Abstract
Background
Plasma amino acid measurements have been extensively investigated in individuals with autism spectrum disorder (ASD). Results thus far have been inconclusive as studies generally disagree on which amino acids are different in individuals with ASD versus their typically developing (TD) peers, due in part to methodological limitations of several studies.
Method
This paper investigates plasma amino acids in children and adults with ASD using data from Arizona State University’s Comprehensive Nutritional and Dietary Intervention Study. Measurements from 64 individuals with ASD and 49 TD controls were analyzed using univariate and multivariate statistical techniques.
Results
Univariate analysis indicated increased median levels of glutamate (+21%, p=0.014) and serine (+8%, p=0.043), and increased mean levels of hydroxyproline (+17%, p=0.018) for the ASD cohort, although these differences were insignificant after correcting for multiple comparisons. A multivariate approach was used to classify study participants into ASD/TD cohorts using Fisher discriminant analysis (FDA) and its nonlinear extension, kernel Fisher discriminant analysis (KFDA). Model fitting with FDA using all available measurements produced Type I and Type II errors of 27.0% and 27.8%, respectively. KFDA was most effective when using hydroxyproline, leucine, and threonine as inputs; however, leave-one-out cross-validation with this nonlinear model only resulted in 70.3% sensitivity and 77.6% specificity.
Conclusions
The finding of elevated glutamate in ASD is in agreement with several other studies. Overall, however, these results suggest that plasma amino acid measurements are of limited use for purposes of ASD classification, which may explain some of the inconsistencies in results presented in the literature.
Keywords: autism spectrum disorder, plasma amino acids, Fisher discriminant analysis, classification, multivariate statistics, cross-validation
1. Introduction
Amino acids, commonly referred to as the building blocks of proteins, have important roles in cell metabolism, neurotransmission, immune system regulation, and cell signaling (Wu, 2009). A number of published studies have measured amino acid concentrations in the plasma of individuals with autism spectrum disorder (ASD) and found differences from measured concentrations in typically developing (TD) controls. While an overall consensus among these studies is that some plasma amino acid levels differ between TD individuals and those on the autism spectrum, there is a general disagreement as to what these differences are. The inconsistencies in results can possibly be attributed to several challenges in study design, primarily small sample sizes, the use of non-age-matched or non-gender-matched controls, a lack of overnight fasting by study participants, and/or failure to correct for multiple hypothesis testing. The use of age- and gender-matched controls is important as both age and gender have been found to influence plasma amino acid composition (Lepage, McDonald, Dallaire, & Lambert, 1997; Proenza, Cresp, Roca, & Palou, 2001; Caballero, Gleason, & Wurtman, 1991). Overnight fasting by participants is also a necessary study design component as it minimizes the short-term influence of diet on blood measurements.
Only two recent studies (after 2011) were found to address all four of these study design components. A previous study in Arizona analyzed concentrations of 41 amino acids and amino acid metabolites in the plasma of 55 children with ASD and 44 TD children, and detected significantly elevated glutamate and significantly decreased tryptophan in the ASD cohort (Adams et al., 2011). Additionally, a study in China measured glutamate in the plasma of 51 children with ASD and 51 TD controls, and found mean glutamate levels to be significantly higher in the children with ASD (Cai, Ding, Zhang, Xue, & Wang, 2016), consistent with the study by Adams et al. (Adams et al., 2011).
A potential physiological connection between amino acids and ASD status could perhaps be found in the investigation of folate-dependent one-carbon metabolism (FOCM) and transsulfuration (TS), two metabolic pathways of particular importance to the study of ASD that are associated with the regulation of epigenetic gene expression and intracellular redox status in the body (James et al., 2006). The interactions of nutritional/environmental factors with genetic irregularities in FOCM and TS are believed to contribute to a decreased capacity for DNA methylation and increased levels of intracellular oxidative stress in individuals with ASD (Deth, Muratore, Benzecry, Power-Charnitsky, & Waly, 2008). Mathematical modeling of these pathways and estimation of model parameters using clinical FOCM/TS metabolite data has revealed significant differences in several metabolic reaction rate parameters between TD individuals and those on the autism spectrum (Vargason, Howsmon, Melnyk, James, & Hahn, 2017). Multivariate statistical modeling of clinical measurements related to DNA methylation and redox status has also suggested a strong ability to distinguish individuals with ASD from their TD peers and predict severity of ASD-related symptoms based on the activity in these pathways (Howsmon, Kruger, Melnyk, James, & Hahn, 2017). Given that the amino acids methionine and glycine feature in FOCM, while serine, cysteine, glutamate, and glycine contribute to the TS pathway, there may be correlations between these amino acids and the metabolic abnormalities that have been observed in ASD.
Amino acids also influence brain activity by serving various roles as neurotransmitters or as precursors to such in the central nervous system (Fernstrom, 1994). There is evidence to suggest that the activity of certain neurotransmitters is abnormal in individuals with ASD, indicating another potential link between amino acids and the disorder. Glutamate and γ-aminobutyric acid (GABA), for example, are the primary excitatory and inhibitory neurotransmitters in the brain, and a reduced GABA/glutamate ratio in the brain has been reported in ASD (Harada et al., 2011). It has also been observed that the synthesis of serotonin, of which tryptophan is a precursor, is regulated differently during early childhood brain development in children with ASD compared to their TD peers (Chugani et al., 1999). While the presence of the blood-brain barrier makes it difficult to directly assess the relationships between amino acid levels in plasma and neurotransmitter activity in the central nervous system, there are some indicators of what these relationships are. Changes in brain serotonin levels, for one, have been suggested to be closely linked to changes in plasma tryptophan levels (Fernstrom & Wurtman, 1971; McDougle et al., 1996). At the same time, tryptophan’s uptake by the brain is affected by its competition for transport with the other large neutral amino acids, i.e., isoleucine, leucine, phenylalanine, tyrosine, and valine (Fernstrom & Wurtman, 1972).
All of this information considered, the relationships between plasma amino acids and ASD pathophysiology are still unclear. The use of multivariate statistical methods for differentiating individuals with ASD from their TD peers using plasma amino acid measurements may elucidate certain aspects of these relationships (Vargason, Howsmon, McGuinness, & Hahn, 2017). Previous work has demonstrated the utility of multivariate methods, namely Fisher discriminant analysis (FDA) and its nonlinear extension kernel Fisher discriminant analysis (KFDA), for performing this ASD/TD classification task using biochemical data (Adams et al., 2017; Howsmon et al., 2017). Therefore, after performing a detailed univariate analysis, the current work will apply these multivariate statistical methods to the analysis of data from the Comprehensive Nutritional and Dietary Intervention Study at Arizona State University (Adams et al., 2017), which addresses the aforementioned challenges associated with previous plasma amino acids studies by featuring a larger sample size, age- and gender-matched controls, overnight fasting, and correction for multiple hypothesis testing.
2. Methods
2.1 Study participants
The plasma amino acid data for this work comes from the Comprehensive Nutritional and Dietary Intervention Study, a 12-month nutritional and dietary intervention study at Arizona State University. Only data from the baseline measurements (i.e. before the start of treatment) are described and analyzed in this work. Enrollment criteria and participant characteristics from the study are described in detail by Adams et al. (Adams et al., 2017). Participants in the study included 67 individuals on the autism spectrum (37 of which received the treatment and 30 that did not) and 50 age- and gender-matched TD controls. As plasma amino acid measurements were unavailable for a few study participants, only 64 and 49 participants from each study group, respectively, were included in the analysis for the present work. The characteristics of this subset of participants are summarized in Table 1.
Table 1.
Characteristics of participants in the Comprehensive Nutritional and Dietary Intervention Study whose plasma amino acid measurements were included in the present work.
| ASD Cohort | TD Cohort | |
|---|---|---|
| Total Participants | 64 | 49 |
| Males | 52 (81%) | 40 (82%) |
| Females | 12 (19%) | 9 (18%) |
| Age (years) | 11.8 ± 8.5 | 12.2 ± 7.6 |
| Children (ages 3–12) | 45 (70%) | 34 (69%) |
| Teens (ages 13–20) | 13 (20%) | 10 (20%) |
| Adults (ages 20+) | 6 (9%) | 5 (10%) |
All participants in the ASD cohort had a previous diagnosis of ASD. The Autism Diagnostic Observation Schedule and/or Childhood Autism Rating Scale were also used by staff at Arizona State University to confirm the ASD diagnosis. A number of additional assessments of ASD-related symptoms and behaviors were also conducted (Adams et al., 2017); these were the Severity of Autism Scale, Aberrant Behavior Checklist, Autism Treatment Evaluation Checklist, Pervasive Developmental Disorders Behavior Inventory, Social Responsiveness Scale, Short Sensory Profile, and Parent Global Impressions. The IQ of participants in the ASD cohort was also evaluated using the Reynolds Intellectual Assessment Scales, consisting of the Verbal Intelligence Index, Nonverbal Intelligence Index, Composite Intelligence Index, and Composite Memory Index. The outcomes of these evaluations for participants in the ASD cohort are available in the supplementary data.
In the ASD cohort, 29 participants (45.3%) were reported to be taking at least one medication at the time of the study. This was also the case for 4 participants (8.2%) in the TD cohort. However, the study’s inclusion criteria (Adams et al., 2017) required that participants did not have any significant changes to their medical treatment in the two months prior to the study, or have any intention to make such changes during the study period.
2.2 Amino acid measurements
Blood samples were collected after an overnight fast, and plasma concentrations of amino acids and amino acid metabolites were measured according to a protocol previously described (Adams et al., 2011). In total, forty-two plasma amino acids and related amino acid metabolites were measured in the clinical study. Concentrations of the nine essential amino acids histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine were measured. Additionally, the concentrations of the eleven non-essential amino acids alanine, arginine, asparagine, aspartate, cystine (the oxidized form of cysteine), glutamate, glutamine, glycine, proline, serine, and tyrosine were quantified. Concentrations of eight secondary amino acids (i.e. those that are not essential or non-essential) were also evaluated: α-amino-N-butyrate (AABA), β-alanine, citrulline, GABA, homocystine (the oxidized form of homocysteine), hydroxyproline, methionine sulfoxide, and ornithine. The fourteen measured amino acid metabolites (hereafter referred to as amino acids, for brevity) were 1-methylhistidine, 3-methylhistidine, α-aminoadipate, ammonia, anserine, β-aminoisobutyrate, carnosine, cystathionine, ethanolamine, phosphoethanolamine, phosphoserine, sarcosine, taurine, and urea. The data for these amino acids are available in the supplementary information.
2.3 Univariate hypothesis testing
Student’s t-test, Welch’s t-test, or the Mann-Whitney U test were used to compare the central tendencies of each plasma amino acid measurement between the ASD and TD cohort. Preliminary statistical tests were performed to evaluate the distributional assumptions of these location tests (i.e. statistical test of mean or median as appropriate) and determine which was most suitable for each measurement. Normality of a measurement’s distribution in each cohort was first assessed with the Anderson-Darling test (Anderson & Darling, 1954). If a measurement was found to be normally distributed in both cohorts, equality of variance of the distributions was evaluated using the two-sample Brown-Forsythe test (Brown & Forsythe, 1974). For measurements in which the two normal distributions were found to have equal variances, the two-sample Student’s t-test (Student, 1908) was used to test for a significant difference in the means of the two distributions. Measurements in which the variances of the two distributions were found to be unequal were instead evaluated with the two-sample Welch’s t-test (Welch, 1938) to test for a significant difference in means.
For measurements whose distributions were non-normal in either or both cohorts, the two-sample Kolmogorov-Smirnov test (Massey, 1951) was used to compare the shapes of the distributions in the ASD and TD cohorts. If the shapes of the two distributions were determined to be similar, the Mann-Whitney U test (Mann & Whitney, 1947) was then used to test for a significant difference in medians between the distributions. Measurements that did not follow identical, but shifted, distributions in the two cohorts were evaluated with Welch’s t-test.
All hypothesis tests were performed at significance level α = 0.05. Two-sample location tests were two-tailed and the outcomes of these tests were adjusted for multiple comparisons using Bonferroni correction. Differences indicated by a p-value less than 0.05 were defined as possibly significant, while differences indicated by a p-value less than the Bonferroni-adjusted significance level were defined as significant.
2.4 Effect sizes and confidence intervals
The effect size for each amino acid was quantified as the median value in the ASD cohort minus the median value in the TD cohort. Bootstrap resampling with 10,000 replications was used to obtain a distribution of the effect size for each amino acid, and the 95% confidence interval (CI) for the effect size was then estimated by measuring the 0.025 and 0.975 quantiles of this bootstrap distribution (Banjanovic & Osborne, 2016).
2.5 Multivariate statistical analysis
The multivariate analytical methods used in the current study were implemented with previously-developed MATLAB code (Adams et al., 2017), but adapted for the measurements of this work. For classification, the data were normalized such that all sample values for each plasma amino acid had a mean of zero and a standard deviation of one.
2.5.1 ASD classification
Classification of individuals as belonging to the ASD or TD cohort was performed using FDA and KFDA. The aim of FDA is to determine the linear combination of plasma amino acid measurements that best separates the samples of the ASD study cohort from the samples of the TD study cohort, where each sample consists of the amino acid concentrations for a given study participant. FDA uses a set of measurements stored in a matrix X, where each row in X corresponds to an individual measurement of x and each column represents a single measurement of all data points. In this matrix, a subset of data points X1 belong to the ASD cohort and the remaining subset X2 belong to the TD cohort. FDA determines the projection vector w such that calculation of discriminant scores t from the dot product t = w · x, for all x, will best separate the values of t for data points in the ASD cohort from the values of t for the points in the TD cohort. This is done by maximizing the difference between the mean value of t for each class while simultaneously minimizing the variance of t within each class (Fisher, 1936). The ratio to be maximized, J, is defined by the objective function
also known as the Fisher criterion (Bishop, 2006). In this ratio, t̄1 and t̄2 are the sample means for t = w · x of the ASD and TD cohorts, respectively, while and are the variances in discriminant scores for the ASD and TD cohorts, respectively.
Linear FDA calculates w to best separate the samples x between the ASD and TD cohorts. KFDA, a nonlinear extension of FDA, first maps each x to a higher-dimensional feature space f through the nonlinear transformation f = φ(x) and then calculates w to maximize separation (quantified by J) of the discriminant scores t between the two cohorts as calculated by t = w · f (Mika, Ratsch, Weston, Scholkopf, & Mullers, 1999). Although linear in the higher-dimensional space, the projection vector w is applied to the feature space. Nonlinear relationships and interactions between variables in the data are then able to be examined to yield a nonlinear classifier. A radial basis function kernel was used for the KFDA analysis performed in this work.
2.5.2 Kernel density estimation
For distinguishing the participants in the ASD and TD cohorts during classification, kernel density estimation was used to estimate the probability density functions (PDFs) of the discriminant scores for both groups. The aim of kernel density estimation is to determine the underlying probability distribution that a set of observed data points were drawn from (Silverman, 1986). Use of density estimates requires the selection of a value for the bandwidth parameter, which determines the smoothness of the PDF. For this work, the mean integrated squared error (Marron & Wand, 1992) was used as the criterion for choosing this parameter. The PDFs were estimated for each cohort’s discriminant scores resulting from FDA/KFDA model evaluation.
2.5.3 Null hypothesis for classification
For classification, the null hypothesis H0 states that an individual is TD and the alternative hypothesis H1 states that an individual has ASD. The Type I error for the null hypothesis is the probability of incorrectly classifying a TD participant as having ASD. Similarly, the Type II error is the probability of incorrectly classifying a participant with ASD as being TD. These errors are calculated based on a threshold chosen to separate the PDFs for the ASD and TD cohorts. Since there is overlap between the two PDFs, adjusting this threshold to reduce one error will typically cause increases in the other error. Selection of the classification threshold is thus dependent on the shapes of the PDFs as well as desired diagnostic outcomes. For this study, the threshold will be chosen so as to minimize the absolute difference between the Type I and Type II errors.
2.5.4 Leave-one-out cross-validation
As the overall goal of classification is to be able to evaluate new samples that were not used during model development, leave-one-out cross validation (Kohavi, 1995) was used to provide an independent assessment of model performance. Leave-one-out cross-validation involves removing the first sample from the data set containing N samples and then training the discriminant model using the remaining N-1 samples. The model is then used to predict the discriminant score for the first sample that was left out of the data set. This prediction is recorded and the first sample is then returned to the data set. The process is repeated such that each sample is individually removed from the data set once and subsequently evaluated with the trained model. The outcome is a set of N discriminant scores resulting from cross-validation that are then analyzed.
To evaluate classification accuracies and misclassification errors resulting from cross-validation, the discriminant scores for each of the N-1 samples included for model training were first used to estimate PDFs for the ASD and TD cohorts. The left-out validation sample was then evaluated with the trained model and its resulting discriminant score was plotted with the estimated ASD and TD distributions from the training samples. The overlap of the validation sample with its own cohort’s PDF (the correct PDF) was then calculated as the proportion of the correct PDF’s area falling between the validation sample and the tail of the correct PDF closest to the other cohort’s PDF (the incorrect PDF); this value was taken to be the sample’s classification accuracy. Similarly, the proportion of the incorrect PDF’s area falling between the validation sample and the tail of the incorrect PDF that was closest to the correct PDF was considered to be the misclassification error for the sample. Samples with a classification accuracy less than or equal to 0.05, or with a misclassification error greater than 0.05, were considered to be poorly predicted by the classifier.
3. Results
3.1 Omission of select measurements
Upon conclusion of the 12-month study period, sixteen amino acid measurements in the subset of participants in the ASD cohort that did not receive any treatment were found to be significantly changed from their baseline values (according to the paired t-test or Wilcoxon signed-rank test, depending on which was appropriate for the distribution of the data; results not shown). As this group of participants did not receive any intervention, the measurements should not have exhibited a large change for these amino acids; therefore, the data for these specific amino acids were deemed unreliable and not considered for further analysis in the present work. These omitted measurements included two essential amino acids (lysine, tryptophan), three non-essential amino acids (alanine, asparagine, aspartate), two secondary amino acids (homocystine, methionine sulfoxide), and nine amino acid metabolites (3-methylhistidine, α-aminoadipate, β-aminoisobutyrate, ethanolamine, phosphoethanolamine, phosphoserine, sarcosine, taurine, urea). Four additional measurements contained at least 90% of values in both the ASD and TD cohorts that were below their respective detection limits (anserine, carnosine, cystathionine, GABA), and therefore were also omitted. The remaining twenty-two measurements did not have any values below the detection limit and were included for all further analyses.
3.2 Univariate analysis of amino acids
Comparisons of the twenty-two included measurements in the ASD and TD cohorts are provided in Table 2; these include the results of the appropriate location tests, along with effect sizes and their 95% confidence intervals. Of these measurements, the Mann-Whitney U test found the median levels of glutamate (+21%, p = 0.014) and serine (+8%, p = 0.043) to be possibly different in the ASD cohort compared to TD controls. Welch’s t-test also showed the mean level of hydroxyproline (+17%, p = 0.018) in the ASD cohort to be possibly different from the mean level in the TD cohort. It should also be reiterated that all measurements in this table had no values that were below the detection limit.
Table 2.
Plasma concentrations (μmol/dL) of included amino acids in the ASD and TD cohorts. Means are reported for measurements when Student’s t-test or Welch’s t-test were used, and medians when the Mann-Whitney U test was used. Instances in which Welch’s t-test was used for non-identical distributions are denoted with an asterisk (*), and p-values greater than α = 0.05 are labeled not significant (n.s.). Percent difference is calculated as 100*(ASD mean or median - TD mean or median)/(TD mean or median). Effect size is defined as a measurement’s median value in the ASD cohort minus the median value in the TD cohort.
| Amino Acids | Location Test | ASD Mean or Median (25th %/75th %) | TD Mean or Median (25th %/75th %) | Percent Difference in ASD | p-value | Effect Size (95% Confidence Interval) |
|---|---|---|---|---|---|---|
| Essential | ||||||
|
| ||||||
| Histidine | Student’s t-test | 7.92 (7.33/8.50) | 8.20 (7.58/8.85) | −3% | n.s. | −0.132 (−0.518, 0.129) |
| Isoleucine | Mann-Whitney U test | 5.97 (4.88/6.72) | 5.43 (4.81/6.38) | +10% | n.s. | 0.539 (−0.194, 0.988) |
| Leucine | Mann-Whitney U test | 11.2 (9.50/13.0) | 10.7 (9.77/12.2) | +4% | n.s. | 0.475 (−0.982, 1.54) |
| Methionine | Student’s t-test | 2.24 (1.94/2.60) | 2.29 (1.92/2.63) | −2% | n.s. | −0.069 (−0.216, 0.239) |
| Phenylalanine | Mann-Whitney U test | 5.08 (4.50/5.70) | 5.14 (4.77/5.65) | −1% | n.s. | −0.054 (−0.502, 0.301) |
| Threonine | Mann-Whitney U test | 11.0 (9.18/12.7) | 11.2 (8.80/13.9) | −2% | n.s. | −0.235 (−2.15, 1.15) |
| Valine | Mann-Whitney U test | 20.9 (18.6/24.9) | 20.8 (18.5/23.1) | 0% | n.s. | 0.040 (−1.87, 2.40) |
|
| ||||||
| Non-Essential | ||||||
|
| ||||||
| Arginine | Student’s t-test | 7.72 (6.76/8.58) | 8.03 (7.01/9.15) | −4% | n.s. | −0.360 (−0.966, 0.444) |
| Cystine | Mann-Whitney U test | 3.79 (3.39/4.50) | 4.05 (3.60/4.63) | −7% | n.s. | −0.267 (−0.584, 0.157) |
| Glutamate | Mann-Whitney U test | 4.36 (3.24/5.80) | 3.61 (2.84/4.62) | +21% | 0.014 | 0.749 (0.097, 1.44) |
| Glutamine | Mann-Whitney U test | 37.4 (33.3/41.2) | 36.8 (34.0/40.8) | +2% | n.s. | 0.615 (−1.47, 2.60) |
| Glycine | Mann-Whitney U test | 24.5 (21.1/28.7) | 25.2 (21.3/29.4) | −3% | n.s. | −0.650 (−2.67, 2.18) |
| Proline | Mann-Whitney U test | 17.4 (14.6/22.9) | 17.2 (14.5/23.2) | −1% | n.s. | 0.205 (−4.90, 2.62) |
| Serine | Mann-Whitney U test | 7.80 (6.87/9.24) | 7.19 (6.53/8.46) | +8% | 0.043 | 0.613 (−0.282, 1.31) |
| Tyrosine | Mann-Whitney U test | 5.89 (5.22/6.95) | 5.75 (5.17/6.53) | +2% | n.s. | 0.141 (−0.376, 0.700) |
|
| ||||||
| Secondary | ||||||
|
| ||||||
| AABA | Student’s t-test | 2.04 (1.61/2.50) | 2.02 (1.73/2.37) | +1% | n.s. | −0.004 (−0.225, 0.278) |
| β-Alanine | Mann-Whitney U test | 0.396 (0.305/0.545) | 0.359 (0.305/0.439) | +10% | n.s. | 0.037 (−0.030, 0.091) |
| Citrulline | Student’s t-test | 2.83 (2.30/3.26) | 2.99 (2.52/3.45) | −5% | n.s. | −0.120 (−0.501, 0.246) |
| Hydroxyproline | Welch’s t-test* | 1.83 (1.22/2.21) | 1.56 (1.41/1.77) | +17% | 0.018 | 0.185 (−0.017, 0.429) |
| Ornithine | Mann-Whitney U test | 5.93 (5.05/7.06) | 5.71 (5.19/6.71) | +4% | n.s. | 0.221 (−0.550, 0.657) |
|
| ||||||
| Metabolites | ||||||
|
| ||||||
| 1-Methylhistidine | Mann-Whitney U test | 0.280 (0.213/0.372) | 0.269 (0.222/0.352) | +4% | n.s. | 0.011 (−0.051, 0.076) |
| Ammonia | Mann-Whitney U test | 7.90 (6.90/9.70) | 8.30 (6.90/12.0) | −5% | n.s. | −0.400 (−3.10, 0.950) |
After correcting for multiple comparisons (α/22 = 0.002), none of the measurements presented a statistically significant p-value. Glutamate was the only measurement with a 95% confidence interval for effect size that did not contain zero, indicating it to have the only significant effect size by our definition. However, the overlap between cohorts in these measurements demonstrates that univariate statistics are not useful for differentiating the ASD and TD cohorts in these data.
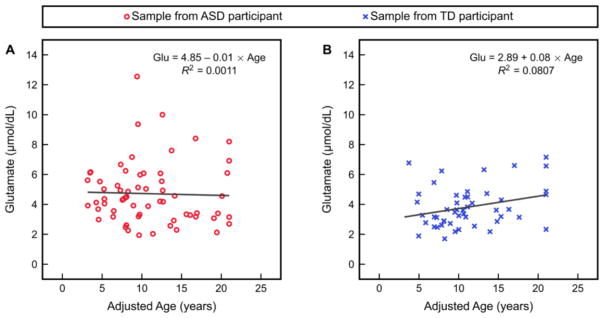
Given the observation of elevated glutamate in the ASD cohort, as well as the observation that plasma amino acid levels vary with age (Lepage et al., 1997), glutamate was then plotted as a function of age separately in each study cohort (Figure 1). The age of each individual used for this analysis was adjusted such that any individual with a clinical age greater than 21 years was assigned an age of 21 years. This adjusted age was used because most participants were under age 21, and amino acid levels are highly age-dependent for children while levels generally become more stable at older ages (Lepage et al., 1997; Caballero et al., 1991); therefore, compressing the data for those over age 21 allowed for plots that were less sparse. Linear regression of this glutamate-versus-age relationship revealed no significant association between glutamate and age for either cohort, accompanied by a poor fit for each regression model.
Figure 1.
Glutamate as a function of adjusted age (i.e. any individual with a clinical age greater than 21 years is assigned an age of 21 years) and the results of linear regression of this relationship in (A) the ASD study cohort and (B) the TD study cohort.
3.3 Classification with FDA
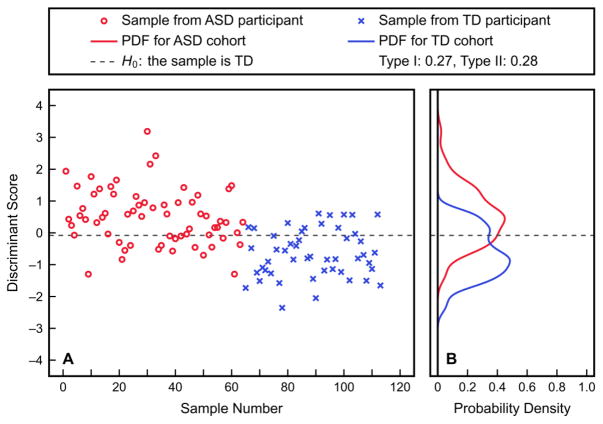
To next explore the possibility of distinguishing the two study cohorts using multiple plasma amino acid measurements, an FDA model was first fit to all twenty-two included measurements under consideration. The fitted discriminant scores output by this analysis and the estimated PDFs of these scores in the two cohorts are shown in Figure 2; the Type I and Type II errors associated with the shown threshold are 27.0% and 27.8%, respectively. Table 3 demonstrates the trade-off between the Type I and Type II errors with these PDFs. The large errors suggest only weak linear relationships between measurements that are useful for distinguishing the ASD and TD cohorts. Cross-validation results are not reported for this linear analysis as the results for fitting already show poor separation between the ASD and TD cohorts that would be even worse after cross-validation.
Figure 2.
Results of fitting an FDA model to all twenty-two included amino acid measurements. Discriminant scores for each sample (A), as well as PDFs of these fitted scores for the ASD and TD study cohorts (B), are presented. The shown threshold corresponds to a Type I error of 27.0% and a Type II error of 27.8%.
Table 3.
Type I and Type II errors obtained from the fitted FDA model using all twenty-two included amino acid measurements as inputs. Errors were calculated using the estimated PDFs and are with respect to the null hypothesis H0 that a sample belongs to the TD cohort.
| Fitted FDA Model | |||||||
|---|---|---|---|---|---|---|---|
| Type I error | 0.40 | 0.35 | 0.30 | 0.25 | 0.20 | 0.15 | 0.10 |
| Type II error | 0.15 | 0.19 | 0.25 | 0.30 | 0.36 | 0.43 | 0.50 |
3.4 Classification with KFDA
Nonlinear KFDA was then employed to assess whether any nonlinear relationships between plasma amino acids are present that could offer improvements to classification performance. Two factors that must be considered when using KFDA are that its computational cost is significantly greater than that of FDA, and that it is more prone to model overfitting when a greater number of input variables are used. To address both of these concerns while identifying the subset of measurements with the best KFDA classification performance, all combinations of up to five plasma amino acids were analyzed; in other words, variable sets containing more than five plasma amino acids were not considered.
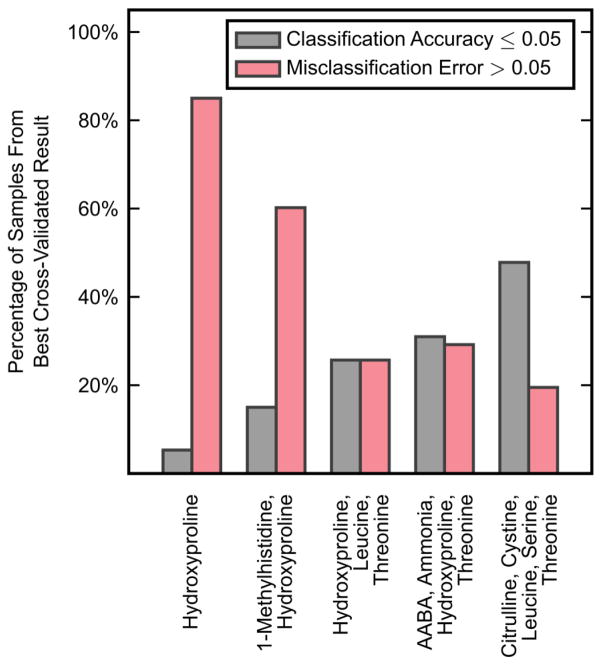
A KFDA model was first fit to each of these candidate subsets of measurements, with the output value of J, the Fisher criterion, being recorded for each combination. For each subset size, the 1000 measurement combinations that produced the greatest value of J were then evaluated with leave-one-out cross-validation; in the cases with fewer than 1000 total combinations, all combinations were tested with cross-validation. Figure 3 shows the combination of measurements for each subset size that performed best under cross-validation, presenting both the percentage of samples that had poor classification accuracy and the percentage of samples that had high misclassification error. Each of these combinations minimized the sum of these percentages for its respective subset size. Using three input variables (hydroxyproline, leucine, threonine) produced the best overall results with both of these percentages at 25.7% of samples. These three variables were thus analyzed further with KFDA.
Figure 3.
The best leave-one-out cross-validation results obtained across all possible combinations of different numbers of KFDA input variables. Cross-validation results are expressed as the percentage of samples that had a classification accuracy less than or equal to 0.05 and the percentage of samples that had a misclassification error greater than 0.05. The best combinations of variables were those that minimized the sum of these two percentages.
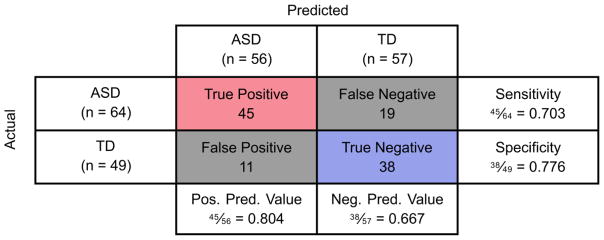
Fitted discriminant scores and PDFs resulting from the aforementioned KFDA model, as well as the predicted discriminant scores resulting from cross-validation, are presented in Figure 4. It is clear from these plots that the Type I error (4.2%) and Type II error (5.0%) from KFDA fitting are small compared to the fitting errors observed with FDA; however; these smaller fitting errors are not representative of the classification performance observed after model cross-validation. Additionally, it should be noted that some cross-validated discriminant scores had very large positive or negative values and thus could not be appropriately presented; the samples with these extreme values are shown as darker points at the bounds of the plot, but were not manipulated for the analysis. Figure 5 then provides the individual samples’ classification accuracies and misclassification errors resulting from cross-validation with this KFDA model. 25.7% of samples had a classification accuracy less than or equal to 0.05 and 25.7% of samples also had a misclassification error greater than 0.05. Finally, the confusion matrix for classification into the ASD and TD cohorts after cross-validation is given in Figure 6. Despite these results being for the best-performing KFDA model, the sensitivity was only 70.3% and the specificity was marginally better at 77.6%. Combined, these results indicate that the classifier can correctly predict 73.5% of samples as belonging either to the ASD or TD cohort, which makes these amino acid measurements insufficient as a biochemical marker for ASD by themselves.
Figure 4.
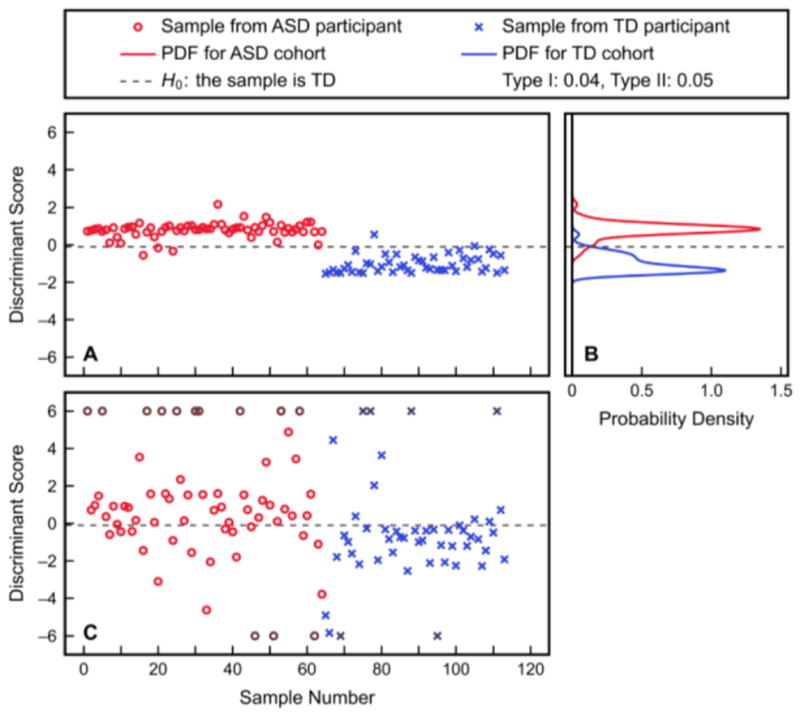
Classification with KFDA using hydroxyproline, leucine, and threonine as inputs. Shown are (A) the discriminant scores from fitting, (B) the PDFs of these fitted scores for the ASD and TD study cohorts, and (C) the predicted discriminant scores resulting from leave-one-out cross-validation. The Type I and Type II errors from the fitted PDFs with the shown classification threshold are 4.2% and 5.0%, respectively. Discriminant scores outside the range of [-6, 6] are plotted as darker points at the respective bound, for visualization purposes only. Misclassification errors of the fitted scores are significantly smaller than those of the cross-validated results.
Figure 5.
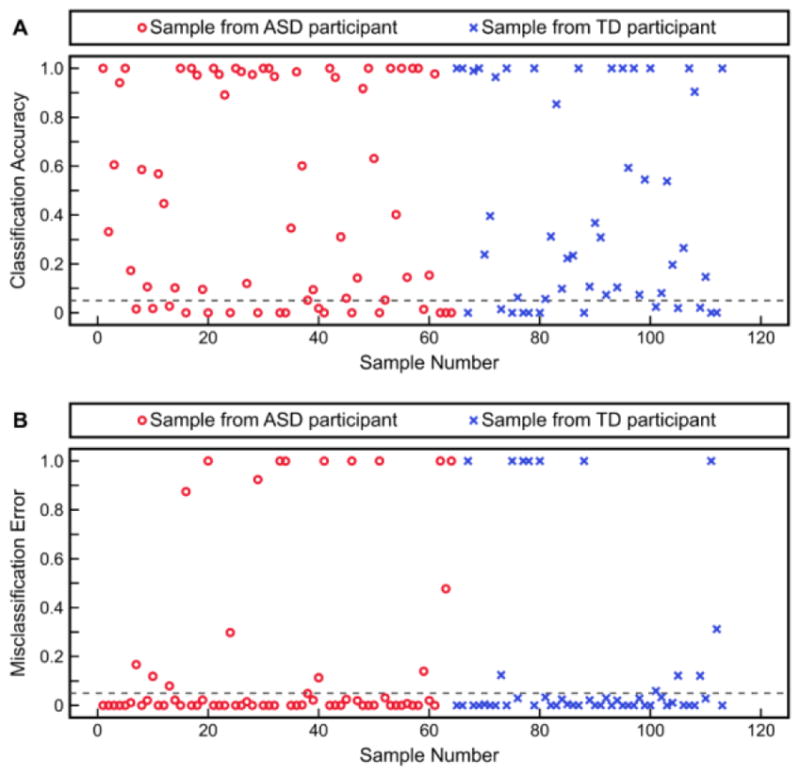
Leave-one-out cross-validation results from classification with KFDA using hydroxyproline, leucine, and threonine as inputs. Shown are each individual sample’s (A) classification accuracy and (B) misclassification error. Samples with a classification accuracy less than or equal to 0.05, or with a misclassification error greater than 0.05, are considered to be poorly predicted.
Figure 6.
Confusion matrix showing the results of leave-one-out cross-validation with KFDA using hydroxyproline, leucine, and threonine as inputs. The sensitivity is calculated as the number of true positives divided by the total number of individuals with ASD, while specificity is the number of true negatives divided by the total number of TD individuals. Positive predictive value is equal to the number of true positives divided by the total number of individuals predicted to have ASD, and negative predictive value is defined as the number of true negatives divided by the total number of predicted TD individuals.
4. Discussion
Measurements from the Comprehensive Nutritional and Dietary Intervention Study indicate possibly elevated concentrations of glutamate, hydroxyproline, and serine in the plasma of individuals with ASD. The finding of elevated glutamate, in particular, is consistent with findings from other recent studies (Adams et al., 2011; Shimmura et al., 2011; Tirouvanziam et al., 2011; Tu, Chen, & He, 2012; Hassan et al., 2013; Naushad, Jain, Prasad, Naik, & Akella, 2013; El-Ansary & Al-Ayadhi, 2014; Cai et al., 2016; El-Ansary, 2016; Zaki, Abdel-Al, & Al-Sawi, 2017), suggesting a potential role for glutamate in ASD pathophysiology. Glutamate is the most abundant neurotransmitter and the primary excitatory neurotransmitter in the human nervous system (Meldrum, 2000). It contributes to synaptic plasticity with significant roles in learning and memory formation (Nakanishi, 1992). Overstimulation of glutamate receptors can lead to excitotoxicity and subsequent neuronal damage or death, which are thought to be linked to a number of chronic neurologic disorders such as Huntington’s disease, Parkinson’s disease, and epilepsy (Sheldon & Robinson, 2007); elevated plasma glutamate in ASD may indicate a similar condition of excitotoxicity. Some studies have also found increased plasma glutamate levels to be correlated with the severity of certain ASD-related behaviors and symptoms (Adams et al., 2011; Cai et al., 2016). Our finding of glutamate being independent of age (in both the ASD and the TD cohort) may be indicative of a lifelong elevation of plasma glutamate in individuals with ASD. However, this finding is in disagreement with that of Lepage et al. (Lepage et al., 1997), who reported plasma glutamate concentrations to decrease notably between 0 and 18 years of age.
Aside from glutamate, our findings conflict with some of the studies in the literature that focus on plasma amino acid measurements in individuals with ASD. As mentioned previously, a likely explanation for this discrepancy is the varying degree to which these studies addressed the study design challenges associated with small sample size, use of age- and gender-matched controls, overnight fasting, and corrections for multiple hypothesis testing. One or more of these methodological limitations are present in most recent studies (after 2011) that have compared the plasma amino acid measurements of individuals on the autism spectrum to those of their TD peers. Two studies were identified (El-Ansary & Al-Ayadhi, 2014; El-Ansary, 2016) where the only limitation was an insufficient sample size (i.e. less than 25 participants in each study cohort). Two other studies were found to only lack correction for multiple hypothesis testing (Naushad et al., 2013; Zaki et al., 2017). Five identified studies were affected by two of the aforementioned limitations (Shimmura et al., 2011; Tu et al., 2012; Hassan et al., 2013; Kuwabara et al., 2013; Bugajska, Berska, Wojtyto, Bik-Multanowski, & Sztefko, 2017), and three others were limited by three of these study design challenges (Tirouvanziam et al., 2011; ElBaz, Zaki, Youssef, ElDorry, & Elalfy, 2014; Bala et al., 2016).
While it is difficult to compare the results of studies that do not account for these factors, a comparison to the two studies that did address all four components will be offered. In their previous study in Arizona, Adams et al. (Adams et al., 2011) detected significantly elevated mean levels of glutamate (+18%, p = 0.001), an insignificant increase in hydroxyproline (+9.6%, p-value not significant), and a possibly significant increase in serine (+10%, p = 0.04) in the ASD cohort, along with a significantly decreased mean concentration of tryptophan (−19%, p = 0.001) that was not seen in the current data. That study did not use measurements for classification and so it is unknown whether those data would be useful for predicting ASD. In the second study, Cai et al. (Cai et al., 2016) measured only glutamate levels and found them to be significantly higher (+53%, p < 0.0001) in the cohort of ASD patients relative to healthy controls; that difference was sufficiently significant to achieve an area under the receiver operating characteristic curve of 0.92 using glutamate as the only predictor variable. Based on this finding, glutamate may potentially offer stronger predictive ability than was indicated with our data.
It is also worth discussing the two studies that lacked only an adjustment for multiple comparisons, as they still provide insight into which plasma measurements may be elevated or reduced in individuals with ASD. The first of these studies, by Naushad et al. (Naushad et al., 2013), detected significantly increased mean plasma levels of glutamate (+45%, p = 0.01) and asparagine (+81%, p = 0.0001) in their ASD cohort, along with significant decreases in the mean levels of four amino acids that included tryptophan (−41%, p = 0.0001). In the other study, Zaki et al. (Zaki et al., 2017) reported significantly elevated median levels of plasma glutamate (+517%, p = 0.031) and hydroxyproline (+900%, p = 0.022), plus three other amino acids, in individuals with ASD; their study also found median levels of thirteen amino acids, including serine (−48%, p = 0.032) and tryptophan (−92%, p = 0.03), to be significantly lower in their ASD cohort.
Despite our finding of possible differences in glutamate, hydroxyproline, and serine measurements between the ASD and TD cohorts, clear discrimination of the cohorts was not possible using these data. Fitting an FDA model to all available measurements yielded large Type I and Type II errors, indicating that a linear classifier is not adequate for uncovering meaningful patterns in the data. Further nonlinear analysis with KFDA was also unable to reveal a robust set of measurements for predicting which individuals had ASD, as indicated by the model’s poor predictive performance under cross-validation. Together, these results suggest that any differences in plasma amino acid measurements in ASD are not sufficiently significant by themselves to accurately distinguish individuals with the disorder from their TD peers. However, it is possible that they could be used as part of a larger biomarker set.
Although glutamate and other amino acids, such as methionine, have been found by other studies to be important in ASD etiology, these measurements did not appear in our best model for the classification task. While a model using hydroxyproline, leucine, and threonine produced the best overall results for KFDA, the model predictions were still poor when cross-validation was involved. This suggests that these amino acids likely do not serve any significant biochemical roles in the pathophysiology of ASD, but further investigation of their relevance may still be warranted. It is also worth highlighting that some studies have reported significant associations between certain ASD-related symptom/behavior assessments (Adams et al., 2011; Kuwabara et al., 2013; ElBaz et al., 2014; Cai et al., 2016) and individual plasma amino acids while others have reported these relationships to be insignificant (Shimmura et al., 2011; Tirouvanziam et al., 2011). In the interest of evaluating any such associations in our data, linear regression was used to model the relationships between each ASD assessment (as detailed in Section 2.1) and each individual amino acid in the individuals with ASD. The strongest relationship identified by linear regression was between the Reynolds Intellectual Assessment Scales Verbal Intelligence Index and the amino acid isoleucine (R2 = 0.096). Given the generally poor fit of these regression models, there do not appear to be any strong relationships.
Considering the roles of amino acids in FOCM/TS and the highly accurate prediction of individuals with ASD previously achieved using plasma measurements from these pathways, it at first seems surprising that the current classification results were so poor. However, the reason for this disagreement is actually quite clear. The accurate classification presented by Howsmon et al. (Howsmon et al., 2017) was achieved using a set of epigenetic and oxidative stress markers derived from FOCM/TS measurements, with minimal contribution from the amino acid measurements themselves; only the ratio of oxidized free cysteine to reduced free cysteine (a measure of extracellular redox status) was determined to be in the final set of predictor variables, and even then this does not rely on a single measured quantity. This suggests that the amino acid measurements themselves are not strong predictors of ASD. Much greater value is offered by instead taking into account the physiological contexts of the amino acids and perhaps by incorporating other physiologically-relevant quantities, e.g., FOCM biomarkers, into the classification procedure.
4.1 Limitations
The current study is not without limitations. For one, all study participants were from Arizona, so observed plasma amino acid profiles may be affected by the geographic region. A study including participants from other regions would provide a more general description of these profiles and help to address this potential bias. Another limitation is the broad age range, which is a problem since some amino acid levels are reported to change greatly with age; a narrower age range may yield more significant results. The heterogeneity of ASD may also make it more difficult to identify a biomarker for the disorder that is applicable to all individuals. Additionally, omitting twenty out of forty-two amino acid measurements from multivariate analysis may have removed potentially important information that could have helped to classify the ASD and TD cohorts with greater accuracy. However, it could also be argued that finding so many amino acids to be undetectable or significantly changed from baseline in a non-treatment ASD group is reflective of the large variability in plasma amino acid levels at the individual level, and highlights the general caution with which amino acid measurements should be interpreted by themselves.
The sample size of our study also might not be regarded as a substantial improvement over those studies that have been previously discussed. However, our sample size is still more than double that of most similar studies. For example, other recent studies have used cohort sample sizes of 23 ASD and 22 TD (Shimmura et al., 2011); 27 ASD and 20 TD (Tirouvanziam et al., 2011); 20 ASD and 20 TD (Tu et al., 2012); 10 ASD and 10 TD (Hassan et al., 2013); 25 ASD and 28 TD (Kuwabara et al., 2013); 20 ASD and 20 TD (ElBaz et al., 2014); 20 ASD and 19 TD (El-Ansary & Al-Ayadhi, 2014); 21 ASD and 21 TD (Bala et al., 2016); 20 ASD and 20 TD (El-Ansary, 2016); 42 ASD and 26 TD (Zaki et al., 2017); and 27 ASD and 13 TD (Bugajska et al., 2017). While some studies do have similar or larger sample sizes, such as with 55 ASD and 44 TD (Adams et al., 2011); 138 ASD and 138 TD (Naushad et al., 2013); or 51 ASD and 51 TD (Cai et al., 2016), there are many more studies with small sample sizes than with comparable ones. Thus it is worth highlighting that our study features a larger sample size than is commonly seen in studies of plasma amino acids in ASD.
In addition, the use of medications by 45.3% of the ASD cohort and 8.2% of the TD cohort may have influenced the concentrations of certain plasma amino acids in these individuals. That being said, requiring participants to maintain the same medication regimen throughout the course of the study was intended to minimize any potential variability that may have arisen from the use of medications.
4.2 Implications
The results of this study suggest that some plasma amino acid measurements may be different in individuals with ASD compared to TD controls. However, this difference is not sufficient for clear discrimination between our two study cohorts. This may offer an explanation for the general disagreement across studies that have investigated plasma amino acid profiles in ASD (Zheng, Wang, Li, Rauw, & Baker, 2017); there may not be any consistent patterns in the measurements that are truly correlated to ASD incidence. Diverse roles for amino acids in the body may make it difficult to assess what a deficiency or excess of a particular amino acid in the plasma truly indicates in the context of ASD.
Supplementary Material
Highlights.
Plasma amino acids from 64 children with ASD and 49 typically-developing (TD) peers were analyzed.
No significant univariate differences were found between the ASD and TD cohorts.
Multivariate analysis revealed poor ability to classify as ASD/TD with these data.
Our findings suggest plasma amino acids are not a good predictor for ASD status.
Acknowledgments
The authors gratefully acknowledge partial financial support from the National Institutes of Health (Grant 1R01AI110642-01A1). The funders had no role in study design; in collection, analysis, or interpretation of data; in writing the report; or in the decision to submit the article for publication.
Footnotes
Conflicts of interest
The authors declare that they have no conflicts of interest.
Data availability
The de-identified data used in this study are available in the supplementary information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JB, Audhya T, McDonough-Means S, Rubin RA, Quig D, Geis E, … Lee W. Nutritional and metabolic status of children with autism vs. neurotypical children, and the association with autism severity. Nutrition & Metabolism. 2011;8:34. doi: 10.1186/1743-7075-8-34. https://doi.org/10.1186/1743-7075-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JB, Howsmon DP, Kruger U, Geis E, Gehn E, Fimbres V, … Hahn J. Significant association of urinary toxic metals and autism-related symptoms—a nonlinear statistical analysis with cross validation. PLOS ONE. 2017;12(1):e0169526. doi: 10.1371/journal.pone.0169526. https://doi.org/10.1371/journal.pone.0169526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TW, Darling DA. A test of goodness of fit. Journal of the American Statistical Association. 1954;49(268):765. https://doi.org/10.2307/2281537. [Google Scholar]
- Bala KA, Doğan M, Mutluer T, Kaba S, Aslan O, Balahoroğlu R, … Kocaman S. Plasma amino acid profile in autism spectrum disorder (ASD) European Review for Medical and Pharmacological Sciences. 2016;20(5):923–929. [PubMed] [Google Scholar]
- Banjanovic ES, Osborne JW. Confidence intervals for effect sizes: applying bootstrap resampling. Practical Assessment, Research & Evaluation. 2016;21(5) [Google Scholar]
- Bishop CM. Pattern Recognition and Machine Learning. New York, NY: Springer-Verlag New York, Inc; 2006. [Google Scholar]
- Brown MB, Forsythe AB. Robust tests for the equality of variances. Journal of the American Statistical Association. 1974;69(346):364–367. https://doi.org/10.2307/2285659. [Google Scholar]
- Bugajska J, Berska J, Wojtyto T, Bik-Multanowski M, Sztefko K. The amino acid profile in blood plasma of young boys with autism. Psychiatria Polska. 2017;51(2):359–368. doi: 10.12740/PP/65046. https://doi.org/10.12740/PP/65046. [DOI] [PubMed] [Google Scholar]
- Caballero B, Gleason RE, Wurtman RJ. Plasma amino acid concentrations in healthy elderly men and women. The American Journal of Clinical Nutrition. 1991;53(5):1249–1252. doi: 10.1093/ajcn/53.5.1249. [DOI] [PubMed] [Google Scholar]
- Cai J, Ding L, Zhang JS, Xue J, Wang LZ. Elevated plasma levels of glutamate in children with autism spectrum disorders. Neuroreport. 2016;27(4):272–276. doi: 10.1097/WNR.0000000000000532. https://doi.org/10.1097/WNR.0000000000000532. [DOI] [PubMed] [Google Scholar]
- Chugani DC, Muzik O, Behen M, Rothermel R, Janisse JJ, Lee J, Chugani HT. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Annals of Neurology. 1999;45(3):287–295. doi: 10.1002/1531-8249(199903)45:3<287::aid-ana3>3.0.co;2-9. https://doi.org/10.1002/1531-8249(199903)45:3<287::AID-ANA3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Deth R, Muratore C, Benzecry J, Power-Charnitsky VA, Waly M. How environmental and genetic factors combine to cause autism: A redox/methylation hypothesis. NeuroToxicology. 2008;29(1):190–201. doi: 10.1016/j.neuro.2007.09.010. https://doi.org/10.1016/j.neuro.2007.09.010. [DOI] [PubMed] [Google Scholar]
- El-Ansary A. Data of multiple regressions analysis between selected biomarkers related to glutamate excitotoxicity and oxidative stress in Saudi autistic patients. Data in Brief. 2016;7:111–116. doi: 10.1016/j.dib.2016.02.025. https://doi.org/10.1016/j.dib.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ansary A, Al-Ayadhi L. GABAergic/glutamatergic imbalance relative to excessive neuroinflammation in autism spectrum disorders. Journal of Neuroinflammation. 2014;11:189. doi: 10.1186/s12974-014-0189-0. https://doi.org/10.1186/s12974-014-0189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElBaz FM, Zaki MM, Youssef AM, ElDorry GF, Elalfy DY. Study of plasma amino acid levels in children with autism: An Egyptian sample. Egyptian Journal of Medical Human Genetics. 2014;15(2):181–186. https://doi.org/10.1016/j.ejmhg.2014.02.002. [Google Scholar]
- Fernstrom JD. Dietary amino acids and brain function. Journal of the American Dietetic Association. 1994;94(1):71–77. doi: 10.1016/0002-8223(94)92045-1. https://doi.org/10.1016/0002-8223(94)92045-1. [DOI] [PubMed] [Google Scholar]
- Fernstrom JD, Wurtman RJ. Brain serotonin content: physiological dependence on plasma tryptophan levels. Science (New York, NY ) 1971;173(3992):149–152. doi: 10.1126/science.173.3992.149. [DOI] [PubMed] [Google Scholar]
- Fernstrom JD, Wurtman RJ. Brain serotonin content: physiological regulation by plasma neutral amino acids. Science (New York, NY ) 1972;178(4059):414–416. doi: 10.1126/science.178.4059.414. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The use of multiple measurements in taxonomic problems. Annals of Eugenics. 1936;7(2):179–188. https://doi.org/10.1111/j.1469-1809.1936.tb02137.x. [Google Scholar]
- Harada M, Taki MM, Nose A, Kubo H, Mori K, Nishitani H, Matsuda T. Non-invasive evaluation of the GABAergic/glutamatergic system in autistic patients observed by MEGA-editing proton MR spectroscopy using a clinical 3 tesla instrument. Journal of Autism and Developmental Disorders. 2011;41(4):447–454. doi: 10.1007/s10803-010-1065-0. https://doi.org/10.1007/s10803-010-1065-0. [DOI] [PubMed] [Google Scholar]
- Hassan TH, Abdelrahman HM, Abdel Fattah NR, El-Masry NM, Hashim HM, El-Gerby KM, Abdel Fattah NR. Blood and brain glutamate levels in children with autistic disorder. Research in Autism Spectrum Disorders. 2013;7(4):541–548. https://doi.org/10.1016/j.rasd.2012.12.005. [Google Scholar]
- Howsmon DP, Kruger U, Melnyk S, James SJ, Hahn J. Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLOS Computational Biology. 2017;13(3):e1005385. doi: 10.1371/journal.pcbi.1005385. https://doi.org/10.1371/journal.pcbi.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, … Gaylor DW. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2006;141B(8):947–956. doi: 10.1002/ajmg.b.30366. https://doi.org/10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of the 14th International Joint Conference on Artificial Intelligence; San Francisco, CA, USA: Morgan Kaufmann Publishers Inc; 1995. pp. 1137–1143. [Google Scholar]
- Kuwabara H, Yamasue H, Koike S, Inoue H, Kawakubo Y, Kuroda M, … Kasai K. Altered metabolites in the plasma of autism spectrum disorder: a capillary electrophoresis time-of-flight mass spectroscopy study. PLOS ONE. 2013;8(9):e73814. doi: 10.1371/journal.pone.0073814. https://doi.org/10.1371/journal.pone.0073814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage N, McDonald N, Dallaire L, Lambert M. Age-specific distribution of plasma amino acid concentrations in a healthy pediatric population. Clinical Chemistry. 1997;43(12):2397–2402. [PubMed] [Google Scholar]
- Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. The Annals of Mathematical Statistics. 1947;18(1):50–60. [Google Scholar]
- Marron JS, Wand MP. Exact mean integrated squared error. The Annals of Statistics. 1992;20(2):712–736. [Google Scholar]
- Massey FJ. The Kolmogorov-Smirnov test for goodness of fit. Journal of the American Statistical Association. 1951;46(253):68. https://doi.org/10.2307/2280095. [Google Scholar]
- McDougle C, Naylor ST, Cohen DJ, Aghajanian GK, Heninger GR, Price LH. Effects of tryptophan depletion in drug-free adults with autistic disorder. Archives of General Psychiatry. 1996;53(11):993–1000. doi: 10.1001/archpsyc.1996.01830110029004. https://doi.org/10.1001/archpsyc.1996.01830110029004. [DOI] [PubMed] [Google Scholar]
- Meldrum BS. Glutamate as a neurotransmitter in the brain: review of physiology and pathology. The Journal of Nutrition. 2000;130(4):1007–1007. doi: 10.1093/jn/130.4.1007S. [DOI] [PubMed] [Google Scholar]
- Mika S, Ratsch G, Weston J, Scholkopf B, Mullers KR. Fisher discriminant analysis with kernels. Proceedings of the Neural Networks for Signal Processing IX Workshop; 1999. pp. 41–48. https://doi.org/10.1109/NNSP.1999.788121. [Google Scholar]
- Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258(5082):597–603. doi: 10.1126/science.1329206. https://doi.org/10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- Naushad SM, Jain JMN, Prasad CK, Naik U, Akella RRD. Autistic children exhibit distinct plasma amino acid profile. Indian Journal of Biochemistry & Biophysics. 2013;50(5):474–478. [PubMed] [Google Scholar]
- Proenza AM, Cresp C, Roca P, Palou A. Gender related differences in the effect of aging on blood amino acid compartmentation. The Journal of Nutritional Biochemistry. 2001;12(7):431–440. doi: 10.1016/s0955-2863(01)00157-7. https://doi.org/10.1016/S0955-2863(01)00157-7. [DOI] [PubMed] [Google Scholar]
- Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochemistry International. 2007;51(6–7):333–355. doi: 10.1016/j.neuint.2007.03.012. https://doi.org/10.1016/j.neuint.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmura C, Suda S, Tsuchiya KJ, Hashimoto K, Ohno K, Matsuzaki H, … Mori N. Alteration of plasma glutamate and glutamine levels in children with high-functioning autism. PLOS ONE. 2011;6(10):e25340. doi: 10.1371/journal.pone.0025340. https://doi.org/10.1371/journal.pone.0025340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman BW. Density Estimation for Statistics and Data Analysis. New York: CRC Press; 1986. [Google Scholar]
- Student. The probable error of a mean. Biometrika. 1908;6(1):1–25. https://doi.org/10.2307/2331554. [Google Scholar]
- Tirouvanziam R, Obukhanych TV, Laval J, Aronov PA, Libove R, Banerjee AG, … Hardan AY. Distinct plasma profile of polar neutral amino acids, leucine, and glutamate in children with autism spectrum disorders. Journal of Autism and Developmental Disorders. 2011;42(5):827–836. doi: 10.1007/s10803-011-1314-x. https://doi.org/10.1007/s10803-011-1314-x. [DOI] [PubMed] [Google Scholar]
- Tu WJ, Chen H, He J. Application of LC-MS/MS analysis of plasma amino acids profiles in children with autism. Journal of Clinical Biochemistry and Nutrition. 2012;51(3):248–249. doi: 10.3164/jcbn.12-45. https://doi.org/10.3164/jcbn.12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason T, Howsmon DP, McGuinness DL, Hahn J. On the use of multivariate methods for analysis of data from biological networks. Processes. 2017;5(3):36. doi: 10.3390/pr5030036. https://doi.org/10.3390/pr5030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason T, Howsmon DP, Melnyk S, James SJ, Hahn J. Mathematical modeling of the methionine cycle and transsulfuration pathway in individuals with autism spectrum disorder. Journal of Theoretical Biology. 2017;416:28–37. doi: 10.1016/j.jtbi.2016.12.021. https://doi.org/10.1016/j.jtbi.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch BL. The significance of the difference between two means when the population variances are unequal. Biometrika. 1938;29(3/4):350–362. https://doi.org/10.2307/2332010. [Google Scholar]
- Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37(1):1–17. doi: 10.1007/s00726-009-0269-0. https://doi.org/10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- Zaki MM, Abdel-Al H, Al-Sawi M. Assessment of plasma amino acid profile in autism using cation-exchange chromatography with postcolumn derivatization by ninhydrin. Turkish Journal of Medical Sciences. 2017;47(1):260–267. doi: 10.3906/sag-1506-105. https://doi.org/10.3906/sag-1506-105. [DOI] [PubMed] [Google Scholar]
- Zheng HF, Wang WQ, Li XM, Rauw G, Baker GB. Body fluid levels of neuroactive amino acids in autism spectrum disorders: a review of the literature. Amino Acids. 2017;49(1):57–65. doi: 10.1007/s00726-016-2332-y. https://doi.org/10.1007/s00726-016-2332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.