Abstract
The thioredoxin system plays a central role in the intracellular redox maintenance in the majority of cells. The canonical system consists of an NADPH-dependent thioredoxin reductase (TrxR) and thioredoxin (Trx), a disulfide reductase. Although Trx is encoded in almost all sequenced genomes of methanogens, its incorporation into their unique physiology is not well understood. Methanosarcina acetivorans contains a single TrxR (MaTrxR) and seven Trx (MaTrx1–MaTrx7) homologues. We previously showed that MaTrxR and at least MaTrx7 compose a functional NADPH-dependent thioredoxin system. Here, we report the characterization of all seven recombinant MaTrxs. MaTrx1, MaTrx3, MaTrx4 and MaTrx5 lack appreciable disulfide reductase activity, unlike previously characterized MaTrx2, MaTrx6 and MaTrx7. Enzyme assays demonstrated that, of the MaTrxs, only the reduction of disulfide-containing MaTrx7 is linked to the oxidation of reduced coenzymes. NADPH is shown to be supplied to the MaTrxR–MaTrx7 system through the oxidation of the primary methanogen electron carriers F420H2 and ferredoxin, indicating that it serves as a primary intracellular reducing system in M. acetivorans. Bioinformatic analyses also indicate that the majority of methanogens likely utilize an NADPH-dependent thioredoxin system. The remaining MaTrxs may have specialized functions. MaTrx1 and MaTrx3 exhibited thiol oxidase activity. MaTrx3 and MaTrx6 are targeted to the membrane of M. acetivorans and likely function in the formation and the reduction of disulfides in membrane and/or extracellular proteins, respectively. This work provides insight into the incorporation of Trx into the metabolism of methanogens, and this reveals that methanogens contain Trx homologues with alternative properties and activities.
Keywords: Methanosarcina, methanogens, anaerobes, archaea, thioredoxin–thioredoxin reductase, cytochromes
Introduction
The strictly anaerobic, methane-producing archaea (methanogens) are the only cellular organisms capable of biological methane production (methanogenesis), an important step in the global carbon cycle [1, 2]. Methanogens are ubiquitous microbes, found in diverse environments ranging from the human gastrointestinal tract to the Antarctic [2, 3]. No matter the environment, significant methane production by methanogens only occurs under strictly anaerobic conditions due to the requirement of a large number of redox-sensitive enzymes, coenzymes and cofactors for methanogenesis [4]. Thus, methanogens require an intracellular electron transfer system(s) to maintain a reduced intracellular environment. Although the enzymes and factors involved in energy-conserving electron transfer reactions during methanogenesis have been fairly well-characterized [2, 4, 5], the enzymes and factors involved in intracellular electron transfer for redox maintenance, biosynthesis and protection from oxidative stress are less understood.
Thiol-disulfide oxidoreductases play a central role in the intracellular redox maintenance of cells. In particular, the vast majority of cells rely on thioredoxin (Trx), a small (~12 kDa) thiol-disulfide oxidoreductase, to maintain a reduced intracellular environment [6]. The canonical thioredoxin system consists of a thioredoxin reductase (TrxR), which uses reducing equivalents from NADPH, generated from metabolism, to reduce the active site cysteines within a CXXC motif of Trx. Trx primarily catalyses the reduction of disulfides, but it can also provide reductant for other enzymes. As such, Trxs are typically capable of reducing disulfides in a diverse number of proteins and are involved in physiological processes, in addition to general redox maintenance and protection during oxidative stress [7]. For example, Trx provides reducing equivalents to biosynthetic enzymes, such as ribonucleotide reductase [7]. There are a number of more complex and specialized Trx-related systems. In particular, many cells also contain glutaredoxin (Grx), in addition to Trx. Grx is structurally and functionally similar to Trx, but it receives reducing equivalents from glutathione that is reduced by an NADPH-dependent glutathione reductase [8]. However, the glutaredoxin system is primarily found in aerobes and is typically not present in strict anaerobes [8]. Methanogens do not contain glutathione, indicating the lack of a functional glutaredoxin system [9–11]. Recent evidence showed that Trx homologues are present in almost every sequenced methanogen genome, indicating that Trx is likely the primary thiol-disulfide oxidoreductase involved in redox maintenance in methanogens [12, 13]. Indeed, Trx was shown to target a large number of proteins, including those involved in methanogenesis, in the methanogen Methanocaldococcus jannaschii [13]. Yet, how Trx is assimilated into the metabolism of methanogens, in particular, the enzyme(s) and coenzyme(s) involved in providing reducing equivalents to Trx, is less understood. Importantly, NADPH is not directly generated by methanogenesis. Instead, reduced coenzyme F420H2, a 5′-deazaflavin derivative, and reduced ferredoxin are produced during methanogenesis [2, 4]. Methanogens may therefore directly use F420H2 and/or reduced ferredoxin to provide reducing equivalents to Trx or alternatively generate NADPH from the oxidation of F420H2 and ferredoxin.
Previous work by our group has demonstrated that the methanogen, Methanosarcina acetivorans, contains seven Trx homologues (MaTrx1–MaTrx7) and a single TrxR (MaTrxR). Recombinant MaTrx2, MaTrx6 and MaTrx7 have catalytic disulfide reductase activity, and recombinant MaTrxR is specific for NADPH as an electron donor [12]. Of the three characterized MaTrxs, only MaTrx7 was reduced by MaTrxR, indicating that M. acetivorans possesses a canonical Trx system composed of NADPH, MaTrxR and at least MaTrx7. Here, we report the characterization of all seven MaTrxs, including reduction by MaTrxR, analyses of electron donors and alternative activities. Results support that the NADPH-dependent thioredoxin system, composed of MaTrxR–MaTrx7, is likely the general intracellular reducing system in M. acetivorans. Bioinformatic analyses indicate that the majority of methanogens contain an NADPH-dependent TrxR, suggesting that the canonical Trx system is used by the majority of extant methanogens. The remaining MaTrxs likely have specialized functions, including two (MaTrx3 and MaTrx6) that are associated with the membrane of M. acetivorans.
Methods
Cloning of M. acetivorans Trx homologue genes
The genes encoding MA_RS05020 (MaTrx1), MA_RS19290 (MaTrx3), MaTrx3ΔSp (deletion of signal peptide amino acids 1–34), MA_RS20550 (MaTrx4) and MA_RS20570 (MaTrx5) were cloned into the Escherichia coli expression vector pET28a as previously described for MaTrx2, MaTrx6, MaTrx6Δsp and MaTrx7 [12]. Plasmids containing matrx1, matrx3, matrx3ΔSp, matrx4 and matrx5 were verified by DNA sequencing and named pDL342, pDL343, pDL344, pDL345 and pDL346, respectively.
Purification of recombinant proteins
Proteins were expressed in E. coli Rosetta DE3 (pLacI) transformed with pDL342, pDL343, pDL344, pDL345 or pDL346. Each E. coli expression strain was grown in Luria broth medium and protein expression was induced with 500 µM IPTG at OD600 of 0.5–0.7. The induced cultures were incubated at 25 °C for 16 h. The cells were harvested, and recombinant protein was purified as described previously [12], except that the His6 tag could not be cleaved from MaTrx4. Purified recombinant protein was stored in buffer A [50 mM Tris and 150 mM NaCl (pH 7.2)] at −80 °C.
Generation of oxidized and reduced MaTrxs
MaTrxs were incubated anaerobically at 25 °C for 20 min in buffer A containing a 10 : 1 molar excess of either H2O2 or DTT, to generate oxidized MaTrx (MaTrxox) and reduced MaTrx (MaTrxred), respectively. After incubation, residual H2O2 or DTT was removed by buffer exchange with an NAP-5 column (GE Healthcare). The number of thiols were quantified in each MaTrx sample using 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). The standard assay contained 175 µM DTNB in buffer A and MaTrx (5–60 µM). After 15 min of anaerobic incubation, the absorbance at 412 nm was used to calculate the thiol concentration using ε412=14 150 M−1 cm−1 [14]. MaTrxox samples were analysed by non-reducing 15 % SDS-PAGE for the presence of oligomers due to the formation of intermolecular disulfides.
Enzyme activity assays with M. acetivorans cell-free lysate
M. acetivorans was grown in high-salt medium supplemented with 125 mM methanol and 0.025 % Na2S (w/v) to an OD600 of 0.75 [15]. Unless stated otherwise, all subsequent manipulations were done inside an anaerobic chamber (Coy Laboratories). Cells were harvested by centrifugation for 10 min at 16 000 g and 10 °C. The cell pellet was resuspended in buffer A supplemented with protease inhibitors (1 mM benzamidine and 1 mM PMSF). Cell suspensions were stored at −80 °C in anaerobic vials. Cell suspensions were thawed on ice and sonicated to lyse cells. Cell lysate was clarified by centrifugation at 16 000 g for 10 min at 25 °C and the soluble fraction was stored at −80 °C in anaerobic vials.
F420H2 : NADP oxidoreductase (Fno) activity in M. acetivorans cell lysate was determined by measuring the NADP-dependent oxidation of F420H2. F420, provided by Dr Lacy Daniels (Texas A&M University, Kingsville, TX, USA), was chemically reduced to F420H2 using sodium borohydride as previously described [16]. Fno activity assays were performed in buffer A containing M. acetivorans lysate (100 µg total protein) and 70 µM F420H2. After equilibration, reactions were initiated by the addition of 1 mM NADP or an equivalent volume of buffer A as a control. The oxidation of F420H2 was monitored at 420 nm for 10 min. The rate of F420H2 oxidation was determined using ε420=40 000 M−1 cm−1 [17].
To test for the presence of a functional ferredoxin : NADP reductase (Fnr), CO-dependent reduction of NADP by M. acetivorans was assayed. Lysate was prepared from acetate-grown cells as described above. To exclude the possibility of F420 mediating electron transfer between ferredoxin and NADP [18], low molecular weight compounds were removed from the lysate by two consecutive sixfold concentration and dilution steps using buffer A with a 10 kDa cutoff Nanosep spin column (Pall Corporation). Cell lysate was pre-incubated with either CO or N2 by transferring lysate to a 2 ml serum vial and flushing the headspace with CO or N2 for 2 min, followed by incubation on ice for 30 min. Assays were performed in a sealed quartz cuvette containing 400 µl of buffer A with a headspace of either N2 or CO. Lysate (100 µg) was added to the cuvettes and reactions were initiated by the addition of 500 µM NADP to each sealed cuvette. The amount of NADPH produced over time was determined using ε340=6220 M−1 cm−1.
MaTrxox-dependent oxidation of NADH, NADPH and F420H2 by M. acetivorans cell lysate was measured spectrophotometrically by monitoring the change in absorbance at 340 nm for NAD(P)H or 420 nm for F420H2. Assays were performed under anaerobic conditions in cuvettes containing 300 µg cell lysate, 70–100 µM MaTrx1–7ox and either 100 µM NAD(P)H or 70 µM F420H2 in buffer A (total volume of 100 µl). Assay mixtures were incubated in the absence of MaTrxox until a stable baseline was obtained, then reactions were initiated by the addition of MaTrxox. The amount of NAD(P)H or F420H2 consumed over time was determined using ε340=6220 M−1 cm−1 and ε420=40 000 M−1 cm−1, respectively.
MaTrx activity assays
Disulfide reductase activity of each MaTrx was measured with DTT as an electron donor and insulin as the substrate as previously described [12]. Specific activity is reported as ΔA650 min−2 mg−1 ×10−3 after subtraction of the background rate of insulin reduction by DTT alone.
Disulfide isomerase and thiol oxidase activity of MaTrxs was determined using bovine pancreatic RNase A (Amresco) as a substrate. Reduced RNase A (rRNaseA) was specifically used as the substrate to measure thiol oxidase activity, and it was generated as previously described [19], except that 20 mg of RNase A was brought into an anaerobic chamber and dissolved in 2 ml of 6 M guanidine HCl, 2 mM EDTA, 50 mM Tris/HCl (pH 9.0) and 0.2 M DTT. After 2 h of incubation, the rRNaseA was buffer-exchanged into buffer A using a PD-10 column (GE Healthcare) and stored in sealed vials at −80 °C. Scrambled RNase A (scRNaseA) was used as the substrate to measure disulfide isomerase activity and was generated as described previously [20]. The in vitro refolding of rRNaseA and scRNaseA to generate active RNase A was performed as previously described [19]. Briefly, refolding buffer contained 5 µM rRNaseA or scRNaseA, 1 mM GSH and 1.11 mM GSSG yielding a redox potential of −150 mV based on the Nernst equation. E. coli DsbA was used as a positive control and was provided as a gift by Dr James Bardwell (University of Michigan, Ann Arbor, MI, USA). RNase refolding assays contained 20 µM DsbA or MaTrx1–MaTrx7. All refolding assays were performed in triplicate for 3 min at 15 °C and RNase A activity after incubation with DsbA or MaTrx1–MaTrx7 was compared to activity recovered in refolding buffer alone to account for non-enzymatic disulfide formation or rearrangement due to GSH/GSSG. RNase A activity was determined by monitoring the amount of RNA degraded over time using the methylene blue assay as previously described [21]. The RNA substrate was prepared by dissolving 100 mg of Torula yeast RNA (Sigma) in 10 ml of anaerobic 0.1 M MOPS/HCl (pH 7.5) and 2 mM EDTA (buffer B). Preparation of methylene blue binding buffer was done by adding 1 mg of methylene blue to 100 ml of anaerobic buffer B and the absorbance at 688 nm was adjusted to 0.5±0.02 with buffer B. A standard curve of methylene blue bound to RNA was generated using a range of 0–1000 µg of RNA, and the absorbance at 688 nm was determined. The Excel kinetics modelling add-in was used to obtain the maximum absorbance change (Vmax) and the concentration of RNA needed to obtain ½Vmax (Km). The Michaelis–Menten equation was used to calculate the concentration of RNA (S) at a given absorbance (V).
Generation of M. acetivorans strains expressing FLAG-tagged MaTrx3 or MaTrx6 for localization analysis
PCR was used to amplify matrx3, maTrx3ΔSp, matrx6 and maTrx6ΔSp. Each forward primer contained a 5′ NdeI site and each reverse primer encoded a C-terminal FLAG tag followed by a HindIII site. The PCR product was digested with NdeI and HindIII and ligated with similarly digested pJK027A [22]. The resulting plasmids containing matrx3-FLAG, maTrx3ΔSp-FLAG, matrx6-FLAG and maTrx6ΔSp-FLAG were named pDL350, pDL353, pDL348 and pDL349 respectively. M. acetivorans strain WWM73 was transformed with pDL350, pDL353, pDL348 and pDL349 as described previously [23]. Successful integration of the plasmid into the chromosome of each strain was determined as previously described [22], and the resulting strains were named DJL80 (MaTrx6-FLAG), DJL81 (MaTrx6ΔSp-FLAG), DJL82 (MaTrx3-FLAG) and DJL83 (MaTrx6ΔSp-FLAG). These strains allow the tetracycline-inducible chromosomal expression of each MaTrx-FLAG.
Immunodetection of FLAG-tagged MaTrx3 and MaTrx6 in membrane and soluble fractions of M. acetivorans strains
Cultures of strains DJL80–DJL83 were grown in high-salt medium (100 ml) supplemented with 125 mM methanol and 0.025 % Na2S (w/v). Tetracycline (100 µg ml−1) was added where indicated to induce expression of the MaTrx-FLAG in each strain. The cells were harvested by centrifugation when the OD600 reached 0.5–0.75. Pelleted cells were resuspended in a volume of buffer A containing 1 mM benzamidine and 1 mM PMSF, and they were normalized based on OD600. Cell suspensions were stored in microfuge tubes at −80 °C.
Methanogen cell membrane and soluble (cytoplasmic) fractions were separated in a manner similar to that described previously [24]. Frozen cells of strains DJL80–DJL83 were lysed by five freeze/thaw cycles. After lysis, 10 units of RQ1 DNase 1 (Promega) was added to each tube and incubated at 37 °C for 25 min, followed by centrifugation at 10 500 g for 5 min at 10 °C. The supernatant was removed and centrifuged a second time. The supernatant (soluble fraction) was removed and membranes were pelleted by centrifugation at 70 000 g for 1 h at 10 °C in 1.5 ml safe-lock Eppendorf tubes. The pellet (membrane fraction) was washed in 500 µl of 25 mM Tris (pH 7.5). Both the membrane fraction and the soluble fraction were separately centrifuged a second time at 70 000 g for 1 h at 10 °C to further remove contaminating proteins from each fraction. The supernatant was removed from the centrifuged soluble fractions and was used as the final soluble fraction for each strain. The pellet from each centrifuged membrane fraction was resuspended in a small volume of 25 mM Tris, 8 M urea (pH 7.5), then diluted to 150 µl with 25 mM Tris (pH 7.5) and was used as the final membrane fraction for each strain. Total protein was quantified for all fractions using the Bradford assay with BSA as a standard. For detection of each MaTrx-FLAG by Western blot, identical amounts of protein for the membrane fractions and soluble fractions of strains DJL80/DJL81 and DJL82/DJL83 were analysed by 15 % SDS-PAGE. Protein was transferred to a PVDF membrane and Western blotting was performed using standard protocols using an α-FLAG tag primary antibody (Rockland Immunochemicals) and an HRP-conjugated secondary antibody (Promega). An enhanced chemiluminescent substrate (Thermo Scientific) was used for detection.
Determination of the cytochrome c content of membrane fractions
M. acetivorans strains DJL80–DJL83 were grown with methanol to late log phase, cells were harvested and membrane fractions were generated as described above. The production of the previously detected 25 kDa cytochrome c in M. acetivorans [25] was quantified by SDS-PAGE analysis followed by densitometry of bands in haem-stained gels, similar to that previously described [26]. Loaded samples (26 µg) were normalized based on total protein. SDS-PAGE analysis was carried out on 15 % polyacrylamide gels run at 90 V at 10 °C. Gels were stained for covalently bound haem using o-dianisidine as previously described [27]. Due to observed differences in staining efficiencies between gels, the cytochrome c content of membrane fractions from each strain grown under non-inducing and inducing conditions was compared in a single gel. Each gel was loaded with duplicate samples of membrane fractions generated from two independent cultures for each condition (total samples=4). Gels were imaged using a UMAX Powerlook 2100XL tri-linear CCD scanner at 400 dpi. Intensity of the band corresponding to the 25 kDa cytochrome c was determined using ImageJ. Using the method described by Gassmann et al. [28], only the central 30 % of the band was used to calculate intensity. For comparison, the intensity of cytochrome c determined in membrane fractions of each non-induced culture was set to 100 arbitrary units.
Results
Analysis of MaTrx disulfide reductase activity and reduction by MaTrxR
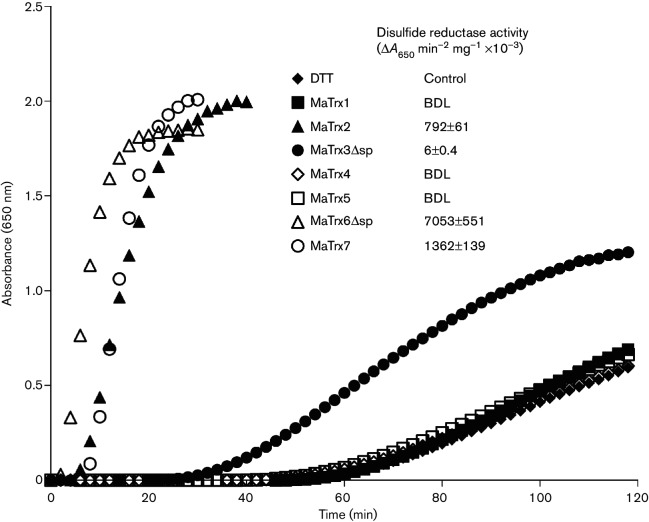
Recombinant MaTrx1, MaTrx3Δsp, MaTrx4 and MaTrx5 were each purified to homogeneity as described for recombinant MaTrx2, MaTrx6Δsp and MaTrx7 [12]. Each MaTrx was assayed for disulfide reductase activity with insulin as a substrate. Unlike MaTrx2, MaTrx6Δsp and MaTrx7, none of the additional MaTrxs exhibited significant insulin disulfide reductase activity (Fig. 1). Although MaTrx3Δsp exhibited low activity, it is likely not physiologically relevant when compared to the activities of MaTrx2, MaTrx6Δsp and MaTrx7.
Fig. 1.
Insulin disulfide reductase activity of MaTrxs. Assays were performed in anaerobic buffer containing DTT (1 mM) alone or with the following: MaTrx1 (20 µM), MaTrx2 (10 µM), MaTrx3Δsp (20 µM), MaTrx4 (20 µM), MaTrx5 (20 µM), MaTrx6Δsp (5 µM) or MaTrx7 (5 µM). BDL, below detection limit.
Using insulin disulfide reduction assays, MaTrxR was previously shown to reduce MaTrx7, but not MaTrx2 or MaTrx6Δsp [12]. The lack of insulin disulfide reduction by MaTrx1, MaTrx3Δsp, MaTrx4 and MaTrx5 occludes using the insulin reduction assay. Thus, MaTrxs with active site disulfides were generated to test as substrates for MaTrxR. H2O2 was used to generate MaTrxs with oxidized active site cysteines (MaTrxox). With the exception of MaTrx1, the number of thiols per monomer for each MaTrxox was close to zero (Table 1), consistent with complete thiol oxidation. MaTrx1 contains four cysteines, unlike the other MaTrxs, and the data indicate that at least two of these cysteines cannot be oxidized by H2O2. This is possibly due to these cysteines being inaccessible (i.e. buried in the protein) to H2O2. Importantly, non-reducing SDS-PAGE of each MaTrxox showed that each protein was monomeric (Fig. 2), consistent with the presence of intramolecular active site disulfides after oxidation with H2O2. If intermolecular disulfides were formed during oxidation with H2O2, then higher molecular weight species (e.g. dimers) would have been observed by non-reducing SDS-PAGE. Moreover, similar levels of disulfide reductase activity were observed with MaTrx2ox, MaTrx6Δspox and MaTrx7ox compared to non-oxidized samples (data not shown), indicating that the proteins were not damaged beyond active site oxidation by incubation with H2O2. Incubation of each MaTrx with DTT resulted in an increase in the number of thiols detected (Table 1), revealing that each protein contains cysteines capable of thiol-disulfide exchange. Each MaTrxox was then tested for reduction by MaTrxR. Only the addition of MaTrx7ox to MaTrxR resulted in significant NADPH oxidation (Table 1). These results suggest that MaTrx1, MaTrx3Δsp, MaTrx4 and MaTrx5 are capable of thiol-disulfide exchange, but they are likely not disulfide reductases. Moreover, MaTrxR is specific for MaTrx7 and is incapable of reducing any of the additional MaTrxs.
Table 1. MaTrx thiol-disulfide exchange ability and specificity of MaTrxR.
| MaTrx | Total cysteines | MaTrxox thiols* | MaTrxred thiols† | NADPH oxidation‡ (MaTrxR + MaTrxox) |
|---|---|---|---|---|
| MaTrx1 | 4 | 3.20±0.35 | 4.58±0.30 | BDL |
| MaTrx2 | 2 | 0.05±0.02 | 1.66±0.77 | nd |
| MaTrx3Δsp | 2 | 0.05±0.03 | 2.04±0.31 | BDL |
| MaTrx4 | 2 | 0.06±0.14 | 1.04±0.07 | 20±2 |
| MaTrx5 | 2 | 0.11±0.03 | 1.83±0.17 | BDL |
| MaTrx6Δsp | 2 | 0.21±0.05 | 1.14±0.31 | nd |
| MaTrx7 | 2 | 0.13±0.04 | 1.4±0.08 | 829±134 |
BDL, below detection limit; nd, not determined.
*Thiols per MaTrx monomer after incubation with H2O2 as determined by the DTNB assay.
†Thiols per MaTrx monomer after incubation with DTT as determined by the DTNB assay.
‡nmol NADPH oxidized min−1 (mg oxidized MaTrx)−1.
Fig. 2.
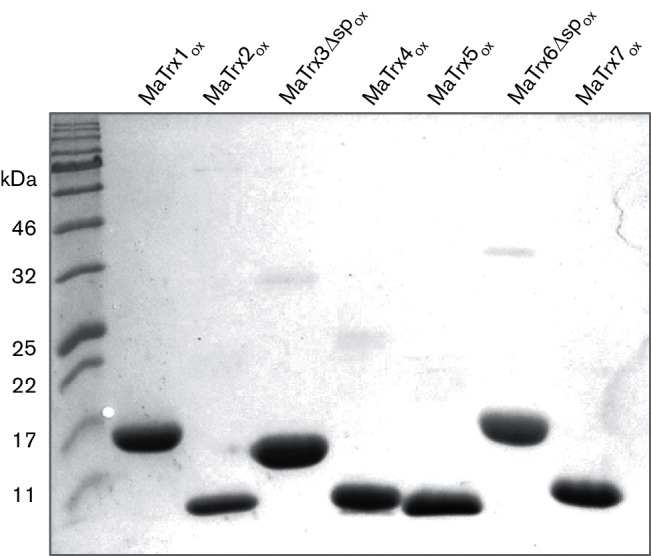
Non-reducing SDS-PAGE of H2O2-oxidized MaTrxs. Each MaTrxox (10 µg) was separated by 15 % SDS-PAGE in the absence of a reducing agent.
Examination of the ability of NADH, NADPH and F420H2 to supply electrons for the reduction of MaTrxs
Of the seven MaTrxs, only MaTrx7ox was reduced by MaTrxR, indicating that, if the other MaTrxs function as specific disulfide reductases, they must have a different redox partner(s) and/or electron donor(s). Thus, to test for the presence of enzymes that mediate the oxidation of the electron carrier NADH, NADPH or F420H2 and reduction of the disulfide in MaTrxsox, oxidation assays with cell lysates were performed. Only the addition of MaTrx7ox to M. acetivorans cell lysate resulted in the statistically significant oxidation of both NADPH (331±18 pmol min−1 mg−1) and F420H2 (138±12 pmol min−1 mg−1). All assays with other oxidized MaTrxs did not result in activity that was statistically significantly above the background. The addition of MaTrx7red to cell lysate also did not result in the significant oxidation of either NADPH or F420H2, confirming that oxidation of both electron donors was due to the reduction of the disulfide in MaTrx7ox. These data indicate that reduction of the other MaTrxs is not linked to the oxidation of NADH, NADPH or F420H2. Since both NADPH and F420H2 were oxidized by the addition of MaTrx7ox to cell lysate, it is possible that MaTrx7ox is reduced by an unknown enzyme that directly oxidizes F420H2. More likely, the oxidation of F420H2 to generate NADPH needed by MaTrxR is mediated by the Fno homologue (MA_RS22115) encoded in the genome of M. acetivorans. The addition of recombinant MaTrxR to cell lysate resulted in a fivefold increase in the rate of MaTrx7ox-dependent F420H2 oxidation (787±60 pmol min−1 mg−1), consistent with the transfer of reducing equivalents from F420H2 to MaTrx7ox involving MaTrxR and therefore NADPH. Overall, these results confirm that the in vivo reduction of MaTrx7ox is dependent on MaTrxR, and link the production of NADPH to the oxidation of F420H2, through the activity of Fno, presumably the product of the MA_RS22115 gene.
M. acetivorans can generate NADPH for MaTrxR from the activities of Fno and Fnr
The results from the incubation of MaTrx7ox with cell lysates indicate that NADPH is produced by the oxidation of F420H2, consistent with the activity of Fno. To confirm that M. acetivorans contains a functional Fno, cell lysates were examined for NADP-dependent F420H2 oxidation. Lysate from M. acetivorans cells exhibited NADP-dependent F420H2 oxidation at a rate of 6.8±1 nmol min−1 mg−1, revealing the presence of a functional Fno. During growth of M. acetivorans with acetate and CO, ferredoxin is the primary electron carrier [1, 29]. Thus, M. acetivorans would likely need to generate NADPH for MaTrxR from the oxidation of ferredoxin. MA_RS19715–MA_RS19720 in the genome of M. acetivorans encode a homologue of a two-subunit Fnr, similar to NfnAB from clostridia [30]. CO-dependent reduction of NADP by cell lysates, as described in Methods, was used to examine for the presence of Fnr activity in M. acetivorans. Carbon monoxide dehydrogenase from Methanosarcina oxidizes CO to CO2 with the concomitant reduction of ferredoxin [31, 32]. An approximately threefold higher rate of NADP reduction was observed when M. acetivorans cell lysates were provided CO compared to N2 as a control (Fig. 3). These data reveal that M. acetivorans can generate NADPH by the oxidation of F420H2 or reduced ferredoxin.
Fig. 3.
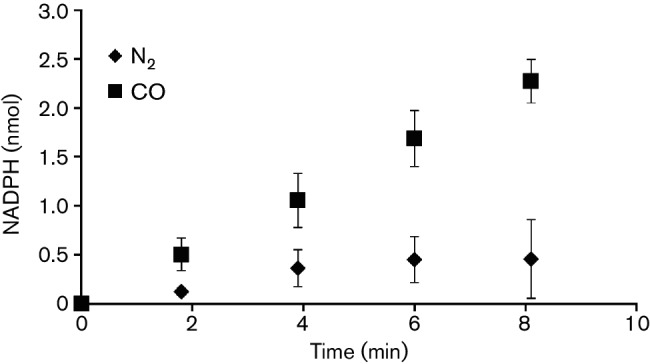
CO-dependent reduction of NADP by M. acetivorans cell lysate. Assays were performed in sealed anaerobic cuvettes containing lysate (100 µg), NADP (0.5 mM) and a headspace of either N2 or CO. Data points are the mean (±sd) of triplicate assays.
The majority of sequenced methanogens encode NADPH-dependent TrxR, Fno and Fnr
To assess the prevalence of an NADPH-dependent thioredoxin system in methanogens, we analysed sequenced methanogen genomes currently available in the NCBI database for the presence of TrxR, Fno and Fnr. Using MaTrxR as a blast search query, a TrxR homologue with >30 % identity and >70 % coverage was found in 64 of the 75 analysed methanogen genomes (83 %). Table S1 (available in the online Supplementary Material) shows the prevalence of TrxR among methanogens at the genus level. We next assessed the electron donor preference of the methanogen TrxR homologues by aligning the active site region of the 64 sequences (Fig. S1) and examining them for the presence of the NADPH-binding motifs GXGXXA [33] and VXXXHRRRDXXRA, an arginine-rich sequence found in E. coli TrxR [34]. Characterized archaeal, bacterial and eukaryotic TrxRs which have the consensus GXGXXA motif can accept reducing equivalents from NADPH [35–39]. However, the TrxR from the archaeon Thermoplasma acidophilum lacks the consensus GXGXXA motif and cannot accept reducing equivalents from NADPH even though it can reduce T. acidophilum Trx [34]. The physiological electron donor to T. acidophilum TrxR is unknown. Thus, conservation of the GXGXXA motif appears critical to the ability of TrxR to use NADPH as an electron donor. The GXGXXA motif is present in 55 of the 64 methanogen TrxR homologues (86 %) (Fig. S1), indicating that the majority of methanogens contain an NADPH-dependent TrxR. However, the GXGXXA motif is not conserved in TrxR homologues from certain methanococci. Thus, NADPH is likely not the electron donor to TrxR in a small subset of the methanogens.
Using M. acetivorans MA_RS22115 (Fno), a blast search of methanogen genomes revealed that all TrxR-containing methanogens encode a homologue of Fno (Table S1). Likewise, using M. acetivorans MA_RS19715 (Fnr), a blast search of methanogen genomes revealed that all TrxR-containing methanogens, with the exception of species of the genera Methanothermus and Methermicoccus, encode a homologue of Fnr (Table S1). These data suggest that all methanogens have the ability to direct reducing equivalents generated from methanogenesis (F420H2 and reduced ferredoxin) to an NADPH-dependent thioredoxin system.
MaTrx1 and MaTrx3 have thiol oxidase activity similar to E. coli DsbA
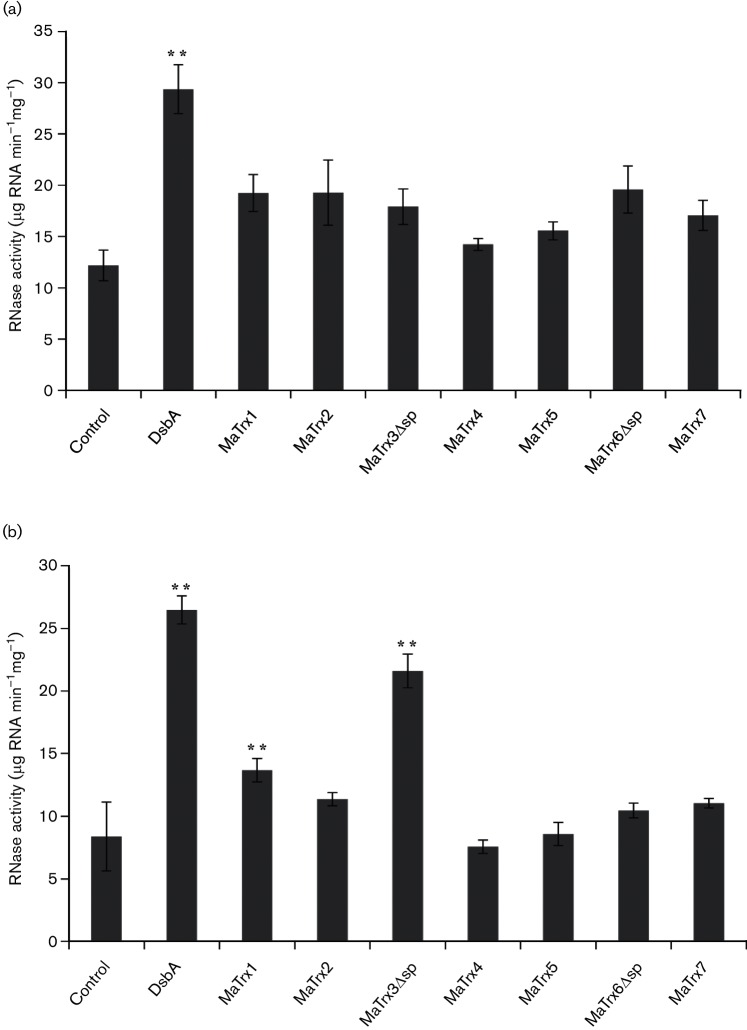
A number of Trx-like proteins are involved in activities other than disulfide reduction, including protein disulfide isomerase and disulfide-forming (thiol oxidase) activities [40]. For example, DsbA is a Trx-like protein found in the periplasm of E. coli that is capable of both rearranging (disulfide isomerization) and forming disulfides by the oxidation of thiols in proteins [41]. Thus, since four of the seven MaTrxs lack insulin disulfide reductase activity but appear capable of thiol-disulfide exchange, the MaTrxs were tested for disulfide isomerase and thiol oxidase activities using RNase as a substrate. RNase requires disulfides in the correct configuration for activity; thus, RNase with incorrect disulfides (scrambled) and RNase with thiols (reduced) can be used as substrates to measure the disulfide isomerase and thiol oxidase activities, respectively [19]. In comparison to E. coli DsbA, which was included as a positive control, none of the MaTrxs exhibited statistically significant disulfide isomerase activity with scrambled RNase above the control level determined with buffered GSH/GSSG alone (Fig. 4a). However, both MaTrx1 and MaTrx3Δsp showed statistically significant thiol oxidase activity with reduced RNase, with the activity of MaTrx3Δsp comparable to that of DsbA (Fig. 4b). These data reveal that MaTrx1 and MaTrx3 are thiol-disulfide oxidoreductases, but they likely serve as thiol oxidases to form disulfides rather than reduce disulfides, similar to DsbA. The enzymatic activities of MaTrx4 and MaTrx5 are unknown.
Fig. 4.
Analysis of disulfide isomerase and disulfide-forming activities of MaTrxs using scrambled and reduced RNase. (a) RNase activity of disulfide-scrambled RNase after incubation in GSH/GSSG redox buffer alone (control), with DsbA or with the indicated MaTrx. (b) RNase activity of reduced RNase after incubation in GSH/GSSG redox buffer alone (control), with DsbA or with the indicated MaTrx. Data points are the mean (±sd) of triplicate assays. **P≤0.002 (t-test).
MaTrx3 and MaTrx6 contain a signal peptide that localizes each protein to the membrane of M. acetivorans
Both MaTrx3 and MaTrx6 contain an N-terminal signal peptide predicted to target each protein across the membrane of M. acetivorans [12]. Consistent with this prediction, expression of recombinant MaTrx3 and MaTrx6 in E. coli results in accumulation of each protein primarily in the insoluble fraction, whereas expressions of recombinant MaTrx3Δsp and MaTrx6Δsp are found in the soluble fraction of E. coli lysates (data not shown). To examine the importance of the signal peptide to the localization of MaTrx3 and MaTrx6 directly in M. acetivorans, strains were generated capable of expressing MaTrx3, MATrx3Δsp, MaTrx6 or MaTrx6Δsp with a C-terminal FLAG tag (MaTrx-FLAG) to allow immunodetection by Western blot. Strain DJL80 contains MaTrx6-FLAG, strain DJL81 contains MaTrx6Δsp-FLAG, strain DJL82 contains MaTrx3-FLAG and strain DJL83 contains MaTrx3Δsp-FLAG. Growth of each strain under inducing conditions (+ tetracycline) did not alter growth rate or yield but led to the immunodetection of a protein consistent with the predicted molecular weight of each MaTrx-FLAG that was absent in cells grown under non-inducing conditions (data not shown). These data indicate that each MaTrx-FLAG is expressed and does not alter the general growth of each strain. Therefore, lysate from induced cells of each strain was separated by centrifugation into soluble and membrane fractions, followed by Western blot analysis using anti-FLAG antibodies. MaTrx6Δsp-FLAG was only detected in the soluble fraction of DJL81 cells, whereas MaTrx6-FLAG was detected in both the membrane and soluble fractions of DJL80 cells (Fig. 5a). Similarly, MaTrx3Δsp-FLAG was only detected in the soluble fraction of DJL83 cells, whereas MaTrx3-FLAG was detected in both the membrane and soluble fractions of DJL82 cells (Fig. 5b). These results demonstrate that the N-terminal signal peptide of MaTrx6 and MaTrx3 directs each protein to the membrane of M. acetivorans. This is the first evidence of membrane-localized Trx homologues in a methanogen.
Fig. 5.
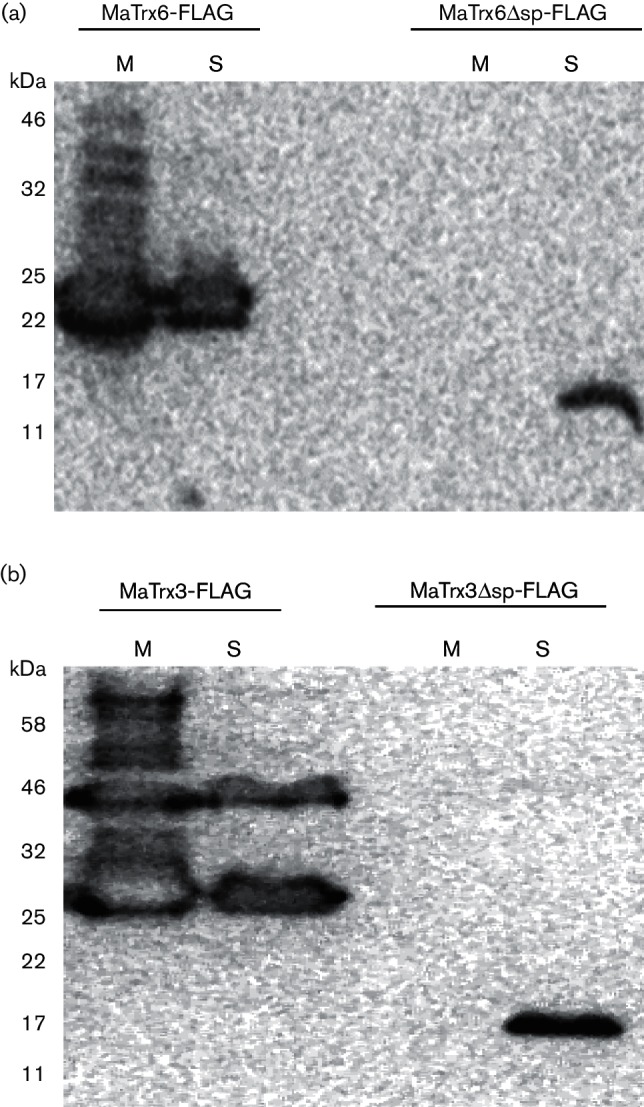
Western blot analysis of fractionated cell lysates from M. acetivorans strains expressing FLAG-tagged MaTrx3 and MaTrx6. (a) Western blot analysis using anti-FLAG antibodies of membrane (M) and soluble (S) fractions of DJL80 cells containing MaTrx6-FLAG (18.8 kDa) and DJL81 cells containing MaTrx6Δsp-FLAG (15.8 kDa). Total protein in samples: M, 7.5 µg; S, 50 µg. (b) Western blot analysis using anti-FLAG antibodies of membrane (M) and soluble (S) fractions of DJL82 cells containing MaTrx3-FLAG (20.3 kDa) and DJL83 cells containing MaTrx3Δsp-FLAG (16.7 kDa). Total protein in samples: M, 5 µg; S, 35 µg.
Expression of FLAG-tagged MaTrx3 and MaTrx6 alters the level of cytochrome c in M. acetivorans
Among methanogens, homologues of MaTrx3 and MaTrx6 are restricted to Methanomicrobia [12], the only methanogens that contain cytochromes, including cytochrome c [2]. In other organisms, extracellular and membrane-associated Trx-like proteins serve key roles in the maturation of cytochrome c, which has haem covalently attached to thiols of cysteines typically within a CXXCH motif [26, 42]. Thus, to provide initial insight into the potential for MaTrx3 and MaTrx6 to play a similar role in M. acetivorans, the effect of the increased expression of FLAG-tagged MaTrx3 and MaTrx6 on the level of cytochrome c was examined in M. acetivorans strains DJL80–DJL83. The genome of M. acetivorans encodes three predicted cytochrome c proteins, two of which have been experimentally detected [25]. A 25 kDa cytochrome c is produced during growth with methanol or acetate and a 55 kDa cytochrome c is produced only during growth with acetate [25]. The effect of the expression of FLAG-tagged MaTrx3 and MaTrx6 on the level of the 25 kDa cytochrome c in methanol-grown cells was determined by densitometry of bands in haem-stained SDS-PAGE gels. Gels were loaded with the membrane fraction of cells grown under conditions that do not induce (− tetracycline) or induce (+ tetracycline) expression of each FLAG-tagged MaTrx. A similar level of cytochrome c was detected in the membrane fractions from induced and non-induced cells of strains DJL81 and DJL83 (Table 2), which express MaTrx6Δsp-FLAG or MaTrx3Δsp-FLAG in the cytoplasm, respectively. In contrast, the membrane fraction of induced DJL80 cells that express MaTrx6-FLAG, shown to localize to the membrane (Fig. 5), contains approximately 50 % less cytochrome c than non-induced cells of DJL80. Furthermore, the membrane fraction of induced cells of DJL82 that express MaTrx3-FLAG, also shown to localize to the membrane (Fig. 5), contains approximately 50 % more cytochrome c than non-induced DJL82 cells. Thus, the expression of membrane-localized MaTrx3-FLAG and MaTrx6-FLAG alters the level of the haem-containing 25 kDa cytochrome c in M. acetivorans. However, altered cytochrome c content as a result of indirect effects, such as changes in membrane protein content due to the presence of MaTrx3-FLAG and MaTrx6-FLAG, cannot be ruled out though it is important to note that expression of MaTrx6-FLAG had an opposite effect on the level of cytochrome c (decreased), compared to expression of MaTrx3-FLAG, which resulted in an increase in cytochrome c. This difference indicates that the catalytic activities of MaTrx6 (disulfide reductase) and MaTrx3 (thiol oxidase) may play a role in the altered cytochrome c levels.
Table 2. The effect of the expression of FLAG-tagged MaTrx3 and MaTrx6 on the levels of the 25 kDa cytochrome c in membrane fractions of methanol-grown M. acetivorans.
| Strain | Tetracycline | Level of cyt c* |
|---|---|---|
| DJL80 (MaTrx6-FLAG) | − | 100±10 |
| + | 57±16† | |
| DJL81 (MaTrx6Δsp-FLAG) | − | 100±13 |
| + | 123±37 | |
| DJL82 (MaTrx3-FLAG) | − | 100±25 |
| + | 170±18† | |
| DJL83 (MaTrx3Δsp-FLAG) | − | 100±30 |
| + | 138±13 |
cyt c, Cytochrome c.
*The levels of cyt c were determined by densitometry of haem-stained SDS-PAGE gels. Gels were loaded with replicate samples of normalized membranes purified from two independent cultures. The amount of cyt c produced in non-induced cells (−tetracycline) is set to 100 arbitrary units.
†Significantly different from the level of cyt c in non-induced cells (− tetracycline), P≤0.05 (t-test).
Discussion
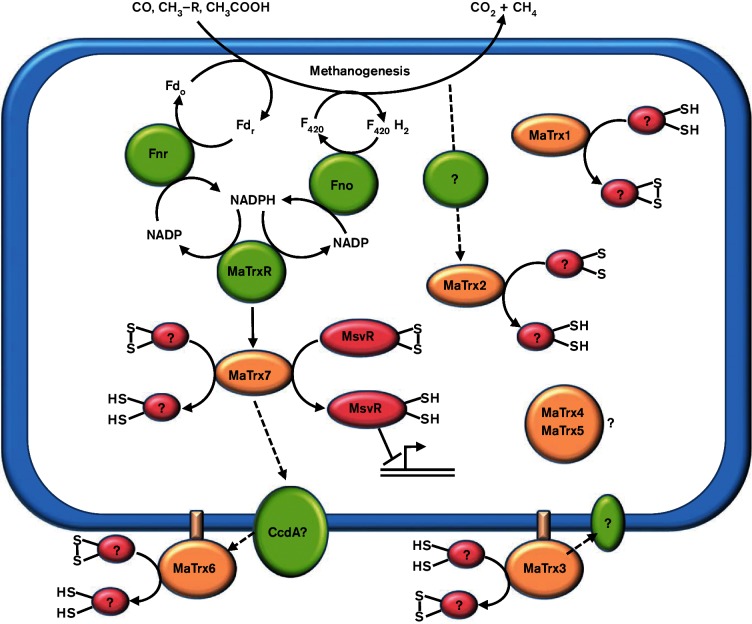
The majority of sequenced methanogens encode a TrxR homologue, and all methanogens, with the exception of Methanopyrus kandleri, contain at least one Trx homologue, underscoring the importance of the thioredoxin system to methanogen physiology [12, 13]. Among methanogens, Methanosarcina species encode the highest number of Trx homologues (six to eight) yet typically encode a single TrxR homologue [12]. Results from the biochemical characterization of MaTrxR and MaTrx1–MaTrx7 in this study confirm that NADPH-dependent MaTrxR is specific for MaTrx7 [12] and reveal that the remaining MaTrxs have distinct properties and, thus, likely different functions. The model shown in Fig. 6 illustrates the proposed role(s) of the MaTrxs in M. acetivorans. MaTrx7 appears to be the primary intracellular reducing Trx in M. acetivorans. MaTrx7 is the only MaTrx reduced by MaTrxR, and experiments with cell lysates support the in vivo reduction of MaTrx7 by MaTrxR. Reducing equivalents can be provided during methanogenesis with all growth substrates used by M. acetivorans through Fno and/or Fnr activities (Fig. 6), supporting the assimilation of an NADPH-dependent thioredoxin system into the physiology of M. acetivorans. The proteins targeted for reduction by MaTrx7 in M. acetivorans are largely unknown. However, MaTrx7 was shown to reduce the disulfides in the redox-sensitive transcription repressor MsvR, which activates DNA binding [43]. Moreover, recent results from a MaTrx7 pull-down experiment reveal that MaTrx7 is capable of reducing disulfides in several hundred M. acetivorans proteins (unpublished results), consistent with MaTrx7 serving as a general disulfide reductase. Thus, the NADPH-dependent MaTrxR–MaTrx7 system likely serves as a general reducing system in M. acetivorans.
Fig. 6.
Model showing the location and proposed function(s) of MaTrxs in M. acetivorans. Solid lines indicate detected activities and dashed lines indicate putative activities. Question marks denote unknown or proposed protein(s) or factor(s). MaTrx7 was previously shown to reduce disulfides in oxidized MsvR, activating binding of DNA [43]. Fdo, oxidized ferredoxin; Fdr, reduced ferredoxin; Fnr, ferredoxin : NADP oxidoreductase; Fno, F420H2 : NADP oxidoreductase.
Results from comparative bioinformatic analyses also indicate that the TrxR homologue in the majority of methanogens, with the exception of a subset of methanococci, is likely specific for NADPH. Thus, an NADPH-dependent thioredoxin system is likely used by the majority of methanogens. One potential benefit of using NADPH to directly reduce TrxR, instead of the primary methanogenesis electron carrier F420 or ferredoxin, is to provide specificity and to minimize competition for reducing equivalents needed for energy conservation. This separation of electron donors is similar to that used by the vast majority of cells, which use NADH for catabolism and NADPH for anabolism. However, some species from deeper methanogen lineages (e.g. Methanocaldococcus jannaschii) may use F420H2 or ferredoxin directly to reduce Trx, as these methanogens lack the conserved NADPH-binding site in the encoded TrxR homologue. Indeed, during review of this manuscript, it was demonstrated that F420H2 serves as the electron donor to M. jannaschii TrxR, which in turn reduces functional Trx1 [44].
Recent evidence has also revealed that methanogens contain additional Trx-related proteins that function in intracellular redox physiology. In M. acetivorans, ferredoxin : disulfide reductase (Fdr), a protein homologous to ferredoxin : thioredoxin reductase (Ftr) from plants, can reduce protein disulfides with reducing equivalents provided directly from ferredoxin [45]. In the same study, Fdr was shown to be incapable of reducing MaTrx2, the only intracellular MaTrx other than MaTrx7 with disulfide reductase activity. More recently, Fdr from Methanosarcina barkeri was shown to be specific for NrdH, a Trx-like protein, which reduces the active site disulfide in the unusual anaerobic ribonucleotide reductase NrdD found in some methanogens [46]. The Fdr–NrdH system is not ubiquitous in methanogens, and it appears to be an intracellular reducing system that is specific for a subtype of anaerobic ribonucleotide reductase restricted to methanogens from the orders Methanomicrobiales and Methanosarcinales [46]. In addition, the genome of some methanogens encodes Grx-like proteins, even though methanogens lack glutathione. A Grx-like protein from M. acetivorans was named methanoredoxin (Mrx) based on the ability to use coenzyme M, as well as glutathione, as a direct source of reductant [47]. Coenzyme M is a low molecular weight thiol found in all methanogens where it is directly involved in methanogenesis [2]. However, Mrx homologues are only found in roughly 50 % of methanogen species with sequenced genomes [47], indicating that disulfide reduction by Mrx may also serve a more specialized rather than general function. The disulfide-containing targets of Mrx have not been identified. It is common for organisms to have more than one intracellular reducing system. For example, E. coli contains both Trx and Grx [8]. This appears true of at least some methanogens as well, in particular, members of Methanosarcinales. Nonetheless, despite the unique physiology of methanogens, one that relies heavily on ferredoxin and coenzyme F420 as electron carriers, rather than NAD(P), the accumulated results indicate that the majority of methanogens likely use a canonical NADPH-dependent thioredoxin system.
In addition to a canonical thioredoxin system composed of MaTrxR–MaTrx7, M. acetivorans contains four additional intracellular Trx homologues. MaTrx1 is unique among the MaTrxs as it contains two additional cysteines [12] and an active site motif (CPYC) similar to Grx [8]. Also, unlike the other MaTrxs, not all of the cysteines in MaTrx1 were capable of thiol-disulfide exchange. MaTrx1 also lacked disulfide reductase activity, instead having low, but detectable, thiol oxidase activity. These results indicate that MaTrx1 is possibly an intracellular disulfide-forming enzyme (Fig. 6), but the importance of such an activity to methanogens is unclear. MaTrx2 has disulfide reductase activity but is not reduced by MaTrxR, and experiments with cell lysates did not link the reduction of disulfide-containing MaTrx2 to the oxidation of NADH, NADPH or F420H2. Thus, the redox partner(s) to MaTrx2 is unknown (Fig. 6). MaTrx2 homologues appear restricted to Methanosarcina [12] and may have a specialized function in these methanogens. MaTrx4 and MaTrx5 are similar to one another and have the same CAKC active site motif [12]. Although the cysteines of both proteins could be oxidized and reduced, consistent with thiol-disulfide exchange activity, neither protein exhibited disulfide reductase, disulfide isomerase or thiol oxidase activities. Thus, the function and role(s) of MaTrx4 and MaTrx5 are also unclear and may be unrelated to known functions of Trx-related proteins.
Results from the expression of FLAG-tagged MaTrx3 and MaTrx6 demonstrate that the N-terminal signal sequence targets both proteins to the membrane of M. acetivorans, providing the first experimental evidence that methanogens possess membrane-associated Trx proteins. The signal peptide is also likely retained, as evidenced by the presence of full-length product detected by Western analysis in membrane fractions (Fig. 5). Thus, the N-terminal sequence of both MaTrx3 and MaTrx6 likely serves to anchor each protein to the membrane, most likely on the extracellular surface. Although, MaTrx3 and MaTrx6 are homologous proteins and are within the same methanogen Trx clade [12], they are clearly not functionally equivalent. Experimental results support that each protein is capable of thiol-disulfide exchange; however, MaTrx6 is a disulfide reductase, whereas MaTrx3 is a thiol oxidase. The cellular location and activities indicate that MaTrx3 and MaTrx6 function in the formation and reduction of disulfides in membrane and/or extracellular proteins, respectively. M. acetivorans contains several membrane-associated and extracellular proteins with cysteines that are required for growth. For example, there are multiple CXXCH motifs in the 55 kDa cytochrome c encoded by MA_RS03460, which is involved in electron transfer by the Rnf complex, which is required for growth of M. acetivorans with acetate [48]. In bacteria and eukaryotes, extracellular thiol oxidases and disulfide reductases are documented to oxidize and reduce the CXXCH cysteines in apo-cytochrome c to stabilize and prepare the protein for haem insertion [26, 42, 49]. The involvement of these enzymes in the maturation of cytochrome c in methanogens has not been demonstrated. However, the altered levels of cytochrome c in the membranes of M. acetivorans as a result of the expression of FLAG-tagged MaTrx3 and MaTrx6 provide indirect evidence that MaTrx3 and MaTrx6 may play a role in the steps leading to haem insertion in apo-cytochrome c in this methanogen.
For MaTrx3 and MaTrx6 to function at the membrane of M. acetivorans, a membrane-associated redox partner(s) would be required to serve as an electron donor to MaTrx6 and an electron acceptor to MaTrx3. One probable redox partner to MaTrx6 is MA_RS22215, which encodes a homologue of CcdA. MaTrx6 is encoded upstream of, and possibly cotranscribed with, MA_RS22215. This gene arrangement is conserved in all Methanosarcinales [12]. In bacteria, CcdA functions to transfer reducing equivalents from intracellular Trx across the membrane to support the catalytic disulfide reductase activity of an extracellular Trx homologue. For example, in Bacillus subtilis, CcdA provides reductant to membrane-anchored ResA, a Trx homologue that is responsible for reducing the CXXCH disulfide in apo-cytochrome c [50]. Thus, it seems reasonable to propose that the CcdA homologue in M. acetivorans serves a similar role, supplying reductant to MaTrx6 from MaTrx7, the primary intracellular Trx (Fig. 6). For redox partners to MaTrx3, there are no obvious candidates encoded in the genome. A few systems have been characterized that use a thiol oxidase to catalyse the specific formation of disulfides in extracellular and/or periplasmic proteins. For example, DsbA is a Trx homologue that oxidizes thiols to disulfides in proteins, including the CXXCH motif of apo-cytochrome c, to increase protein stability in the periplasm of E. coli [26]. DsbA is re-oxidized by the cytoplasmic membrane protein DsbB, which then transfers electrons to the membrane-bound quinone pool [51]. The genome of M. acetivorans lacks genes for homologues of proteins known to serve a redox partners to extracellular thiol oxidases, such as DsbB. Determining the importance of MaTrx3 and MaTrx6 to the physiology of M. acetivorans, including the identification of redox partners and target proteins, will require additional experimentation and is currently underway.
Conclusions
The results presented here reveal that M. acetivorans contains seven Trx homologues with different functional properties and cellular locations. NADPH-dependent MaTrxR is specific for MaTrx7, and MaTrxR–MaTrx7 likely constitutes the general intracellular reducing system in M. acetivorans. Reducing equivalents are provided to the MaTrxR–MaTrx7 system through the oxidation of the primary methanogen electron carriers, F420H2 and ferredoxin. Bioinformatic analyses indicate that the majority of methanogens also likely use an NADPH-dependent thioredoxin system. MaTrx3 and MaTrx6 are localized to the membrane of M. acetivorans, and they function to generate or reduce membrane and/or extracellular proteins, respectively. The physiological function(s) of MaTrx1, MaTrx2, MaTrx4 and MaTrx5 are unclear and will require additional experimentation to elucidate what roles these Trx homologues serve in the physiology of M. acetivorans.
Funding information
This work was supported in part by grant number P30 GM103450 from the National Institute of General Medical Sciences of the National Institutes of Health (D. J. L.), National Science Foundation grant number MCB1121292 (D. J. L.), NASA Exobiology grant number NNX12AR60G (D. J. L.) and the Arkansas Biosciences Institute (D. J. L.), the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
Acknowledgements
We thank Dr Lacy Daniels (Texas A&M University-Kingsville) for providing F420 and Dr James Bardwell (University of Michigan) for providing purified E. coli DsbA.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); Fdr, ferredoxin : disulfide reductase; Fno, F420H2 : NADP oxidoreductase; Fnr, ferredoxin : NADP oxidoreductase; Ftr, ferredoxin : thioredoxin reductase; Grx, glutaredoxin; rRNaseA, reduced RNase A; scRNaseA, scrambled RNase A; Trx, thioredoxin.
One supplementary figure and one supplementary table are available with the online Supplementary Material.
Edited by: S. Gribaldo and W. Achouak
References
- 1.Ferry JG. One-carbon metabolism in methanogenic anaerobes. In: Ljungdahl LL, Adams MW, Barton LL, Ferry JG, Johnson MK, editors. Biochemistry and Physiology of Anaerobic Bacteria. New York: Springer-Verlag; 2003. pp. 143–156. (editors) [Google Scholar]
- 2.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 3.Zinder S. Physiological ecology of methanogens. In: Jerry JG, editor. Methanogenesis. New York, NY: Chapman and Hall; 1993. pp. 128–206. (editor) [Google Scholar]
- 4.Ferry JG. Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol Rev. 1999;23:13–38. doi: 10.1111/j.1574-6976.1999.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 5.Deppenmeier U. The membrane-bound electron transport system of Methanosarcina species. J Bioenerg Biomembr. 2004;36:55–64. doi: 10.1023/b:jobb.0000019598.64642.97. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 7.Arnér ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 8.Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 9.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol. 2001;55:333–356. doi: 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 10.Mcfarlan SC, Terrell CA, Hogenkamp HP. The purification, characterization, and primary structure of a small redox protein from Methanobacterium thermoautotrophicum, an archaebacterium. J Biol Chem. 1992;267:10561–10569. [PubMed] [Google Scholar]
- 11.Ondarza RN, Rendón JL, Ondarza M. Glutathione reductase in evolution. J Mol Evol. 1983;19:371–375. doi: 10.1007/BF02101641. [DOI] [PubMed] [Google Scholar]
- 12.Mccarver AC, Lessner DJ. Molecular characterization of the thioredoxin system from Methanosarcina acetivorans. FEBS J. 2014;281:4598–4611. doi: 10.1111/febs.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Susanti D, Wong JH, Vensel WH, Loganathan U, Desantis R, et al. Thioredoxin targets fundamental processes in a methane-producing archaeon, Methanocaldococcus jannaschii. Proc Natl Acad Sci USA. 2014;111:2608–2613. doi: 10.1073/pnas.1324240111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 15.Sowers KR, Boone JE, Gunsalus RP. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl Environ Microbiol. 1993;59:3832–3839. doi: 10.1128/aem.59.11.3832-3839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deppenmeier U, Blaut M, Mahlmann A, Gottschalk G. Reduced coenzyme F420 : heterodisulfide oxidoreductase, a proton-translocating redox system in methanogenic bacteria. Proc Natl Acad Sci USA. 1990;87:9449–9453. doi: 10.1073/pnas.87.23.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berk H, Thauer RK. Function of coenzyme F420-dependent NADP reductase in methanogenic archaea containing an NADP-dependent alcohol dehydrogenase. Arch Microbiol. 1997;168:396–402. doi: 10.1007/s002030050514. [DOI] [PubMed] [Google Scholar]
- 18.Welte C, Deppenmeier U. Re-evaluation of the function of the F420 dehydrogenase in electron transport of Methanosarcina mazei. FEBS J. 2011;278:1277–1287. doi: 10.1111/j.1742-4658.2011.08048.x. [DOI] [PubMed] [Google Scholar]
- 19.Messens J, Collet JF, van Belle K, Brosens E, Loris R, et al. The oxidase DsbA folds a protein with a nonconsecutive disulfide. J Biol Chem. 2007;282:31302–31307. doi: 10.1074/jbc.M705236200. [DOI] [PubMed] [Google Scholar]
- 20.Akiyama Y, Kamitani S, Kusukawa N, Ito K. In vitro catalysis of oxidative folding of disulfide-bonded proteins by the Escherichia coli dsbA (ppfA) gene product. J Biol Chem. 1992;267:22440–22445. [PubMed] [Google Scholar]
- 21.Greiner-Stoeffele T, Grunow M, Hahn U. A general ribonuclease assay using methylene blue. Anal Biochem. 1996;240:24–28. doi: 10.1006/abio.1996.0326. [DOI] [PubMed] [Google Scholar]
- 22.Guss AM, Rother M, Zhang JK, Kulkarni G, Metcalf WW. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea. 2008;2:193–203. doi: 10.1155/2008/534081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci USA. 1997;94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn W, Fiebig K, Hippe H, Mah RA, Huser BA, et al. Distribution of cytochromes in methanogenic bacteria. FEMS Microbiol Lett. 1983;20:407–410. [Google Scholar]
- 25.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, et al. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J Bacteriol. 2006;188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavridou DA, Ferguson SJ, Stevens JM. The interplay between the disulfide bond formation pathway and cytochrome c maturation in Escherichia coli. FEBS Lett. 2012;586:1702–1707. doi: 10.1016/j.febslet.2012.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz H, Pellicioli EC, Thöny-Meyer L. New insights into the role of CcmC, CcmD and CcmE in the haem delivery pathway during cytochrome c maturation by a complete mutational analysis of the conserved tryptophan-rich motif of CcmC. Mol Microbiol. 2000;37:1379–1388. doi: 10.1046/j.1365-2958.2000.02083.x. [DOI] [PubMed] [Google Scholar]
- 28.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis. 2009;30:1845–1855. doi: 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- 29.Ferry JG. Acetate-based methane production. In: Wall J, Harwood CS, Demain A, editors. Bioenergy. Washington, DC: ASM press: 2008. pp. 155–170. (editors) [Google Scholar]
- 30.Wang S, Huang H, Moll J, Thauer RK. NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J Bacteriol. 2010;192:5115–5123. doi: 10.1128/JB.00612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terlesky KC, Nelson MJ, Ferry JG. Isolation of an enzyme complex with carbon monoxide dehydrogenase activity containing corrinoid and nickel from acetate-grown Methanosarcina thermophila. J Bacteriol. 1986;168:1053–1058. doi: 10.1128/jb.168.3.1053-1058.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terlesky KC, Ferry JG. Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J Biol Chem. 1988;263:4075–4079. [PubMed] [Google Scholar]
- 33.Hanukoglu I, Gutfinger T. cDNA sequence of adrenodoxin reductase. identification of NADP-binding sites in oxidoreductases. Eur J Biochem. 1989;180:479–484. doi: 10.1111/j.1432-1033.1989.tb14671.x. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez HH, Jaquez OA, Hamill MJ, Elliott SJ, Drennan CL. Thioredoxin reductase from Thermoplasma acidophilum: a new twist on redox regulation. Biochemistry. 2008;47:9728–9737. doi: 10.1021/bi8006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berglund O, Holmgren A. Thioredoxin reductase-mediated hydrogen transfer from Escherichia coli thioredoxin-(SH)2 to phage T4 thioredoxin-S2. J Biol Chem. 1975;250:2778–2782. [PubMed] [Google Scholar]
- 36.Grimaldi P, Ruocco MR, Lanzotti MA, Ruggiero A, Ruggiero I, et al. Characterisation of the components of the thioredoxin system in the archaeon Sulfolobus solfataricus. Extremophiles. 2008;12:553–562. doi: 10.1007/s00792-008-0161-y. [DOI] [PubMed] [Google Scholar]
- 37.Kashima Y, Ishikawa K. A hyperthermostable novel protein-disulfide oxidoreductase is reduced by thioredoxin reductase from hyperthermophilic archaeon Pyrococcus horikoshii. Arch Biochem Biophys. 2003;418:179–185. doi: 10.1016/j.abb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira MA, Discola KF, Alves SV, Medrano FJ, Guimarães BG, et al. Insights into the specificity of thioredoxin reductase–thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system. Biochemistry. 2010;49:3317–3326. doi: 10.1021/bi901962p. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Ma K. Characterization of a thioredoxin–thioredoxin reductase system from the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol. 2010;192:1370–1376. doi: 10.1128/JB.01035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, Holmgren A. The thioredoxin superfamily in oxidative protein folding. Antioxid Redox Signal. 2014;21:457–470. doi: 10.1089/ars.2014.5849. [DOI] [PubMed] [Google Scholar]
- 41.Chim N, Harmston CA, Guzman DJ, Goulding CW. Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis. BMC Struct Biol. 2013;13:23. doi: 10.1186/1472-6807-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens JM, Mavridou DA, Hamer R, Kritsiligkou P, Goddard AD, et al. Cytochrome c biogenesis system I. FEBS J. 2011;278:4170–4178. doi: 10.1111/j.1742-4658.2011.08376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheehan R, Mccarver AC, Isom CE, Karr EA, Lessner DJ. The Methanosarcina acetivorans thioredoxin system activates DNA binding of the redox-sensitive transcriptional regulator MsvR. J Ind Microbiol Biotechnol. 2015;42:965–969. doi: 10.1007/s10295-015-1592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Susanti D, Loganathan U, Mukhopadhyay B. A novel F420-dependent thioredoxin reductase gated by low potential FAD: a tool for redox regulation in an anaerobe. J Biol Chem. 2016;291:23084–23100. doi: 10.1074/jbc.M116.750208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar AK, Kumar RS, Yennawar NH, Yennawar HP, Ferry JG. Structural and biochemical characterization of a ferredoxin : thioredoxin reductase-like enzyme from Methanosarcina acetivorans. Biochemistry. 2015;54:3122–3128. doi: 10.1021/acs.biochem.5b00137. [DOI] [PubMed] [Google Scholar]
- 46.Wei Y, Li B, Prakash D, Ferry JG, Elliott SJ, et al. A ferredoxin disulfide reductase delivers electrons to the Methanosarcina barkeri class III ribonucleotide reductase. Biochemistry. 2015;54:7019–7028. doi: 10.1021/acs.biochem.5b01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yenugudhati D, Prakash D, Kumar AK, Kumar RS, Yennawar NH, et al. Structural and biochemical characterizations of methanoredoxin from Methanosarcina acetivorans, a glutaredoxin-like enzyme with coenzyme M-dependent protein disulfide reductase activity. Biochemistry. 2016;55:313–321. doi: 10.1021/acs.biochem.5b00823. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Tomb JF, Ferry JG. Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 2011;11:165. doi: 10.1186/1471-2180-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen JW. Cytochrome c biogenesis in mitochondria–systems III and V. FEBS J. 2011;278:4198–4216. doi: 10.1111/j.1742-4658.2011.08231.x. [DOI] [PubMed] [Google Scholar]
- 50.Simon J, Hederstedt L. Composition and function of cytochrome c biogenesis system II. FEBS J. 2011;278:4179–4188. doi: 10.1111/j.1742-4658.2011.08374.x. [DOI] [PubMed] [Google Scholar]
- 51.Hatahet F, Boyd D, Beckwith J. Disulfide bond formation in prokaryotes: history, diversity and design. Biochim Biophys Acta. 2014;1844:1402–1414. doi: 10.1016/j.bbapap.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.