Abstract
Both in mammals and plants, excess lysine (Lys) is catabolized via saccharopine into α-amino adipic semialdehyde and glutamate by two consecutive enzymes, Lys-ketoglutarate reductase (LKR) and saccharopine dehydrogenase (SDH), which are linked on a single bifunctional polypeptide. To study the control of metabolite flux via this bifunctional enzyme, we have purified it from developing soybean (Glycine max) seeds. LKR activity of the bifunctional LKR/SDH possessed relatively high Km for its substrates, Lys and α-ketoglutarate, suggesting that this activity may serve as a rate-limiting step in Lys catabolism. Despite their linkage, the LKR and SDH enzymes possessed significantly different pH optima, suggesting that SDH activity of the bifunctional enzyme may also be rate-limiting in vivo. We have previously shown that Arabidopsis plants contain both a bifunctional LKR/SDH and a monofunctional SDH enzymes (G. Tang, D. Miron, J.X. Zhu-Shimoni, G. Galili [1997] Plant Cell 9: 1–13). In the present study, we found no evidence for the presence of such a monofunctional SDH enzyme in soybean seeds. These results may provide a plausible regulatory explanation as to why various plant species accumulate different catabolic products of Lys.
The cellular level of the essential amino acid Lys is subject to tight regulation both in mammals and plants. In both types of organisms, excess Lys is catabolized via the α-amino adipic acid pathway, also named as the saccharopine pathway (Moeller, 1976; Arruda et al., 1982; Arruda and Da Silva, 1983; Markovitz et al., 1984; Brochetto-Braga et al., 1992; Galili, 1995; Goncalves-Butruille et al., 1996; Azevedo et al., 1997). The first enzyme in the Lys catabolic pathway is Lys-ketoglutarate reductase (LKR), also named as Lys 2-oxoglutarate reductase, which condenses Lys and α-ketoglutarate into saccharopine and uses the co-factor NADPH. The second enzyme, saccharopine dehydrogenase (SDH), converts saccharopine into α-amino adipic semialdehyde and Glu. This enzyme uses NAD′, or much less efficiently NADP′as a co-factor (Markovitz et al., 1984; Goncalves-Butruille et al., 1996). α-Amino adipic semialdehyde is further converted via α-amino adipic acid to acetyl-coenzyme A (Lehninger, 1975).
The molecular and biochemical regulation of Lys catabolism is still not clearly understood. Feeding of Lys to rat liver or developing tobacco seeds stimulated the activity of LKR (Foster et al., 1993; Karchi et al., 1994). A similar stimulation of this enzyme was also observed in transgenic tobacco (Nicotiana tabaccum) seeds that overproduced Lys due to expression of a bacterial feedback-insensitive dihydrodipicolinate synthase (Karchi et al., 1995). This suggested that both in animal and plant cells, Lys may auto regulate its own catabolism in vivo. Recent studies using developing tobacco and soybean (Glycine max) seeds have also suggested that the activity of LKR is modulated by an intracellular signaling cascade requiring Ca2+ and protein phosphorylation (Karchi et al., 1995; Miron et al., 1997). The control of LKR activity in plants may even be more complex. Various plant species were shown to accumulate different catabolic products of Lys, implying differential metabolite flux via LKR and SDH enzymes (Falco et al., 1995). Moreover, in developing maize seeds, LKR activity was found to be significantly reduced in the high-Lys opaque-2 mutant, as compared with wild-type plants (Brochetto-Braga et al., 1992; Gaziola et al., 1999; Kemper et al., 1999). Opaque-2 is a βZIP transcription factor that regulates the expression of seed storage proteins (Shotwell and Larkins, 1988). In addition, expression of an Arabidopsis LKR/SDH gene was shown to be significantly up-regulated in floral organs and developing seeds (Tang et al., 1997).
Biochemical studies (Markovitz and Chuang, 1987; Goncalves-Butruille et al., 1996; Gaziola et al., 1997; Miron et al., 1997) and gene cloning (Epelbaum et al., 1997; Tang et al., 1997; Kemper et al., 1999) have shown that both in mammals and plants LKR is linked to the second enzyme of Lys catabolism, SDH, on a single bifunctional polypeptide. The molecular basis for this linkage was not elucidated, but suggested that the structure of this bifunctional enzyme may also have an important regulatory role (Kemper et al., 1998). In the present report, we have purified the bifunctional LKR/SDH to near homogeneity from developing soybean seeds to study its enzymatic properties. We found that the linked LKR and SDH activities contained significantly different pH optima, similarly to LKR and SDH enzymes from other plant species (Goncalves-Butruille et al., 1996; Gaziola et al., 1997, 1999) as well as significantly different Kms for their specific substrates at their optimal pH. In addition, our results suggest that soybean may not contain an additional monofunctional SDH isozyme as has previously been observed in Arabidopsis and canola (Tang et al., 1997; Deleu et al., 1999). This may explain previous observations (Falco et al., 1995) showing that various plant species accumulate different catabolic intermediates of Lys.
RESULTS
Purification of Soybean LKR and SDH
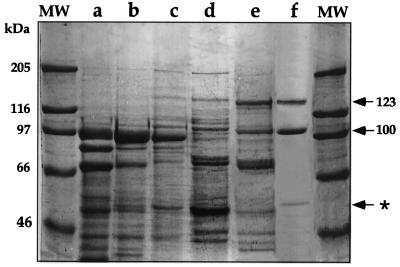
To study the biochemical properties of the soybean bifunctional LKR/SDH, this enzyme was purified to near homogeneity from developing soybean seeds. The degrees of purification after the various steps, as described in “Materials and Methods,” are shown in Table I and Figure 1. During all purification steps the LKR activity co-fractionated with the SDH activity and the ratios between the activities of the two enzymes remained constant. The final degree of purification for both enzymes as compared to the polyethylene glycol (PEG) precipitation stage is approximately 284-fold. The degree of purification as calculated from the crude extract is not accurate because of the large variation in the activity measurements due to a high background in the crude extract. The purified LKR/SDH fraction, following the fast Blue Sepharose column (Pharmacia Biotech, Piscataway, NJ), contained three major Coomassie Blue stained bands with estimated sizes of 123, 100, and 60 kD. The 123-kD protein band is the expected size for the bifunctional LKR/SDH polypeptide and is similar to previously reported LKR/SDH polypeptides from maize (125 kD), Arabidopsis (118 kD), and human (115 kD) (Markovitz et al., 1984; Goncalves-Butruille et al., 1996; Tang et al., 1997). To verify that the 123-kD polypeptide is indeed the LKR/SDH, it was subjected to partial proteolysis, and N-terminal amino acid sequence was obtained from an internal peptide. This amino acid sequence [(R/K)(A/S) GLIDFLHGLGQ] was nearly identical to a RAGLVDFLHGLGQ sequence of an Arabidopsis LKR/SDH (Tang et al., 1997).
Table I.
Purification steps of LKR and SDH from soybean seeds
| Purification Step | Protein | LKR Activity | SDH Activity | Purification
|
|
|---|---|---|---|---|---|
| LKR | SDH | ||||
| mg | units/mg protein | -fold | |||
| Crude | 8,135 | 2 | 1.5 | ||
| PEG | 1,179 | 11.5 | 4.6 | 5.8 | 3.1 |
| DEAE Sepharose | 159 | 77.5 | 32.0 | 38.8 | 21.3 |
| Fractogel | 49 | 113.0 | 47.3 | 56.5 | 31.5 |
| Superdex | 4 | 1,299.0 | 528.0 | 649.5 | 352.0 |
| Blue Sepharose | 0.8 | 3,261.0 | 2,608.0 | 1,630.5 | 1,738.6 |
Figure 1.
Purification of LKR/SDH from developing soybean seeds. Thirty micrograms of protein from the different purification steps were fractionated on a 7.5% (w/v) polyacrylamide gel. The gel was stained with Coomassie Blue. a, Crude extract; b, PEG 8000 7% to 14% fractionation; c, DEAE Sepharose column; d, Fractogel EMD DEAE column; e, Superdex S-200 column; f, Blue Sepharose column. The bifunctional LKR/SDH protein bands of 100 and 123 kD are pointed out by arrows. MW, Molecular mass markers with sizes indicated on the left. The purified LKR/SDH polypeptides are indicated by arrows, whereas the non-related 60-kD protein is indicated by an asterisk on the right.
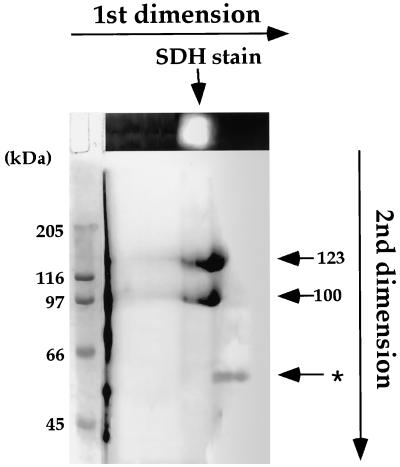
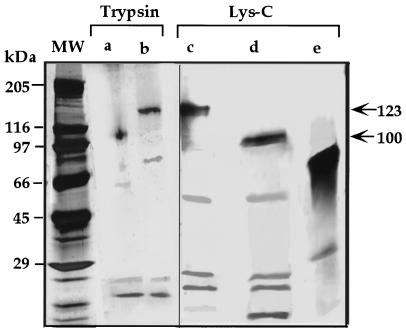
To further study whether the 100-kD and the 60-kD polypeptides are related to the 123-kD polypeptide, the purified proteins eluted from the Blue Sepharose column were subjected to two-dimensional PAGE. In the first dimension, proteins were separated on a native gel and stained for SDH activity. The SDH stained lane was then separated on a second-dimensional SDS PAGE, and the gel was stained with Coomassie Blue. The 123- and 100-kD polypeptides co-migrated with the SDH staining band in the first-dimensional gel, suggesting that they were related to LKR/SDH, whereas the 60-kD polypeptide did not, implying that it represented a different protein (Fig. 2). The 60-kD protein was subsequently identified as Glu dehydrogenase based on N-terminal sequence analysis of several internal peptides (data not shown). The sequence relationships between the 100- and the 123-kD polypeptides was further confirmed by a limited proteolysis with either trypsin or Lys-C, in which the two bands generated similar proteolysis patterns (Fig. 3).
Figure 2.
Two-dimensional separation the purified soybean LKR/SDH. The purified fraction followed the Blue Sepharose column (Fig. 1f) was separated on a first-dimensional native gel and the gel was stained for SDH activity (top gel). The first-dimensional gel was separated on a second-dimensional SDS gel and the gel was stained with Coomassie Blue R-250. The positions of the 100-, 123-, and 60-kD (asterisk) polypeptides in the second-dimensional gel are indicated by arrows on the right. Sizes of protein markers are indicated on the left.
Figure 3.
Limited proteolysis analysis of the 100- and 123-kD polypeptides. The 100- (a and d) and 123-kD (b and c) bands were excised from a SDS gel, treated with trypsin (a and b) or Lys-C (c and d), and separated on a second-dimensional SDS gel. The second-dimensional gel was silver stained. The non-related 60-kD Glu dehydrogenase band, co-fractionated with the LKR/SDH bands, was also digested with Lys-C as a control (e). Sizes of protein markers are indicated on the left and the positions of the intact 100- and 123-kD polypeptides are indicated by arrows on the right.
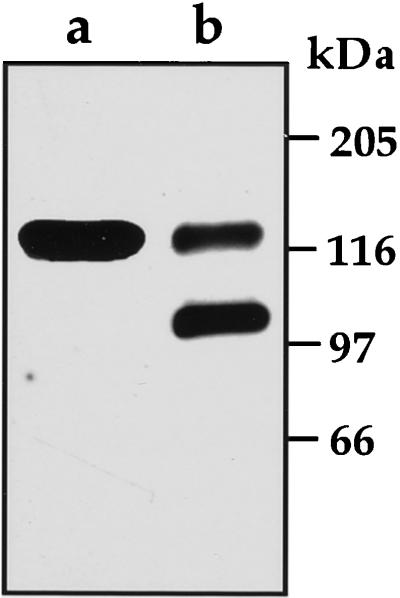
The ratios between the 123- and the 100-kD LKR/SDH polypeptides varied between different purifications, suggesting that the 100-kD polypeptide may have represented a specific processing product of the 123-kD polypeptide rather then being a different isozyme. To address this, developing soybean seeds were extracted either in the absence or presence of a cocktail of protease inhibitors and following PEG precipitation, the proteins were reacted in a western blot with antibodies produced against a synthetic peptide derived from the N-terminal end of the Arabidopsis LKR/SDH. This peptide is highly conserved between the Arabidopsis and maize enzymes (Tang et al., 1997; Kemper et al., 1999), and was therefore expected also to recognize the soybean LKR/SDH. As shown in Figure 4, although these antibodies recognized both the 123- and 100-kD polypeptide in extracts lacking the cocktail of protease inhibitors, only the 123-kD band appeared following extraction with the protease inhibitor mixture. This suggested that the 100-kD polypeptide was derived from the 123-kD polypeptide by a specific proteolytic cleavage at an exposed site in the C-terminal SDH domain, which occurred during the extraction. Unfortunately, due to the high volume and extended processing time, we have not been able to completely prevent the proteolytic cleavage of the 123-kD LKR/SDH in any of the large-scale purifications.
Figure 4.
Effect of extraction in the presence of a cocktail of protease inhibitors on the processing of the bifunctional LKR/SDH 123-kD polypeptide. Developing soybean seeds were extracted either in the presence (a) or absence (b) of a cocktail of protease inhibitors. Following PEG precipitation, the proteins were fractionated on a 7.5% (w/v) polyacrylamide gel, transferred to a PVDF membrane, and reacted in a western blot with antibodies produced against a synthetic peptide derived from the N-terminal end of the Arabidopsis LKR/SDH sequence. Sizes of molecular mass protein markers are indicated on the right.
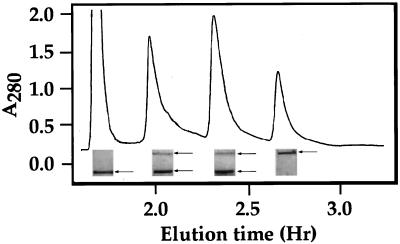
We also tested whether the truncated 100-kD polypeptide maintained SDH and LKR activities comparable to those of the 123-kD protein. The fraction containing these two polypeptides (after the Blue Sepharose column) was further fractionated on a Mono-Q column (Pharmacia Biotech) and individual fractions were initially tested for LKR activity. As shown in Figure 5, this fractionation resulted in four peaks containing LKR activity. Subsequent analysis of the same fractions for SDH activity showed that all of these peaks contained also SDH activity. The subunit composition of these four peaks was tested further by fractionation on a SDS gel. This fractionation showed that one of the peaks contained only the 100-kD truncated LKR/SDH polypeptide, one contained only the 123-kD polypeptide, and two contained a mixture of both (Fig. 5). LKR and SDH activities in all four peaks were comparable to each other and to the activity of LKR and SDH in the fraction following the Blue Sepharose column (data not shown). This suggested that the approximately 23-kD deletion (apparently at the C-terminal domain) played little or no role in SDH activity.
Figure 5.
Fractionation of LKR/SDH on a Mono-Q column. The purified fraction following the Blue Sepharose column (Fig. 1f) was separated on a Mono-Q column and individual fractions were tested for LKR activity. Pooled fractions from each peak were fractionated on SDS gel and the gel were stained with Coomassie Blue R-250. The sections of the gel in the region of the purified LKR/SDH bands are illustrated below each peak, and the bands are indicated by arrows on the right.
We have previously shown that LKR, but not SDH activity in the soybean LKR/SDH isozyme was reduced upon alkaline phosphatase treatment (Miron et al., 1997). It was therefore interesting to test whether LKR activity in the four peaks, obtained by the Mono-Q column would show identical response to alkaline phosphatase. Treatment with alkaline phosphatase decreased LKR, but not SDH activity in all of the peaks (data not shown).
Biochemical Properties of Soybean LKR and SDH
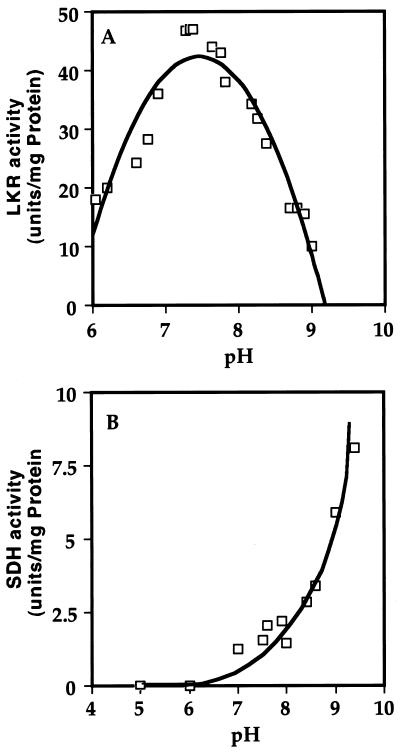
The biochemical properties of LKR/SDH were tested either with the nearly purified protein following the Blue Sepharose column (Table I; Fig. 1f), or with a partially purified preparation, after PEG precipitation, obtained from seeds that were homogenized in the presence of the cocktail of protease inhibitors. In both cases background levels in controls lacking the substrates Lys or saccharopine were minimal, and the results obtained with the two fractions were comparable. Because of this similarity, only the results obtained with the nearly purified LKR/SDH are presented here. Since LKR and SDH are linked on a single polypeptide and hence are expected to function in the same subcellular compartment, it was of interest to test the pH optimum of activity of these two enzymes. As shown in Figure 6, despite their linkage, the two enzymes possessed a markedly different pH optimum. LKR activity was highest at pH approximately 7.2 (Fig. 6A), whereas SDH activity was very low at this pH and its activity was increased with increasing pH up to pH 9 that was the highest pH tested. Due to this observation, the biochemical properties of LKR and SDH were tested at pH 7.2 and 8.5, respectively, as has previously been used for studying the activities of LKR and SDH in monocotyledonous plant species (Moeller, 1976; Brochetto-Braga et al., 1992; Gaziola et al., 1997).
Figure 6.
Determination of the optimal pH for LKR and SDH activity. The kinetics of LKR (A) and SDH (B) activities were assayed spectrophotometrically in reaction mixtures with increasing pH levels as indicated in the figures.
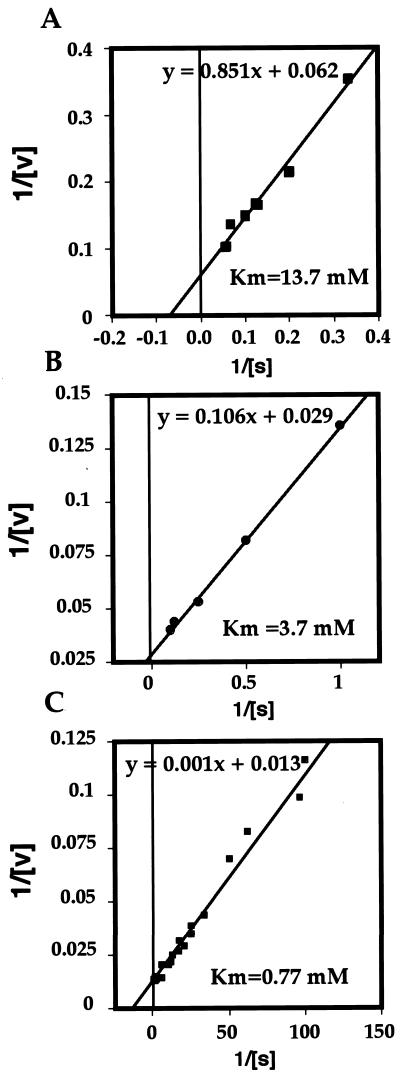
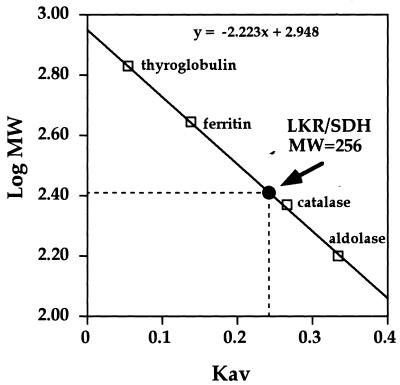
At the next stage, we determined the Km of LKR to Lys and α-ketoglutarate, as well as the Km of SDH to its substrate saccharopine, using a double reciprocal Linweaver-Burk plot. As shown in Figure 7, LKR possessed a relatively high Km of 13.7 mm to Lys, and a lower Km of 3.7 mm to α-ketoglutarate (Fig. 7, A and B). The Km of SDH to its substrate saccharopine was calculated to be 0.77 mm (Fig. 7C). The Mr of the native LKR/SDH was also tested in the Superdex column (SE-751 84; Amersham Pharmacia Biotech, Uppsala; Table I), as compared to a calibration curve, using four Mr markers. As shown in Figure 8, LKR/SDH was eluted with an estimated molecular mass of approximately 256 kD, suggesting that the native enzyme is a dimer.
Figure 7.
Determination of Km values for LKR and SDH toward their substrates. Km values were obtained by double reciprocal plots of the initial velocities of LKR with Lys as the variable substrate (A), LKR with α-ketoglutarate as the variable substrate (B), and SDH with saccharopine as the variable substrate (C). The calculated Km values are given in each panel. These experiments were replicated 12 times with similar results.
Figure 8.
Determination of the Mr of the soybean LKR/SDH. The Mr of the soybean LKR/SDH was determined by fractionation of a partially purified LKR/SDH preparation on a gel filtration-Superdex 200 column. The elution volume calculated as Kav of the LKR/SDH protein was compared to a calibration curve made using standard Mr protein markers (Pharmacia Biotech). The void volume was determined using dextran blue 2000. The calculated Mr is given in the figure.
Do Developing Soybean Seeds Contain a Monofunctional SDH Isozyme?
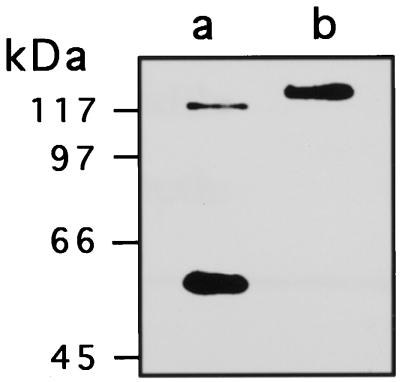
A previous report (Falco et al., 1995) has shown that seeds of Lys overproducing transgenic soybean and canola accumulated saccharopine (a product of LKR and a substrate of SDH) or a down-stream metabolite α-amino adipic acid, respectively. The reason for this is still not clear, but suggests that metabolites flux through the Lys catabolic pathway is apparently regulated in a species-specific manner. We have also recently shown that Arabidopsis plants contain two peaks of SDH activity; one is a bifunctional LKR/SDH and the second containing only SDH activity (Tang et al., 1997). Northern and Southern blots also indicated that these two Arabidopsis proteins are likely to be produced from a single gene (Tang et al., 1997). Thus, since soybean seeds accumulate saccharopine, it was of interest to test whether in soybean, the LKR/SDH gene may also produce two polypeptides (a bifunctional LKR/SDH and a monofunctional SDH), similarly to Arabidopsis. To address this, antibodies were generated against a synthetic peptide resembling the C terminus of the Arabidopsis LKR/SDH polypeptide. This peptide was shown to possess significant homology with the maize enzyme (Tang et al., 1997; Kemper et al., 1999), suggesting that the antibodies should recognize SDH domains in a wide range of plant species. Developing soybean seeds, as well as developing pods of Arabidopsis were extracted in the presence of a cocktail of protease inhibitors, separated on SDS PAGE and reacted in a western blot with the antibodies raised against the Arabidopsis LKR/SDH C- terminal peptide. As shown in Figure 9, lane a, Arabidopsis contains two immunoreactive polypeptide bands with estimated molecular mass of 118 and 54 kD. These correspond to the expected bifunctional LKR/SDH and monofunctional SDH, previously identified in this species. In contrast, in extracts from soybean seeds, only a single band of approximately 123 kD was detected corresponding to the bifunctional LKR/SDH (Fig. 9, lane b).
Figure 9.
Detection of the bifunctional and monofunctional SDH using a western-blot analysis. One to 30 μg of either crude extract of Arabidopsis inflorescence (a) or preparations of developing soybean seeds following PEG precipitation (b) was separated on a 7.5% (w/v) polyacrylamide gel, transferred to a PVDF membrane, and reacted with antibodies produced against a synthetic peptide derived from Arabidopsis C-terminal end of the SDH domain. Sizes of Mr markers are indicated on the left.
DISCUSSION
LKR Is a Rate-Limiting Enzyme in Lys Catabolism
So far, LKR/SDH has been purified and characterized only from the monocotyledonous plants maize and rice (Brochetto-Braga et al., 1992; Goncalves-Butruille et al., 1996; Gaziola et al., 1997; Kemper et al., 1998). In the present report, we describe the first purification and characterization of LKR/SDH from a dicot plant, soybean. The maize, rice, and soybean enzymes are all homodimers, and possess comparable Km for their substrates, as well as comparable pH optima. However, the soybean LKR possesses a slightly higher Km to both of its substrates Lys and α-ketoglutarate than the maize and rice enzymes (13.7 and 3.7 mm, respectively). Assuming that the cellular levels of Lys are much lower than the Km of LKR for this amino acid, it is reasonable to conclude that LKR represents a rate-limiting step in Lys catabolism in vivo, and that Lys catabolism may be regulated by modulations of LKR activity. This is in accord with previous observations showing that LKR activity in different plant species is modulated by Ca2+ and protein phosphorylation/dephosphorylation, and that this modulation may be controlled by sensing the cellular levels of free Lys (Karchi et al., 1995; Miron et al., 1997; Kemper et al., 1998).
Metabolite Flux through LKR/SDH Suggests That SDH Activity of This Bifunctional Enzyme Is Also Rate Limiting
The functional significance of bifunctional enzymes, like LKR/SDH, is still unknown. Such a linkage is common in metabolic pathways with a close example being the bifunctional Asp kinase/homo-Ser dehydrogenase comprising two enzymes in the biosynthesis Thr, Met, Ile, and Lys (Galili, 1995; Azevedo et al., 1997). It has been suggested that linkage of enzymes, such as LKR/SDH, may enable channeling of the product of the first enzyme directly into the catalytic site of the second enzyme (Traut and Jones, 1977; Wahl et al., 1979; Goncalves-Butruille et al., 1996). Such channeling may result in efficient flux of intermediates through the two linked enzymes. However, whether substrate channeling occur in the LKR/SDH enzyme is still questionable since Falco et al. (1995) have shown that transgenic Lys-overproducing soybean seeds accumulate saccharopine, the product of LKR and the substrate of SDH. The results of Falco and associates further suggest that SDH activity of the bifunctional LKR/SDH enzyme may also represent a rate-limiting step in metabolite flux via this bifunctional enzyme. The reason for the limited flux via the SDH activity of LKR/SDH is still not clear. Yet, since plant LKR/SDH enzymes are likely to be localized in the cytosol (Epelbaum et al., 1997; Tang et al., 1997; Kemper et al., 1999), it is possible that SDH operates in a nonoptimal physiological environment having a neutral pH.
Metabolite Flux through SDH May Be Differentially Regulated in Distinct Plant Species
Not all plant seeds accumulate saccharopine as the intermediate catabolic product of Lys. Seeds of transgenic canola plants, expressing the same bacterial dihydrodipicolimate synthase enzyme as was used for the soybean transformation, were shown to accumulate the downstream metabolite α-amino adipic acid (Falco et al., 1995). The reason for the more efficient flux via the SDH step in canola than in soybean is still not known. Yet, this may result from the presence of an additional monofunctional SDH enzyme in this plant species (Deleu et al., 1999), as opposed to soybean, which lacks a monofunctional SDH (this report).
The physiological significance of the species-specific differences in metabolite flux via SDH is also not understood. The final substrate of the α-amino adipic pathway is acetyl-coenzyme A, which serves as a substrate for various macromolecules like lipids (Lehninger, 1975). It is thus possible that efficient flux of Lys catabolism is specifically advantageous in cruciferae seeds, which accumulate high levels of reserve oils.
MATERIALS AND METHODS
Plant Material
Soybean (Glycine max) plants were grown during summer season April through October in a green house. Developing pods were harvested when seeds reached a full-size green stage, approximately 40 d after flowering. Pods were kept on ice until seeds were separated, immediately frozen in liquid nitrogen, and kept in −80°C until used.
Purification of LKR and SDH
Developing soybean seeds were homogenized in 4 volumes of buffer A (25 mm phosphate buffer, pH 7.2, containing 1 mm EDTA, 1 mm DTT, and 10 μg mL−1 leupeptin). After centrifugation for 30 min at 27,000g the supernatant was precipitated by PEG as previously described (Arruda et al., 1982; Goncalves-Butruille et al., 1996). For PEG precipitation the supernatant was brought to pH 5.7 with KH2PO4 and then PEG 8000 was added to a final concentration of 7% (v/v), and after 30-min incubation at 4°C the extract was centrifuged for 10 min at 27,000g. The pellet was discarded and PEG was added to a final concentration of 14% (v/v). After another 30-min incubation and centrifugation in the same conditions, the pellet was resuspended in buffer A at one-tenth of the initial volume. The sample was loaded on a 30-mL anion-exchange column-DEAE Sepharose (Pharmacia Biotech) that was equilibrated with buffer A. After washing out the unbound protein, the column was eluted with a gradient of 170 to 250 mm KCl. The remaining protein was eluted with 1 m KCl. LKR and SDH activities and protein level were determined in the eluted fractions, as well as in the unbound protein and in the protein eluted from the column with 1 m KCl. The fractions containing enzyme activities were pooled and brought to pH 5.7 with KH2PO4, and the protein was precipitated with 14% (w/v) PEG. The PEG pellet was resuspended in a minimal volume of buffer A and applied on a 8-mL DEAE Sepharose fast flow column (Pharmacia Biotech). The unbound protein was washed out and the column was eluted with a gradient of 200 to 230 mm KCl. The fractions containing LKR and SDH activities were pooled, and concentrated to 2 mL using Centriprep 30 concentrators (Amicon, Beverly, MA). The concentrated protein was further fractionated on a Superdex 200 column with buffer A at a flow rate of 0.3 mL min−1. The pooled active fractions were loaded on a 1-mL Blue Sepharose fast flow column. LKR/SDH was eluted with a gradient of 0 to 12 mm NADPH. LKR and SDH activities as well as protein level were determined in all fractions. In the experiment shown in Figure 4, pooled fractions from the Blue Sepharose column, containing LKR and SDH activities, were loaded on a 1-mL Mono-Q column previously calibrated with buffer A. After washing out the unbound protein, the column was eluted with a step gradient of buffer A and buffer A with 1 m KCl. LKR and SDH activity, as well as protein levels, were determined in all fractions.
In-Gel Limited Proteolysis of the 123- and 100-kD Polypeptides
The protein eluted from the Mono-Q column was subjected to a SDS PAGE and staining with Coomassie Blue R-250. The 123- and 100-kD bands, each one containing approximately 1 μg of protein were excised from the gel and applied on different lanes of a gradient, 7% to 15% (w/v) polyacrylamide gel. To each of the wells containing a protein band, 0.4 μg of either Trypsin or Lys-C were added. The gel was run until the protein reached the middle of the stacking gel. The current was stopped for 30 min to allow proteolysis, and renewed until the protein reached the end of the gel. The gel was stained with a silver stain. As a control, the non-related 60-kD Glu dehydrogenase band co-eluted with the LKR/SDH bands after the Blue Sepharose column was run in parallel.
SDH Activity Staining on a Native Gel
SDH activity staining was performed as previously described (Goncalves-Butruille et al., 1996; Gaziola et al., 1997) with some modifications. The purified LKR/SDH, following the Blue Sepharose column, was fractionated on a 7.5% (v/v) polyacrylamide native gel. At the end of the run, the gel was washed three times in 0.1 m Tris buffer, pH 8.5, for 10 min each wash, at 4°C. Following the wash, the gel was incubated for 2 h at 30°C in the staining solution containing: 0.1 m Tris HCl buffer, pH 8.5, 1 mm saccharopine, and 1 mm NAD. The activity band was visualized in UV light as a bright band against a dark background.
Protein Determination and Analysis of LKR and SDH Activities
Protein level was determined by the Bradford method (Bradford, 1976). The kinetics of LKR activity was measured spectrophotometrically by determining the rate of NADPH oxidation at 340 nm for 10 min at 30°C, as previously described (Miron et al., 1997). The kinetics of SDH activity was measured spectrophotometrically by determining the rate of NAD′ reduction at 340 nm for 10 min at 30°C, as previously described (Miron et al., 1997). One unit of LKR activity was defined as the amount of enzyme that catalyzes the oxidation of 1 nmol of NADPH per minute at 30°C. One unit of SDH activity was defined as the amount of enzyme that catalyzes the reduction of 1 nmol of NAD′ per minute at 30°C.
SDS PAGE and Western-Blot Analysis
A protein sample of 1 to 30 μg was fractionated on a 7.5% (w/v) polyacrylamide SDS gel. The protein was transferred to a PVDF membrane using a Protein Trans-Blot apparatus (Bio-Rad Laboratories, Hercules, CA). The membrane was completely dried, stained with Coomassie Blue R, and blocked over-night in a solution of 10% (w/v) milk powder. Following blocking, the membrane was reacted with polyclonal antibodies prepared against a synthetic polypeptide from either the N-terminal end of the LKR domain (AETVNKWERRTPLTPSHC), or the C-terminal end of the SDH domain (CEVYLPALDILQAYGIKLMEKAE) of the Arabidopsis LKR/SDH protein sequence. Both antibodies were diluted 1:500 with the blocking solution. The membrane was incubated with the antibodies at room temperature for 2 h, washed four times for 10 min with phosphate-buffered saline and reacted with goat anti-rabbit peroxidase-linked antibodies at a dilution of 1:4,000 for 1 h at room temperature. Following five 10-min washes with phosphate-buffered saline. Antibodies were detected using ECL western-blotting analysis system (Amersham-Pharmacia Biotech, Uppsala).
ACKNOWLEDGMENTS
We thank Hanna Levanony for excellent technical assistance. G.G. is an incumbent of the Bronfman Chair of Plant Sciences.
Footnotes
This work was supported by grants from the Israel Academy of Sciences and Humanities, National Council for Research and Development, Israel, and by the Leo and Julia Forchheimer Center for Molecular Genetics.
LITERATURE CITED
- Arruda P, Da Silva WJ. Lysine-ketoglutarate reductase activity in developing maize endosperm. Phytochemistry. 1983;22:206–208. doi: 10.1104/pp.69.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda P, Sodek L, Da Silva WJ. Lysine-ketoglutarate reductase activity in developing maize endosperm. Plant Physiol. 1982;69:988–989. doi: 10.1104/pp.69.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo RA, Arruda P, Turner WL, Lea PJ. The biosynthesis and metabolism of the aspartate derived amino acids in higher plants. Phytochemistry. 1997;46:395–419. doi: 10.1016/s0031-9422(97)00319-1. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brochetto-Braga M, Leite A, Arruda P. Partial purification and characterization of lysine-ketoglutarate reductase in normal and opaque-2 maize endosperms. Plant Physiol. 1992;98:1139–1147. doi: 10.1104/pp.98.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleu C, Coustaut M, Niogret M, Larher F. Three new osmotic stress-regulated cDNAs identified by differential display polymerase chain reaction in rapeseed leaf discs. Plant Cell Environ. 1999;22:979–988. [Google Scholar]
- Epelbaum S, McDevitt R, Falco SC. Lysine-ketoglutarate reductase and saccharopine dehydrogenase from Arabidopsis thaliana: nucleotide sequence and characterization. Plant Mol Biol. 1997;35:735–748. doi: 10.1023/a:1005808923191. [DOI] [PubMed] [Google Scholar]
- Falco SC, Guida T, Locke M, Mauvais J, Sandres C, Ward RT, Webber P. Transgenic canola and soybean seeds with increased lysine. Biotechnology. 1995;13:577–582. doi: 10.1038/nbt0695-577. [DOI] [PubMed] [Google Scholar]
- Foster AR, Scislowski PWD, Harris CI, Fuller MF. Metabolic response of liver lysine α-ketoglutarate reductase activity in rats fed lysine limiting or lysine excessive diets. Nutr Res. 1993;13:1433–1443. [Google Scholar]
- Galili G. Regulation of lysine and threonine synthesis. Plant Cell. 1995;7:899–906. doi: 10.1105/tpc.7.7.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziola SA, Alessi ES, Guimaraes PEO, Damerval C, Azevedo RA. Quality protein maize: a biochemical study of enzymes involved in lysine metabolism. J Agric Food Chem. 1999;47:1268–1275. doi: 10.1021/jf980940r. [DOI] [PubMed] [Google Scholar]
- Gaziola SA, Teixeira CMG, Lugli J, Sodek L, Azevedo RA. The enzymology of lysine catabolism in rice seeds: isolation, characterization, and regulatory properties of a lysine 2-oxoglutarate reductase/saccharopine dehydrogenase bifunctional polypeptide. Eur J Biochem. 1997;247:364–371. doi: 10.1111/j.1432-1033.1997.00364.x. [DOI] [PubMed] [Google Scholar]
- Goncalves-Butruille M, Szajner P, Torigoi E, Leite A, Arruda P. Purification of characterization of the bifunctional enzyme lysine-ketoglutarate reductase-saccharopine dehydrogenase from maize. Plant Physiol. 1996;110:765–771. doi: 10.1104/pp.110.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Miron D, Ben-Yaacov S, Galili G. The lysine-dependent stimulation of lysine catabolism in tobacco seeds requires calcium and protein phosphorylation. Plant Cell. 1995;7:1963–1970. doi: 10.1105/tpc.7.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G. Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci USA. 1994;91:2577–2581. doi: 10.1073/pnas.91.7.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper EL, Cord-Neto GC, Capella AN, Goncalves-Butruile M, Azevedo RA, Arruda P. Structure and regulation of the bifunctional enzyme lysine-oxoglutarate reductase-saccharopine dehydrogenase in maize. Eur J Biochem. 1998;253:720–729. doi: 10.1046/j.1432-1327.1998.2530720.x. [DOI] [PubMed] [Google Scholar]
- Kemper EL, Cord-Neto GC, Papes F, Moraes KC, Leite A, Arruda P. The role of opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell. 1999;11:1981–1994. doi: 10.1105/tpc.11.10.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger AL. Biochemistry. New York: Worth Publishers; 1975. p. 572. [Google Scholar]
- Markovitz PJ, Chuang DT. The bifunctional aminoadipic semialdehyde synthase in lysine degradation-separation of reductase and dehydrogenase domains by limited proteolysis and column chromatographgy. J Biol Chem. 1987;262:9353–9358. [PubMed] [Google Scholar]
- Markovitz PJ, Chuang DT, Cox RP. Familial hyperlysinemias: purification and characterization of the bifunctional aminoadipic semialdehyde synthase with lysine-ketoglutarate reductase and saccharopine dehydrogenase activities. J Biol Chem. 1984;259:11643–11646. [PubMed] [Google Scholar]
- Miron D, Ben-Yaacov S, Karchi H, Galili G. In vitro dephosphorylation inhibits the activity of soybean lysine-ketoglutarate reductase in a lysine-regulated manner. Plant J. 1997;12:1453–1458. [Google Scholar]
- Moeller BL. Lysine catabolism in barley (Hordeum vulgare L.) Plant Physiol. 1976;57:687–692. doi: 10.1104/pp.57.5.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotwell MA, Larkins BA. The biochemistry and molecular biology of seed storage proteins. In: Miflin BJ, editor. The Biochemisrty of Plants. Vol. 15. New York: Academic Press; 1988. pp. 297–345. [Google Scholar]
- Tang G, Miron D, Zhu-Shimoni JX, Galili G. Regulation of lysine catabolism through lysine-ketoglutarate reductase and saccharopine dehydrogenase in Arabidopsis. Plant Cell. 1997;9:1–13. doi: 10.1105/tpc.9.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut TW, Jones ME. Kinetic of conformation studies of the orotate phosphoribosyltransferase:orotidine-5′-phosphate decarboxilase enzyme complex from mouse Erlich ascites cells. J Biol Chem. 1977;252:8374–8381. [PubMed] [Google Scholar]
- Wahl GM, Padgett RA, Stark GR. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphoacetyl)-l-aspartate-resistant hamster cells. J Biol Chem. 1979;254:8679–8689. [PubMed] [Google Scholar]