Abstract
Background
Human mitochondrial aldehyde dehydrogenase 2 (ALDH2) plays a critical role in the detoxification of the ethanol metabolite acetaldehyde. The ALDH2*2 (rs671) gene variant is mainly absent among Europeans but is prevalent in populations in East Asia. The aim of this study was to investigate ALDH2*2 mutant alleles and genotype frequencies in the Hakka population of China.
Material/Methods
Between January 2016 and June 2017, 7,966 unrelated individuals were recruited into the study from the Hakka ethnic population residing in the Meizhou area of Guangdong Province, China, who provided venous blood samples. Genotyping of ALDH2 genotypes were determined using a gene chip platform and confirmed by DNA sequencing.
Results
In the 7,966 individuals from the Hakka population of China in this study, the frequencies of the ALDH2 genotypes *1/*1, *1/*2 and *2/*2 were 52.03%, 39.67%, and 8.30%, respectively; 47.97% of the individuals were found to carry the ALDH2*2 genotype, which was associated with a deficiency in the aldehyde dehydrogenase (ALDH2) enzyme activity. The frequency of the ALDH2*2 allele was lower than that previously reported in the Japanese population but higher than that reported in other Oriental populations.
Conclusions
The findings of this study have provided new information on the ALDH2 gene polymorphisms in the Hakka ethnic population residing in the Meizhou area of Guangdong Province, China, including an understanding of the origin of the atypical ALDH2*2 allele. Also, the study findings may be relevant to the primary care of patients in China.
MeSH Keywords: Aldehyde Dehydrogenase; Pharmacogenetics; Polymorphism, Genetic
Background
Human mitochondrial aldehyde dehydrogenase 2 (ALDH2) is a group of enzymes responsible ethanol metabolism and aldehyde detoxification [1,2]. Also, the mitochondrial ALDH2 plays a critical role as an antioxidative by metabolizing 4-hydroxynonenal (4-HNE) and malondialdehyde. The ALDH2 gene is mapped to chromosome 12 in the region of q24.2 [3]. A single nucleotide polymorphism (SNP) in ALDH2, identified as ALDH2*2 at position 504 (rs671) located in exon 12, results in the transition of guanine (G) to adenine (A) and then an amino acid change from glutamic acid to lysine in the ALDH2 protein [3].
Although polymorphisms in the ALDH2 gene are known to contribute to alcohol-induced facial flushing, variation in this response to alcohol and susceptibility to alcoholism due to alterations in ALDH2 activity caused by genetic variation have only recently been recognized as having both a positive and negative effect on human health and disease [3,4]. Large-scale epidemiological and experimental studies have demonstrated that the ALDH2*2 allele is associated with an increased risk for cardiovascular disease, Alzheimer’s disease, alcoholic cirrhosis, and a series of alcohol-related cancers [5–8]. However, data from epidemiological studies have suggested that the ALDH2*2 allele might have a protective effect against chronic diseases, such essential hypertension and psychiatric disorders [9–11].
Currently, a deficiency of the enzyme, ALDH2, is believed to be one of the most significant enzymopathies in humans, affecting an estimated 560 million people of East Asian descent [12–14]. ALDH2 enzyme deficiency is caused by a base mutation in ALDH2*2 at nucleotide 1459, which leads to a single structural polymorphism at amino acid position 487 of the mature protein; this results in a transition of G to A and a dramatic reduction in the enzyme’s activity [15,16]. Following decades of research involving this single point mutation and its widespread effects, various techniques have been developed to determine an individual’s ALDH2 genotype, with recent studies having investigated the underlying mechanisms for the effects of this polymorphism. Depending on the number of ALDH2*2 monomers present in a tetramer, the aldehyde dehydrogenase enzyme encoded by homozygous genotype ALDH2*1/*1 is the catalytically active subunit, whereas the aldehyde dehydrogenase enzyme encoded by the heterozygote ALDH2*1/*2 or the homozygote ALDH2*2/*2 is the partially or completely inactive subunit [3,17,18]. For heterozygous ALDH2*1/*2 individuals and individuals who are homozygous for ALDH2*2/*2, the ALDH2 enzymatic activity has approximately 16% of the effectiveness of the wild-type homozygous ALDH2*1/*1 in individuals [19].
Previously published studies have demonstrated that there are clear differences in the frequencies of the ALDH2 alleles in different geographical regions, nationalities, and races. For example, the Glu504Lys SNP in the ALDH2 gene is rarely found in Caucasians and individuals of African descent [20–22]. However, the ALDH2*2 mutation is has a high prevalence in Asians of between 30–50% [23]. Also, there are variations in the distribution of the ALDH2*2 allele observed among East Asian populations, including between the Chinese, Korean, and Japanese populations [23,24].
Although the prevalence of the ALDH2 gene polymorphisms has been studied in several major populations of the world, there have been few studies on the Hakka Chinese populations [6,10]. Therefore, the aims of this study were to analyze the frequency of the ALDH2*2 allele and genotype in a large cohort of the Hakka Chinese population and to compare this with previously published data of other ethnic groups. It is hoped that these findings may be of practical value for the potential use in implementing diagnosis and disease risk stratification strategies in primary health care.
Material and Methods
Study population
Between January 2016 and June 2017, 7,966 unrelated individuals were recruited into the study from the Hakka ethnic population residing in the Meizhou area of Guangdong Province, China, who provided venous blood samples. The ethical approval for the study protocol was obtained from the Human Ethics Committees of the Meizhou Peoples’ Hospital (Huangtang Hospital), and Meizhou Hospital Affiliated to Sun Yat-sen University, Guangdong Province, China. Written informed consent was obtained from all participants before entering the study.
DNA extraction and genotyping
Two milliliters of venous blood was collected in EDTA tubes from each volunteer. DNA extraction was carried out using the TIANamp Blood DNA Kit (Tiangen, Beijing, China), according to the manufacturer’s instructions, and quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Genotyping of ALDH2 Glu504Lys (rs671) polymorphisms was performed by polymerase chain reaction (PCR), and the hybridization reactions used a commercially available kit (BaiO Technology Co, Ltd., Shanghai, China). PCR was performed with 25 μL of reaction mixture containing 25 ng of genomic DNA, 0.5 pM of each oligonucleotide primer, 250 μM deoxynucleotide, 2 U Taq DNA polymerase and PCR buffer solution. The PCR cycling conditions were as follows: an initial denaturation step at 94°C for 5 min., followed by 35 cycles of denaturation at 94°C for 25 sec., annealing at 56°C for 25 sec., and extension at 72°C for 25 sec., followed by a final extension step at 72°C for 5 min. The amplification products were then dispensed into a hybridization reaction chamber for the hybridization reactions. The genotypes of ALDH2 were analyzed using the BaiO Array Doctor Version 2.0 software (BaiO Technology Co, Ltd., Shanghai, China) and the BaiO® BE-2.0 software (BaiO Technology Co, Ltd., Shanghai, China), according to the manufacturer’s instructions.
To confirm the quality and accuracy of genotyping data from the gene-chip assay, sequencing analysis was also randomly carried out in the 300 duplicate samples by using the sequencing kit, according to the manufacturer’s instructions (SinoMDgene Technology Co., Ltd., Beijing, China).
Statistical analysis
Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS), version 13.0. Categorical data were expressed as percentages, and continuous data were expressed as the mean ± standard deviation (SD). The genotype frequencies were tested for deviation from the Hardy–Weinberg equilibrium using the Chi-squared test. The Chi-squared and Fisher’s exact tests were also used to compare the allele and genotype frequencies between the Hakka population and published data for other ethnic groups. A value of P<0.05 was considered to be statistically significant.
Results
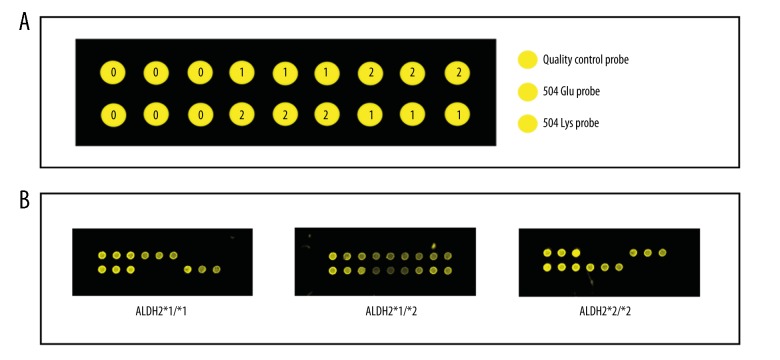
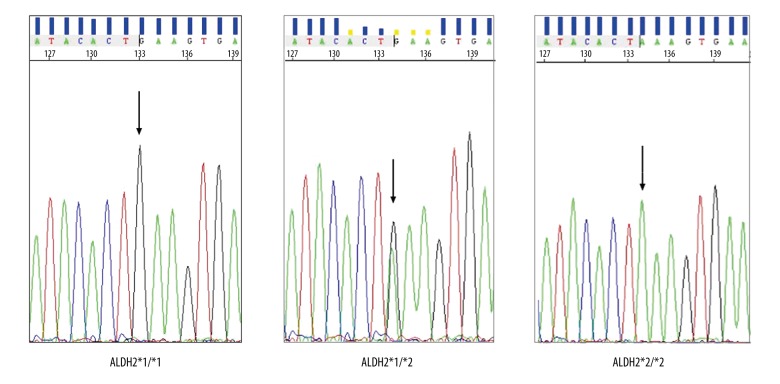
From January 2016 to June 2017, a total of 7,966 unrelated individuals were recruited into the study at our hospital. The study group consisted of 5,453 male individuals and 2,513 female individuals, aged between 10–101 years. As shown in Figure 1, three genotypes of ALDH2 rs671were present: (ALDH2 *1/*1 homozygotes; ALDH2 *1/*2 heterozygotes; and ALDH2 *2/*2 homozygotes. Validation by sequence analysis demonstrated that the gene chip platform used had high sensitivity and accuracy in the genotyping of ALDH2, which were all in concordance with the requirements for DNA sequencing (Figure 2).
Figure 1.
Schematic diagrams of the ALDH2 (Glu504Lys) gene chip and results for the different genotypes. (A) Schematic diagram of the ALDH2 (Glu504Lys) gene chip. (B) Schematic representation of results obtained from the different genotypes.
Figure 2.
Sequencing chromatograms of the ALDH2 (Glu504Lys) genotypes. The presence of base G corresponds to allele ALDH2*1, and the arrow shows the mutation.
The genotype distributions and allele frequencies for the tested ALDH2 variants in the study population are presented in detail in Tables 1 and 2, respectively. The ALDH2 polymorphism frequencies satisfied the Hardy-Weinberg equilibrium (χ2=2.8681, P=0.09). The allele frequency of ALDH2*1 was 71.87%, and ALDH2*2 was 28.13%, respectively. The results showed that a total number of 4,145 individuals (52.03%) were heterozygous (*1/*1) for the ALDH2 polymorphism, and 3,160 individuals (39.67%) were heterozygous (*1/*2), whereas 661 (8.30%) individuals had a homozygous (*2/*2) genotype.
Table 1.
Allele frequencies of CYP2C19 gene among Hakka ethnic groups (n=7,966).
| Variant allele | 2n=15,932 | Frequency (%) |
|---|---|---|
| ALDH2*1 | 11,450 | 71.87% |
| ALDH2*2 | 4,482 | 28.13% |
Table 2.
Genotype frequencies of CYP2C19 gene among Hakka ethnic groups (n=7,966).
| ALDH2*1/*1 | ALDH2*1/*2 | ALDH2*2/*2 | |
|---|---|---|---|
| Age | 59.82±16.31 | 60.00±16.31 | 61.02±16.31 |
| Male (%) | 2,828 (68.23) | 2,180 (68.99) | 445 (67.32) |
| Observed frequency (%) | 4,145 (52.03) | 3,160 (39.67) | 661 (8.30) |
| Expected frequency (%) (Hardy-Weinberg law) | 4,114 (51.65) | 3,221 (40.44) | 630 (7.91) |
Further analysis showed the distribution and frequencies of ALDH2 polymorphism across various populations (Table 3). The Hakka ethnic population had a very similar allele frequency to the Japanese population, but a slightly different frequency to that of the Chinese Han, Korean, and Mongolian populations, and a much higher frequency than for other Asian populations. The results of this study and previous data show that the combined genotypic frequencies of ALDH2*2 was highly prevalent in Eastern Asia populations but is essentially absent among Europeans and other racial populations.
Table 3.
Allele frequencies of ALDH2*2 polymorphisms among the Hakka ethic population and other previously studied populations (n=7,966).
| Population | n | Variant allele (%) | Genotype (%) | Reference | |||
|---|---|---|---|---|---|---|---|
| G | A | *1/*1 | *1/*2 | *2/*2 | |||
| Asian | |||||||
| Hakka | 7,966 | 71.87 | 28.13 | 52.03 | 39.67 | 8.30 | Present study |
| Chinese | 648 | 82.10 | 17.90 | 68.05 | 28.09 | 3.86 | [12] |
| Japanese | 2,299 | 70.09 | 29.90 | 49.63 | 40.93 | 9.44 | [13] |
| Korean | 815 | 81.29 | 18.71 | 66.38 | 29.82 | 3.80 | [14] |
| Thai | 463 | 89.85 | 10.15 | 81.21 | 17.28 | 1.51 | [16] |
| Mongolian | 206 | 74.76 | 25.24 | 57.77 | 33.98 | 8.25 | [23] |
| Uzbek | 161 | 98.45 | 1.55 | 96.89 | 3.11 | 0.00 | [24] |
| Filipino | 86 | 99.42 | 5.81 | 98.84 | 1.16 | 0.00 | [21] |
| Malaysian | 73 | 96.58 | 3.42 | 93.15 | 6.84 | 0.00 | [21] |
| Turkish | 211 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [29] |
| Indian | 87 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [28] |
| European | |||||||
| German | 193 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [21] |
| Polish | 198 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [22] |
| Spanish | 220 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [30] |
| Swedes | 99 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [21] |
| American | |||||||
| Mexican | 101 | 99.51 | 0.49 | 99.01 | 0.99 | 0.00 | [31] |
| Mexican Indian | 118 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [27] |
| Mexican American | 108 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [32] |
| Other | |||||||
| African | 49 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [21] |
| Australian | 37 | 100.00 | 0.00 | 100.00 | 0.00 | 0.00 | [21] |
Discussion
Human mitochondrial aldehyde dehydrogenase 2 (ALDH2) is encoded by the ALDH2 gene, located at exon 12 on chromosome 12q24.2, and is believed to be the key enzyme that degrades and detoxifies acetaldehyde in the liver [18]. Previously published studies have shown that ALDH2 also plays a key role in the removal of other toxic aldehydes, including acrolein malondialdehyde and 4-hydroxynonenal (4-HNE) derived from lipid peroxidation, and thereby protects tissues and cells from oxidative damage [25].
Previously published studies have shown that a mutation in ALDH2 rs671 (Glu504Lys) results in a reduction in the activity of the aldehyde dehydrogenase enzyme, and therefore the clearance of acetaldehyde, a substrate of ALDH2, is limited in individuals who are mutant-type ALDH2 heterozygotes *1/*2 and ALDH2 homozygotes *2/*2 [13]. Not surprisingly, individuals carrying the ALDH2*2 gene polymorphism suffer from various symptoms including facial flushing, headache, drowsiness, and breathlessness, after the consumption of alcohol that may be attributed to the accumulation of acetaldehyde over time, as shown in previous studies [14,15]. Clinical studies have also shown that carriers of ALDH2*2 genotypes are at an increased risk for several diseases, including myocardial infarction, Parkinson’s disease, alcoholic liver disease, and a series of alcohol-related cancers [7,8]. Recent clinical studies have shown that carriers of the ALDH2*2 genotype are protected against chronic diseases, such essential hypertension and psychiatric disorders [10,26].
The inter-individual and inter-ethnic differences in the frequency of ALDH2*2 mutant alleles continues to be a significant topic for clinical research [26]. There have been several published reports on the worldwide genetic polymorphisms of ALDH2*2, which have shown differences in frequency among Asian populations of different geographic areas. The Hakka ethnic group is a unique population who speak the Hakka dialect and mostly inhabit the Meizhou area of Guangdong Province in China. The Hakka people are characterized by their culture, language, lifestyles, and customs, but show some similarities to the people of the Han population in northern China, including their architecture [27]. The Meizhou region is located in the northeast of Guangdong province with a total area of 15.87 km2 and a population of 5.43 million and is bordered by the Jiangxi Province to the northwest and the Fujian Province to the northeast, with approximately 95% of the population of the Meizhou region being Hakka [28]. The Hakka population were considered to be particularly important as a study population to investigate the allele frequencies and genotype distributions of variants of the ALDH2 gene. To our knowledge, this is the first report that has examined the prevalence of alleles of known functional polymorphisms ALDH2*2 in a large study population sample from the Hakka population in the Meizhou region, with all genotype distributions being in Hardy–Weinberg equilibrium.
Several previously published epidemiological studies have shown that a different prevalence of ALDH2*2 variant alleles to be associated with racial origin and geographical distribution [29–31]. In this current study, we assessed the distribution of ALDH2*2 variants in the Hakka ethnic group and compared the data with data from other populations. The frequency of the ALDH2*2 variant was 28.13% in the present study. The frequencies of the ALDH2 genotypes *1/*1, *1/*2 and *2/*2 were 52.03%, 39.67% and 8.30%, respectively. The results demonstrate that Hakka Chinese population have an extremely high allele frequency of ALDH2 *2, which is associated with reduced enzyme activity.
Comparison of the Hakka ethnic group with other ethnic populations indicates differences and similarities in the distribution of the ALDH2*2 allele and genotype (Table 3). The ALDH2*2 allele is not seen among West Asians (absent among Turkish and Indian populations) [30,31]; is minor finding among Central and Southeast Asian populations (10.15% among 463 Thai populations;, 1.55% among 161 Uzbek people; 5.81% among 86 Filipino people; and 3.42% among 73 Malaysian people) [16,21,24]; and is major finding among East Asians (17.90% among 648 Chinese; 29.90% among 2,299 Japanese; 18.71% among 815 Korean; and 25.24% among 206 Mongolian individuals) [12–14,23]. The allelic frequencies of ALDH2*2 observed in the Hakka ethnic group (28.13%) is relatively close to that of the Japanese (29.90%) and Mongolian (25.24%) populations, but showing a large difference from the other oriental population. Furthermore, the ALDH2*2 allele was almost absent among Europeans, including Germans, Polish people, the Spanish, and Swedes, as well as Americans, Australians and Africans [21,22,32–34].
Differences in the frequency of ALDH2 polymorphism in different populations have epidemiologic importance, as many ethnic populations and many genetic variations exist. To date, the global distribution of the prevalence of the ALDH2*2 allele shows a clear east-to-west decrease, where it is dominant in East Asia, rare in Southeast Asia, and absent in West Asia and other parts of the world [21,31–33]. The results of this study showed that the ALDH2*2 allele is virtually exclusive to northeast Asian populations. The differences may be associated with the racial origin and geographical distribution. Some studies have postulated that the ALDH2*2 allele is likely to have dispersed from an origin toward East Asia with the high frequencies in Southeastern coastal regions of China associated with the historical Han migrations thousands of years ago, to Japan, Korea, and Taiwan [35]. Because subjects with the ALDH*2 variant in Asian populations have an abnormality in the metabolism of acetaldehyde, with adverse symptoms, extra clinical care must be taken with this Asian population.
Conclusions
In conclusion, the findings of this study have shown that the determined allelic variants of ALDH2*2 in the Hakka ethnic group in China is 28.13% and is similar to that of East Asian countries. This study confirms the ethnic differences in the ALDH2 allele and genotype frequencies. The high prevalence of the ALDH2*2 allele in this East Asian population may have important implications for public health. In particular, the ALDH2*2 variant is associated with an increased risk for several diseases, including cardiovascular diseases, Alzheimer’s disease, and alcohol-related cancers. Implementing a primary healthcare approach based on the detection of an ALDH2*2 genotype may improve the health of individuals assessing potential disease risk and medication efficacy. A suggested strategy could involve the identification of ALDH2*2 variants within a population such as the Hakka ethnic population and to establish education and intervention in primary healthcare facilities for individuals who may be at increased risk of disease. Future studies are required to determine the clinical consequences of this genetic polymorphism in carrier individuals.
Footnotes
Source of support: This study was supported by The National Key Research and Development Program of China (Grant No: 2016YFD0050405; Dr. Pingsen Zhao), the National Key Research and Development Program of China (Grant No: 2017YFD0501705; Dr. Pingsen Zhao), Natural Science Foundation of Guangdong Province, China (Grant No: 2016A030307031; Dr. Pingsen Zhao), Natural Science Foundation of Guangdong Province, China (Grant No: 2014A030307042; Dr. Pingsen Zhao), Medical Scientific Research Foundation of Guangdong Province, China (Grant No: A2016306; Pingsen Zhao) and Key Scientific and Technological Project of Meizhou People’s Hospital, Guangdong Province, China (Grant No: MPHKSTP-20170102; Pingsen Zhao) and Medical Scientific Research Foundation of Guangdong Province, China (Grant No: A2017404; Dr. Jingyuan Hou)
Conflicts of interest
None.
References
- 1.Agarwal DP. Genetic polymorphisms of alcohol metabolizing enzymes. Pathol Biol. 2001;49(9):703–9. doi: 10.1016/s0369-8114(01)00242-5. [DOI] [PubMed] [Google Scholar]
- 2.Higuchi S, Matsushita S, Murayama M, et al. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry. 1995;52(8):1219–21. doi: 10.1176/ajp.152.8.1219. [DOI] [PubMed] [Google Scholar]
- 3.Kim JS, Kim YJ, Kim TY, et al. Association of ALDH2 polymorphism with sensitivity to acetaldehyde-induced micronuclei and facial flushing after alcohol intake. Toxicology. 2005;210(2):169–74. doi: 10.1016/j.tox.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto T, Hara M, Higaki Y, et al. Influence of alcohol consumption and gene polymorphisms of ADH2 and ALDH2 on hepatocellular carcinoma in a Japanese population. Int J Cancer. 2006;118(6):1501–7. doi: 10.1002/ijc.21505. [DOI] [PubMed] [Google Scholar]
- 5.Hui P, Nakayama T, Morita A, et al. Common single nucleotide polymorphisms in japanese patients with essential hypertension: Aldehyde dehydrogenase 2 gene as a risk factor independent of alcohol consumption. Hypertens Res. 2007;30(7):585–92. doi: 10.1291/hypres.30.585. [DOI] [PubMed] [Google Scholar]
- 6.Chang JS, Hsiao JR, Chen CH. ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective. J Biomed Sci. 2017;24(1):19. doi: 10.1186/s12929-017-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamino K, Nagasaka K, Imagawa M, et al. Deficiency in mitochondrial aldehyde dehydrogenase increases the risk for late-onset Alzheimer’s disease in the Japanese population. Biochem Biophys Res Commun. 2000;273(1):192–96. doi: 10.1006/bbrc.2000.2923. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto Y, Nakayama T, Futanura A, et al. Relationship between genetic polymorphisms of alcohol-metabolizing enzymes and changes in risk factors for coronary heart disease associated with alcohol consumption. Clin Chem. 2002;48(7):1043–48. [PubMed] [Google Scholar]
- 9.Yang S, Lee J, Choi IJ, et al. Effects of alcohol consumption, ALDH2 rs671 polymorphism, and Helicobacter pylori infection on the gastric cancer risk in a Korean population. Oncotarget. 2017;8(4):6630–41. doi: 10.18632/oncotarget.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang SY, Chan SW, Zhou X, et al. Meta-analysis of association between ALDH2 rs671 polymorphism and essential hypertension in Asian populations. Herz. 2015;40(S2):203–8. doi: 10.1007/s00059-014-4166-2. [DOI] [PubMed] [Google Scholar]
- 11.Li D, Zhao H, Gelernter J. Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Human Genetics. 2012;131(5):725–37. doi: 10.1007/s00439-011-1116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo H-R, Israel Y, Tu G-C, et al. Genetic polymorphism of aldehyde dehydrogenase 2 (ALDH2) in a Chinese population: Gender, age, culture, and genotypes of ALDH2. Biochemical Genetics. 2005;43:223–27. doi: 10.1007/s10528-005-5213-8. [DOI] [PubMed] [Google Scholar]
- 13.Matsuo K, Wakai K, Hirose K, et al. Alcohol dehydrogenase 2 His47Arg polymorphism influences drinking habit independently of aldehyde dehydrogenase 2 Glu487Lys polymorphism: Analysis of 2,299 Japanese subjects. Cancer Epidemiol Biomarkers Prev. 2006;15(5):1009–13. doi: 10.1158/1055-9965.EPI-05-0911. [DOI] [PubMed] [Google Scholar]
- 14.Shin CM, Kim N, Cho SI, et al. Association between alcohol intake and risk for gastric cancer with regard to ALDH2 genotype in the Korean population. Int J Epidemiol. 2011;40:1047–55. doi: 10.1093/ije/dyr067. [DOI] [PubMed] [Google Scholar]
- 15.Edenberg HJ. The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- 16.Boonyaphiphat P, Thongsukai P, Sriplung H, et al. Lifestyle habits and genetic susceptibility and the risk of esophageal cancer in the Thai population. Cancer Lett. 2002;186:193–99. doi: 10.1016/s0304-3835(02)00354-3. [DOI] [PubMed] [Google Scholar]
- 17.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83(1):314–16. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai CL, Yao CT, Chau GY, et al. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol Clin Exp Res. 2014;38(1):44–50. doi: 10.1111/acer.12215. [DOI] [PubMed] [Google Scholar]
- 19.Kang TS, Woo SW, Park HJ, et al. Comparison of genetic polymorphisms of CYP2E1, ADH2, and ALDH2 genes involved in alcohol metabolism in Koreans and four other ethnic groups. J Clin Pharm Ther. 2009;34(2):225–30. doi: 10.1111/j.1365-2710.2008.00986.x. [DOI] [PubMed] [Google Scholar]
- 20.Bhaskar LVSK, Thangaraj K, Osier M, et al. Single nucleotide polymorphisms of the ALDH2 gene in six Indian populations. Ann Hum Biol. 2007;34(6):607–19. doi: 10.1080/03014460701581419. [DOI] [PubMed] [Google Scholar]
- 21.Goedde HW, Agarwal DP, Fritze G, et al. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88(3):44–46. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- 22.Cichoz-Lach H, Partycka J, Nesina I, et al. Alcohol dehydrogenase and aldehyde dehydrogenase gene polymorphism in alcohol liver cirrhosis and alcohol chronic pancreatitis among Polish individuals. Scand J Gastroenterol. 2007;42(4):493–98. doi: 10.1080/00365520600965723. [DOI] [PubMed] [Google Scholar]
- 23.Zhou S, Huriletemuer J, Wang J, et al. Absence of association on aldehyde dehydrogenase 2 (ALDH2) polymorphism with Mongolian Alzheimer patients. Neurosci Lett. 2010;468(3):312–15. doi: 10.1016/j.neulet.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Ahn KS, Abdiev S, Rahimov B, et al. Alcohol dehydrogenase 1B and aldehyde dehydrogenase 2 polymorphisms in Uzbekistan. Asian Pac J Cancer Prev. 2009;10(1):17–20. [PubMed] [Google Scholar]
- 25.Singh S, Brocker C, Koppaka V, et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56(3):89–101. doi: 10.1016/j.freeradbiomed.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Ye YL, Wang YN, et al. Aldehyde dehydrogenase 2 genetic variations may increase susceptibility to Parkinson’s disease in Han Chinese population. Neurobiol Aging. 2015;36(9):2660.e9–13. doi: 10.1016/j.neurobiolaging.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Wang WZ, Wang CY, Cheng YT, et al. Tracing the origins of Hakka and Chaoshanese by mitochondrial DNA analysis. Am J Phys Anthropol. 2010;141(1):124–30. doi: 10.1002/ajpa.21124. [DOI] [PubMed] [Google Scholar]
- 28.Lin M, Wang Q, Zheng L, et al. Prevalence and molecular characterization of abnormal hemoglobin in eastern Guangdong of southern China. Clin Genet. 2012;81(2):65–171. doi: 10.1111/j.1399-0004.2011.01627.x. [DOI] [PubMed] [Google Scholar]
- 29.Montano Loza AJ, Ramirez Iglesias MT, Perez Diaz I, et al. Association of alcohol-metabolizing genes with alcoholism in a Mexican Indian (Otomi) population. Alcohol. 2006;39(2):73–79. doi: 10.1016/j.alcohol.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Seng KY, Limenta LM, Heng D, Lee EJ. Population pharmacokinetics and pharmacogenetics of alcohol in Chinese and Indians in Singapore. J Clin Pharm Ther. 2013;38(2):141–49. doi: 10.1111/jcpt.12003. [DOI] [PubMed] [Google Scholar]
- 31.Kayaaltı Z, Söylemezoğlu T. Distribution of ADH1B, ALDH2, CYP2E1 *6, and CYP2E1 *7B genotypes in Turkish population. Alcohol. 2010;44(5):415–23. doi: 10.1016/j.alcohol.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Lorenzo A, Auguet T, Vidal F, et al. Polymorphisms of alcohol-metabolizing enzymes and the risk for alcoholism and alcoholic liver disease in Caucasian Spanish women. Drug Alcohol Depend. 2006;84(2):195–200. doi: 10.1016/j.drugalcdep.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Gordillo-Bastidas E, Panduro A, Gordillo-Bastidas D, et al. Polymorphisms of alcohol metabolizing enzymes in indigenous Mexican population: Unusual high frequency of CYP2E1*c2 allele. Alcoholism Clin Exp Res. 2010;34(1):142–49. doi: 10.1111/j.1530-0277.2009.01075.x. [DOI] [PubMed] [Google Scholar]
- 34.Wan YJ, Poland RE, Lin KM. Genetic polymorphism of CYP2E1, ADH2, and ALDH2 in Mexican-Americans. Genet Test. 1998;2(1):79–83. doi: 10.1089/gte.1998.2.79. [DOI] [PubMed] [Google Scholar]
- 35.Luo HR, Wu GS, Pakstis AJ, et al. Origin and dispersal of atypical aldehyde dehydrogenase ALDH2487Lys. Gene. 2009;435(1–2):96–103. doi: 10.1016/j.gene.2008.12.021. [DOI] [PubMed] [Google Scholar]