Abstract
Benign ovarian Brenner tumors often are associated with mucinous cystic neoplasms, which are hypothesized to share a histogenic origin and progression, however, supporting molecular characterization is limited. Our goal was to identify molecular mechanisms linking these tumors. DNA from six Brenner tumors with paired mucinous tumors, two Brenner tumors not associated with a mucinous neoplasm, and two atypical proliferative (borderline) Brenner tumors was extracted from formalin-fixed, paraffin-embedded tumor samples and sequenced using a 358-gene next-generation sequencing assay. Variant calls were compared within tumor groups to assess somatic mutation profiles. There was high concordance of the variants between paired samples (40% to 75%; P < 0.0001). Four of the six tumor pairs showed KRAS hotspot driver mutations specifically in the mucinous tumor. In the two paired samples that lacked KRAS mutations, MYC amplification was detected in both of the mucinous and the Brenner components; MYC amplification also was detected in a third Brenner tumor. Five of the Brenner tumors had no reportable potential driver alterations. The two atypical proliferative (borderline) Brenner tumors both had RAS mutations. The high degree of coordinate variants between paired Brenner and mucinous tumors supports a shared origin or progression. Differences observed in affected genes and pathways, particularly involving RAS and MYC, may point to molecular drivers of a divergent phenotype and progression of these tumors.
Ovarian Brenner tumors are relatively rare, comprising 1% to 3% of all ovarian neoplasms.1 Although most are benign, approximately 2% are atypical proliferative (borderline) and malignant Brenner tumors.1, 2 Benign Brenner tumors are biphasic, composed of nests of transitional cell epithelium, resembling urothelium, within a fibrous stroma. It is well recognized that many (up to 30%) Brenner tumors arise in association with an ovarian mucinous neoplasm and are hypothesized to share a histogenic origin and progression, however, limited molecular characterization has been conducted that might support this.
Several theories have been proposed to explain the histogenesis of these tumors. It currently is suggested that Brenner tumors have a tubal origin, arising from the tuboperitoneal junction cells (the junction between the tubal fimbria and the mesothelial serosa). It is postulated that these cells undergo transitional cell metaplasia and invaginate into the paratubal or ovarian surface, forming nests termed a Walthard cell nest.3, 4 These Walthard cell nests are frequently the origin of paratubal cysts and it is theorized that when they implant and seed the ovary, they may develop into Brenner tumors.
Mucinous epithelium is not native to the ovary and mucinous epithelial ovarian tumors do not currently have a known cell of origin in the ovary. It is not unusual to find mucinous metaplasia and cystic change in Brenner nests and one hypothesis is that mucinous ovarian tumors arise from these changes in Brenner tumors. However, evidence of transitional cell nests is not evident within a majority of mucinous neoplasms, although the Brenner component of mixed tumors typically is significantly smaller than the mucinous component and without extensive sampling it simply may be missed.4
Morphologic and immunohistochemistry studies have been used to explore the relationship between Brenner tumors and mucinous cystic neoplasms of the ovary.5 More recently, molecular studies evaluating a limited number of genes have been performed in a few cases of Brenner, atypical proliferative (borderline) Brenner, and ovarian mucinous tumors independently.6, 7, 8 Our aim was to investigate the genetic profiles of Brenner tumors and the association with paired ipsilateral mucinous tumors using a 358-gene targeted next-generation sequencing panel. We believe that there is an important molecular relationship between Brenner tumors and ovarian mucinous epithelial tumors that may provide insight into the pathogenesis and progression of these tumors.
Materials and Methods
This study was approved by the Committee for the Protection of Human Subjects at the Dartmouth-Hitchcock Medical Center. The Department of Pathology (Dartmouth-Hitchcock Medical Center) archive was searched from 2004 to 2014 to identify all ovarian benign and atypical proliferative (borderline) Brenner tumors. Six Brenner tumors with paired mucinous cystadenomas (MCA; n = 4) or mucinous borderline tumor (MBT, n = 2), two Brenner tumors not associated with a mucinous neoplasm, and two atypical proliferative (borderline) Brenner tumors that had an adequate amount of tissue to complete the genetic analysis were included in this study.
Sequencing
The histopathology was reviewed by an attending pathologist (L.J.T.) and tumor blocks for each case were selected for analysis. The tissue areas of interest were marked on a hematoxylin and eosin-stained slide, and 10 unstained formalin-fixed paraffin-embedded tissue sections (4 μm) were macrodissected and used for DNA extraction. Macrodissection allowed for separation of the Brenner and mucinous components in the six paired samples (for the Brenner components, stroma and epithelium were dissected together). Sequencing was performed as previously described.9 Briefly, DNA was extracted with the QIAamp DNA Formalin-Fixed Paraffin-Embedded Tissue Kit (Qiagen, Valencia, CA). The DNA quality was evaluated using the NanoDrop 2000 (Thermo Scientific, Waltham, MA). The DNA quantity was analyzed with the Qubit Fluorometer (Life Technologies, Carlsbad, CA). The sample libraries were constructed using the Agilent (Santa Clara, CA) 1-μg DNA sample preparation method, the SureSelectXT Target Enrichment System. Next-generation sequencing was performed at the Jackson Laboratory's Clinical Genomics facility (Farmington, CT) using the JAX Cancer Treatment Profile assay consisting of a 358-gene targeted panel on a HiSeq 2500 (Illumina, San Diego, CA) with 150-bp paired end sequencing. The assay has complete exon coverage of the 358 genes and covers 10 bp of intragenic sequence on either side of each exon.
Data Analysis
Data analysis was performed with the Clinical Genomics Analytical bioinformatics pipeline as follows. FASTQ read files from the Illumina HiSeq instrument were quality-filtered to remove low-quality reads (reads with an average Phred quality score <70) and/or to trim out low-quality nucleotides (with an average Phred quality score <70). The resulting high-quality reads were aligned to the human reference genome [hg19; Genome Reference Consortium Human Reference 37 (GRCh37; http://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13)] using the Burrows-Wheeler Aligner version 0.7.9a,10 and alignments were processed afterward to remove duplicates, realign around insertions and deletions, and recalibrate base quality scores using Picard software version 1.95 (http://broadinstitute.github.io/picard) and the GATK tool suite version 3.1-1.11, 12 Single-nucleotide variants and insertions and deletions (up to 50 bp in length) were called using the GATK Unified Genotyper tool and Pindel version 0.2.5a3.13 Copy number variations were called with CONTRA (Copy Number Analysis for Targeted Resequencing) software version 2.0.4.14 Finally, genomic and functional annotations (gene name, transcript, type of change at the nucleotide and protein levels, Catalogue of Somatic Mutation in Cancer (COSMIC) idenfication numbers, population frequencies, conservation scores, and so forth) were assigned to variants using the SnpEff and SnpSift tool suite version 4.0.e.15 SnpEff/SnpSift variant call files were filtered to retain only high-/medium-impact variants (including missense, nonsense, frameshifts, codon insertions and deletions, stop codon loss or gain, and start codon loss) that met a number of filtering criteria as outlined later. Variants not passing the established quality control metrics (coverage, ≤140×; allelic frequency, ≤5%) were removed from the analysis. Amplification was defined as >6 copies. Germline variants (defined as variants with an allele frequency between 40% and 60% and a 1000 Genomes Population16 frequency >1%, or an allele frequency >90% and a 1000 Genomes Population frequency >1%) and noncanonical transcripts were filtered out. The resulting high-quality, high-/medium-impact somatic variants were compared within tumor pairs and among tumors to assess variant overlap.
Statistical Analysis
All called variants in paired samples sets were compared for statistical significance using the binomial exact test. Statistical analysis was performed in R version 3.0.2 (http://www.R-project.org).17 Results were considered statistically significant at P < 0.05.
Results
Clinicopathologic Features
The patients were 28 to 88 years of age (median age, 60 years) at the time of diagnosis. The ovarian tumors all were unilateral and ranged in size from 9.0 to 32.0 cm. The mixed Brenner and mucinous tumors contained variable amounts of cystic and solid components. Associated findings included a contralateral cystadenofibroma in one patient. The two patients with atypical proliferative (borderline) Brenner tumors both had clinical evidence of virilization.
Histologic Review
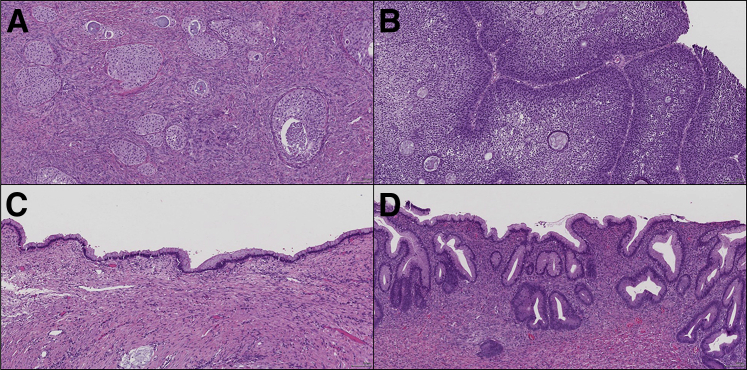
The benign Brenner tumors were composed of nests of transitional epithelium within a dense stroma. At times, the Brenner tumors showed dilatation of the epithelial nests and were associated with mucinous epithelial cells in the center of the nests as well as spiculated calcifications. The MCAs were multicystic and lined by intestinal-type mucinous epithelium containing goblet cells and lacked significant cytologic atypia. The MBTs consisted of similar mucinous epithelium with a more complex architecture, increased cytologic atypia, hyperchromasia, nuclear stratification, and mitotic activity. Only cases in which the Brenner and mucinous components could be macroscopically dissected separately were included in this study. The two atypical proliferative (borderline) Brenner tumors both were associated with a benign Brenner component and showed confluent nests and papillae of transitional cells with intraepithelial microcystic spaces (Figure 1).
Figure 1.
A: Brenner tumor. B: Atypical proliferative Brenner tumor. C: Mucinous cystadenoma. D: Mucinous borderline tumor. Original magnification, ×10.
Pairwise Comparisons and Reportable Mutations
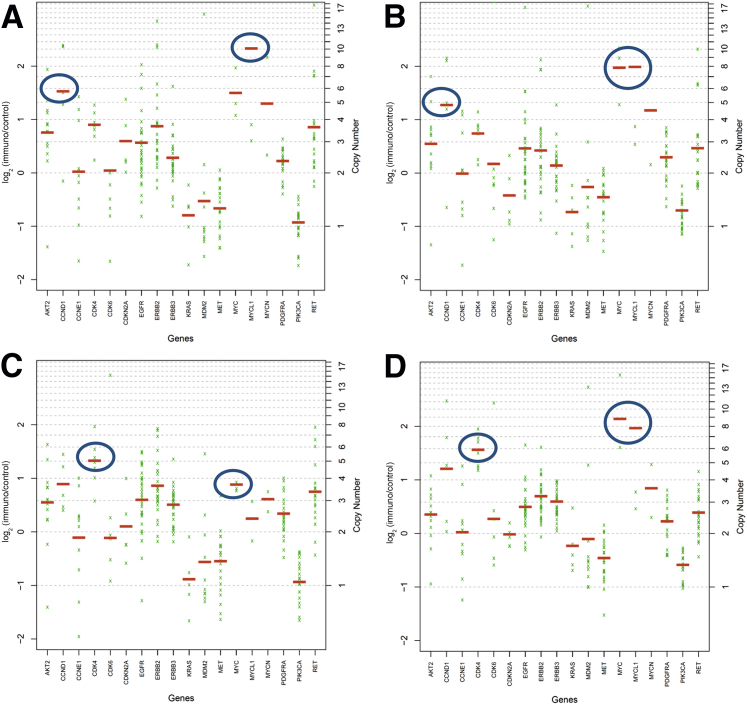
After filtering variants, 122 different high- or medium-impact variants were detected in 70 genes across Brenner, atypical proliferative (borderline) Brenner, and mucinous tumors. Individual tumors had 6 to 18 variants. No characteristic genetic profile across all Brenner or all mucinous tumors was identified. However, pairwise comparisons of filtered genetic variants within the six sets of paired Brenner and mucinous neoplasm samples showed 40% to 75% overlap of variants between paired samples. Concordance of variants was found to be significant (P < 0.0001) for each pair using the binomial exact test. The reportable mutations by tumor are listed in Table 1. Four of the six tumor pairs showed KRAS hotspot driver mutations (KRAS p.G12X, 2; KRAS p.Q61H, 2) specifically in the mucinous tumor, and the KRAS hotspot variant allele frequencies for two of four of these pairs was less than 10%, suggesting a potential early driver event. In the two paired samples that lacked KRAS activating mutations, MYC amplification was detected in both of the MCA and the Brenner components [one Brenner had a low-level amplification with a MYC (alias c-myc) copy number of 3.6] and a PIK3CA mutation in one of the MCAs (Figure 2). MYC amplification also was detected in a third Brenner tumor (Brenner 7). Of the five tumors with MYC amplification, three also had amplification of CDK4 and two had amplification of CCND1 (Figure 2). Five of the Brenner tumors had no reportable potential driver alterations. The two atypical proliferative (borderline) Brenner tumors both had RAS mutations (HRAS p.G12S and KRAS p.G12V), and one also contained a common PIK3CA mutation (p.H1047R). None of the benign Brenner tumors (0 of 7) had RAS mutations. These distinct results also confirm the successful isolation of the two components from one mixed tumor (Table 1).
Table 1.
Reportable Potential Driver Alterations Identified in All Tumor Types
| Brenner tumors | Genetic alterations (allelic frequency) | Mucinous tumors | Genetic alterations (allelic frequency) | Percentage of variant overlap of paired tumors (%) |
|---|---|---|---|---|
| Brenner 1 | None detected | MBT 1 | KRAS p.G12C (18%) | 46 |
| Brenner 2 | c-MYC amplification | MCA 1 | PIK3CA p.C420R (16%) | 40 |
| CCND1 amplification | c-MYC amplification | |||
| CCND1 amplification (4.6 copies)∗ | ||||
| Brenner 3 | c-MYC amplification (3.6 copies)∗ | MCA 2 | c-MYC amplification | 44 |
| CDK4 amplification (5.0 copies)∗ | CDK4 amplification | |||
| Brenner 4 | None detected | MCA 3 | KRAS p.G12R (8%) | 75 |
| Brenner 5 | None detected | MCA 4 | KRAS p.Q61H (6%) | 57 |
| Brenner 6 | None detected | MBT 2 | KRAS p.Q61H (43%) | 47 |
| Brenner 7 | c-MYC amplification | |||
| CDK4 amplification | ||||
| Brenner 8 | None detected | |||
| AP Brenner 1 | HRAS p.G12S (31%) | |||
| AP Brenner 2 | KRAS p.G12V (43%) | |||
| PIK3CA p.H1047R (46%) |
Brenner tumors 1 to 6 are paired with ipsilateral mucinous tumors. The percentage of variant overlap was calculated based on a comparison of the variants remaining after data filtration.
AP Brenner, atypical proliferative Brenner; MBT, mucinous borderline tumor; MCA, mucinous cystadenoma.
Low-level amplification below the data analysis pipeline cut-off value of ≥6 copies.
Figure 2.
Brenner 2 with MYC and CCND1 amplification (A) and paired MCA 1 (B) also showing MYC amplification and 4.6 copies of CCND1. Brenner 3 with CDK4 copy number 5.0 and MYC copy number 3.6 (C) and paired MCA 2 with MYC and CDK4 (D) amplification. The blue circles indicate the areas of increased copy number; with this assay, amplification is defined as more than six copies of a gene per cell.
Discussion
In this study we performed next-generation sequencing analysis of a 358-gene panel to detect single-nucleotide variants and copy number variations in eight benign ovarian Brenner tumors, six associated mucinous neoplasms, and two atypical proliferative (borderline) Brenner tumors. The co-occurrence of a mucinous tumor with a Brenner tumor has been well recognized since the 1920s. The reported incidence of Brenner tumors with a mucinous component is 14% to 30%.5, 18 This is the first study to perform such a comprehensive molecular analysis of Brenner tumors and their associated ipsilateral mucinous neoplasms as well as atypical proliferative (borderline) Brenner tumors.
Several immunohistochemical (IHC)-based studies have been performed to evaluate the association of Brenner tumors with Walthard cell nests, fallopian tube epithelium, and germ cells. The epithelial component of Brenner tumors and most Walthard cell nests are diffusely positive for GATA3, AKR1C3, androgen receptor, and lack staining for PAX8 and PAX2 (two Mullerian markers), calretinin or inhibin (these two markers are positive in the stromal component), and SALL4 (a germ cell tumor marker).3, 5, 19 In an immunohistochemistry study of Brenner tumors and associated mucinous tumors, Roma and Masand5 reported that the mucinous tumors lacked PAX8, PAX2, and SLL4, but, unlike the Brenner component, also lacked GATA3 expression. Taken together, these IHC analyses support that the Brenner and associated mucinous neoplasms are not of a Mullerian phenotype (Table 2).
Table 2.
| Histology | GATA3 | AKR1C3 | AR | PAX8 | PAX2 | SALL4 |
|---|---|---|---|---|---|---|
| Brenner | + | + | + | − | − | − |
| Mucinous metaplasia associated with Brenner | − | UK | UK | − | − | − |
| MCA/MBT | − | UK | UK | − | − | − |
AR, androgen receptor; MBT, mucinous borderline tumor; MCA, mucinous cystadenoma; UK, unknown at this time.
One limitation of the JAX Cancer Treatment Profile assay and current data analysis pipeline used in this study is that it is not validated to detect gene deletions, which may be of significance. For instance, alternations in CDKN2A, the gene that encodes for p16INK4a (also known as p16), have been described in atypical proliferative (borderline) Brenner tumors. Several studies have shown expression of p16 by IHC in benign Brenner tumors with loss of staining in atypical proliferative (borderline) Brenner tumors. Kuhn et al6 found that loss of p16 expression in seven of seven atypical proliferative (borderline) Brenner tumors (but none of the benign Brenner tumors) was the result of a homozygous deletion of the CDKN2A gene detected by fluorescence in situ hybridization analysis. Silencing of CDKN2A via either homozygous deletion or loss of heterozygosity with loss of p16 expression also has been described in 40% to 68% of MCAs and MBTs.8, 20
Reports of RAS pathway aberrations in MCAs/MBTs and atypical proliferative (borderline) Brenner tumors have been reported previously but separately. In a study of 20 MCAs and 22 MBTs, KRAS mutations were detected in 60% and 64% of tumors, respectively.8 Similarly, Ryland et al20 reported a KRAS mutation rate of 55% (12 of 22) in MCAs and 62% (18 of 29) in MBTs. Kuhn et al6 reported the identification of one KRAS and two PIK3CA somatic mutations in a study of seven atypical proliferative (borderline) Brenner tumors. Similar to prior reports, in our series four of the six (67%) MCAs/MBTs harbored KRAS mutations and the two MCAs that lacked KRAS mutations both showed MYC amplification; one also had a PIK3CA mutation. In addition, RAS mutations were identified in both atypical proliferative (borderline) Brenner tumors (a PIK3CA mutation also was identified in one Brenner tumor), however, none were detected in the benign Brenner tumors. RAS mutations were identified in both MBTs and atypical proliferative (borderline) Brenner tumors, suggesting that a more proliferative phenotype is associated with alterations in RAS genes. The two PIK3CA mutations detected (p.H1047R and p.C420R) both are considered activating mutations.21 These findings (along with the high concordance of gene variant overlap) suggest that RAS mutations, and possibly to a lesser extent PIK3CA mutations, have a role in the progression and pathogenesis of ovarian MCAs/MBTs and atypical proliferative (borderline) Brenner tumors from a benign Brenner tumor. Table 3 summarizes the molecular findings of this and prior studies.
Table 3.
Molecular Findings in Brenner Tumors, Atypical Proliferative Brenner, and MCA/MBT as Seen in Our Study and Reported in the Literature
| Reference/mutation | Benign Brenner | Atypical proliferative Brenner | MCA/MBT |
|---|---|---|---|
| Current study | MCA/MBT | ||
| KRAS | 0/8 | 1/2 | 4/6 |
| PIK3CA | 0/8 | 1/2 | 1/6 |
| MYC amplification | 3/8 | 2/6 | |
| CDKN2A-HD | NT | NT | NT |
| HRAS | 1/2 | ||
| Kuhn et al6 | |||
| KRAS | 0/20 | 1/7 | |
| PIK3CA | 1/20 | 2/7 | |
| CDKN2A-HD | 0/13 | 7/7 | |
| Cuatrecasas et al7 | |||
| KRAS | 0/5 | 0/5 | |
| PIK3CA | 0/2 | 1/3 | |
| CDKN2A-HD | NT | NT | |
| Hunter et al8 | MCA; MBT | ||
| KRAS | 12/20; 14/22 | ||
| PIK3CA | NT; NT | ||
| CDKN2A-HD | 7/20; 15/22 | ||
| Ryland et al20 | MCA; MBT | ||
| KRAS | 12/22; 18/29 | ||
| PIK3CA | ND; ND | ||
| CDKN2A-HD | 4/22; 12/29 |
CDKN2A-HD, CDKN2A homozygous deletion; MBT, mucinous borderline tumor; MCA, mucinous cystadenoma; ND, not detected; NT, not tested.
A novel, previously unreported finding in this study was the amplification of MYC in five samples (two pairs of Brenner and MCAs, and a separate Brenner tumor). MYC is a proto-oncogene and the encoded protein is considered a pleiotropic transcription factor that plays roles in multiple pathways that regulate cellular proliferation, metabolism, differentiation, and apoptosis.22 MYC amplification has been reported in up to 25% of epithelial ovarian cancers, although no Brenner tumors were studied.23 In our study, MYC amplification appeared to co-occur with amplification of either CDK4 (three cases) or CCND1 (two cases). MYC amplification was mutually exclusive with RAS mutations, suggesting an alternate mechanism of pathway activation. It is known that activation of the extracellular signal-regulated kinases/mitogen-activated protein kinases pathway by KRAS mutants leads to downstream MYC activation and tumor dependency.24, 25 Therefore, in the tumors lacking RAS mutations, MYC amplification likely serves the same purpose.
Cyclin D1 (encoded by the CCND1 gene) frequently is deregulated in cancers and has the ability to activate cyclin-dependent kinases (CDKs) CDK4 and CDK6. CDKs play a role in regulating the cell cycle and also are key checkpoint proteins to pause replication in response to DNA damage. One very important negative/inhibitory regulator of the CDK proteins is p16 (encoded by CDKN2A). Thus, the deregulation of the cyclin pathway via alterations in these genes, such as CCND1 and CDK4 amplification or homozygous deletion of CDKN2A, can lead to unchecked proliferation, cellular autonomy, and other characteristics of cancer independent from external or internal controls.26, 27
Hormonal, particularly androgen, signaling likely also plays a role in the development of atypical proliferative (borderline) Brenner tumors. Brenner tumors are known to express androgen receptors and the aldo-keto reductase AKR1C3 (HSD17 β5), which is a key enzyme in androgen production,3, 28 and excessive androgen stimulation may be responsible in part for the epithelial proliferation of these tumors. Of importance, both of our patients with atypical proliferative (borderline) Brenner tumors had clinical signs of virilization as a result of excess androgen production.
Wang et al29 recently reported another approach to evaluate the relationship between mixed Brenner and mucinous tumors. In their study, they analyzed the patterns of X chromosome inactivation (methylation) at eight loci, and microsatellite patterns at nine loci, in five paired Brenner and mucinous tumors, respectively. They showed that the two components of the combined tumors shared a clonal relationship that was statistically significant.
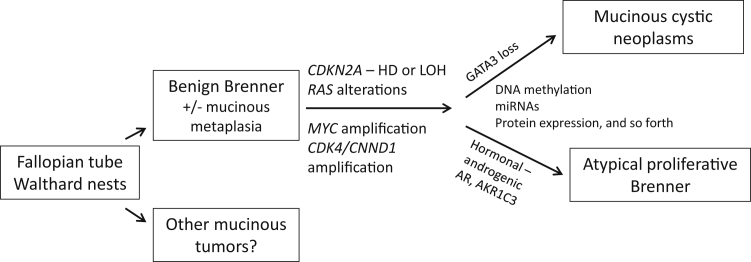
Taking our findings and prior literature together, we propose a model for the relationship between Brenner and mucinous tumors of the ovary (Figure 3). Transitional metaplasia in the form of Walthard rests likely derived from fallopian tube epithelium, may seed the ovary, and develop into a Brenner tumor. Walthard rests also may undergo cystic change to form paratubal cysts and possibly mucinous metaplasia to form other mucinous tumors. Brenner epithelial components may develop mucinous metaplasia with cyst formation. At this point, accumulation of genetic alterations including RAS mutations and CDKN2A aberrations or MYC and CDK4/CNND1 amplification are necessary to further drive histogenesis. Although the RAS alterations are common to both mucinous neoplasms and atypical proliferative (borderline) Brenner tumors, other alterations likely are necessary for the development of these neoplasms. Although the expression of androgen receptors and AKR1C3 have not been well characterized in mucinous tumors at this time, they are known to be overexpressed in atypical proliferative (borderline) Brenner tumors, which, as in our cases, may manifest as virilization. In addition, other differences in gene expression, methylation, and miRNA regulation all are possible mechanisms to differentiate the phenotypes of these tumors. In summary, our findings suggest that RAS mutations, MYC amplification, and, possibly to a lesser extent, PIK3CA mutations, have a role in the progression and pathogenesis of ovarian MCAs/MBTs and atypical proliferative (borderline) Brenner tumors from a benign Brenner tumor.
Figure 3.
Proposed histogenesis of Brenner, atypical proliferative Brenner, and mucinous cystic neoplasms of the ovary and important associated molecular alterations. HD, homozygous deletion; LOH, loss of heterozygosity.
Acknowledgments
We thank the staff of the Dartmouth-Hitchcock Medical Center Molecular Pathology Laboratory, the Translational Research Program (Dartmouth-Hitchcock Medical Center), and the Jackson Clinical Genomics Laboratory. We also thank Dr. Jiang Gui (Geisel School of Medicine at Dartmouth) for statistical advice.
Footnotes
Supported by the Department of Pathology Translational Research Shared Resource Laboratory of the Geisel School of Medicine at Dartmouth, the Dartmouth Hitchcock Medical Center, the Norris Cotton Cancer Center, and the Jackson Laboratory for Genomic Medicine.
Disclosures: Next-generation sequencing was performed at the Jackson Laboratory's Clinical Genomics facility (Farmington, CT) using the JAX Cancer Treatment Profile assay.
A portion of these data were presented as a poster presentation at the 2015 College of American Pathologists Annual Meeting, October 4 to 7, 2015, Nashville, TN (poster 117; Arch Pathol Lab Med 2015;139:e159).
References
- 1.Kondi-Pafiti A., Kairi-Vassilatou E., Iavazzo C.H., Vouza E., Mavrigiannaki P., Kleanthis C.H., Vlahodimitropoulos D., Liapis A. Clinicopathological features and immunoprofile of 30 cases of Brenner ovarian tumors. Arch Gynecol Obstet. 2012;285:1699–1702. doi: 10.1007/s00404-011-2182-5. [DOI] [PubMed] [Google Scholar]
- 2.Gilks C.B., Carinelli S.G., Liu A., Prat J., Seidman J.D., Soslow R. Brenner tumors. In: Kurman R.J., Carcangiu M.L., Herrington C.S., Young R.H., editors. WHO Classification of Tumours of Female Reproductive Organs. IARC; Lyon: 2014. pp. 35–37. [Google Scholar]
- 3.Kuhn E., Ayhan A., Shih IeM., Seidman J.D., Kurman R.J. Ovarian Brenner tumour: a morphologic and immunohistochemical analysis suggesting an origin from fallopian tube epithelium. Eur J Cancer. 2013;49:3839–3849. doi: 10.1016/j.ejca.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Seidman J.D., Khedmati F. Exploring the histogenesis of ovarian mucinous and transitional cell (Brenner) neoplasms and their relationship with Walthard cell nests: a study of 120 tumors. Arch Pathol Lab Med. 2008;132:1753–1760. doi: 10.5858/132.11.1753. [DOI] [PubMed] [Google Scholar]
- 5.Roma A.A., Masand R.P. Different staining patterns of ovarian Brenner tumor and the associated mucinous tumor. Ann Diagn Pathol. 2015;19:29–32. doi: 10.1016/j.anndiagpath.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn E., Ayhan A., Shih IeM., Seidman J.D., Kurman R.J. The pathogenesis of atypical proliferative Brenner tumor: an immunohistochemical and molecular genetic analysis. Mod Pathol. 2014;27:231–237. doi: 10.1038/modpathol.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuatrecasas M., Catasus L., Palacios J., Prat J. Transitional cell tumors of the ovary: a comparative clinicopathologic, immunohistochemical, and molecular genetic analysis of Brenner tumors and transitional cell carcinomas. Am J Surg Pathol. 2009;33:556–567. doi: 10.1097/PAS.0b013e318188b84c. [DOI] [PubMed] [Google Scholar]
- 8.Hunter S.M., Gorringe K.L., Christie M., Rowley S.M., Bowtell D.D., Australian Ovarian Cancer Study Group. Campbell I.G. Pre-invasive ovarian mucinous tumors are characterized by CDKN2A and RAS pathway aberrations. Clin Cancer Res. 2012;18:5267–5277. doi: 10.1158/1078-0432.CCR-12-1103. [DOI] [PubMed] [Google Scholar]
- 9.Ananda G., Mockus S., Lundquist M., Spotlow V., Simons A., Mitchell T., Stafford G., Philip V., Stearns T., Srivastava A., Barter M., Rowe L., Malcolm J., Bult C., Karuturi R.K., Rasmussen K., Hinerfeld D. Development and validation of the JAX Cancer Treatment Profile™ for detection of clinically actionable mutations in solid tumors. Exp Mol Pathol. 2015;98:106–112. doi: 10.1016/j.yexmp.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Lupat R., Amarasinghe K.C., Thompson E.R., Doyle M.A., Ryland G.L., Tothill R.W., Halgamuge S.K., Campbell I.G., Gorringe K.L. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–1313. doi: 10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.1000 Genomes Project Consortium. Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Core Team . R Foundation for Statistical Computing. Austria; Vienna: 2013. R: A language and environment for statistical computing. [Google Scholar]
- 18.Waxman M. Pure and mixed Brenner tumors of the ovary: clinicopathologic and histogenetic observations. Cancer. 1979;43:1830–1839. doi: 10.1002/1097-0142(197905)43:5<1830::aid-cncr2820430536>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 19.Roma A.A., Masand R.P. Ovarian Brenner tumors and Walthard nests: a histologic and immunohistochemical study. Hum Pathol. 2014;45:2417–2422. doi: 10.1016/j.humpath.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Ryland G.L., Hunter S.M., Doyle M.A., Caramia F., Li J., Rowley S.M., Christie M., Allan P.E., Stephens A.N., Bowtell D.D., Australian Ovarian Cancer Study Group. Campbell I.G., Gorringe K.L. Mutational landscape of mucinous ovarian carcinoma and its neoplastic precursors. Genome Med. 2015;7:87. doi: 10.1186/s13073-015-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gymnopoulos M., Elsliger M.A., Vogt P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross J.S., Ali S.M., Wang K., Palmer G., Yelensky R., Lipson D., Miller V.A., Zajchowski D., Shawver L.K., Stephens P.J. Comprehensive genomic profiling of epithelial ovarian cancer by next generation sequencing-based diagnostic assay reveals new routes to targeted therapies. Gynecol Oncol. 2013;130:554–559. doi: 10.1016/j.ygyno.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Bachireddy P., Bendapudi P.K., Felsher D.W. Getting at MYC through RAS. Clin Cancer Res. 2005;11:4278–4281. doi: 10.1158/1078-0432.CCR-05-0534. [DOI] [PubMed] [Google Scholar]
- 25.Huang S., Ren X., Wang L., Zhang L., Wu X. Lung-cancer chemoprevention by induction of synthetic lethality in mutant KRAS premalignant cells in vitro and in vivo. Cancer Prev Res (Phila) 2011;4:666–673. doi: 10.1158/1940-6207.CAPR-10-0235. [DOI] [PubMed] [Google Scholar]
- 26.Musgrove E.A., Caldon C.E., Barraclough J., Stone A., Sutherland R.L. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 27.Asghar U., Witkiewicz A.K., Turner N.C., Knudsen E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji Q., Aoyama C., Chen P.K., Stolz A., Liu P. Localization and altered expression of AKR1C family members in human ovarian tissues. Mol Cell Probes. 2005;19:261–266. doi: 10.1016/j.mcp.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Wu R.C., Shwartz L.E., Haley L., Lin M.T., Shih I.M., Kurman R.J. Clonality analysis of combined Brenner and mucinous tumours of the ovary reveals their monoclonal origin. J Pathol. 2015 doi: 10.1002/path.4572. [Epub ahead of print], doi: 10.1002/path.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]