genotypes for those taking diuretics were assessed by ANOVA (P = 0.0183) and by t‐test, i.e. GG vs. GT (P = 0.0432), GG vs. TT (P = 0.0188), and, GT vs. TT (P = 0.63). Differences in prediction error for those with and without concomitant diuretic therapy stratified by ABCG2 genotype was assessed by t‐test as follows; GG genotype no diuretic vs. diuretic (P < 0.0001), GT genotype no diuretic vs. diuretic (P < 0.001) and TT genotype no diuretic vs. diuretic (P = 0.0107)
The MPE was significantly influenced by ABCG2 (rs2231142) genotype (MPE 201, 107, 15 mg day−1, GG, GT and TT respectively, P < 0.0001) (Table 2, Figure 1C). This genotype effect appears to have been largely retained in those with and without concomitant diuretics (Table 2, Figure 1D). There was no identifiable influence of the other genotypes tested on dose predictions.
The multilinear regression results are presented in the Supplementary Material. ABCG2 genotype and diuretic use explained 53% of the variability in PE (adjusted R 2 = 0.53, P = 0.0008). Diuretic use was estimated to overpredict the dose by about 170 mg day−1 (P = 0.0084) while the GG genotype overpredicted the dose by 94 mg day−1 (P = 0.036). No other patient factors were found to significantly influence dose predictions in the multilinear regression analysis (see Supplementary Material). Inclusion of interaction terms in the regression to account for correlation between patient factors did not improve the model fit and was not explored further.
Discussion
We have observed that the Otago dosing tool overestimated dose requirements by about 150 mg daily. Concomitant diuretic therapy appears to be a major source of the overprediction. For those patients not taking diuretics, the dosing tool showed reasonable accuracy and precision. The residual overprediction of about 63 mg daily in those not taking diuretics is less than the smallest available tablet size (100 mg) and likely to be of minimal clinical importance. When the diuretic effect in the published dosing table is ignored by using the non‐diuretic dose predictions for all patients, regardless of diuretic use, the overprediction was found to be similar at about 70 mg daily (see Figure S3 in the Supplemental Material). We propose that using the non‐diuretic dose predictions for all patients is a pragmatic solution until the diuretic effect on allopurinol dose requirements can be clarified.
The reason for the overprediction in allopurinol doses by the Otago tool for individuals taking diuretics is unclear. The diuretic effect estimated in the original PKPD model used to develop the tool was a 25% reduction in oxypurinol clearance and 14% increase in baseline SU concentrations on average 8. This is broadly similar to the diuretic effect reported elsewhere for allopurinol therapy 9, 22. The original PKPD model was developed using a dataset of 134 patients. Of these, 24 had pretreatment SU concentrations available and, of those, only 10 were taking a diuretic. This means that the impact of diuretic therapy on urate‐lowering was estimated in the model‐building analysis from only 10 individuals. This introduces the risk of type 1 statistical error and may have resulted in an inflated diuretic effect. Re‐estimation of the diuretic effect using the additional data from the RCT will be needed to clarify the magnitude of the diuretic effect.
ABCG2 genotype had an important impact on dose predictions. Our results align with published work where the T‐allele has also been associated with poor allopurinol response 14, hyperuricemia 15, and the presence of tophi 16. The inference is that patients carrying the T‐allele will have a reduced efflux of urate and will therefore require higher allopurinol doses to achieve the target urate response. Examination of the observed doses required to achieve the urate target in the RCT (evaluation) data used for this analysis found a significant difference between the three genotypes (344, 427 and 489 mg daily for the GG, GT and TT genotype, respectively, P < 0.0001). In the current study, model‐based dose predictions were unbiased for the TT genotype, but were overpredicted for the GT and GG genotype. This suggests that adjustment for the genotype effect, resulting in lower predicted doses for those with the GG and GT genotype, should significantly improve the predictive performance of the model‐based dosing tool.
The small residual overprediction in those subjects not taking diuretics (about 63 mg day−1) could speculatively be attributed to differences in adherence between the cohorts used for model building and model evaluation. The model‐building data included some small, intensively sampled, studies using selected cohorts of patients where good adherence might be expected. To limit the impact of nonadherence in the evaluation data, the first dose to achieve the target urate after the start of the study was used, rather than the dose observed at the end of the study. It is also acknowledged that we cannot rule out type 1 statistical error in the dose predictions in those with different ABCG2 genotypes, given the low numbers with the TT genotype.
In summary, the allopurinol dosing tool produced reasonably accurate and precise maintenance dose predictions for patients not taking diuretics. Concomitant diuretic therapy resulted in an overprediction of allopurinol dose and, for this reason, we recommend that the non‐diuretic dose predictions from the published dosing tool be used for all patients, regardless of diuretic use. Inclusion of ABCG2 genotype and a revised adjustment for diuretics would further improve the performance of the Otago dosing tool.
Competing Interests
All authors have competed the Unified Competing Interest form at http://www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author). D.F.B.W., J.D., A.J.P.‐G., P.T. and A.H. declare no support from any organization for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years; no other relationships or activities that could appear to have influenced the submitted work. N.D. discloses consulting fees, speaker fees or grants from Takeda, Menarini, Teijin, Pfizer, Ardea Biosciences, AstraZeneca, Amgen, Horizon, and Cymabay. L.K.S. declares speaker fees from Amgen and grants from Ardea Biosciences. TRM declares research funding from Ardea Biosciences and consulting fees from Ironwood Pharmaceuticals.
We wish to thank Stephen Duffull and Chihiro Hasegawa, School of Pharmacy, University of Otago for advice on the data analysis. We thank Dr Mei Zhang, Canterbury Health Laboratories for her work with the oxypurinol assay. The RCT and genetics data were originally collected as part of research supported by the Health Research Council of New Zealand.
Supporting information
Table S1 Allopurinol maintenance dose predictions to achieve plasma urate concentrations of <0.36 mmol l−1 (with >75% probability). Reproduced with permission (John Wiley & Sons licence 4 186 130 656 587)
Table S2 Genotype details for the evaluation dataset
Table S3 Results of the multilinear regression analysis (note that the dependent variable is prediction error in mg day−1)
Figure S1 Relationship between prediction error and creatinine clearance (A) and weight (A) stratified by diuretic use
Figure S2 Relationship between the observed and predicted allopurinol maintenance doses where predicted doses of the diuretic effect in the published dosing table is ignored and the non‐diuretic dose predictions used for all patients, regardless of diuretic use
Wright, D. F. B. , Dalbeth, N. , Phipps‐Green, A. J. , Merriman, T. R. , Barclay, M. L. , Drake, J. , Tan, P. , Horne, A. , and Stamp, L. K. (2018) The impact of diuretic use and ABCG2 genotype on the predictive performance of a published allopurinol dosing tool. Br J Clin Pharmacol, 84: 937–943. doi: 10.1111/bcp.13516.
References
- 1. Khanna D, Khanna PP, Fitzgerald JD, Singh MK, Bae S, Neogi T, et al American College of Rheumatology Guidelines for the Management of Gout. Part 1: systematic nonpharmacologic and pharmacologic therapeutic approaches to hyperuricaemia. Arthritis Care Res 2012; 64: 1431–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richette P, Doherty M, Pascual E, Barskova V, Becce F, Castaneda‐Sanabria J, et al 2016 updated EULAR evidence‐based recommendations for the management of gout. Ann Rheum Dis 2017; 76: 29–42. [DOI] [PubMed] [Google Scholar]
- 3. Stamp LK, O'Donnell JL, Zhang M, James J, Frampton C, Barclay ML, et al Using allopurinol above the dose based on creatinine clearance is effective and safe in patients with chronic gout, including those with renal impairment. Arthritis Rheum 2011; 63: 412–421. [DOI] [PubMed] [Google Scholar]
- 4. Stamp LK, Barclay ML, O'Donnell JL, Zhang M, Drake J, Frampton C, et al Relationship between serum urate and plasma oxypurinol in the management of gout: determination of minimum plasma oxypurinol concentration to achieve a target serum urate level. Clin Pharmacol Ther 2011; 90: 392–398. [DOI] [PubMed] [Google Scholar]
- 5. Dalbeth N, Kumar S, Stamp L, Gow P. Dose adjustment of allopurinol according to creatinine clearance does not provide adequate control of hyperuricemia in patients with gout. J Rheumatol 2006; 33: 1646–1650. [PubMed] [Google Scholar]
- 6. Stamp LK, Chapman PT, Barclay ML, Horne A, Frampton C, Tan P, et al A randomised controlled trial of the efficacy and safety of allopurinol dose escalation to achieve target serum urate in people with gout. Ann Rheum Dis 2017; 76: 1522–1528. [DOI] [PubMed] [Google Scholar]
- 7. Stamp LK, Chapman PT, Barclay ML, Horne A, Frampton C, Tan P, et al Allopurinol dose escalation to achieve serum urate below 6 mg/dL: an open‐label extension study. Ann Rheum Dis 2017; 76: 2065–2070. [DOI] [PubMed] [Google Scholar]
- 8. Wright DFB, Duffull SB, Merriman TR, Dalbeth N, Barclay ML, Stamp LK. Predicting allopurinol response in patients with gout. Br J Clin Pharmacol 2016; 81: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stocker SL, McLachlan AJ, Savic RM, Kirkpatrick CM, Graham GG, Williams KM, et al The pharmacokinetics of oxypurinol in people with gout. Br J Clin Pharmacol 2012; 74: 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wright DFB, Stamp LK, Merriman TR, Barclay ML, Duffull SB, Holford NH. The population pharmacokinetics of allopurinol and oxypurinol in patients with gout. Eur J Clin Pharmacol 2013; 69: 1411–1421. [DOI] [PubMed] [Google Scholar]
- 11. Reyes AJ. Cardiovascular drugs and serum uric acid. Cardiovasc Drugs Ther 2003; 17: 397–414. [DOI] [PubMed] [Google Scholar]
- 12. Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al Genome‐wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 2013; 45: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merriman TR, Dalbeth N. The genetic basis of hyperuricaemia and gout. Joint Bone Spine 2011; 78: 35–40. [DOI] [PubMed] [Google Scholar]
- 14. Wen CC, Yee SW, Liang X, Hoffman TJ, Kvale MN, Banda Y, et al Genome‐wide association study identifies ABCG2 (BCRP) as an allopurinol transporter and a determinant of drug response. Clin Pharmacol Ther 2015; 97: 518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kannangara DRW, Phipps‐Green AJ, Dalbeth N, Stamp LK, Williams KM, Graham GG, et al Hyperuricaemia: contributions of urate transporter ABCG2 and the fractional renal clearance of urate. Arthritis Care Res 2016; 75: 1363–1366. [DOI] [PubMed] [Google Scholar]
- 16. He W, Phipps‐Green AJ, Stamp LK, Merriman TR, Dalbeth N. Population‐specific association between ABCG2 variants and tophaceous disease in people with gout. Arthritis Res Ther 2017; 19: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron 1976; 16: 31–41. [DOI] [PubMed] [Google Scholar]
- 18. Hande KR, Noone RM, Stone WJ. Severe allopurinol toxicity. Description and guidelines for prevention in patients with renal insufficiency. Am J Med 1984; 76: 47–56. [DOI] [PubMed] [Google Scholar]
- 19. Sheiner LB, Beal SL. Some suggestions for measuring predictive performance. J Pharmacokinet Biopharm 1981; 9: 503–512. [DOI] [PubMed] [Google Scholar]
- 20. Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S, et al The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acid Res 2018; 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alexander SPH, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD, et al The Concise Guide to PHARMACOLOGY 2017/18: Transporters. Br J Pharmacol 2017; 174: S360–S446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stamp LK, Barclay M, O'Donnell JL, Zhang M, Drake J, Frampton C, et al Furosemide increases plasma oxypurinol without lowering serum urate – a complex drug interaction: implications for clinical practice. Rheumatology 2012; 51: 1670–1676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Allopurinol maintenance dose predictions to achieve plasma urate concentrations of <0.36 mmol l−1 (with >75% probability). Reproduced with permission (John Wiley & Sons licence 4 186 130 656 587)
Table S2 Genotype details for the evaluation dataset
Table S3 Results of the multilinear regression analysis (note that the dependent variable is prediction error in mg day−1)
Figure S1 Relationship between prediction error and creatinine clearance (A) and weight (A) stratified by diuretic use
Figure S2 Relationship between the observed and predicted allopurinol maintenance doses where predicted doses of the diuretic effect in the published dosing table is ignored and the non‐diuretic dose predictions used for all patients, regardless of diuretic use


