Abstract
Human pluripotent stem cells (hPSCs) have great potential for use in regenerative medicine and cell replacement therapies; however, prior to clinical application, cultured cell populations need to be screened to ensure the quality of the culture, as well as the capacity of these pluripotent cells to differentiate into desired cell types. Flow cytometry, utilizing antibodies recognizing targets restricted to the hPSC surfaceome, offers an invaluable tool for high-throughput validation of hPSC lines. Here we describe the immunophenotyping of live human embryonic stem cell (hESC, H9) and human induced pluripotent stem cell (hiPSC, KB3) lines by flow cytometry using a panel of antibodies identified as either stem cell reference markers (CD90, EpCam) or reported as being prevalent or restricted (c-Kit, HPI-1, Integrin α6, Semaphorin-6A) to these cells. The protocols described here with hPSCs are also applicable to differentiated hPSC progeny and should be instrumental in the immunophenotyping and isolation of well-defined homogeneous cell populations useful in regenerative medicine.
Keywords: Embryonic stem cells, Induced pluripotent stem cells, Surfaceome, Immunophenotyping, Flow cytometry, Antibody
1 Introduction
Embryonic stem (ES) cells are characterized by their pluripotency and self-renewing properties. It is because of these properties that the establishment of the first human embryonic stem cell (hESC) lines in 1998 [1] garnered much enthusiasm for their significant potential use in research and regenerative medicine. Clinical application of these cells, however, is limited by concerns surrounding the immunogenicity of the donor cells, and while the more recent development of hESCs derived by somatic cell nuclear transfer (ntES cells) overcomes the potential for immune rejection [2], there remains both the ethical controversy surrounding the use of human embryos as well as the issue regarding the limited supply of donor embryos.
Human induced pluripotent stem cells (hiPSC), which are derived from somatic cells through the ectopic expression of a specific set of transcription factors [3–5] and share the ESC properties of self-renewal and pluripotency, have overcome many of the limitations associated with hESC. Despite the advantages, there remain many obstacles to overcome prior to using hiPSC in the treatment of disease. Variability in the differentiation potential of hiPSC compared to hESC has been reported [6], and could be related to “epigenetic memory” from the somatic cell of origin [7] or the result of residual transgene expression [8]. Furthermore, recent comparisons of hESC and hiPSC transcriptional profiles at the single cell level demonstrate greater heterogeneity in gene expression levels in hiPSC, which may ultimately underlie the differentiation capabilities of these cells [9]. These factors likely contribute to the heterogeneity of differentiating human pluripotent stem cells (hPSCs) or the presence of contaminating undifferentiated cells found in many hPSC cultures. Therefore, before any hPSC line can be deemed acceptable for use in clinical settings, it is critical that cells be evaluated for their ability to differentiate into desired cells types, as well as assessing the overall quality and purity of differentiated cultures. Several methods exist for identification and characterization of PSCs, including single-nucleotide polymorphism (SNP) analysis, epigenetic profiling, immunocytochemistry, PCR, western blotting, in vitro differentiation, and teratoma formation; however, the majority of these methods require either the destruction or alteration (fixation) of the cell, or take days to complete, precluding the ability to use identified populations in downstream applications.
Immunophenotyping of live cells by flow cytometry, utilizing targets found on the cell surface, provides a high-throughput, nonmutagenic, and reproducible method for validation of hESC and hiPSC cultures, and eventually for the removal of potential contaminating hPSCs that would potentially lead to tumor formation after transplantation. Current state-of-the-art cytometric evaluations allow one to define distinct cell population using two physical parameters. The first parameter includes both forward scattered (FSC) and side scatter light (SSC), while the second parameter involves fluorescence, usually following some sort of cell labeling through the use of a dye or use of a fluorescently labeled antibody. The number of fluorescence parameters can range from 0 to >12 depending on the equipment available, with the latter requiring high-end multicolor flow cytometry. Each fluorescence parameter can be used to independently measure a function or the presence of a protein. When these parameters are combined, the measurements can be used to immunophenotype and sort selected cell populations with known functions.
Although a number of kits are available to characterize undifferentiated hPSC, most rely on antibodies against SSEA-1 (mouse), SSEA-3 (rat), and TRA-1-81 (human). SSEA-1 is absent from hPSCs, while SSEA-3 (or SSEA-4) is not wholly specific to hPSC. TRA-1-81 (or TRA-1-60) requires fixation, which limits their use for sorting and selecting authentic hiPSCs for cultivation and expansion. SSEA-5 has also been reported as useful for removal of teratoma-forming cells as part of a surface antibody panel [10]. A recent publication of a human pluripotent stem cell surface proteome (surfaceome), which compared hESCs and hiPSC against human fibroblasts and 50 additional cell types, identified >30 positive and negative markers for hPSCs and found an additional >100 proteins of interest for hPSCs [11]. This resource allows for the selection of a panel of markers for the identification of newly derived hPSC populations with greater purity, potency or with enhanced lineage specific differentiation potential.
The protocols described here demonstrate the immunophenotyping of KB3 hiPS cells cultured in feeder free, defined media conditions, in comparison to an established hESC line (H9) using live cell antibody labeling and analysis by flow cytometry. Cells were probed for the reference stem cell markers CD90 and EpCam (CD326), as well as for stem cell prevalent/restricted markers c-Kit (CD117) (hematopoietic), HPI-1 (neural progenitor cell) [12], Semaphorin-6A [11] and Integrin α6 (CD49f) (hematopoietic [13] and mesenchymal stem cells [14] multipotency markers). Although polychromatic flow cytometry is not described, the antibodies used, if conjugated with appropriate fluorophores, can be employed with multicolor flow cytometric technologies. The approaches described here are valid for hPSCs, but these protocols can be adapted for the analysis of any mammalian cell line or any differentiated hPSC progeny, provided that informative epitopes are known and antibodies are available that are suitable for the species and cell type being analyzed.
2 Materials
All solutions should be prepared using cell culture grade reagents and sterile supplies. All procedures should be performed using aseptic techniques in a Biological Safety Cabinet/Tissue Culture Hood. If analyses of cells without sorting are performed, then cell preparation after cultivation and flow cytometry can be performed using nonsterile techniques.
2.1 Coating Cell Culture Plates with hESC Qualified Matrix
Microcentrifuge tubes, 1.5 mL, sterile.
DMEM/F-12 (with L-Glutamine and HEPES) culture medium (ThermoFisher Scientific, Waltham, MA, USA) chilled to 4 °C.
hESC qualified matrix coating stock solution: Corning® Matrigel® hESC-qualified Matrix (catalog number 354277) (Corning Incorporated, Corning, NY, USA) (see Note 1). Thaw vial of hESC-qualified matrix overnight in 4 °C refrigerator on ice. Swirl vial to ensure material is evenly dispersed and dispense into single use aliquots (see Note 2) in chilled 1.5 mL microcentrifuge tubes on ice, switching pipette tips frequently to prevent clogging. Store the aliquots at −20 or −80 °C. Alternatively use Geltrex® LDEV-Free hESC-qualified matrix (Catalog number A1413202; ThermoFisher Scientific, Waltham, MA, USA). Allow a bottle of growth factor-reduced Geltrex® to thaw at 4 °C overnight. Aliquot (see Note 2) and store at −20 °C. Thaw at 4 °C, thawed vials may be stored at 4 °C until needed.
Culture dishes: 6-well, 100 mm, sterile (Corning).
Conical centrifuge tubes: 50 mL, sterile (Corning, Falcon Brand).
Serological Pipettes: 5 and 10 mL.
P10, P20, P200, and P1000 Pipettes (e.g., Gilman).
Pipette tips.
2.2 Passaging and Maintenance of Undifferentiated hPSCs in Monolayer Culture
H9 (WA09) (WiCell, Madison, WI, USA) hESCs cultured in 6-well plates. Any established hESC line can be used.
KB-3 hiPSCs cultured in 6-well plates. Any established hiPSC line can be used.
hESC qualified matrix coated 100 mm culture dishes.
Conical centrifuge tubes: 15 mL.
FACS (Fluorescence-activated cell sorting) tubes or equivalent: Polystyrene round bottom tube (5 mL) with 35 μm nylon mesh cell strainer cap (catalog number 352235) (Corning Incorporated, Corning, NY, USA).
StemPro® Accutase® Cell Dissociation Solution (Catalog number A1110501) (ThermoFisher Scientific, Waltham, MA, USA). Thaw Accutase® at 4 °C overnight. Use Accutase® within 2 months, if stored at 4 °C. Otherwise, aliquot 10 mL into 15 mL conical centrifuge tubes and store at −20 °C (see Note 3).
Trypan Blue Stain (0.4%) (catalog number T8154) (Sigma- Aldrich Corp, St. Louis, MO, USA).
Ultrapure water.
Dulbecco’s PBS (DPBS) without Calcium and Magnesium, pH 7.4 (catalog number 14190250) (ThermoFisher Scientific, Waltham, MA, USA).
Borosilicate glass disposable 9″ pipette.
Cotton-plugged borosilicate glass disposable 9″ pipette with bulb.
Rho kinase (ROCK) inhibitor Y-27632 stock solution (10 mM) (STEMCELL Technologies, Vancouver, Canada): In 1.5 mL microcentrifuge tube, dissolve 1 mg ROCK inhibitor in 312.5 μL DPBS. Store in 250 μL aliquots at −20 °C.
Essential 8 Medium (E8™ Medium) (catalog number A1517001) (ThermoFisher Scientific, Waltham, MA, USA) (see Note 4).
E8™ Medium with 1× ROCK Inhibitor: Add 1.25 μL ROCK Inhibitor stock solution per mL of E8™ Media (see Note 5). Mix well.
Vacuum Aspiration System.
Automated cell counter or manual hemocytometer.
Centrifuge for 15 mL conical tubes and 5 mL FACS tubes with cooling (4 °C) capability.
5% CO2, 37 °C, humidified incubator.
2.3 Harvesting hPSCs for Antibody Labeling
100 mm plate of hESC/hiPSC grown to 70–80% confluency.
Phosphate Buffered Saline (PBS), pH 7.4, without calcium, without magnesium.
Fetal Bovine Serum (FBS) (Atlanta Biologicals, Flowery Branch, GA, USA).
Cell Wash Solution: 1× PBS containing 1% FBS Cell Wash Solution. Add 5 mL FBS to 500 mL PBS. Store solution at 4 °C for up to 2 weeks.
Enzyme Free Cell Dissociation Solution (catalog number S-014-B; EMD Millipore, Billerica, MA, USA).
Falcon® Cell Strainers: 40 μm nylon mesh cell strainer (catalog number 352340; Corning Incorporated, Corning, NY, USA).
Conical Centrifuge Tubes: 50 mL, sterile (Corning, Fisher Brand)
2.4 Titration of Antibodies
Stock Solution of antibody to be titrated (fluorochrome-conjugated or unconjugated) (see Note 6).
Stock Solution of fluorochrome-conjugated Secondary Antibody.
Isotype Control antibodies (see Note 7).
Target Cells for antibody titration (H9 (WA09) hESCs or KB-3 hiPSCs).
Cell Staining Solution for Flow Cytometry: 1× PBS containing 5% FBS. Add 5 mL FBS to 500 mL PBS. Store at 4 °C for up to 2 weeks.
Round-Bottom polystyrene tubes: 5 mL.
2.5 Antibody Labeling of Cell Surface Antigens
Antibodies. See Table 1 for antibodies used in the current protocol.
Isotype Controls. See Table 1 for Isotype controls used in the current protocol.
Borosilicate glass disposable 9″ pipette.
Phosphate Buffered Saline (PBS), pH 7.4, without calcium, without magnesium.
Cell Staining Solution for Flow Cytometry (see item 5 of Subheading 2.4).
Round-Bottom polystyrene tube: 5 mL.
Serological Pipettes: 5 and 10 mL.
Human Trustain FcX™Fc Blocking Solution (Catalog number 422301; BioLegend, San Diego, CA, USA).
Platform Rocker.
10% normal goat serum in 1× PBS Secondary Antibody Blocking Solution (Catalog number 50062Z; ThermoFisher Scientific, Waltham, MA, USA).
Table 1.
List of antibodies (primary and secondary) used in the current protocols
| Antigen (clone) | Fluorochrome | Company | Isotype control | Amount of primary antibody per 1 × 106 cells in 100 μL | Secondary anti body | Amount of secondary antibody per 1 × 106 cells in 100 μL |
|---|---|---|---|---|---|---|
| CD49f (GoH3) | PE | eBioScience | Rat IgG2a | 0.13 μg | N/A | N/A |
| CD90 | PE | BD Pharmingen | Mouse IgG1 | 4.0 μg | N/A | N/A |
| CD117 (104D2) | PE | Bio Legend | Mouse IgG1 | 0.25 μg | N/A | N/A |
| CD326 (9C4) | Alexa Fluor 647 | Bio Legend | Mouse IgG2b | 0.25 μg | N/A | N/A |
| HAI-1 (9B10) | PE | eBioScience | Mouse IgG2a | 0.25 μg | N/A | N/A |
| SEMA6A | R&D Systems | Goat IgG | 0.5 μg | Donkey anti-goat IgG-PE | 0.125 μg |
2.6 Preparation of Cells for Flow Cytometry
Hanks Balanced Salt Solution (without calcium or magnesium) (HBSS).
Cell Maintenance Solution for Flow Cytometry: Add 5 mL FBS to 95 mL HBSS. Store at 4 °C.
FACS Tubes (see 2.2 above).
FBS.
2.7 Flow Cytometry Analysis
The current protocol used a FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Single peak Rainbow beads (catalog number RFP-30-5A; Spherotech Inc., Green Oaks, IL, USA).
Compensation beads (catalog number 557640; Beckton Dickinson Immunocytometry Systems).
0.5% paraformaldehyde (PFA).
3 Methods
The protocols described here are applicable for both flow cytometry analysis and sorting of hPSC (see Note 8). All procedures should be performed using aseptic techniques in a Biological Safety Cabinet/Tissue Culture Hood unless otherwise noted.
3.1 Coating Cell Culture Plates with hESC Qualified Matrix
Slowly thaw 1 aliquot of hESC qualified matrix (Matrigel®) at 4 °C for 30 min.
Transfer 25 mL of cold DMEM/F12 medium to a 50 mL centrifuge tube on ice.
Add an aliquot of hESC qualified matrix to 25 mL of chilled DMEM/F12 medium and mix well immediately before coating plates (see Note 9).
Add 2 mL hESC-qualified Matrix in culture medium per 9.5 cm2 culture well (one well of a 6-well plate) growth areas.
Swirl plate to ensure even coating, and store at 4 °C for Matrigel or at 37 °C for Geltrex. Before using, allow hESC-qualified Matrix coated plates to set, covered, for 30 min at room temperature (see Notes 10 and 11).
Immediately prior to plating cells, aspirate the excess hESC qualified matrix (see Note 12).
3.2 Passaging Undifferentiated hPSCs/hiPSC in Monolayer Culture
Add 1 mL or 7.5 mL prewarmed E8™ medium containing 2× ROCK inhibitor to the selected number of wells of a 6-well plate or a 100 mm plate that has been coated with hESC qualified matrix (see Note 13).
Thaw sufficient Accutase® (see Note 14) to passage cells and warm to room temperature (see Note 15).
Observe cells under a brightfield microscope. Identify and if necessary remove regions of differentiation by scraping with a pipette tip or by aspiration, using a new tip for each plate to reduce the possibility of cross-contamination (see Note 16).
Aspirate medium from culture plates using sterile Pasteur pipette and vacuum aspiration system.
For a single well or a 100mmculture dish, wash cells twice with 2 or 10 mL of 1× DPBS (room temperature), respectively.
Add 1 or 4 mL of Accutase® to each well or 100 mm culture dish, respectively and leave undisturbed for 3–7 min until colony boundaries appear folded back and show signs of becoming less well packed.
Using a pipetteman (P1000) or a cotton-plugged glass pipette with bulb, dislodge cells with very gentle pipetting and transfer to a 15 or 50 mL conical tube containing DMEM/F-12 medium, prewarmed to room temperature (see Note 17). Gently triturate suspension to generate single cells.
Transfer a 10 μL aliquot of cell solution to a 1.5 mL microcentrifuge tube and mix with 10 μL trypan blue. Count cells (see Note 18). Use total cell counts for all applications going forward.
Collect remainder of cells by centrifugation at 130 × g for 5 min at room temperature (see Note 19).
Aspirate media and resuspend the cell pellet in DMEM/F12 or E8™ medium, prewarmed to 37 °C, to an optimal concentration of 0.1–1.0 × 106 cells per mL (see Note 20).
Add up to 1.0 or 7.5 mL of the cell suspension to each well or 100 mm tissue culture plate containing E8™ medium plus 2x ROCK inhibitor prepared in step 1. Optimally, the volume of the cell suspension being added is less than the volume in the previously prepared plate; therefore, the final volume should be brought to 2.0 mL per well, or 15 mL per 100 mm dish, such that the ROCK inhibitor is at a final concentration of 1×. Before placing the cells in the incubator, gently move in a front-to-back and side-to-side motion to uniformly disperse cells across the well (see Note 21).
Incubate cells at 5% CO2, 37 °C in humidified incubator, and replace the ROCK inhibitor containing medium within 18–24 h of plating with E8™ culture medium without ROCK inhibitor. The volume to add is 2 or 15 mL per well or 100 mm culture dish, respectively.
Replace media daily with E8™ culture medium without ROCK inhibitor.
Harvest cells when plates reach 70–80% confluency.
3.3 Harvesting hESCs and hiPSCs for Antibody Labeling
For antibody labeling, we use primarily 100 mm cultures of hPSCs; however, for simple tests, there are usually enough cells (1–3 × 106) in a single well of a 6-well plate.
Examine cells for morphological signs of differentiation using a brightfield microscope and, as needed, remove differentiated cells as described in step 3 of Subheading 3.2.
Aspirate the growth medium from the culture plates. Wash cells twice with 2 mL cold Cell Wash Solution
Aspirate Cell Wash Solution then add cold Millipore “cell dissociation solution” to cover cells (3–4 mL/100 mm plate) (see Note 22).
Incubate dish at 4 °C for 20 min, on a rocker to maximize cell dissociation (see Note 23).
Using a cotton-plugged borosilicate glass disposable 9″ pipette with bulb, dislodge cells with gentle pipetting and transfer to a 15 mL conical tube on ice.
Collect cells by centrifugation at 200 × g for 5 min at 4 °C.
Aspirate the supernatant.
Resuspend cells in 10.1 mL Cell Wash solution using a 10 mL serological pipette with repeated gentle trituration to break up cell clumps and ensure a single cell suspension.
Using a 10 mL serological pipette, pass cells through a 40 μm nylon mesh cell strainer fitted to the top of a 50 mL conical tube (see Note 24).
Transfer a 100 μL aliquot of cell solution to a 1.5 mL microcentrifuge tube and mix with 100 μL trypan blue. Count cells as described in step 6 of Subheading 3.2. Use total cell counts for all applications going forward.
Collect cells by centrifugation at 200 × g for 5 min at 4 °C.
3.4 Titration of Antibodies for Percent Positive Measurements
All steps should be performed on ice and samples protected from light.
For Fluorochrome-conjugated Primary Antibodies:
Determine the concentration and volume of the antibody stock solutions and recommended antibody concentration for use in flow cytometry analysis from the manufacturer’s product data sheet (see Note 25).
For each antibody and isotype control, number 6–8 microcentrifuge tubes and place on ice.
Using the manufacturer’s recommended antibody concentration as a guide, serial dilute antibody to create several stock antibody concentrations. Begin dilutions at slightly above the recommended concentration. Table 2 provides an example of a twofold serial dilution scheme for a PE conjugated CD90 antibody with a master stock antibody concentration of 0.2 mg/mL (see Note 26).
Prepare the first working stock antibody solution by pipetting 30 μL of master stock antibody into a labeled microcentrifuge tube (Tube No. 1) on ice.
Perform 6–8 twofold serial dilutions from the highest concentration of the working stock antibody. Pipette 20 μL from the working stock into microfuge tube containing 20 μL Cell Staining Solution. Gently vortex the tube followed by a quick spin in a microfuge. Repeat for subsequent dilutions until series is complete.
Label cells with antibody and prepare for flow cytometry analysis as described in Subheadings 3.5 and 3.6. Include tubes for each antibody to be titrated as well as for unstained and isotype controls (see Note 27).
Refer to Subheading 3.8 for determination of the optimal antibody concentration.
Table 2.
Antibody serial dilution scheme
| Tube no. | Cell wash solution volume (μL) | Volume and source of primary Ab (μL) | Working stock dilution | Working stock concentration (μg/mL) | Ab staining reaction volume (μL/106 cells) | Concentration in staining reaction (μg/106 cells) |
|---|---|---|---|---|---|---|
| 1 | 0 | 30 of stock | 0 | 200 | 20 | 4.0 |
| 2 | 20 | 20 of tube 1 dilution | 2 | 100 | 20 | 2.0 |
| 3 | 20 | 20 of tube 2 dilution | 4 | 50 | 20 | 1.0 |
| 4 | 20 | 20 of tube 3 dilution | 8 | 25 | 20 | 0.5 |
| 5 | 20 | 20 of tube 4 dilution | 16 | 12.5 | 20 | 0.25 |
| 6 | 20 | 20 of tube 5 dilution | 32 | 6.25 | 20 | 0.125 |
| 7 | 20 | 20 of tube 6 dilution | 64 | 3.125 | 20 | 0.0625 |
| 8 | 20 | 20 of tube 7 dilution | 128 | 1.5625 | 20 | 0.03125 |
Example of a serial dilution scheme for a PE conjugated CD90 antibody with a master stock concentration of 0.2 mg/mL for staining 1 × 105 cells
3.5 Antibody Labeling of Cell Surface Antigens on Live Cells
All steps should be performed on ice and samples protected from light.
Resuspend cells prepared in Subheading 3.3 in cold Wash Solution so that final concentration of cells is 1 × 106 total cells in 95 μL. Using a P200 pipette, gently triturate to disaggregate cells.
Block cells by adding 5 μL Human Trustain FcX™Fc Blocking Solution for every 1 × 106 cells and mix by gently flicking tube with finger, then incubate for 10 min on ice, gently rocking.
Gently triturate cell solution using P200 pipette to ensure homogeneous mixture of cells then aliquot 1 × 106 cells (100 μL) per tube into 5 mL round bottom polystyrene tubes on ice (see Note 28).
Add primary antibody or isotype control at its optimal concentration (as determined in Subheadings 3.4 and 3.8) then incubate for 60 min on ice with gentle rocking (see Note 29). Include an unstained control.
Add 3 mL coldWash Buffer, then collect cells by centrifugation at 200 × g for 5 min at 4 °C.
Aspirate solution being careful not to disturb cell pellet.
Repeat washing steps 5 and 6 for a total of two washes following antibody labeling.
If a primary antibody directly conjugated to a fluorochrome is used, proceed directly to Subheading 3.6. Continue as follows for labeling with a secondary antibody conjugated to a fluorochrome.
Resuspend cells in 100 μL secondary antibody blocking solution using a P200 pipette with gentle trituration.
Add secondary antibody, gently tap tube to mix, and then incubate for 30 min on ice, gently rocking.
Add 3 mL coldWash Buffer, then collect cells by centrifugation at 200 × g for 5 min at 4 °C.
Aspirate solution being careful not to disturb cell pellet
Repeat washing steps 11 and 12 for a total of two washes after secondary antibody labeling.
All steps should be performed on ice and samples protected from light.
3.6 Preparation of Cells for Flow Cytometry
Resuspend cells prepared in 400 μL cold Cell Maintenance Solution. Using a P1000 pipette, gently triturate to disaggregate cells.
Prewet the 35 μm nylon mesh cell-strainer cap on 5 mL FACS tube with 50 μL cell maintenance solution (see Note 30). Keep tubes on ice.
Transfer cell solution to cell strainer cap and allow to pass across themesh and drop to the bottom of tube by gravity (see Note 31).
Rinse strainer with 250 μL cell maintenance solution.
Keep cells on ice and protected from light until analyzed by flow cytometry.
3.7 Flow Cytometry Analysis
Prior to running samples, alignment beads and calibration standards should be run to ensure that the data are reproducible from day to day. This may be routinely run by an experienced user or in a core facility (if so, go to step 16 below), but if not, then beads should be run to ensure good instrument performance.
Place diluted single peak Rainbow beads onto the sample insertion tube and initiate data acquisition.
While observing FSC versus SSC and all other fluorescence parameter combinations, adjust the instrument according to the manufacturer’s instructions to ensure the narrowest CV with the highest signal intensity. Tolerance ranges are established for the coefficient of variance and fluorescent intensity as well as all instrument parameters.
Adjust the voltages for each photomultiplier tube (PMT) to achieve the predetermined intensity levels for the bead population.
Collect and save all single parameter histograms for subsequent analyses.
The signal-to-background ratio (S/B) is obtained by using the eight peak beads (i.e., the median channel of each peak) divided by the median channel. Each PMT will have a characteristic S/B, and the correct tolerance plotted against time should be in the range of ±10%.
If multiple fluorophores are run, then compensation beads (mouse anti-K beads) will also need to be run as described in https://www.bdbiosciences.com/documents/BD_FACSDiva_setup_system.pdf.
Latex beads coated with anti-mouse K antibody are used with each antibody conjugate to determine the compensation matrix for polychromatic flow cytometry. Into a conical tube add 40 μL of compensation beads and the volume of previously titered antibody conjugate (this is done for each antibody). Dilute to 100 μL with cell wash solution.
Incubate at room temperature for 15 min.
Wash once with cell wash solution.
Remove the supernatant and resuspend in 250 μL of cell wash solution.
Vortex and add 150 μL of 0.5% PFA.
Acquire each compensation control tube and the unstained bead control using the previously defined voltage settings.
Set the automated compensation matrix. Compensation should be rechecked regulatory by acquiring cell samples stained with combinations of antibody conjugates.
Once established, it is inappropriate to alter these voltages during data acquisition among samples.
For experimental data acquisition, gently vortex or triturate each biological sample immediately prior to placement onto sample insertion tube to ensure dispersion of cells and the absence of aggregates.
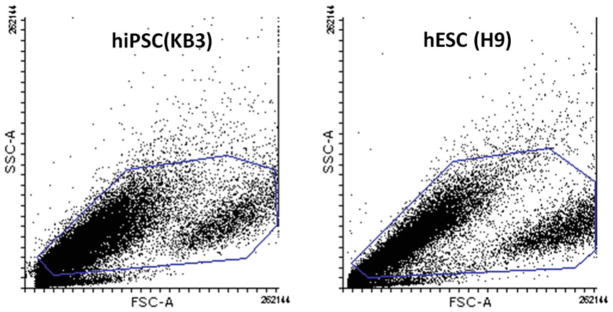
For each cell type, load the unstained control or preferably the isotype control sample onto the flow cytometer and optimize forward and side scatter voltage settings (see Note 32). For statistical analysis, we use forward scatter (FSC) (abscissa) and side scatter (SSC) (ordinate) to gate viable, single cell events and eliminate debris and cell aggregates (see Fig. 1) (see Note 33). If there are multiple isotype controls we begin with unstained controls and then optimize settings to maximize data acquisition of all isotype controls tested.
Ideally, the scatter plots should show an equal distribution of cells distributed along a 45° angle relative to the ordinate and abscissa.
Visualize the cell distribution of isotype controls and fluorochrome-labeled cells to determine threshold values to limit the number of events acquired by the flow cytometer. If using total counts, then the threshold value is low and most events will be acquired; however, population gating may be useful to eliminate events such as cell debris and dead cells (see Note 34).
Adjust the voltages to ensure that the signals with the isotype control can be readily visualized. Ideally, the peak signal will be well defined when viewed as a histogram, and it will have an even distribution, and a signal on the Fluorochrome log scale that is minimal.
Using fluorochrome-labeled cells, check to see if there is a signal above that seen with the isotype control. Normally, we test each antibody singly on a specific cell type before attempting any multilabeling experiments. Establish optimum baseline PMT gains (see Note 35) to maximize resolution. Use the minimum intensity required to achieve a histogram of fluorescence, which clearly displays both left and right edges of the peak(s).
Record settings, as this will be valuable for future repeat experiments. We recommend multiple repeats with an n > 4 for each cell surface protein examined.
Once these thresholds have been determined, maintain the laser voltage settings of each fluorochrome when analyzing each corresponding antibody labeled sample. It is inappropriate to alter these voltages during data acquisition among samples, either when determining optimal antibody dilutions or when performing analyses.
Collect a minimum of 10,000 events; however, a higher acquisition may be needed for multicolor analyses.
Acquire data using flow cytometry for each antibody tested; however, it may be necessary to adjust laser voltage settings for each fluorochrome using the appropriate isotype controls.
If multicolor parameters are assessed, then the voltage settings should be set to maximize data acquisition.
If displaying multiple parameters with multiple fluorochromes, then 2D, 3D, and other plots may be necessary for analyzing data. Quenching must be considered, and settings must be based on the absorption spectra of fluorochromes (see Note 36).
Fig. 1.
Gating strategy for selective live cells. Dot blots showing light scatter profiles of hiPSC and hESC gated for the selection of live cells and exclusion of debris. In this instance, cells have not been stained for viability, and all cells are included, excluding those with a very low SSC value. These likely correspond to dead or damaged cells. 50,000 events were collected and the gated population used for determination of cells positive for marker proteins
3.8 For Antibody Titration Determinations
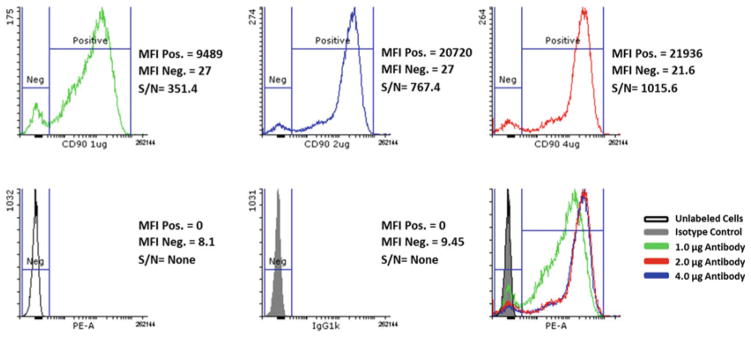
Display histogram for each dilution and negative controls (see Fig. 2).
Determine the total counts or median fluorescent intensity (MFI) of both positive (Signal) and negative (Noise) for all samples.
Calculate the signal to noise ratio by dividing the MFI value for positive cells by that for the negative cells.
In the final evaluations, choose an antibody for all subsequent analyses at the concentration that gives the highest Signal to Noise ratio for the best discrimination between positive and negative cells with the least amount of added antibody.
Alternatively, for quantitation purposes, where saturation of the target protein is necessary to achieve accurate measurements, the antibody should be used at the concentration where antibody saturation is achieved without significantly increasing nonspecific binding, as indicated by shifts in fluorescence in isotypes and negative controls (see Note 37).
Fig. 2.
Titration of CD90-PE antibody using H9 hESC. The data shown here reflect titration results from pilot tests with three independent antibodies. Data from the antibody showing the best results are shown, but at three concentrations. Histograms of CD90 in H9 cells illustrating that the optimal signal to noise ratio (S/N) is achieved using 4.0 μg antibody. MFI median fluorescence intensity, S/N MFI positive/MFI negative
3.9 Analysis of Data
Data will need to be exported and analyzed using software. There are a number of software packages that can be used, and we recommend that you discuss with your Flow Cytometry Core, which package may be apt. Examples include FCS Express, FloJo, and free software from Flowing Software.
For data analyses in the histogram mode, adjust the gates so that less than 2% of the signals/events from the isotype controls are above the negative control gate. All signals falling below this setting/gate will be considered as negatives, while those signals located above this value will be considered positive. When viewed as a histogram (ordinate—counts (total, FSC, SSC); abscissa—Fluorochrome Log scale (channel)), only signals with intensities equal to or greater than the threshold channel value will be processed.
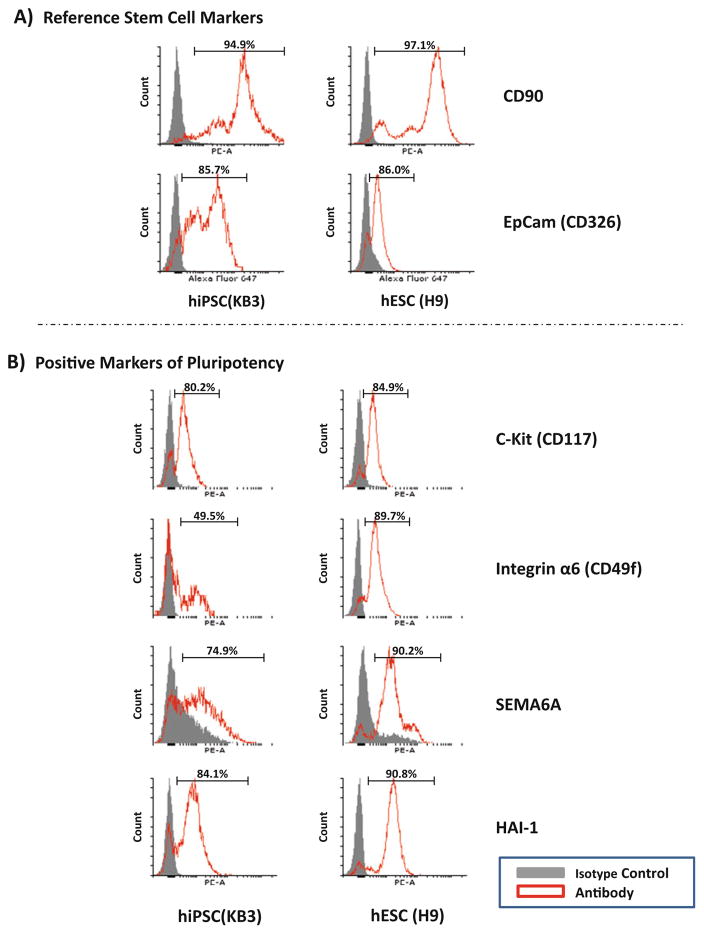
Determine the percent positive cells within the gated population by generating a univariate histogram and using the isotype control to gate the negative population to include >98% of the events, with staining greater than this in the antibody stained sample considered positive (see Fig. 3a, b).
Fig. 3.
Characterization of hiPSC and hESC by flow cytometry. Histograms of live KB3 hiPSC and H9 hESC showing populations positive for (a) Stem cell reference markers and (b) markers of pluripotency
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging, by the Research Grants Council of Hong Kong Theme-based Research Scheme T13-706/11, and the Hong Kong Research Grant Committee General Research Fund (Project number 17100214). We thank Robert Wersto and the NIA FC Core Facility for assistance with flow cytometry.
Footnotes
Although Matrigel® in conjunction with E8™ medium has been the most widely used culture system for hiPSC maintenance, the optimal coating matrix should be tested whenever developing or characterizing new hiPS cell lines. Alternative coating matrices, such as Geltrex®, Vitronectin XF™, or BD™ Laminin/Entactin Complex High Concentration are available if a fully chemical defined system is required.
The dilution factor for creating the correct concentration of hESC-qualified matrix for plate coating is calculated for each lot of Matrigel® or Geltrex® LDEV-Free hESC-qualified matrix based on the product protein concentration, and therefore, the user should refer to the manufacturer’s lot specific Certificate of Analysis for product protein concentrations and follow the supplier’s instructions for generating appropriate dilutions and single use aliquots.
Dispase can also be used (though in our hands, it has proven less good than accutase) or cells can be passaged using EDTA (EDTA dissociation buffer: 500 μL 0.5 M EDTA and 0.9 g NaCl in 500 mL DPBS). Refer to Notes 14 and 22 for further information regarding the use of EDTA dissociation in this protocol.
E8, or Essential 8™ medium, is a simplified medium originally developed by Chen et al. [15] for the culturing of hPSC and hiPSC in a feeder free, chemically defined culture system. TeSR™-E8™ is the commercially manufactured version of the E8 formulation made by STEMCELL Technologies and it can also be used. Other media, like NutriStem, which is both serum free and xenofree, can also be used.
Adjust volume to ensure the final concentration of ROCK inhibitor is 10 μM following the addition of cells/medium.
We routinely use Phycoerythrin (PE)-conjugated antibodies for most simple fluorescence analyses, however, when multiple antibodies are needed, appropriate secondary fluorochrome-conjugated antibodies must be used or primary antibodies should be conjugated in the laboratory. Unconjugated monoclonal antibodies can be purchased in bulk from many manufacturers and subsequently conjugated with selected fluorophores, ranging from fluorescein (FITC) to Texas Red to Cascade Blue. All conjugations should be performed as detailed in (http://www.drmr.com/abcon/). Antibody conjugation is a fairly rapid procedure that can be accomplished in 2–3 h.
An isotype control is an antibody of the same species, class (heavy and light chains), and has the same fluorochrome (e.g., PE) as the target primary antibody, but which lacks specificity. The ideal isotype controls will also have the same number of fluorescent molecules (fluorochrome–protein (F:P) ratio). Isotype controls are necessary negative controls used to determine the specific antibody signal by allowing the subtraction of nonspecific antibody binding (background) from the total positive signal.
This protocol can be readily employed with most standard flow cytometers, including those that permit cell sorting. In the case of the latter, ROCK inhibitor should be added to the cells at all stages to ensure cell viability after plating. Failure to add ROCK inhibitor will adversely affect cell survival.
One aliquot of Matrigel® hESC-qualified Matrix in 25 mL of DMEM/F-12 is sufficient to coat three 100mmdishes (8 mL/dish) or four 6-well plates (1 mL/well). It is important to realize that different batches of Matrigel® or Geltrex® (see Note 11) may require different volumes for coating of plates and plating of cells. This needs to be determined empirically, but we usually start with the recommended dilution; however, we have on occasion been able to use a twofold greater dilution or needed to use up to fourfold greater amounts of the matrix for optimal hPSC growth.
Coating can also be accomplished at 30 min in 37 °C incubator. Make sure incubation surfaces are level to achieve evenly coated plates.
As an alternative to Matrigel®, add 250 μL of Geltrex® to 50 mL of cold (4 °C) DMEM/F-12 (a 1:200 dilution) and mix thoroughly. It is also important to realize that the amount of Geltrex® or Matrigel® required for optimal growth varies from 1:50 to 1:400 dilutions, and must be determined empirically. Plate Geltrex® onto tissue culture plates (as was done for the Matrigel®), allow to polymerize at 37 °C for at least 1 h, and store, sealed with Parafilm™, at 37 °C for up to 2 weeks.
Matrigel®-coated plates can be used immediately, or if desired, stored at 4 °C immediately after coating for up to 1 week with addition of 1 mL per well of a 6-well plate or 7 mL per 100 mm culture plate of DMEM/F-12 media and sealed with Parafilm ® (to prevent dehydration). Previously stored plates should be equilibrated to room temperature for 60 min prior to use. Plates that have uneven coating or where the Matrigel® has evaporated should not be used.
6-Well plates are used for routine passaging and expansion, while 100 mm plates are used for cell sorting following immunostaining. Each 100 mm plate of cells should be sufficient for 10–15 antibody labeling reactions required for flow cytometry analysis, and therefore, the total number of plates prepared should be adjusted to reflect the scope of the experiment. Alternatively, for cell sorting experiments, 1–2 100 mm plates of cells should be prepared for each antibody labeling required to ensure sufficient numbers of live cells for subsequent culturing and any potential downstream experiments.
It is also possible to use an EDTA-based procedure in conjunction with E8™ medium for routine passaging of hPSCs. This method has the advantage of shorter protocol times and may be preferred for high throughput experiments or when an enzyme-free method is necessary. See Beers et al. [16] for a detailed protocol.
Thawed aliquots of Accutase® can be used immediately or stored at 2–8 °C for up to 2 weeks. Do not refreeze.
The current protocol assumes an already established culture of hESCs or hiPSCs. For initial thawing and seeding of hESCs and hiPSCs, see supplier recommended procedures. Cell lines should be well established (>p20) and exhibit a homogeneous morphology with less than 10–20% differentiation in high quality cultures. Also, when cultivating cells in E8™ medium, there is normally very little overt differentiation. If there is obvious differentiation, then it may be better to repeat the experiment and ensure optimal growth conditions.
Use a new pipette or tip for each well/plate to reduce the chance of cross-contamination. For large scale experiments, cells from multiple wells/plates can be combined into single 15 or 50 mL conical centrifuge tube. When combining cells from multiple plates, the ratio of enzyme/cells solution to DMEM/F-12 should remain at a minimum of 1:1 to ensure proper inactivation of the cell detachment solution.
Use either automated or manual hemocytometer. The current protocol has typically yielded 70–85% viability when using an automated hemocytometer.
To save time, cells can be centrifuged during cell counting.
Depending on the cell line, we plate anywhere from 70 to 150,000 cells/well for use in 3–4 days. Higher numbers can be used, however, we recommend cells be left in culture for 3–4 days before harvest; therefore, initial plating numbers will need to be adjusted accordingly to attain the desired cell density prior to harvesting cells for antibody labeling.
Do not use a circular motion as cells will accumulated at the center of the plate.
Use of EDTA dissociation solutions are not optimal for this step as cells will clump together once calcium is added back, affecting downstream labeling of cells.
Cell Dissociation incubation time is dependent on the cell type and should be determined empirically, using the shortest possible time to generate optimal combination of single cell suspension and high cell viability (at least 70–80%). Cells are typically ready to harvest when cell boundaries begin to round-up and colonies become less well packed.
Passing cells through a filter can improve downstream antibody labeling by eliminating cell aggregates and ensuring only single cells are labeled.
Optimizing the antibody concentration is essential for reducing nonspecific antibody binding and allowing for the best discrimination between positive and negative results. For each cell type and antibody used, an antibody titration assay should be performed. Titrations should be performed in the same conditions in which you plan to use the antibody, and an appropriate isotype control should be included for each antibody tested.
Some manufacturers provide the recommended concentration as a volume per number of cells (e.g., 10 μL/106 cells). In such a case, begin the titration using 2–4× the recommended volume and prepare serial (1/2) dilutions in Cell Staining Solution as described.
Isotype controls should be added to samples at the same concentration as that of the test antibody.
It is not necessary to wash the cells between blocking and immunodetection steps.
For optimal results, ensure primary antibody and corresponding isotype controls are run at the same concentrations.
The small volume may take several minutes to wet the entire area of the strainer. This step can be performed during final wash/centrifugation steps.
Straining of cells is necessary to eliminate large aggregates that could clog the flow cytometer. Tubes can be tapped gently on the benchtop to aid the collection of cell solution through strainer and into the tube. Return cells to ice as quickly as possible.
Forward-scattered light (FSC) is proportional to cell surface area or cell size. Side-scattered light (SSC) is proportional to cell granularity or internal complexity.
Dead cells can nonspecifically bind many antibody conjugates. These signals can erroneously be counted as positively labeled cells; therefore, gating strategies should be used to gate out these cells. Intercalated dyes like propidium iodide (PI) and live cell assays like Calcein AM can be useful for identifying and gating dead cells.
Preliminary tests should be run to determine whether a cell type can be effectively labeled using a specific antibody. For this type of analysis, we usually use the cells from a single well of a 6-well plate and test the antibody using the manufacturer’s recommendations. Antibody dilutions must then be performed with more cells to optimize conditions (see Subheadings 3.4 and 3.8).
Optimal baseline PMT gains need to be established empirically at the outset, and should be determined based on the advice of a FC Core facility or with the help of someone who is well acquainted with flow cytometry.
It is possible for multiple parameters/signals to be measured; however, if multiple fluorochromes are being assessed simultaneously, then quenching must be considered. Contact a FC Core or experienced user for assistance.
If using a fluorochrome-conjugated secondary antibody, it is best to titrate both the neat, primary antibody, as well as the fluorochrome-conjugated secondary antibody. Begin by labeling the cells with the primary antibody at the manufacturer’s recommended concentration (typically 1.25–20 ng/mL final dilution) and then perform a series of secondary antibody dilutions similar to the method outlined for the titration of primary antibodies. Once the optimal concentration has been determined for the secondary antibody, perform a titration of the primary antibody as described. The listed concentration of a fluorochrome-conjugated antibody includes both the antibody as well as the fluorochrome; therefore, the concentration of each antibody required will likely be less.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Narsinh KH, Plews J, Wu JC. Comparison of human induced pluripotent and embryonic stem cells: fraternal or identical twins? Mol Ther. 2011;19:635–638. doi: 10.1038/mt.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narsinh KH, Sun N, Sanchez-Freire V, Lee AS, Almeida P, Hu S, Jan T, Wilson KD, Leong D, Rosenberg J, et al. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest. 2011;121:1217–1221. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang C, Lee AS, Volkmer JP, Sahoo D, Nag D, Mosley AR, Inlay MA, Ardehali R, Chavez SL, Pera RR, et al. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boheler KR, Bhattacharya S, Kropp EM, Chuppa S, Riordon DR, Bausch-Fluck D, Burridge PW, Wu JC, Wersto RP, Chan GC, et al. A human pluripotent stem cell surface N-glycoproteome resource reveals markers, extracellular epitopes, and drug targets. Stem Cell Rep. 2014;3:185–203. doi: 10.1016/j.stemcr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koivuniemi R, Makela J, Hokkanen ME, Bruelle C, Ho TH, Ola R, Korhonen L, Schroder J, Kataoka H, Lindholm D. Hepatocyte growth factor activator inhibitor- 1 is induced by bone morphogenetic proteins and regulates proliferation and cell fate of neural progenitor cells. PLoS One. 2013;8:e56117. doi: 10.1371/journal.pone.0056117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE, Xie S, Laurenti E, Hermans K, Eppert K, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 14.Yu KR, Yang SR, Jung JW, Kim H, Ko K, Han DW, Park SB, Choi SW, Kang SK, Scholer H, et al. CD49f enhances multipotency and maintains stemness through the direct regulation of OCT4 and SOX2. Stem Cells. 2012;30:876–887. doi: 10.1002/stem.1052. [DOI] [PubMed] [Google Scholar]
- 15.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]