Abstract
Objective
Single nucleotide polymorphisms predisposing to coronary artery disease (CAD) have been shown to predict cardiovascular risk in healthy individuals when combined into a genetic risk score (GRS). We examined whether the cumulative burden of known genetic risk variants associated with risk of CAD influences the development and progression of coronary atherosclerosis.
Approach and Results
We investigated the combined effects of all known CAD variants in a cross-sectional study of 8,622 Icelandic patients with angiographically significant CAD (≥50% diameter stenosis). We constructed a GRS based on 50 CAD variants and tested for association with the number of diseased coronary arteries on angiography. In models adjusted for traditional cardiovascular risk factors, the GRS associated significantly with CAD extent (difference per SD increase in GRS, 0.076; P=7.3×10−17). Compared to the bottom GRS quintile, patients in the top GRS quintile were roughly 1.67× more likely to have multivessel disease (odds ratio, 1.67; 95% confidence interval, 1.45–1.94). The GRS significantly improved prediction of multivessel disease over traditional cardiovascular risk factors (Χ2 likelihood ratio 48.1, P<0.0001) and modestly improved discrimination, as estimated by the C-statistic (without GRS vs. with GRS, 64.0% vs. 64.8%) and the integrated discrimination improvement (0.52%). Furthermore, the GRS associated with an earlier age at diagnosis of angiographic CAD. These findings were replicated in an independent sample from the Emory Biobank study (n=1,853).
Conclusions
When combined into a single GRS, known genetic risk variants for CAD contribute significantly to the extent of coronary atherosclerosis in patients with significant angiographic disease.
Keywords: Coronary disease, Atherosclerosis, Genetics, Genetic risk score
Coronary artery disease (CAD) is a complex disease with both environmental and heritable contributions.1 To date, genome-wide association studies have yielded common single nucleotide polymorphisms (SNPs) at 50 chromosomal loci associated with risk of CAD.2 Multilocus genetic risk scores (GRS) combining multiple SNPs with modest effects on cardiovascular risk have been shown to predict incident cardiovascular events in several prospective cohorts of European ancestry.3–10 GRSs based on common CAD risk variants have been associated with atherosclerotic phenotypes such as peripheral artery disease11 and carotid intima-media thickness12, and coronary artery calcium5, an indirect measure of atherosclerotic burden.
Coronary angiography remains the ‘gold standard’ in quantifying the extent and severity of CAD and thus atherosclerotic burden. Previous studies have shown that genetic sequence variants at chromosome 9p21 and in the apolipoprotein(a) gene (LPA) not only associate with risk of CAD but also predict the extent of angiographic CAD, suggesting a role for these loci in influencing the development and progression of coronary atherosclerosis.13–15 In this study, we evaluated the effects of all known common genetic variants associated with risk of CAD on the extent of coronary atherosclerosis in patients with significant CAD on coronary angiography, both individually and combined in a GRS.
Materials and Methods
Materials and Methods are available in the online-only Data Supplement.
Results
Characteristics of the Patients
A total of 8,622 Icelandic patients with significant angiographic CAD (≥50% diameter stenosis) were included in the main analysis. Replication was sought in 1,853 patients from the Emory Biobank. All participants were of European ancestry. Characteristics of the study patients are shown in Table 1. Diabetes, hypertension and hyperlipidemia were more common in patients from the Emory Biobank, whereas Icelandic patients tended to be younger and were more likely to be current smokers. On average, patients from the Emory Biobank had more extensive coronary disease and were more likely to have prior history of myocardial infarction and coronary revascularization.
Table 1.
Characteristics of the Patients
| Characteristics | Iceland (n=8,622) | Emory Biobank (n=1,853) | P value* |
|---|---|---|---|
| Age (years) | 64.4 (10.7) | 65.6 (10.6) | <0.001 |
| Male sex (%) | 75.1 | 73.8 | 0.24 |
| Diabetes (%) | 11.4 | 31.5 | <0.001 |
| Hypertension (%) | 54.3 | 70.7 | <0.001 |
| Hyperlipidemia (%) | 50.3 | 74.6 | <0.001 |
| Current smoker (%) | 27.4 | 14.7 | <0.001 |
| Former smoker (%) | 47.6 | 47.9 | 0.83 |
| Prior MI (%) | 29.7 | 49.2 | <0.001 |
| Prior PCI (%) | 4.2 | 57.5 | <0.001 |
| Prior CABG (%) | 7.6 | 31.3 | <0.001 |
| Family history (%) | 43.1 | 44.7 | 0.21 |
| Number of diseased vessels† | 1.94 (0.88) | 2.10 (0.91) | <0.001 |
| One-vessel disease | 39.1 | 32.0 | <0.001 |
| Two-vessel disease | 30.3 | 31.6 | 0.30 |
| Three-vessel disease | 28.1 | 31.3 | 0.007 |
| Four-vessel disease | 2.5 | 5.1 | <0.001 |
Data are presented as percentages or means (standard deviation).
MI indicates myocardial infarction; PCI, percutaneous coronary intervention; CABG, coronary artery bypass grafting.
P values for continuous variables were calculated using Student’s t-test. P values for categorical variables were calculated using the chi-squared test.
Total number of coronary arteries with at least 50% stenosis on coronary angiography (left anterior descending, circumflex, the right and the left main coronary artery).
Association With CAD Extent
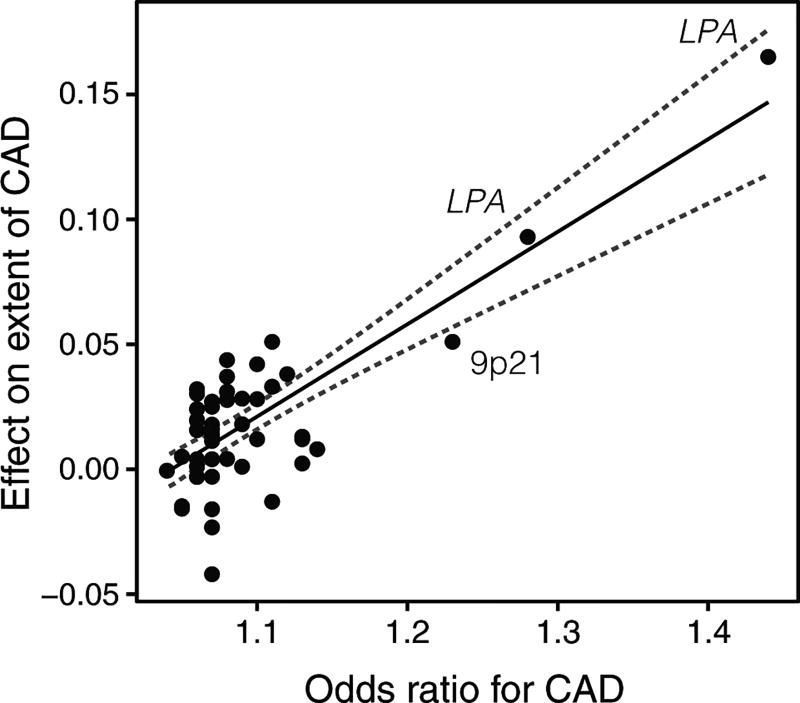
Among the 50 SNPs tested, rs1333049 at chromosome 9p21 and rs10455872 at the apolipoprotein(a) gene (LPA) associated significantly (P<0.001) with CAD extent in a combined analysis of the samples, adjusting for traditional cardiovascular risk factors: age, sex, hyperlipidemia, diabetes, hypertension, current and former smoking (Table I in the online-only Data Supplement). Figure 1 illustrates the linear relationship between the magnitude of the effects of individual SNPs on CAD extent and their respective odds ratio for the risk of CAD, previously reported in meta-analyses of genome-wide association studies (Table I in the online-only Data Supplement).
Figure 1.
The effects of 50 single nucleotide polymorphisms (SNPs) on the extent of coronary artery disease (CAD), expressed as the increase in number of diseased coronary vessels (with at least 50% stenosis) per SNP risk allele, plotted against their respective effect on CAD risk (odds ratio), previously reported in meta-analyses of genome-wide association studies (Table I in the online-only Data Supplement for references). Combined effect sizes in the Icelandic and Emory Biobank samples are presented where available (Table I in the online-only Data Supplement). The solid line denotes best linear fit, the dashed lines indicate 95% confidence limits.
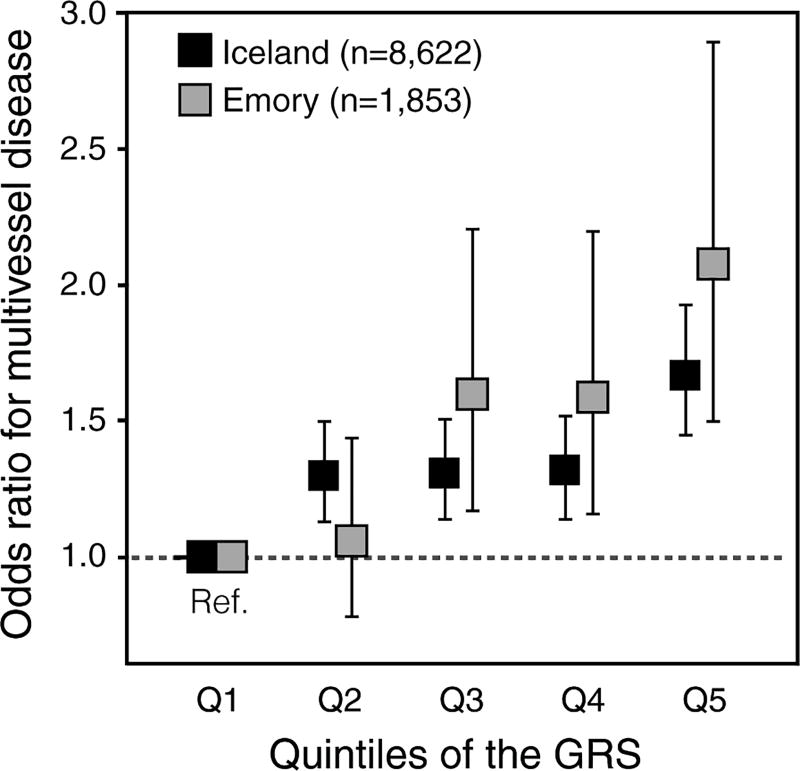
The combined GRS was strongly associated with the number of coronary arteries with at least 50% diameter stenosis when adjusting for traditional cardiovascular risk factors (difference per SD increase in GRS, 0.076; P=7.3×10−17). Estimates in models adjusting for traditional cardiovascular risk factors did not differ substantially from those in models adjusted for age and sex only (Table II in the online-only Data Supplement) and the association remained significant after further consecutive adjustment for family history of premature CAD (difference per SD increase in GRS, 0.072; P=3.0×10−15) (Table III in the online-only Data Supplement). To further illustrate this relationship, we divided patients into quintiles based on the GRS and compared the proportion of patients with multivessel disease (two or more coronary arteries with at least 50% diameter stenosis) between the top and bottom quintiles (Figure 2). Roughly 65% of patients in the top quintile had multivessel disease compared to 56% of patients in the bottom quintile (Table IV in the online-only Data Supplement). Thus, patients who were in the top quintile of the GRS were 1.67× more likely to have multivessel disease compared with patients in the bottom quintile (adjusted odds ratio, 1.67; 95% confidence interval, 1.45–1.94) (Table 2).
Figure 2.
Adjusted odds ratios for multivessel disease by quintiles of the genetic risk score (GRS) in the Icelandic (black) and Emory Biobank (grey) samples. Odds ratios are referenced to the bottom GRS quintile and presented with 95% confidence intervals.
Table 2.
Association of the GRS with the Number of Diseased Coronary Arteries on Coronary Angiography
| No. of SNPs |
Difference per SD increase (95% CI) |
Standard error |
P value | Contrast top versus bottom GRS quintile |
|
|---|---|---|---|---|---|
| OR for multivessel disease*(95% CI) | |||||
| Iceland (n=8,622) | |||||
| Full GRS | 50 | 0.076 (0.058–0.094) | 0.0091 | 7.3×10−17 | 1.67 (1.45–1.94) |
| Full GRS excluding 9p21 and LPA | 47 | 0.049 (0.031–0.066) | 0.0091 | 8.1×10−8 | 1.32 (1.14–1.52) |
| Restricted GRS | 32 | 0.072 (0.054–0.089) | 0.0091 | 3.2×10−15 | 1.55 (1.34–1.78) |
| Emory Biobank (n=1,853) | |||||
| GRS | 32 | 0.115 (0.075–0.155) | 0.021 | 2.6×10−8 | 2.08 (1.50–2.90) |
| GRS excluding 9p21 and LPA | 29 | 0.070 (0.029–0.110) | 0.021 | 7.3×10−4 | 1.50 (1.09–2.08) |
GRS indicates genetic risk score; SD, standard deviation; OR, odds ratio; CI, confidence interval.
Associations were tested using linear and logistic regression models adjusted for traditional cardiovascular risk factors (age, sex, hyperlipidemia, diabetes, hypertension, current and former smoking).
Multivessel disease was defined as having at least two coronary arteries with ≥50% stenosis on coronary angiography.
Since variants at chromosome 9p21 and the LPA loci have previously been reported to associate with the extent of angiographic CAD13–15, we investigated whether the effect of the GRS was dominated by these variants. After excluding variants at chromosome 9p21 (rs1333049) and LPA (rs10455872 and rs3798220) from the GRS, the association remained significant (P=8.1×10−8; Table 2). Similar results were obtained in models additionally adjusted for family history of premature CAD and in models adjusted for age and sex only (Table II and Table III in the online-only Data Supplement).
Model Performance
As shown in Table 3, the GRS significantly improved prediction of multivessel disease over cardiovascular risk factors in models including age and sex only (model 1), traditional cardiovascular risk factors (model 2) and family history of premature CAD (model 3), as evaluated by likelihood-ratio tests (P<0.0001). To estimate the improvement in discrimination, we compared the C-statistics (area under the receiver-operating-characteristic curve) for models with and without the GRS. The C-statistics for the models without the GRS ranged from 62.9% to 64.8% (Table 3). Addition of the GRS to the models resulted in modest increases in the C-statistic, ranging from 0.6% to 0.8% (Table 3). Similarly, the integrated discrimination improvement ranged from 0.46% to 0.53%, indicating a marginal improvement in discrimination for multivessel disease with the addition of the GRS (Table 3).
Table 3.
Model Prediction for Multivessel Disease With and Without the GRS
| Model covariates | C-statistic | IDI | LR Χ2 | P value* | ||
|---|---|---|---|---|---|---|
| Without GRS |
With GRS |
Increase | ||||
| Iceland (n=8,622)† | ||||||
| Model 1: Age and sex only | 62.9% | 63.7% | 0.8% | 0.53% | 48.7 | <0.0001 |
| Model 2: Traditional cardiovascular risk factors‡ | 64.0% | 64.8% | 0.8% | 0.52% | 48.1 | <0.0001 |
| Model 3: Traditional cardiovascular risk factors‡ and family history of premature CAD | 64.8% | 65.4% | 0.6% | 0.46% | 42.7 | <0.0001 |
| Emory Biobank (n=1,853)§ | ||||||
| Model 1: Age and sex only | 60.9% | 62.8% | 1.9% | 1.5% | 28.1 | <0.0001 |
| Model 2: Traditional cardiovascular risk factors‡ | 62.1% | 63.8% | 1.7% | 1.5% | 28.4 | <0.0001 |
| Model 3: Traditional cardiovascular risk factors‡ and family history of CAD | 62.2% | 63.9% | 1.7% | 1.5% | 28.4 | <0.0001 |
GRS indicates genetic risk score; CAD, coronary artery disease; IDI, integrated discrimination improvement; LR, likelihood ratio. C-statistics and the IDI are reported as percentages.
All P values reported are from likelihood ratio Χ2 tests for nested models.
GRS based on 50 SNPs.
Traditional cardiovascular risk factors were defined as age, sex, hyperlipidemia, diabetes, hypertension, current and former smoking.
GRS based on 32 SNPs.
Association with Age at Angiography
The GRS associated significantly with age at angiography when adjusting for sex, hyperlipidemia, diabetes, hypertension, current and former smoking (difference per SD increase in the GRS, −0.90 years; P=7.2×10−17). This association persisted when variants at chromosome 9p21 and LPA were excluded from the GRS (difference per SD increase in the GRS, −0.64 years; P=2.5×10−9). Patients in the top quintile of the GRS were on average 2.4 years younger than patients in the bottom quintile (63.6 years compared to 66.0 years), as shown in Table IV in the online-only Data Supplement.
Replication
In the Emory Biobank sample, the GRS based on 32 SNPs was significantly associated with CAD extent (difference per SD increase in the GRS, 0.115; P=2.6×10−8) (Table 2). In the Emory Biobank sample, 77% of patients in the top quintile of the GRS had multivessel disease compared to 62% of patients in the bottom quintile (Table V in the online-only Data Supplement), corresponding to an adjusted odds ratio of 2.08 (95% confidence interval, 1.50–2.90; Table 2). As shown in Table 3, The GRS significantly improved prediction of multivessel disease over cardiovascular risk factors and modestly improved discrimination, as estimated by the increase in the C-statistics for the models with the addition of the GRS (ranging from 1.7% to 1.9%) and the integrated discrimination improvement (1.5% for all models). The GRS associated with age at angiography when adjusting for sex, hyperlipidemia, diabetes, hypertension, current and former smoking (difference per SD increase in the GRS, −0.60 years; P=0.011) but the association was not significant when variants at 9p21 and LPA were excluded from the GRS (difference per SD increase in the GRS, −0.44 years; P=0.062).
Association With CAD Extent When Including Individuals With Non-Significant CAD
As expected, the association between the GRS and CAD extent was even more pronounced when individuals with non-significant CAD (<50% stenosis) were also included (Table VI and Figure in the online-only Data Supplement).
Discussion
In this study, we demonstrate that a genetic score based on known common CAD risk variants is strongly associated with the extent of coronary atherosclerosis in patients with established angiographic CAD. This association is independent of traditional cardiovascular risk factors and family history of CAD. We found that the GRS significantly improved prediction of multivessel disease over established cardiovascular risk factors, although the improvement in discrimination was modest. Compared to patients in the bottom quintile of the GRS, patients in the top quintile were roughly 1.67× (Iceland) and 2.08× (Emory Biobank) more likely to have multivessel disease. Furthermore, we found that the GRS associated with younger age at angiography, consistent with an earlier disease onset for individuals with a high burden of common genetic risk variants for CAD.
Previously, a genetic variant at chromosome 9p21 and two variants at LPA were shown to influence the extent of coronary atherosclerosis as determined by coronary angiography.13–15 In keeping with these findings, these variants showed the strongest association with CAD extent in the present study. Due to their large effect sizes on the risk of CAD, they were assigned the greatest weights in the GRS. To evaluate whether the GRS was dominated by these loci, we excluded them from the GRS in a separate analysis. We found that the GRS restricted to variants outside chromosome 9p21 and LPA was also significantly associated with CAD extent, despite showing a somewhat weaker effect compared to that of the unrestricted GRS. These results show that currently known genetic risk variants for CAD, not previously associated with CAD extent, collectively associate with extent of coronary atherosclerosis. This suggests that many of these genetic variants influence the development of coronary atherosclerosis, although the effect of a single variant is likely to be small.
Previous studies have suggested that some genetic variants associated with CAD may primarily promote coronary atherosclerosis whereas other variants may predispose to myocardial infarction in the presence of coronary atheroma. For example, chromosome 9p21 has been shown to associate primarily with coronary atherosclerosis but not myocardial infarction per se.16, 17 Reilly et al. showed that 12 genome-wide significant CAD variants did not associate individually with myocardial infarction among patients with angiographic CAD.18 Extending these observations, Patel et al. showed that a GRS based on 11 CAD risk variants associated with prevalent myocardial infarction in individuals undergoing coronary angiography but not when the analysis was restricted to patients with established angiographic CAD.19 These studies suggest that genetic risk variants for CAD, identified in early large-scale GWAS, relate primarily to coronary atherosclerosis and may have a minimal role in plaque rupture or thrombosis leading to acute coronary events. Our findings support the hypothesis that most common CAD variants identified to date influence the development of coronary atherosclerosis. While the extent and overall burden of angiographic CAD unequivocally increase the risk of adverse cardiovascular events20, 21, it remains to be established whether genotype scores based on common CAD variants are predictive of cardiovascular events in patients with established disease. Large prospective studies are warranted to evaluate the potential clinical utility of genomic data as prognostic factors in patients with established CAD.
Our study should be interpreted in the context of several important limitations. Firstly, we used standard coronary angiography to assess and quantify the extent of coronary atherosclerosis as the number of coronary arteries with at least 50% diameter stenosis. While angiography is the most widely used and validated method for CAD assessment, it does not provide information on the volume or composition of the atherosclerotic plaque.22 In the Icelandic sample, angiographic data for calculation of more sophisticated angiographic scoring systems such as the Gensini score or Duke CAD Severity Index were not available. Secondly, the GRS used for replication analyses in the Emory Biobank was constructed from an available 32-SNP subset of the 50 SNPs and was therefore not directly comparable to the GRS used for the main analyses. The main strengths of our study include large sample sizes and an unbiased nationwide coverage for the selection of the larger sample of Icelandic patients.
In summary, we have demonstrated that a combined GRS based on known common genetic risk variants for CAD is associated with the extent of coronary atherosclerosis in two independent populations of patients with established angiographic CAD. These findings show that CAD patients with a high burden of common genetic variants associated with CAD risk are more likely to have extensive coronary disease than those who carry a low burden of such risk variants.
Supplementary Material
Significance.
Prior studies have shown that common genetic risk variants for coronary artery disease at chromosome 9p21 and in the lipoprotein(a) gene associate with angiographic extent of the disease, suggesting a role for these loci in the development of coronary atherosclerosis. In this study, we show that the cumulative burden of currently known genetic risk variants for coronary artery disease associates significantly with the extent of coronary atherosclerosis in two independent populations of patients with established angiographic coronary artery disease. Compared to patients in the bottom quintile of the genetic score, patients in the top quintile were significantly more likely to have multivessel disease.
Acknowledgments
The authors thank all the individuals who participated in this study and whose contribution made this work possible. We also thank our valued colleagues who contributed to the data collection and phenotypic characterization of clinical samples, as well as genotyping and analysis of genome-wide association data.
Sources of Funding:
The work was supported by Landspitali University Hospital Research Fund, Jónína Gísladóttir fund, Bent Scheving Thorsteinsson research fund and Research Fund of the Icelandic Society of Cardiology. Emory Cardiovascular Biobank: This work was supported by the American Heart Association (Postdoctoral Fellowship for RSP), National Institutes of Health R01 HL89650-01, Robert W. Woodruff Health Sciences Center Fund, Emory Heart and Vascular Center Funds and supported in part by NIH Grant UL1 RR025008 from the Clinical and Translational Science Award program and NIH grant R24HL085343.
Nonstandard Abbreviations and Acronyms
- CAD
Coronary artery disease
- GRS
Genetic risk score
- SNP
Single-nucleotide polymorphism
Footnotes
Disclosures:
E.B., A.H., D.F.G., G.T., U.T. and K.S. are employees of deCODE Genetics/Amgen Inc.
References
- 1.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 2.Roberts R. Genetics of Coronary Artery Disease. Circ Res. 2014;114:1890–1903. doi: 10.1161/CIRCRESAHA.114.302692. [DOI] [PubMed] [Google Scholar]
- 3.Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy MJ, Kane JP, Pankow JS, Devlin JJ, Willerson JT, Boerwinkle E. Coronary heart disease risk prediction using a genetic risk score: the Atherosclerosis Risk in Communities (ARIC) Study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 4.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies RW, Dandona S, Stewart AFR, Chen L, Ellis SG, Wilson Tang WH, Hazen SL, Roberts R, McPherson R, Wells GA. Improved prediction of cardiovascular disease based on a panel of SNPs identified through genome wide association studies. Circ Cardiovasc Genet. 2010;3:468–474. doi: 10.1161/CIRCGENETICS.110.946269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D’Agostino RB, Hwang SJ, O’Donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5:113–121. doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes MF, Saarela O, Stritzke J, et al. Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One. 2012;7:e40922. doi: 10.1371/journal.pone.0040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaarhorst AAM, Lu Y, Heijmans BT, et al. Literature-based genetic risk scores for coronary heart disease: the Cardiovascular Registry Maastricht (CAREMA) prospective cohort study. Circ Cardiovasc Genet. 2012;5:202–209. doi: 10.1161/CIRCGENETICS.111.960708. [DOI] [PubMed] [Google Scholar]
- 9.Ganna A, Magnusson PKE, Pedersen NL, de Faire U, Reilly M, Arnlöv J, Sundström J, Hamsten A, Ingelsson E. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol. 2013;33:2267–72. doi: 10.1161/ATVBAHA.113.301218. [DOI] [PubMed] [Google Scholar]
- 10.Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:2261–6. doi: 10.1161/ATVBAHA.112.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tragante V, Doevendans PAFM, Nathoe HM, van der Graaf Y, Spiering W, Algra A, de Borst GJ, de Bakker PIW, Asselbergs FW. The impact of susceptibility loci for coronary artery disease on other vascular domains and recurrence risk. Eur Heart J. 2013;34:2896–2904. doi: 10.1093/eurheartj/eht222. [DOI] [PubMed] [Google Scholar]
- 12.Hamrefors V, Hedblad B, Engström G, Almgren P, Sjögren M, Melander O. A myocardial infarction genetic risk score is associated with markers of carotid atherosclerosis. J Intern Med. 2012;271:271–81. doi: 10.1111/j.1365-2796.2011.02472.x. [DOI] [PubMed] [Google Scholar]
- 13.Dandona S, Stewart AFR, Chen L, Williams K, So D, O’Brien E, Glover C, Lemay M, Assogba O, Vo L, Wang YQ, Labinaz M, Wells GA, McPherson R, Roberts R. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2010;56:479–86. doi: 10.1016/j.jacc.2009.10.092. [DOI] [PubMed] [Google Scholar]
- 14.Patel RS, Su S, Neeland IJ, Ahuja A, Veledar E, Zhao J, Helgadottir A, Holm H, Gulcher JR, Stefansson K, Waddy S, Vaccarino V, Zafari AM, Quyyumi AA. The chromosome 9p21 risk locus is associated with angiographic severity and progression of coronary artery disease. Eur Heart J. 2010;31:3017–3023. doi: 10.1093/eurheartj/ehq272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helgadottir A, Gretarsdottir S, Thorleifsson G, et al. Apolipoprotein(a) genetic sequence variants associated with systemic atherosclerosis and coronary atherosclerotic burden but not with venous thromboembolism. J Am Coll Cardiol. 2012;60:722–729. doi: 10.1016/j.jacc.2012.01.078. [DOI] [PubMed] [Google Scholar]
- 16.Chan K, Patel RS, Newcombe P, et al. Association between the chromosome 9p21 locus and angiographic coronary artery disease burden: a collaborative meta-analysis. J Am Coll Cardiol. 2013;61:957–70. doi: 10.1016/j.jacc.2012.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel RS, Asselbergs FW, Quyyumi AA, Palmer TM, Finan CI, Tragante V, Deanfield J, Hemingway H, Hingorani AD, Holmes MV. Genetic variants at chromosome 9p21 and risk of first versus subsequent coronary heart disease events: A systematic review and meta-analysis. J Am Coll Cardiol. 2014;63:2234–45. doi: 10.1016/j.jacc.2014.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reilly MP, Li M, He J, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel RS, Sun YV, Hartiala J, et al. Association of a genetic risk score with prevalent and incident myocardial infarction in subjects undergoing coronary angiography. Circ Cardiovasc Genet. 2012;5:441–449. doi: 10.1161/CIRCGENETICS.111.960229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ringqvist I, Fisher LD, Mock M, Davis KB, Wedel H, Chaitman BR, Passamani E, Russell RO, Alderman EL, Kouchoukas NT, Kaiser GC, Ryan TJ, Killip T, Fray D. Prognostic value of angiographic indices of coronary artery disease from the Coronary Artery Surgery Study (CASS) J Clin Invest. 1983;71:1854–1866. doi: 10.1172/JCI110941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholls SJ, Hsu A, Wolski K, Hu B, Bayturan O, Lavoie A, Uno K, Tuzcu EM, Nissen SE. Intravascular ultrasound-derived measures of coronary atherosclerotic plaque burden and clinical outcome. J Am Coll Cardiol. 2010;55:2399–2407. doi: 10.1016/j.jacc.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 22.Topol EJ, Nissen SE. Our preoccupation with coronary luminology. The dissociation between clinical and angiographic findings in ischemic heart disease. Circulation. 1995;92:2333–2342. doi: 10.1161/01.cir.92.8.2333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.