Abstract
The microRNA miR-132 serves as a key regulator of a wide range of plasticity-associated processes in the central nervous system. Interestingly, miR-132 expression has also been shown to be under the control of the circadian timing system. This finding, coupled with work showing that miR-132 is expressed in the hippocampus, where it influences neuronal morphology and memory, led us to test the idea that daily rhythms in miR-132 within the forebrain modulate cognition as a function of circadian time. Here, we show that hippocampal miR-132 expression is gated by the time-of-day, with peak levels occurring during the circadian night. Further, in miR-132 knockout mice and in transgenic mice, where miR-132 is constitutively expressed under the control of the tetracycline regulator system, we found that time-of-day dependent memory recall (as assessed via novel object location and contextual fear conditioning paradigms) was suppressed. Given that miRNAs exert their functional effects via the suppression of target gene expression, we examined the effects that transgenic miR-132 manipulations have on MeCP2 and Sirt1—two miR-132 targets that are associated with neuronal plasticity and cognition. In mice where miR-132 was either knocked out, or transgenically expressed, rhythmic expression of MeCP2 and Sirt1 was suppressed. Taken together, these results raise the prospect that miR-132 serves as a key route through which the circadian timing system imparts a daily rhythm on cognitive capacity.
Work in many species has shown that the circadian timing system exerts a potent modulatory influence over both learning and memory (Holloway and Wansley 1973a,b; Wansley and Holloway 1975; Monk et al. 1997; Fernandez et al. 2003; for reviews, see Schmidt et al. 2007; Gerstner and Yin 2010; Lyons 2011; Smarr et al. 2014). For example, in mammals, the efficacy of memory formation, acquisition, and recall vary depending on the time-of-day (Davies et al. 1973; Monk et al. 1997; Valentinuzzi et al. 2001; Chaudhury and Colwell 2002; Eckel-Mahan et al. 2008). Further, disruption of circadian timing in the suprachiasmatic nucleus (SCN), the locus of the master circadian clock, has profound effects on the acquisition and maintenance of learning and memory (Stephan and Kovacevic 1978; Tapp and Holloway 1981; Ruby et al. 2008; Phan et al. 2011).
In addition to the SCN, the role of ancillary clocks located in forebrain circuits has also been shown to exert circadian influence over cognition. For example, Snider et al. (2016) showed deficits in circadian-gated learning in a mouse model where a key circadian gene, Bmal1, was deleted from forebrain excitatory neurons. Likewise, Shimizu et al. (2016) demonstrated that consolidation of long-term recognition memory is abrogated when the hippocampal clock is disrupted. These findings raise interesting questions about the molecular effectors that couple the circadian clock to rhythms of plasticity and memory in the hippocampus.
If one were to posit potential genetic/cellular signaling mechanisms by which the clock modulates cognition, a candidate gene would likely function at the interface of circadian timing and memory; hence this gene would be clock-regulated, and it would serve as a modulator of synaptic plasticity in the hippocampus. One gene that could fit these criteria is the microRNA, miR-132. Vo et al. (2005) first identified miR-132, characterizing its inducible regulation and its marked effects on neurite outgrowth. Since this initial study, a large body of work has shown that miR-132 is inducibly expressed in vivo following a wide range of stimuli (Cheng et al. 2007; Nudelman et al. 2010; Mellios et al. 2011; Hansen et al. 2013; Hernandez-Rapp et al. 2015). miR-132 also plays a role in shaping neuronal morphology and synaptic plasticity/transmission (Impey et al. 2010; Jasińska et al. 2016; for review, see Aten et al. 2016). Consistent with this, deletion of miR-132 attenuates activity-dependent dendritic growth and leads to a reduction in hippocampal dendritic length and spine density (Wayman et al. 2008; Magill et al. 2010) and a decrease in the amplitude of both evoked and spontaneous EPSCs (Luikart et al. 2011; Remenyi et al. 2013), whereas overexpression of miR-132 leads to an increase in spine density (Hansen et al. 2010) and an increase in paired-pulse facilitation and mEPSC amplitude (Edbauer et al. 2010; Lambert et al. 2010). In accordance with these observations, knockout of miR-132 leads to cognitive deficits (Hernandez-Rapp et al. 2015; Hansen et al. 2016), while moderate overexpression of transgenic miR-132 enhances cognition (Hansen et al. 2013).
With respect to circadian physiology, miR-132 expression is under the control of the circadian oscillator in the SCN, and photic entrainment cues trigger a marked increase in miR-132 levels (Cheng et al. 2007). Additionally, miR-132 also varies over the diurnal cycle in the rat brain, with significant time-of-day expression differences found in the somatosensory cortex (Davis et al. 2011).
The noted studies raise the possibility that miR-132 could serve as a signaling intermediate through which the circadian timing system fine tunes learning and memory efficacy. Here, we used transgenic and knockout mouse lines and a series of molecular and behavioral approaches to examine the contribution of miR-132 to time-of-day dependent measures of cognition. We show that both the deletion of miR-132 and the transgenic expression of miR-132 to stable (noncircadian-regulated) levels disrupt the effect of the circadian system on learning and memory efficiency. These data suggest that the rhythmic expression of miR-132 modulates synaptic circuits that facilitate optimal cognitive performance across the circadian day.
Results
Endogenous miR-132 expression is higher during the circadian night
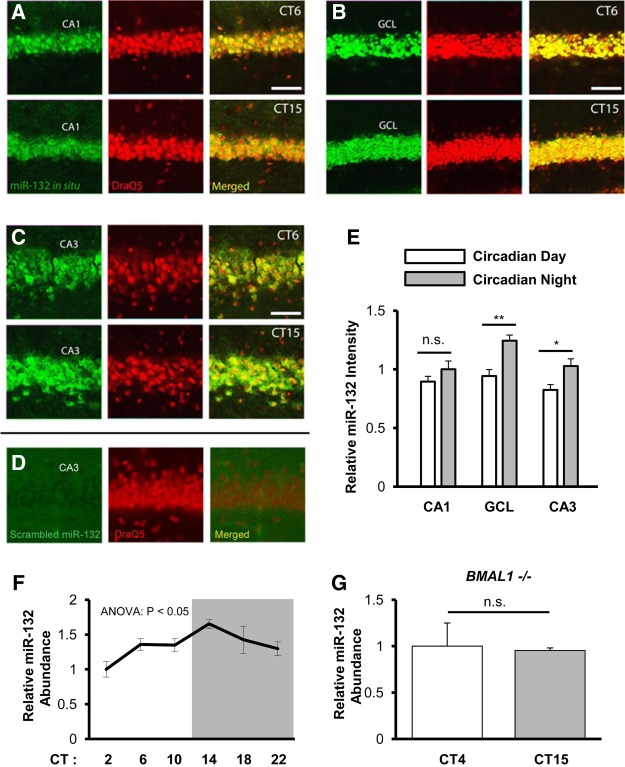
Previous work has demonstrated that miR-132 expression is under the control of the circadian timing system in the SCN, with peak expression occurring during the subjective day (Cheng et al. 2007). Given that circadian rhythms have been detected in the forebrain, we examined whether miR-132 could also be expressed in a time-of-day-dependent manner within the hippocampus. To this end, C57Bl/6 mice were entrained to a 12 h light–dark cycle, and were then dark-adapted for 2 d (to remove the entraining effects of light) before being sacrificed during either the circadian day (CT 6) or the circadian night (CT 15). Subsequently, hippocampal tissue was processed for miR-132 expression using fluorescence in situ hybridization (FISH) (Fig. 1A–E). Representative images (Fig. 1A–C) and quantitative analysis (Fig. 1E) revealed that miR-132 expression was significantly higher during the circadian night (CT15) (Fig. 1E, overall the time-of-day effect on brain regions F(1,32) = 20.107, P < 0.001; interaction F(2,32) = 1.500, P = 0.241; two-way ANOVA). Specifically, miR-132 was significantly higher at night in the GCL and CA3 (Fig. 1E, t(9) = 3.796, P = 0.004 for GCL and t(9) = 2.625, P = 0.028 for CA3; selective Bonferroni post hoc test).
Figure 1.
Time-of-day miR-132 expression in the hippocampus. miR-132 expression in the CA1 (A), GCL (B), CA3 (C), was examined by FISH in WT animals during the circadian day (CT6) and the circadian night (CT15) time points. Scale bar = 50 µm for CA1, GCL, and CA3. (D) FISH profiling of a control, scrambled miRNA probe: note the lack of cellular labeling. (E) Quantification of the relative intensity of miR-132 expression in the noted brain regions during the circadian day and the circadian night. Data were analyzed by two-way ANOVA, n = 5–6 animals per group. CA1 miR-132 mean intensity during the circadian night was set equal to a value of one, and the remaining brain regions were normalized to this condition. Data are displayed as the mean ± standard error (SEM); (*) P < 0.05; (**) P < 0.01; (n.s.) not significant (P > 0.05). (F) Total hippocampal RNA was isolated from WT mice sacrificed every 4 h across the 24-h circadian cycle, and miR-132 cDNA was probed via real-time PCR. Relative miR-132 abundance at CT2 was set equal to a value of one. Significance was examined via Kruskal Wallis Test, n = 7–8 animals per group. Data are displayed as the mean ± SEM for each timepoint. (G) Total hippocampal RNA was isolated from BMAL1−/− animals sacrificed at CT4 and CT15. Relative miR-132 abundance at CT4 was set equal to a value of one, and miR-132 expression at CT15 was normalized to this value. Significance was examined via Student's t-test, n = 3–4 animals per timepoint.
Next, we examined the circadian profile of miR-132 expression in the hippocampus. Thus, WT mice were dark-adapted for 2 d, and tissue was isolated at 4 h intervals over the circadian cycle. Quantitative RT-PCR revealed an oscillation in miR-132 expression, with peak expression occurring during the early subjective night (CT14) and nadir occurring during the early subjective day (CT2) (Fig. 1F; F(5,44) = 12.94, P = 0.024; Kruskal–Wallis one-way ANOVA). Specifically, Dunn's Multiple Comparison Test revealed a significant difference in miR-132 expression between CT2 and CT14 (Fig. 1F, t(15) = −24.39, P < 0.01). Further, to determine whether this oscillation is under the influence of the circadian clock, miR-132 expression was examined in tissue isolated from BMAL1−/− mice. BMAL1 is an essential component of the molecular circadian clock, and as such BMAL1−/− mice do not exhibit clock-gated molecular or behavioral rhythms (Bunger et al. 2000). Using the approximate peak and nadir of miR-132 expression in WT mice as a reference point, we did not detect a time-of-day difference in miR-132 expression in hippocampal tissue from BMAL1−/− mice (Fig. 1G; t(5) = 0.864; Student's t-test). Together these data indicate a clock-driven miR-132 rhythm in the hippocampus.
miR-132 knockout and miR-132 transgenic mice
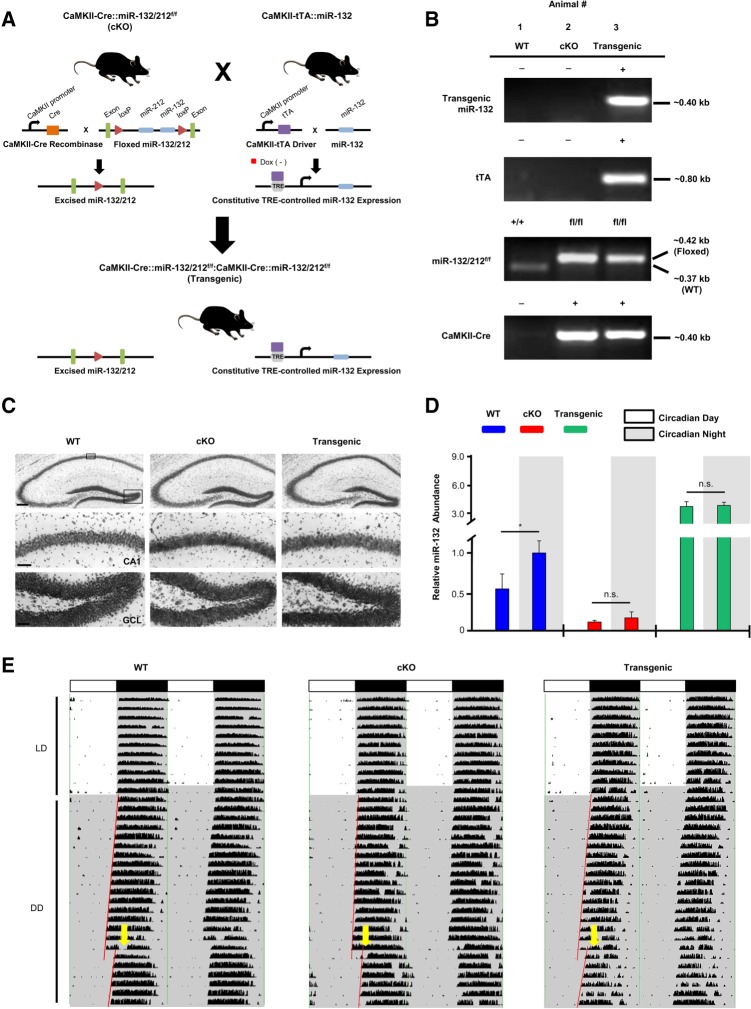
Prior studies from our laboratory demonstrated that cognitive capacity is tightly regulated by miR-132 (Hansen et al. 2010, 2013). This observation, coupled with the observed rhythm in hippocampal miR-132 expression, led us to test the role of miR-132 in time-of-day learning and memory. To this end, we utilized two genetically modified mouse lines: one in which miR-132 is deleted, and a second line, where miR-132 time-of-day expression is suppressed. In the first line, a Cre/lox strategy was used to conditionally delete the miR-132/212 locus from excitatory forebrain neurons, including those of the cortex and hippocampus (miR-132/212f/f conditional knockout; Fig. 2A, hereafter referred to as the “cKO” line: Fig. 2B, Animal #2). Prior work with this line confirmed the selective loss of miR-132 from forebrain excitatory neurons (Hansen et al. 2016). In the second mouse line, we sought to test the cellular and behavioral effects that constitutive, noncircadian-gated, expression of miR-132 would have on time-of-day-dependent learning and memory. Thus, we crossed the miR-132/212f/f conditional knockout (cKO) line to a tetracycline-inducible transgenic miR-132 mouse line. The tetracycline inducible (i.e., “Tet-off”) miR-132 transgenic line (previously described by Hansen et al. 2010) was generated by crossing a CaMKII-tTA driver line with a line expressing miR-132 (and cyan fluorescent protein) under the control of the TRE promoter. Therefore, crossing of the CaMKII-Cre::miR-132/212f/f (cKO) line to the CaMKII-tTA::miR-132-CFP transgenic line generated a four transgene mouse line: CaMKII-tTA::miR-132:CaMKII-CRE::miR-132/212f/f (Fig. 2A, hereafter referred to as the “Transgenic” miR-132 line: Fig. 2B, Animal #3). This unique animal model allowed us to selectively delete endogenous miR-132 and transgenically express miR-132 in the same population of excitatory forebrain neurons. Of note, cKO and transgenic mice were generated in the expected Mendelian distribution with normal sex ratios, and no anatomical abnormalities were detected. Cresyl violet labeling revealed no obvious gross morphological differences in the hippocampus of cKO and Transgenic animals compared to WT animals (Fig. 2C). Control animals (hereafter referred to as “WT”) were either negative for all four noted transgenes (as depicted in Fig. 2B, Animal #1), or they were positive for the driver (CaMKII-Cre or CaMKII-tTA) or the responder transgene (miR-132 or miR-132/212f/f).
Figure 2.
miR-132 knockout and transgenic mouse models and circadian phenotyping. (A) Schematic depiction of CaMKII-Cre::miR-132/212f/f (denoted as “cKO”) and CaMKII-tTA::miR-132:CamKII-Cre::miR-132/212f/f (“Transgenic”) mice. (B) PCR-based genotyping results for the miR-132 cKO and Transgenic mice. Of note, mouse #2 represents a miR-132 cKO animal—positive for both the CaMKII-Cre driver and the miR-132/212f/f transgene. Mouse #3 represents a miR-132 Transgenic animal—positive for both the tTA and miR-132 transgenes, in addition to the CaMKII-Cre driver and the miR-132/212f/f transgene. (C) Representative hippocampal tissue stained with cresyl violet. Of note, no gross morphological differences in cKO and Transgenic animals were detected; scale bar = 150 μm for low-magnification images and 50 μm for high-magnification images. (D) To profile miR-132 expression levels, total hippocampal RNA from WT, cKO, and Transgenic animals was isolated, reverse transcribed and probed via real-time PCR. Relative miR-132 abundance (in WT animals) during the circadian night was set equal to a value of one. Time-of-day comparisons were analyzed by Student's t-test, n = 3–5 animals per group. Data are displayed as the mean ± SEM; (*) P < 0.05; (n.s.) not significant (P > 0.05). (E) Double-plotted wheel running actograms of WT, cKO, and Transgenic mice. White and black horizontal bars across the tops of the plots represent the light and dark (LD) periods, respectively; DD (shown on the Y-axis) refers to the time when mice were housed in constant darkness. Yellow arrows indicate a light pulse (15 min, 40 lux) given at CT15 to induce a phase delay.
In order to assess hippocampal miR-132 expression, quantitative RT-PCR was performed on the three noted mouse lines at CT4 (circadian day) and CT15 (circadian night). Consistent with the data shown in Figure 1, WT animals showed significantly higher miR-132 expression at night (Fig. 2D, t(9) = 2.560, P = 0.031; Student's t-test); in contrast, a time-of-day difference in cKO and Transgenic animals was not detected (Fig. 2D). The low level of miR-132 detected in cKO animals likely results from interneuron and nonneuronal cell populations. Importantly, expression of the reference snRNA that was used for normalization (RNU6B) was not altered between circadian day and night conditions (data not shown). Together, these results suggest that the WT rhythmic profile of miR-132 is eliminated in miR-132 cKO and Transgenic animals.
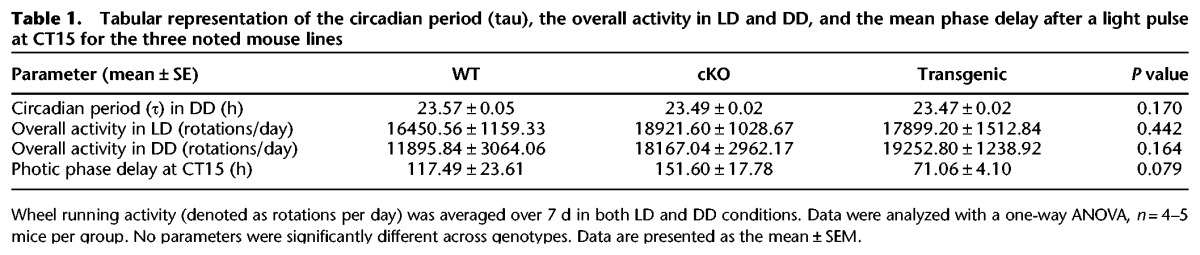
Next, we examined circadian wheel running activity of WT, cKO, and Transgenic mice in order to determine whether the SCN clock machinery was significantly affected by the noted genetic manipulations. Double plotted actograms of WT, cKO, and Transgenic mice are presented (Fig. 2E). All circadian wheel running data were analyzed by one-way ANOVA. The circadian period (tau) under DD conditions was similar for all genotypes (Table 1; F(2,13) = 2.087, P = 0.170), and the overall activity (rotations per day) in LD and in DD was similar for all three genotypes (Table 1; F(2,13) = 0.876, P = 0.442) and (F(2,13) = 2.139, P = 0.164). Additionally, a light pulse (40 lux, 15 min) administered during the early subjective night (CT15), elicited phase delaying responses in all three lines. The Transgenic animals exhibited an attenuated phase shift relative to the two other lines, however this difference did not reach statistical significance (Table 1; F(2,13) = 3.217, P = 0.079). Together, these experiments show that key functional features of the SCN are retained in miR-132 cKO and Transgenic animals.
Table 1.
Tabular representation of the circadian period (tau), the overall activity in LD and DD, and the mean phase delay after a light pulse at CT15 for the three noted mouse lines
Impairments in time-of-day dependent novel object location memory in mice with forebrain miR-132 dysregulation
We next sought to examine the effects of miR-132 dysregulation on time-of-day dependent measures of learning and memory. To this end, we utilized the novel object location (NOL) task—a hippocampal-dependent memory test (Barker and Warburton 2011; Chao et al. 2016) with an efficiency that has previously been shown to be influenced by circadian time (McGowan and Coogan 2013; Takahashi et al. 2013, Snider et al. 2016). The NOL task examines the ability of a mouse to remember the location of an object after moving one of the two objects in the arena. Thus, the more time spent exploring the moved object (compared to the object in the familiar location), the better the memory recall (paradigm shown in Fig. 3A).
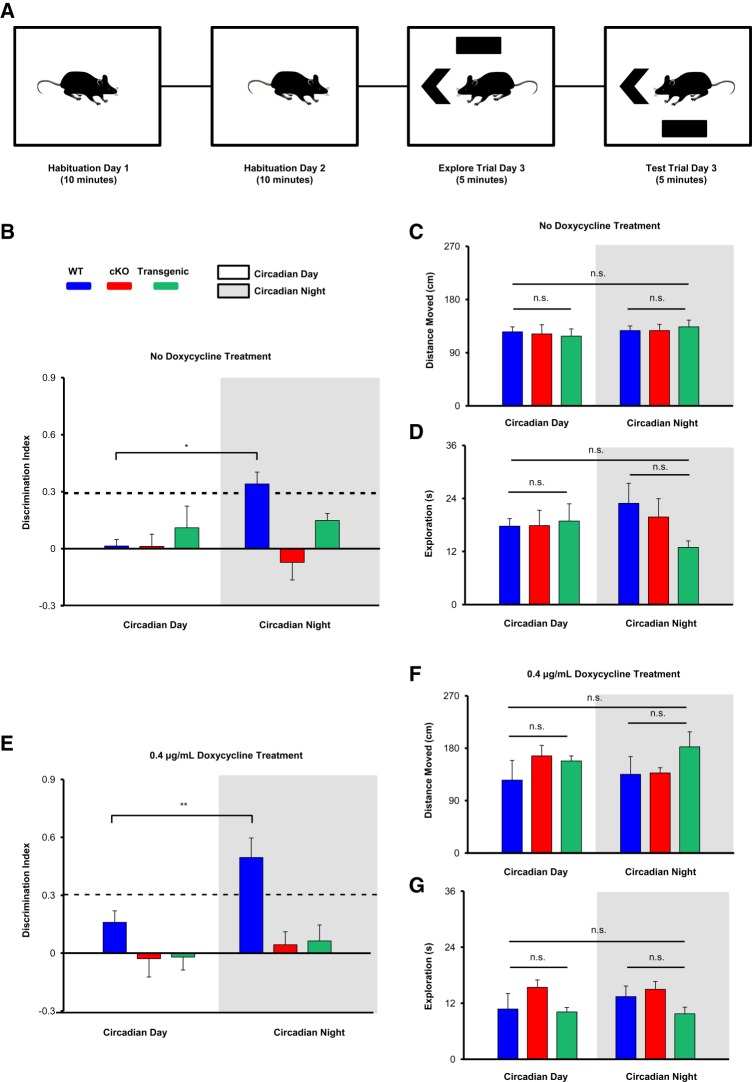
Figure 3.
Time-of-day-dependent learning assayed using the NOL task. (A) Outline of the experimental design. (B) Discrimination indices of WT, cKO, and Transgenic mice were profiled (in the absence of doxycycline treatment) during the circadian day and the circadian night, recorded as [(novel object exploration time-familiar object exploration time)/(total exploration time)]. Significant time-of-day differences were tested using the repeated-measures ANOVA with Bonferroni post hoc correction for within genotype comparisons; n = 8–10 animals per group. The dashed line indicates 30% more time spent exploring the NOL compared to the familiar object location. Only WT animals (tested during the circadian night) demonstrated above-chance NOL discrimination, analyzed with a one-sample t-test. Total distance moved (C) and total exploration time (D) did not differ between WT, cKO, and Transgenic animals, and did not differ in any genotype depending on time-of-day, assessed via Repeated-Measures ANOVA. (E) Discrimination indices of WT, cKO, and Transgenic animals (maintained on 0.4 μg/mL doxycycline water) profiled during both time points, as described in B. Only WT animals (tested at night) demonstrated above-chance NOL discrimination. Total distance moved (F) and total exploration time (G) did not differ between genotypes or time-of-day after doxycycline treatment, assessed via Repeated-Measures ANOVA; n = 6–9 animals per group. Data are presented as mean ± SEM. (*) P < 0.05; (**) P < 0.01; (n.s.) not significant (P > 0.05).
To determine time-of-day performance in the NOL task, mice were tested at circadian day (CT4) and circadian night (CT15) time points. For this experiment, mice on a 12 h/12 h LD schedule were dark-adapted (DD) for 2 d and were subsequently tested under dim red light (∼5 lux). The red light condition allowed animals to use visual/contextual cues, while not affecting the circadian clock timing system (Hattar et al. 2003; Figueiro and Rea 2010; Zhang et al. 2017). Interestingly, WT animals exhibited time-of-day differences in NOL discrimination, while miR-132 cKO and Transgenic animals did not. Indeed, an interaction at the level of genotype that depended on time-of-day was detected (Fig. 3B, interaction F(2,28) = 5.011, P = 0.001; repeated-measures ANOVA). In particular, when comparing within genotype, WT animals displayed a significantly higher discrimination index (DI) when tested at night compared to when tested during the day (Fig. 3B, t(9) = 3.497, P = 0.020; Bonferroni post hoc test). On the other hand, cKO and Transgenic animals showed no such differences (Fig. 3B, t(9) = 0.978, P = 1.000 for cKO mice and t(10) = 0.776, P = 1.000 for Transgenic mice; Bonferroni post hoc test). Additionally, statistical analyses were performed to determine whether the DI for each group (at each time point) was significantly greater than chance levels. Only WT animals demonstrated greater-than-chance discrimination for the novel location at night (Fig. 3B, t(9) = 3.47, P = 0.005; one sample t-test). Finally, no significant effect of time-of-day or genotype on total distance moved (Fig. 3C) or total exploration time (Fig. 3D) was observed (assessed via repeated-measures ANOVA).
Previous work from our laboratory has shown that overexpression of miR-132 (greater than approximately threefold over basal levels) can impair learning and memory (Hansen et al. 2010, 2013). Given that the Transgenic animals displayed an approximately sevenfold increase in miR-132 during the day, and a ∼3.5-fold increase relative to endogenous miR-132 levels at night (Fig. 2D), we wanted to determine whether time-of-day-dependent cognitive deficits revealed in the NOL assay in the miR-132 Transgenic animals could be attributed to the high, supra-physiological, levels of transgenic miR-132. Thus, we administered 0.4 μg/mL doxycycline to the drinking water of a separate cohort of mice, and tested the animals in the NOL task 3 wk later. Of note, 0.4 μg/mL doxycycline treatment was previously shown to reduce transgenic miR-132 expression to a level that is approximately equivalent to WT (i.e., endogenous) miR-132 expression levels in the hippocampus (Hansen et al. 2013). Remarkably, Transgenic animals maintained on doxycycline did not exhibit time-of-day-dependent differences in NOL discrimination (Fig. 3E). We again found an interaction at the level of genotype that depended on the time point of testing (Fig. 3E, interaction F(2,18) = 3.594, P = 0.0176; repeated-measures ANOVA). When comparing within genotype, WT animals displayed a significantly higher DI at night compared to during the day (Fig. 3E, t(5) = 3.974, P = 0.0078; Bonferroni post hoc test). However, cKO and Transgenic animals did not show a significant difference (Fig. 3E, t(8) = 1.512, P = 0.523 for cKO mice and t(5) = 0.272, P = 1.000 for Transgenic mice; Bonferroni post hoc tests). Again, only WT animals demonstrated greater-than-chance discrimination for the novel object at night (Fig. 3E, t(5) = 4.504, P = 0.006; one sample t-test). Finally, no significant effect of time-of-day or genotype on total distance moved or total exploration time (assessed via repeated-measures ANOVA) was observed (Fig. 3F,G). Together, these results indicate that the daily rhythm in miR-132 facilitates both the formation of NOL memory and regulates its strength as a function of time-of-day.
Disruption of circadian-gating of contextual fear memory recall in miR-132 cKO and Transgenic mice
Next, we tested whether miR-132 influences time-of-day-dependent memory recall. This approach is in line with previous studies showing that the strength of memory recall is influenced by the time-of-day (Chaudhury and Colwell 2002; Eckel-Mahan et al. 2008). For this experiment, mice were initially handled once a day under LD conditions (for two consecutive days). Next, they were transferred to darkness (DD), where they were handled once per day for an additional 2 d. The 2 d of handling in DD, the training trial, and all retrieval trials were conducted under dim red light (∼5 lux), allowing the animals to use visual/contextual cues without influencing the circadian timing system, as described in the NOL section above.
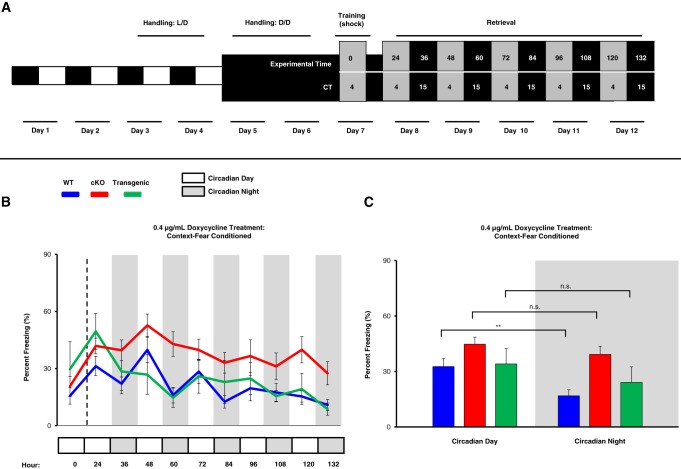
On the day of training, mice received a conditioning footshock during the early subjective day (CT4; Fig. 4A, Day 7). Memory retrieval (based on the percent time an animal was immobile/freezing) was then tested 24 h after conditioning and then probed every 12 h thereafter for 5 d (Fig. 4A; days 8 through 12). Mice were maintained on drinking water supplemented with 0.4 μg/mL doxycycline (from 3 wk prior to the beginning of the experiment until the completion of the experiment). A graphical representation of the daily recall profile of contextual fear memory for all three genotypes is shown in Figure 4B. We report that WT mice showed a daily oscillation in contextual fear memory recall; thus, over the first 3 d of testing, WT mice consistently exhibited more efficient recall during the subjective day. In contrast, daily oscillations in contextual fear memory recall were not consistently observed in miR-132 cKO and Transgenic animals. Specifically, a significant effect of genotype (Fig. 4C, F(2,24) = 3.609, P = 0.043; repeated-measures ANOVA) and a trend between genotype and freezing level that is dependent on time-of-day was observed (Fig. 4C, interaction F(2,24) = 3.156, P = 0.061; repeated-measures ANOVA). WT mice showed higher daytime freezing compared to nighttime freezing (Fig. 4C, t(9) = 5.476, P < 0.001; Bonferroni post hoc test), whereas miR-132 cKO and Transgenic mice showed no significant time-of-day-dependent differences in memory recall (Fig. 4C, t(10) = 2.122, P = 0.180 for cKO mice and t(5) = 2.559, P = 0.153 for Transgenic mice, Bonferroni post hoc tests). The abrogation of a circadian rhythm in memory recall in the knockout and transgenic animals supports the idea that the rhythmic expression of miR-132 gates time-of-day- dependent contextual fear memory recall.
Figure 4.
Time-of-day-dependent contextual fear memory recall. (A) Outline of the experimental design. After 2 d of dark-adaptation (days 5–6) mice were trained to associate a context box with a mild footshock. Beginning 24 h after training, mice were placed in the box every 12 h during circadian day and circadian night time points to profile their freezing behavior (i.e., memory retrieval). Of note, from day 5 onward, experimental manipulations were conducted under dim red light. Gray boxes represent circadian day time points (retrieval at CT4) and black boxes represent circadian night time points (retrieval at CT15). Mice were maintained on doxycycline drinking water (0.4 μg/mL) for 3 wk prior to fear conditioning (and throughout the fear conditioning experiment). (B) Graphical representation of the percentage of time spent freezing for WT, cKO, and Transgenic mice. 24, 48, 72, 96, and 120 h (post-training) represent day contextual fear retrieval time points, while 36, 60, 84, 108, and 132 h represent night retrieval time points. (C) Comparison of the freezing percentage (averaged over the first three respective day time points and the first three night time points) revealed a significant difference in WT animals, but not in cKO or Transgenic animals; analyzed using repeated-measures ANOVA with Bonferroni post hoc correction for within genotype comparison by TOD; n = 6–10 mice per group. Data are presented as mean ± SEM. (**) P < 0.01; (n.s.) not significant (P > 0.05).
To control for the effects of doxycycline treatment, we also examined time-of-day contextual fear memory recall in mice that were not treated with doxycycline; consistent with the doxycycline-treated paradigm, we found that only WT mice exhibited a significant time-of-day contextual fear memory recall (data not shown). Further, to test whether the observed reduction in freezing during subjective night in WT animals was a result of a circadian rhythm in fear memory, or a result of an innate increase in an animal's activity, we subjected a second cohort of animals to a context-only paradigm (data not shown). In this experiment, the animals were “trained” and tested in the same shock-box as the mice that underwent contextual fear conditioning; however, the context-only mice did not receive a footshock when they were exposed to the arena. As expected, all groups (WT, cKO, and Transgenic) displayed a mild, nonsignificant, overall increase in average freezing percentage during subjective day time points compared to subjective night time points, and no significant interaction was observed between genotypes that was dependent on the time-of-day (interaction F(2,13) = 2.32, P = 0.0814; repeated-measures ANOVA). Additionally, while context-only WT animals froze an average of 1.8 sec more during the day than the night, fear conditioned WT animals froze an average of 12.7 sec more during the day than the night. These data suggest that the increase in freezing observed in WT animals during the subjective day is due to a circadian rhythm in fear memory retrieval and is not simply the result of the animals’ increase in activity during the night time point domain.
Hippocampal time-of-day-dependent expression changes in miR-132 target genes
MicroRNAs function via the suppression of translation and/or the degradation of target mRNA. Given the marked effects on time-of-day-dependent memory recall in the miR-132 cKO and Transgenic lines, we examined whether miR-132 affects the time-of-day expression of putative target genes that affect synaptic plasticity and/or cognition. To this end, we profiled the expression of MeCP2 and Sirt1.
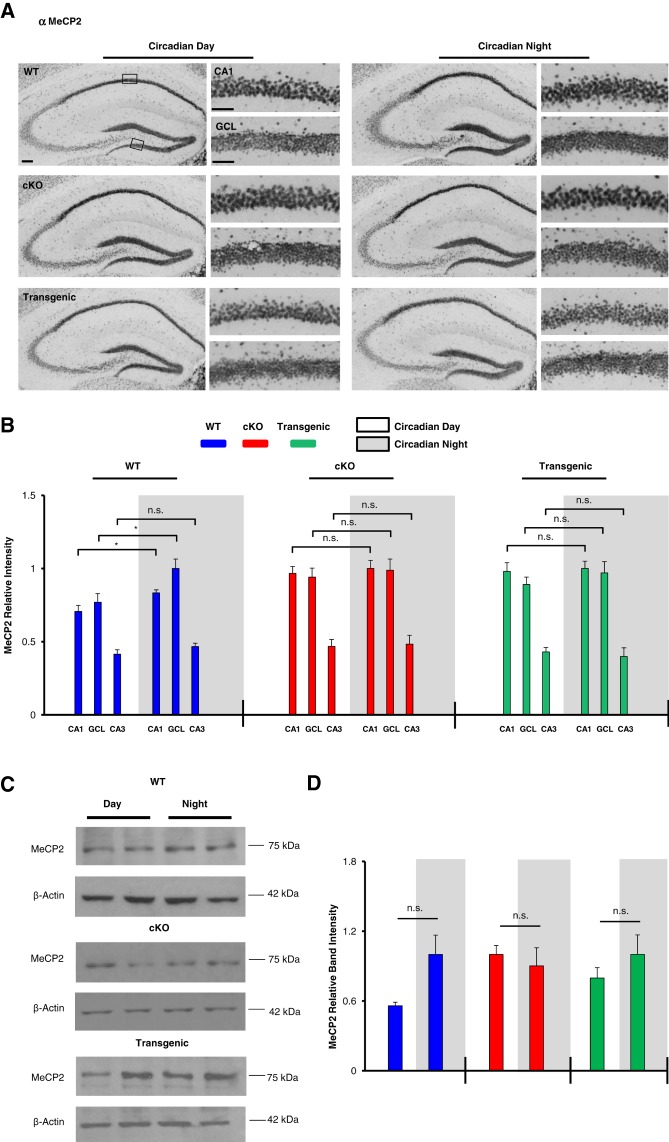
For the analysis of MeCP2, hippocampal tissue was profiled at CT4 and CT15 via immunohistochemical labeling and Western blotting. Quantitative immunohistochemical analysis revealed a significant time-of-day variation in MeCP2 expression within the CA1 (Fig. 5A,B, t(10) = 2.762, P = 0.017; Student's t-test) and the GCL (Fig. 5B, t(10) = 2.309, P = 0.044; Student's t-test) of WT mice, with higher levels observed during the circadian night. Interestingly, time-of-day-dependent differences in MeCP2 expression within the CA1, GCL, and CA3 were not detected in miR-132 cKO or Transgenic mice (Fig. 5B, P > 0.3 for all brain regions in both genotypes, Student's t-tests). Western analysis of hippocampal lysates from WT animals showed a mean increase in MeCP2 expression during the subjective night, although significance was not reached (Fig. 5D, t(4) = 1.229, P = 0.287; Student's t-test). Significant time-of-day differences in band intensity were not detected in miR-132 cKO or Transgenic mice (Fig. 5D, P > 0.30 for both genotypes, Student's t-tests). As a loading control, all Western blots were probed for β-actin. Importantly, previous studies have reported that β-actin expression is not under the control of the circadian clock (Eckel-Mahan et al. 2008; Gerstner et al. 2014; Jang et al. 2015).
Figure 5.
Clock-gated and miR-132-regulated MeCP2 expression in the hippocampus. (A) Low and high magnification representative images of MeCP2 immunohistochemical labeling in WT, cKO, and Transgenic animals sacrificed during the circadian day and the circadian night; scale bar = 50 μm for low-magnification images and 30 μm for high-magnification images. Boxed regions in the low magnification images approximate the locations from which the high magnification images were acquired. (B) Immunolabeling quantification for the CA1, GCL, and CA3. Within-genotype time-of-day differences were statistically analyzed using the Student's t-test, n = 5–6 mice per condition. For each genotype, MeCP2 expression is presented as relative intensity values, with the highest of the six expression levels set to a value of one. (C) Representative Western blot images of hippocampal MeCP2 expression in WT, cKO, and Transgenic animals sacrificed during the circadian day and the circadian night. Each time point was run as an experimental replicate (one mouse/lane, two lanes per condition). For each genotype, the time point (day or night) with the highest relative band intensity was set equal to a value of one. (D) Quantitative densitometric analysis of MeCP2 hippocampal protein levels relative to the control protein, β-actin; significance was assessed via the Student's t-test, n = 3–4 animals per group. The experiment was replicated at least one time per condition. Of note, immunolabeling and Western blotting for each genotype was performed on separate occasions, thus comparisons were only made within genotype (i.e., WT day versus WT night), and not across genotypes. Data are presented as mean ± SEM. (*)P < 0.05; (n.s.) not significant (P > 0.05).
Here, it is also worth noting that expression level comparisons for the noted target genes were only made within a genotype (e.g., WT circadian day versus WT circadian night), as immunohistochemical labeling and Western blotting experiments were run independently for each genotype. Hence, though one would expect for MeCP2 (and Sirt1) levels to be highest in the miR-132 cKO animals and lowest in the miR-132 Transgenic animals (given that they are miR-132 targets), we did not make across-genotype comparisons.
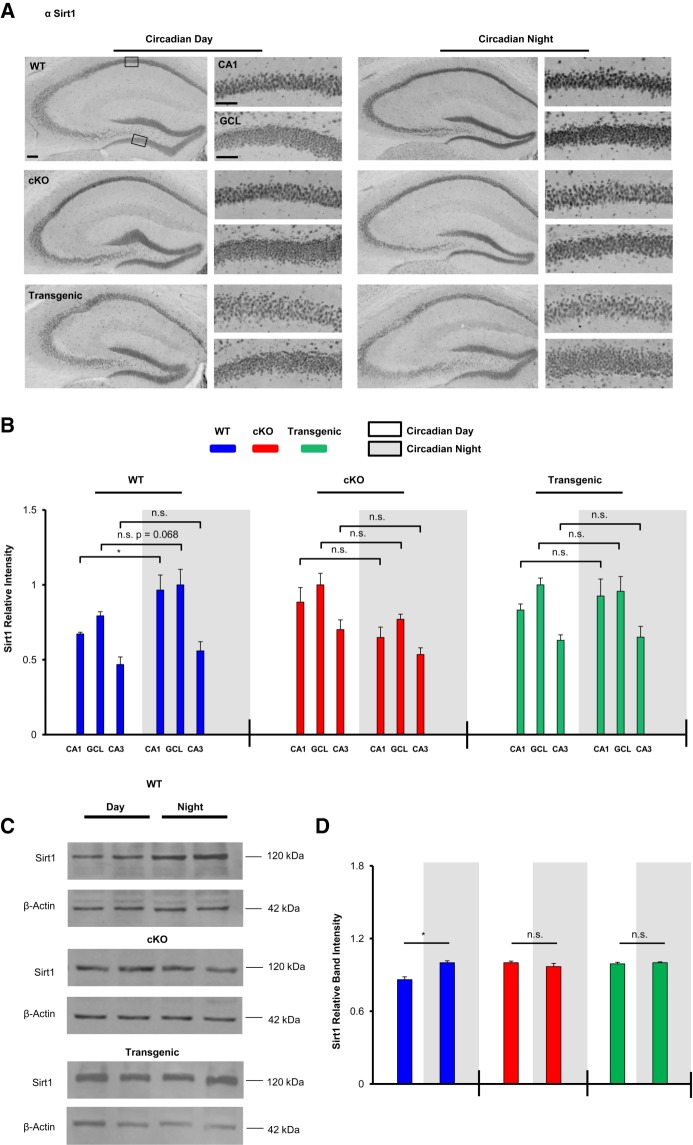
Next we examined Sirt1, an NAD-dependent deacetylase that has been reported to modulate synaptic plasticity and memory (Gao et al. 2010). In WT mice, immunohistochemical labeling detected higher Sirt1 expression within the CA1 cell layer during the circadian night compared to during the circadian day (Fig. 6A,B, t(6) = 2.973, P = 0.025; Student's t-test). In contrast, significant time-of-day expression within the CA1 cell layer (or CA3 and GCL) was not detected in cKO or Transgenic mice (Fig. 6B, P > 0.30 for all brain regions analyzed in both genotypes, Student's t-tests). As a complement to the IHC analysis, Western blotting of hippocampal lysates revealed significant time-of-day expression of Sirt1, with higher expression during the circadian night (Fig. 6D, t(5) = 3.907, P = 0.011; Student's t-test). In contrast, significant time-of-day expression in the miR-132 cKO or Transgenic animals was not observed (Fig. 6D; P > 0.30, Student's t-tests).
Figure 6.
Clock-gated and miR-132-regulated Sirt1 expression in the hippocampus. (A) Representative low and high magnification images of hippocampal Sirt1 immunostaining in WT, cKO, and Transgenic animals sacrificed during the circadian day and the circadian night. Scale bar = 50 μm for low-magnification images and 30 μm for high-magnification images. Boxed regions in the low magnification images approximate the locations from which the high magnification images were acquired. (B) Quantification of Sirt1 expression in the CA1, GCL, and CA3. Statistical comparisons of time-of-day expression were performed using the Student's t-test, n = 4–6 animals per condition. For each genotype, Sirt1 expression is presented as relative intensity values, with the highest of the six expression levels set to a value of one. (C) Representative Western blot images of hippocampal Sirt1 expression in WT, cKO, and Transgenic animals sacrificed during the circadian day or night. Each time point was run as an experimental replicate (one mouse/lane, two lanes per condition). For each genotype, the time point (day or night) with the highest relative band intensity was set equal to a value of one. (D) Quantitative ratiometric densitometric analysis of Sirt1/β-actin protein levels; significance was assessed using the Student's t-test, n = 3–4 animals per group. The experiment was replicated at least one time per condition. A significant time-of-day difference in band intensity was observed for the WT animals, but no significant differences were observed in cKO or Transgenic animals. Expression level comparisons for immunolabeling and Western blotting were only made within genotype. Data are presented as mean ± SEM. (*) P < 0.05. (n.s.) not significant (P > 0.05).
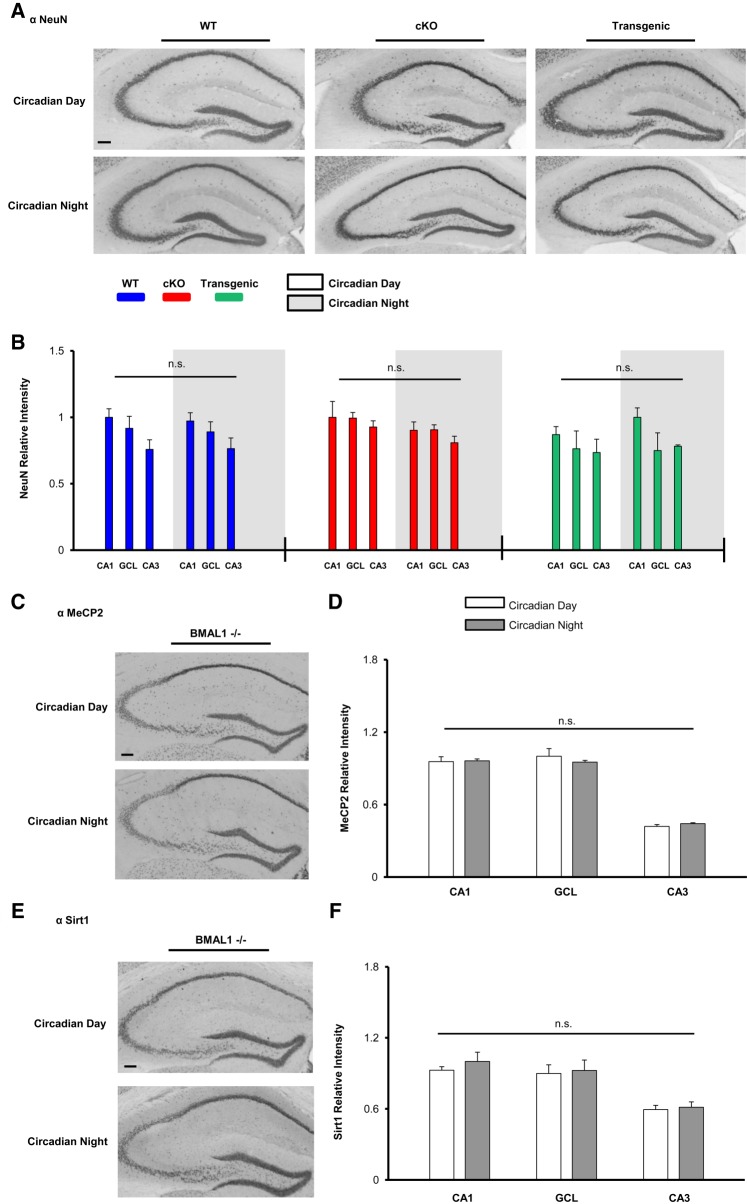
As a control, we profiled the time-of-day expression of the neuronal-enriched protein NeuN (Fig. 7A; Mullen et al. 1992). Given that NeuN is not a target of miR-132 and has not been reported to be regulated by the circadian clock (Campos et al. 2015), we did not anticipate time-of-day-gated NeuN expression. In WT mice, the expression of NeuN was not significantly different between CT4 and CT15 in the CA1, CA3, or GCL (Fig. 7B). Further, the expression of NeuN was not significantly affected in either the miR-132 cKO or the Transgenic lines (Fig. 7B). Together these data indicate that the effects of miR-132 are specific, and as such, add support to the idea that the rhythmic expression of miR-132 plays a key role in shaping the functionality of forebrain circuits that underlie cognition.
Figure 7.
Profiling hippocampal TOD expression: NeuN, and MeCP2/Sirt1 in BMAL1−/− mice. (A) Representative immunohistochemical labeling for NeuN in WT, cKO, and Transgenic animals sacrificed during the circadian day and the circadian night. Scale bar = 30 μm. (B) Immunolabeling quantification; no time-of-day differences in NeuN expression were observed in any genotype. Significance assayed using the Student's t-test; n = 4–6 mice per group. In each of the three noted mouse lines, the hippocampal region with the highest NeuN intensity was set equal to a value of one. (C) Representative immunolabeling for MeCP2 in BMAL1−/− mice; animals were profiled during the circadian day and the circadian night. (D) Quantification of MeCP2 expression did not detect a time-of-day difference in any hippocampal region. Data were analyzed by Student's t-test; n = 4 mice per time point. (E) Representative Sirt1 immunolabeling in BMAL1−/− animals. (F) Quantification of Sirt1 expression did not detect a time-of-day difference in any hippocampal region. Data were analyzed by Student's t-test; n = 4 mice per time point. Data are presented as mean ± SEM. (n.s.) not significant; P > 0.05.
Finally, we examined time-of-day expression of MecP2 and Sirt1 and BMAL1−/− animals (Fig. 7C,E). BMAL1−/− is a critical component of the circadian clock timing mechanism, and its germline deletion results in a complete loss of circadian timekeeping capacity (Bunger et al. 2000). Interestingly, significant time-of-day differences in MeCP2, and Sirt1 in the CA1, GCL, and CA3 were not observed in the BMAL−/− background (P > 0.30 for all three brain regions profiled for both proteins, Student's t-tests). These data provide support for the idea that the circadian timing system drives a rhythm of miR-132, which in turn imparts rhythmicity to target genes and cognition.
Discussion
Here, we provide evidence that miR-132 serves as a conduit through which the biological clock confers daily rhythms on cellular plasticity and cognition in forebrain circuits. With respect to the miR-132 rhythm in the hippocampus, our data are in line with several studies that have found time-of-day expression differences in miR-132 within the murine cortex (Davis et al. 2011) and the SCN (Cheng et al. 2007). Interestingly, a time-of-day difference in hippocampal miR-132 expression was not detected in BMAL1−/− mice, indicating that miR-132 is a clock-regulated gene. Consistent with this idea, Cheng et al. found that the disruption of the circadian clock transcription/translation feedback led to a suppression of rhythmic miR-132 expression in the SCN. However, it should be noted that our assessment of miR-132 levels in the BMAL1−/− mice was conducted at two time points (CT4 and CT15), approximating the time points in which miR-132 was low or high in WT animals. Though unlikely, it is possible that there may be a shift in the phase of the rhythm in miR-132 in the BMAL1−/− mouse lines that evaded our detection; as such, more extensive circadian profiling would be needed to confirm a total loss of the miR-132 hippocampal rhythm in BMAL1−/− animals.
It is worth noting that miR-132 does not appear to be under the direct control of the E-Box dependent core clock timing mechanism wherein heterodimers of CLOCK and BMAL1 promote the transcription of cryptochrome and period genes (Cheng et al. 2007). Consistent with this, the promoter/regulatory region of miR-132 does not contain an E-box element (Cheng et al. 2007). Rather, the rhythmic expression of miR-132 is likely to be driven by the CREB/CRE transcriptional pathway. In support of this hypothesis, multiple CRE-motifs have been identified within the regulatory region of the miR-132/212 gene cluster (Vo et al. 2005; Remenyi et al. 2010). Further, the disruption of CREB-mediated transcription has been shown to suppress inducible miR-132 expression in neurons (Vo et al. 2005; Remenyi et al. 2010).
Interestingly, within the SCN, circadian-gated, CRE-mediated, gene expression has been reported by several laboratories (Obrietan et al. 1999; O'Neill et al. 2008). Further, time-of-day changes in CREB activity have been reported in multiple forebrain regions (Cirelli and Tononi 2000; Graves et al. 2003; Guzman-Marin et al. 2006), thus raising the prospect that rhythmic miR-132 expression in the forebrain is mediated via a circadian clock-driven, CREB-dependent, transcriptional pathway.
In the NOL task, we found that cKO and Transgenic animals exhibited time-of-day-dependent learning deficits, suggesting that peak cognitive performance requires rhythmic miR-132 expression. It should be noted that the NOL memory impairments observed in miR-132 cKO animals are similar to recent studies showing that both forebrain miR-132 cKO animals and miR-132 germline knockout animals exhibit marked deficits in Novel Object Recognition (Hernandez-Rapp et al. 2015; Hansen et al. 2016). Further, with regard to the time domain-specific efficacy observed in the NOL task in WT animals, studies from several laboratories have shown that rodents display better discrimination at night and very low to negligible discrimination during the day (Takahashi et al. 2013; Snider et al. 2016). It was somewhat unexpected that both constitutive expression and targeted deletion of miR-132 disrupted discriminatory capacity; hence neither line scored better than chance during either the circadian day or circadian night. The reasons for these similar effects resulting from two profoundly different transgenic manipulations are not clear. It is worth noting, however, that Transgenic animals displayed increased orientation toward the NOL (during both the subjective day and night; Fig. 3B), but these values did not reach statistical significance; a larger sample size may have been needed to disentangle a potential phenotypic effect.
Turning to the contextual fear conditioning paradigm, we found that both the loss of miR-132 expression in the knockout animals and constitutive expression in the Transgenic animals led to a disruption of time-of-day gated memory recall. This finding contrasts with the recall rhythm in WT mice, which persisted for 4 d after training. Of note, this rhythm is consistent with work from several papers which found that fear memory recall varies as a function of the time-of-day, with better retrieval occurring during the day (Chaudhury and Colwell 2002; Eckel-Mahan et al. 2008). Here, however, it should be noted that we cannot definitively conclude that the superior day-time retrieval in WT mice (or lack thereof in cKO and Transgenic mice) is strictly a time-of-day effect, as we have not controlled for the potential influence of time-stamping in our experiments. Along these lines, several groups (using a variety of memory retention paradigms) have shown that regardless of the time-of-day in which training occurs, memory retention is optimal 24 h later (or multiples of 24 h after training) (Holloway and Wansley 1973a,b; Wansley and Holloway 1975; Holloway and Sturgis 1976; Holloway 1978; Chaudhury and Colwell 2002; Ralph et al. 2002). Hence, since we trained our mice at only one timepoint (CT4), it is possible that our behavioral findings may also be attributed to a miR-132-dependent time-stamping phenomenon. Indeed, additional experiments aiming to tease apart the role of miR-132 and its function in time-of-day memory retrieval and/or time-stamping would be of merit. Finally, one interesting observation from our contextual fear conditioning studies was that miR-132 cKO mice exhibited a higher level of freezing than WT mice, which could be interpreted as more efficient memory retrieval. However, our NOL data reported here, as well as prior work from our laboratory and the Hébert laboratory, found that the disruption of miR-132 results in diminished cognitive capacity (Hansen et al. 2010, 2013; Hernandez-Rapp et al. 2015). Potential explanations for this discord may be related to the differences inherent to the contextual fear conditioning assay compared to spatial learning assays used to detect cognitive deficits (i.e., novel object recognition, NOL and Barnes maze). Along these lines, contextual fear training would be expected to require the involvement of fear circuits to a much greater extent than the novel object recognition, NOL and Barnes maze assays (Fendt and Fanselow, 1999; Moser et al. 2008). As such, the relatively high level of cKO freezing detected in the contextual fear conditioning assay may be related to an elevated anxiety response: a topic that we are currently pursuing.
Prior hippocampal gene profiling work from our laboratory found that miR-132 affects the expression of hundreds of functionally diverse gene transcripts (Hansen et al. 2016). Hence, it is reasonable to expect that the behavioral effects that we report here for cKO and Transgenic mice may result from a complex interplay of genes (i.e., direct targets) and gene networks (i.e., indirect targets) that are affected by miR-132. Given this complexity, attempts to identify a target gene(s) through which miR-132 disrupts time-of-day learning would have been technically challenging. Thus, rather than attempting to identify a behavioral link between miR-132 and a target, we chose to focus on the cellular functional effects of miR-132 rhythms by analyzing two genes that are (1) well-characterized miR-132 targets, (2) reported to exhibit diurnal oscillations, and that (3) affect cognition. To this end, we examined MeCP2 and Sirt1.
MeCP2 is a chromatin-associated protein targeted by miR-132 that functions as a potent transcriptional regulator (Klein et al. 2007; Chahrour et al. 2008; Alvarez-Saavedra et al. 2011; Hansen et al. 2010). Loss-of-function mutations in MeCP2 have been shown to result in Rett syndrome (Amir et al. 1999), a developmental disorder that results in marked deficits in learning and memory tasks (Moretti et al. 2006; Pelka et al. 2006; for reviews, see McGowan 2006; Na et al. 2012). Initially, we detected a significant time-of-day-dependent MeCP2 expression profile within the hippocampus of WT mice. Consistent with this finding, a number of profiling studies performed on both brain and peripheral tissues have reported an oscillatory profile at the level of mRNA and protein (Hoogerwerf et al. 2007; Mouse 1.OST Lung Affymetrix: Circadian Expression Profiles Data Base (CircaDB) Pizarro et al. 2013; Martínez de Paz et al. 2015). Given the profound effects that MeCP2 has on synaptic plasticity and dendritic complexity and cognition (Na et al. 2013), it is reasonable to postulate that MeCP2 could contribute to the circadian clock-mediated, miR-132-dependent, modulation of learning and memory.
Interest in the NAD-deacetylase Sirt1 was piqued by work showing that Sirt1 knockout mice display impairments in both short- and long-term associative memory tasks and show deficits in synaptic plasticity and dendritic branching/arborization (Michán et al. 2010; Codocedo et al. 2012). Further, regarding its post-transcriptional regulation, Strum et al. (2009) and Zhang et al. (2014) identified Sirt1 as a direct target of miR-132, and both the MicroRNA.org and miRanda algorithms detect robust hybridization capacity between miR-132 and Sirt1 (Betel et al. 2008; Peterson et al. 2014).
Our analysis indicates that rhythmic Sirt1 hippocampal expression is driven by miR-132. The temporal profile of Sirt1 expression in WT mice reported here is consistent with the work of Rawashdeh et al. (2014) who reported peak protein levels of hippocampal Sirt1 during the night domain and Asher et al. (2008) who reported peak levels of Sirt1 protein from the mouse liver at ZT16. Interestingly, CREB protein levels have been shown to be significantly reduced in mutant mice lacking brain-specific Sirt1 catalytic activity (Gao et al. 2010). Given the tight regulation of miR-132 by CREB, one could propose a complex interplay wherein Sirt1 regulates plasticity and cognition across the circadian day, in part, through a miR-132-CREB-mediated mechanism.
With respect to the peak in the time-of-day protein expression profiles of MeCP2 and Sirt1 reported here, one may have expected for these gene products to be lower at night (given that they are direct targets of miR-132—which was highest at night). However, at the level of protein expression/regulation, the kinetics of a wide array of processes impacts the functional effects of microRNA. Along these lines, in addition to the kinetic rates of miRNA-mediated mRNA decay and translational repression (Morozova et al. 2012), simple mRNA stability and translation rates, as well as protein half-lives, impact the temporally delimited effects of miRNAs. Hence, within a circadian timeframe, it is conceivable for the peak expression times of MeCP2 and Sirt1 to temporally overlap with the peak in miR-132 expression.
Together, these data suggest that the rhythm of miR-132 expression regulates learning and memory as a function of the time-of-day. In addition, the results presented here raise interesting questions regarding the potential role of miR-132 in an array of time-of-day-delineated processes, including synaptic scaling and homeostatic plasticity; to these ends, additional studies are highly merited.
Materials and Methods
miR-132 transgenic and knockout mouse lines
Generation of the CaMKII-Cre::miR-132/212f/f conditional forebrain neuron knockout (referred to as “cKO”) mouse line was previously described by Hansen et al. (2016). The miR-132/212f/f animals were provided to us by Dr. Simon Arthur, and the CaMKII-Cre line (Mayford et al. 1996) was acquired from Jackson Laboratory (Bar Harbor, ME, USA). To generate the CaMKII-tTA::miR-132:CaMKII-Cre::miR-132/212f/f mouse line (referred to as “Transgenic”), homozygous CaMKII-Cre::miR-132/212f/f mice were crossed to a tetracycline-regulated bidirectional miR-132/cyan fluorescent protein (CFP) transgenic mouse line (driven by CaMKII::tTA); the details of this mouse line are provided in Hansen et al. (2010). Female and male experimental CaMKII-tTA::miR-132:CaMKII-Cre::miR-132/212f/f animals were homozygous for the miR-132/212f/f locus and positive for Cre, tTA, and miR-132. Littermates negative for either the driver(s) (CaMKII-tTA and/or CaMKII-Cre) and/or the responder(s) (miR-132 and/or miR-132/212f/f) served as control, WT-like mice (referred to as “WT”). All genotyping was performed as described previously (Hansen et al. 2010, 2016). Additionally, BMAL1−/− mice from the C57/Bl6 background were purchased from Jackson Laboratory (RRID:SCR_004633B6.129; Arntltm1Bra/J). BMAL−/− line genotyping was performed as described by Bunger et al. (2000). Animal care protocols and methods were approved by the Ohio State University's Institutional Animal Care and Use Committee.
Experimental animals were bred, housed, and maintained under standard 12 h/12 h light–dark (LD) conditions. Experiments performed under circadian time (CT) conditions are noted with the terminology “Circadian Day” or “Circadian Night.” For these experiments, mice were dark-adapted (kept in constant darkness beginning at the normal lights-off time) for the noted periods. Any animal manipulations were performed under dim red light to avoid perturbation of the circadian clock. Under these conditions, CT0 denotes the beginning of the circadian day (when lights would have been turned on) and CT12 (when lights would have been turned off).
Fluorescence in situ hybridization
Tissue isolation and FISH against miR-132 was carried out as described previously (Hansen et al. 2013). Initially, brains were removed from mice at least 8 wk of age, tissue was cut into 500 μm coronal sections using a vibratome, and sections containing the dorsal hippocampus were fixed in 4% paraformaldehyde (PFA) for 4 h at 4°C. After fixing, tissue was cryoprotected overnight in 30% sucrose in PBS, and then thin cut (40 μm thick) using a freezing microtome. Next, free-floating sections were incubated with the nuclear/DNA stain DRAQ5 (1:10,000 dilution; BioStatus Limited, Cat# DR50050 RRID:AB_2314341) and then probed for miR-132 expression using fluorescein-conjugated locked nucleic acid (LNA) probes to mouse miR-132, or to a “scrambled” negative control probe that does not correspond to any known murine miRNA (Exiqon Corp). An anti-fluorescein Alexa 488 signal detection kit (Millipore, Cat# MAB045X RRID:AB_11214450) was used to amplify the fluorescein signal. Of note, 40× images were taken using a Zeiss 510 confocal microscope with LSM Software (LSM Image Examiner, RRID:SCR_014344) and MetaMorph analysis software (MetaMorph Microscopy Automation and Image Analysis Software, RRID:SCR_002368) was used to quantify the signals.
RT-PCR quantification of miR-132 expression levels
Total hippocampal RNA was isolated during the noted day and night time points, using methods described previously (Hansen et al. 2013). In brief, after RNA isolation, hippocampal cDNA was prepared using the miScript II Reverse Transcription kit (Qiagen). Amplification of cDNA was carried out using QuantiFast SYBR Green (Qiagen), and the miScript Primer System (Qiagen) was used to quantify miR-132 levels. The following miR-132 primer sequence was utilized: 5′ UAACAGUCUACAGCCAUGGUCG (Qiagen, Cat# MS00001561). QuantiFast SYBR Green thermocycling conditions were previously described by Alemayehu et al. (2013). Data from both time points were normalized to RNU6B_2 cDNA levels, and Double Delta CT was used for analysis.
Tissue processing and cresyl violet staining
A group of WT, cKO, and Transgenic animals was sacrificed, and brains were cut into 600 μm sections using a vibratome, fixed in 4% PFA (6 h at 4°C), and cryoprotected overnight in 30% sucrose in 1× PBS. After thin sectioning to 40 μm, sections were mounted onto gelatin-coated slides and dehydrated in alcohol (100%), incubated in a 0.1% cresyl violet solution in dH2O (5 min), destained with 0.1% glacial acetic acid in 95% ethanol, cleared with xylenes, and coverslipped with DPX (Electron Microscopy Sciences).
Immunohistochemical labeling
Animals were sacrificed during day and night time points (CT4 and CT15), and tissue was fixed and cut using the methods noted above. Forty micrometer sections were washed in 1% Triton X-100 in PBST (3 times, 5 min each) and then incubated in 0.3% hydrogen peroxide in 1× PBST (20 min). Next, sections were blocked in 10% normal goat serum (NGS) in 1× PBST for 1 h and incubated in primary antibody overnight at 4°C in rabbit polyclonal anti-Sirt1/Sir2α (1:1000 dilution; Millipore, Cat# 09-845 RRID:AB_1587512), rabbit monoclonal anti-MeCP2 (1:3000 dilution; Cell Signaling Technology, Cat# 3456S RRID:AB_2143849), or rabbit polyclonal anti-NeuN (1: 2000 dilution; Millipore, Cat# MAB377 RRID:AB_2298772). The next day, sections were washed in PBST and incubated in biotin-conjugated goat anti-rabbit IgG secondary antibody (1:500 dilution; Vector Laboratories, Cat# BA-1000 RRID:AB_2313606) for 2 h at room-temperature. Next, tissue was processed using the ABC labeling method (Vector Laboratories Cat# PK-6100 RRID:AB_2336819) and the signal was visualized using nickel intensified diaminobenzidine labeling (Vector Laboratories Cat# SK-4100 RRID: AB_2336382). Finally, sections were mounted on gelatin-coated slides, washed in dH2O (2×, 5 min each), cleared using xylene, and coverslipped with Permount Mounting Medium (Fisher Chemical). Bright field images were captured with a 16-bit digital camera (Micromax YHS 1300; Princeton Instruments) on a Leica DMIR microscope with Metamorph software (MetaMorph Microscopy Automation and Image Analysis Software, RRID:SCR_002368). For quantification of MeCP2, Sirt1 and NeuN labeling, images of the CA1, CA3, and GCL were traced digitally from 2 to 4 hippocampi per animal. Intensity levels for each section were background subtracted and analyzed using ImageJ software (ImageJ, RRID:SCR_003070). Values for each animal were averaged and displayed as the mean ± SEM for each noted region of the hippocampus.
Western blotting
The Western blotting protocol has previously been described (Hansen et al. 2010). Animals were sacrificed at CT4 and CT15 as noted above. Hippocampal tissue was collected and lysed in 125 μL of radioimmunoprecipitation assay buffer. Next, protein (10 μg/lane) was loaded into a 12% SDS–Page gel and transferred onto polyvinylidene difluoride membranes (Immobilon-P; EMD Millipore). Upon transfer, membranes were blocked in 10% milk (in PBST or TBST) and incubated overnight at 4°C with the rabbit MeCP2 antibody (1:3000; Cell Signaling Technology, Cat# 2507 RRID:AB_561221) and rabbit Sirt1 (1:1000; Millipore, Cat# 09-845 RRID:AB_1587512). All antibodies were diluted in 5% NGS in PBST or 5% BSA in TBST. On the following day, membranes were incubated in 10% milk (in PBST or TBST) with an anti-rabbit IgG (goat) horseradish peroxidase-conjugated antibody (1:2000; PerkinElmer). Of note, since comparisons were only made within genotype (i.e., density of WT bands at CT4 versus CT15), each membrane contained protein for one specific genotype (WT, cKO, or Transgenic) for profiling of both timepoints (CT4 and CT15). Additionally, all membranes were also probed for mouse β-actin (1:200,000; PhosphoSolutions Cat# 125-ACT RRID:AB_2492035). The following day, membranes were incubated in 10% milk (in PBST or TBST) with an anti-mouse IgG (goat) horseradish peroxidase-conjugated antibody (1:2000; PerkinElmer Cat# NEL750001EA RRID:AB_2617185). A luminescent signal was generated using the Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate (PerkinElmer), and captured using BioBlue Lite Western Blot film (Alkali Scientific). PBST or TBST washes (three times, 5 min each) were carried out between each antibody incubation step. Photoshop CS6 (Adobe Photoshop, RRID:SCR_014199) was used for densitometric band analysis. To this end, the band intensity of MeCP2 and/or Sirt1 bands were digitally traced, background subtracted, and divided by the β-actin signal from the same lane. The mean signal for each genotype/time point was averaged from 3 to 4 animals, and the experiment was replicated at least one time per condition.
Behavioral assays: vision assessment
Animals used in each of the following behavioral assays were screened for proper vision by reaching for the surface before their vibrissae made contact with the table. Based on this assay, no vision issues were noted with any of the mouse lines.
Circadian activity analysis
Circadian activity rhythm analysis was carried out as described previously by Snider et al. (2016). In brief, WT, cKO, and Transgenic animals were singly housed in cages with running wheels, and locomotor activity was collected via a magnet-actuated sensor. Animals were entrained to a 12 h/12 h LD light cycle for 10 d. Mice were then transitioned to total dark conditions (DD) for 17 d to assess free-running rhythms. Finally, to profile photic resetting abilities, mice were exposed to white light (40 lux, 15 min) at CT15 and then were allowed to free-run for six more days. VitalView software (VitalView Software, RRID:SCR_014497) was used to collect wheel-running data. The overall activity, circadian period, and phase delay were calculated based on readouts from ActiView software (Respironics Corp. Bend OR).
Novel object location
The NOL task was adapted from Takahashi et al. (2013) and has been recently described by our laboratory (Snider et al. 2016). In short, animals on a 12 h/12 h LD cycle were dark-adapted. From this point on, all habituation, exploration, and testing trials were conducted under dim red light (∼5 lux). The following day, animals were habituated (two consecutive days) to an arena with shapes serving as contextual cues. Object shapes are described as in Snider et al. (2016) and were randomly distributed among cohorts. Exploration of the two objects and testing of the NOL occurred at CT4 or CT15 (24 h after the second day of habituation). During exploration, mice were allowed to explore two objects in the arena for a total of 5 min. Animals were then returned to their cages for 30 min (70% ethanol was used to clean the chamber in order to extinguish the odor from the previous mouse). During the test trial (which began 30 min after the exploration trial), mice explored the objects in the familiar and NOL. These objects were the same objects as those used in the initial exploration trial; however, one object (the familiar location) was in the same location as the exploration trial, while the other object (the novel location) was moved to a different location in the arena. Animals were then returned to their original 12 h/12 h LD cycle for 8 d. Next, animals were again dark-adapted and underwent 2 d of habituation. The following day, exploration and testing occurred as described above, except at the opposite time of day (i.e., an animal was tested for the first time at CT4 and tested a second time at CT15). A different set of arena contextual cues and objects was used to avoid any confounding effects of the mice having previously been exposed to a particular object. For analysis, the amount of time exploring each object was manually scored. “Exploration” was defined as the animals’ nose being within 2 cm of the object, and the movement of the animal had to represent a distinct deflection from the original path. If an animal was touching or standing on the object, it must also be actively sniffing or exploring the object to constitute “exploration.” Of note, animals that spent less than five total seconds of exploration during the test phase of the experiment were excluded from formal analysis (a total of two WT, one cKO, and two Transgenic animals in the no-doxycycline experiment and a total of four WT, one cKO, and three Transgenic animals in the doxycycline experiment). Total distance moved and total seconds immobile were scored using Noldus Ethovision XT version 11.5 (EthoVision XT, RRID:SCR_000441).
For the NOL assay, half of the animals were tested first during the circadian day and half were tested first during the circadian night, with a 10 d interval separating the two testing trials. Here it is important to note that prior work from our laboratory and from other laboratories has shown that NOL can be repeated with the same subjects with no confounding effects of prior exposure to the arena and objects during the second test (Besheer et al. 1999; Graciarena et al. 2010; Snider et al. 2016). Further, a regression analysis of our NOL data confirmed this. To this this end, we used regression analysis of the DI, which validated that no significant confounding effect (in any genotype) existed when animals were tested at both timepoints (R2 = 0.032, P = 0.450 for WT animals, R2 = 0.102, P = 0.227 for cKO animals, and R2 = 0.109, P = 0.182 for Transgenic animals; data not shown). Thus, NOL performance was not dependent on the sequential nature or the temporal order of testing.
Contextual fear conditioning
Contextual fear conditioning experiments were adapted from Chaudhury and Colwell (2002) and Eckel-Mahan et al. (2008). Animals were individually housed in ventilated cages 1 wk before the start of the experiment. Four days prior to training, animals were handled each day (1 min per day) during different, randomly chosen times of the 24 h period, as described in detail by Chaudhury and Colwell (2002). Two days prior to training, animals were dark-adapted, and from this point on, animals were only exposed to dim red light (∼5 lux) during the training and twice daily recall paradigms. The Passive/Active Avoidance Box Pacs-30 (Columbus Instruments) was used to administer the mild footshock. Contextual patterns and shapes were placed on three of the four walls of the chamber so that the animals could associate the context with the shock. For these experiments, we chose to train (shock) all of the animals at CT4. The decision to train during the early subjective day is based on the work of Eckel-Mahan et al. (2008) who showed that contextual fear memory is impaired when animals are trained at night. Thus, on the day of training (at CT4) mice were placed in the chamber and were given 3 min to acclimate to the surroundings. Two consecutive 0.1 mA footshocks (each lasting 1 sec in duration) were given to the animals, and freezing behavior was monitored for an additional 2 min after the shock. Of note, we chose to administer a 0.1 mA shock, as pilot data revealed that the diurnal rhythm of contextual fear recall was abolished with higher-intensity shocks (data not shown). This 0.1 mA intensity was similar to the intensity reported in similar contextual fear conditioning paradigms (Chaudhury and Colwell 2002; Alexander et al. 2009). After the 5-min trial was completed, mice were placed back into their cages. Seventy percent ethanol was used to clean the chamber in order to extinguish the odor from the previous mouse. The first retrieval test was carried out 24 h after training, and all animals were repeatedly tested every 12 h for five continuous days. For the context-only control experiment, the same protocol, as described above, was used, except the animals did not receive a footshock. Freezing behavior and distance moved were scored by Noldus Ethovision software (EthoVision XT, RRID:SCR_000441). The same freezing parameters were used for both the contextual fear experimental paradigm and for the context-only control paradigm. The percent freezing for each trial was determined by dividing the total cumulative freezing time (generated by Noldus software) by the total amount of time the mouse was allowed to explore the arena.
Regulation of transgenic miR-132 expression via doxycycline treatment
To reduce transgenic miR132 expression, doxycycline (0.40 μg/mL) was administered to the drinking water of a subset of the mice assayed in the NOL paradigm (Fig. 3E–G) and to all mice profiled using the contextual fear training paradigm (Fig. 4). Doxycyline dosing and the effects on transgenic miR-132 expression are described in our prior study (Hansen et al. 2013). Doxycycline water was changed every 3 d. Of note, doxycycline supplemented water was only administered for behavioral assays; animals sacrificed for immunolabeling/immunoblotting, and/or RT-PCR experiments did not receive doxycycline.
Experimental design and statistical analysis
All statistics were done using IBM SPSS Statistics 22 (SPSS, RRID:SCR_002865) and GraphPad Prism 3.0 (Graphpad Prism, RRID: SCR_002798), and all data are represented as the mean ± the SEM. For all data sets, statistical significance was set at *P < 0.05, as denoted in the figures and figure legends. Comparisons between two groups were performed using Student's two-tailed t-tests (unless otherwise stated), while comparisons (not dependent on the time-of-day) between the three noted genotypes were made using a one-way ANOVA. Further, Two-way ANOVA analysis was used to analyze differences between genotype and the time-of-day, as noted in the text. Selective Bonferroni post hoc tests were conducted in PRISM 3.0 in order to examine the time-of-day difference of miR-132 in each noted brain region (see Fig. 1E). Repeated-measures ANOVA analysis was used for experiments in which the same mouse was run through a behavioral assay for more than one trial (i.e., the NOL and Contextual Fear Conditioning paradigms). For these experiments, CT4 and CT15 were used as the repeated measures and genotype was noted as the between-subjects factor. Bonferroni post hoc corrections were made for within-genotype time-of-day comparisons. Grubb's test was conducted on data sets within each group, and animals that were found to be statistically significant outliers (P < 0.05) were removed from analysis.
miR-132 expression levels probed during the circadian day and the circadian night in WT, cKO, and Transgenic animals were not normally distributed (P < 0.02 for both timepoints; Shapiro–Wilks test of normality). Thus, fold changes obtained from qPCR data were log transformed before analysis (as in Snider et al. 2016).
For the NOL paradigm, the DI was calculated as described previously by Snider et al. (2016). Time-of-day differences were analyzed by repeated-measures ANOVA with Bonferroni post hoc analysis. Additionally, regression analysis was conducted in PRISM to ensure that prior exposure did not confound the results of the experiment (i.e., testing each mouse in the NOL paradigm once at CT4 or CT15, and a second time—10 d later—at the opposite timepoint). Further, to determine whether animals displayed above-chance discrimination, a one sample t-test was conducted for each genotype, at each separate time point (CT4 and CT15).
For all immunohistochemical labeling, differences between time points in each region of the brain were statistically analyzed using the Student's t-tests. Of note, expression level comparisons were only made within genotype (i.e., WT CT4 versus WT CT15) and not across genotypes (i.e., WT CT4 versus Transgenic CT4). Similarly, for Western blotting, relative band intensity was only measured within genotype; thus, Student's t-tests were used for analysis.
Acknowledgments
This study was funded by National Institutes of Health (grant nos. MH103361, NS066345, NS091302, F31-MH096460) and National Science Foundation (grant no. 1354612).
Competing interest statement: The authors declare no competing financial interests.
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.047191.117.
References
- Alemayehu S, Feghali KC, Cowden J, Komisar J, Ockenhouse CF, Kamau E. 2013. Comparative evaluation of published real-time PCR assays for the detection of malaria following MIQE guidelines. Malar J 12: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JC, McDermott CM, Tunur T, Rands V, Stelly C, Karhson D, Bowlby MR, An WF, Sweatt JD, Schrader LA. 2009. The role of calsenilin/DREAM/KChIP3 in contextual fear conditioning. Learn Mem 16: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. 2011. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet 20: 731–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 134: 317–328. [DOI] [PubMed] [Google Scholar]
- Aten S, Hansen KF, Hoyt KR, Obrietan K. 2016. The miR-132/212 locus: a complex regulator of neuronal plasticity, gene expression and cognition. RNA Dis 3: e1375. [PMC free article] [PubMed] [Google Scholar]
- Barker GR, Warburton EC. 2011. When is the hippocampus involved in recognition memory? J Neurosci 31: 10721–10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besheer J, Jensen HC, Bevins RA. 1999. Dopamine antagonism in a novel-object recognition and a novel-object place conditioning preparation with rats. Behav Brain Res 103: 35–44. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res 36(Database issue): D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield C. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos LM, Osório EC, Santos GL, Nogueira MI, Cruz-Rizzolo RJ, Pinato L. 2015. Temporal changes in calcium-binding proteins in the medial geniculate nucleus of the monkey Sapajus apella. J Chem Neuroanat 68: 45–54. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320: 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao OY, Huston JP, Nikolaus S, de Souza Silva MA. 2016. Concurrent assessment of memory for object and place: Evidence for different preferential importance of perirhinal cortex and hippocampus and for promnestic effect of a neurokinin-3 R agonist. Neurobiol Learn Mem 130: 149–158. [DOI] [PubMed] [Google Scholar]
- Chaudhury D, Colwell CS. 2002. Circadian modulation of learning and memory in fear-conditioned mice. Behav Brain Res 133: 95–108. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S. 2007. microRNA modulation of circadian-clock period and entrainment. Neuron 54: 813–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. 2000. Differential expression of plasticity-related genes in waking and sleep and their regulation by the noradrenergic system. J Neurosci 20: 9187–9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codocedo JF, Allard C, Godoy JA, Varela-Nallar L, Inestrosa NC. 2012. SIRT1 regulates dendritic development in hippocampal neurons. PLoS One 7: e47073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JA, Navaratnam V, Redfern PH. 1973. A 24-hour rhythm in passive-avoidance behaviour in rats. Psychopharmacologia 32: 211–214. [DOI] [PubMed] [Google Scholar]
- Davis CJ, Clinton JM, Taishi P, Bohnet SG, Honn KA, Krueger JM. 2011. MicroRNA 132 alters sleep and varies with time in brain. J Appl Physiol (1985) 111: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GC-K, Scheiner ZS, Storm DR. 2008. Circadian oscillation of hippocampal MAPK activity and cAmp: implications for memory persistence. Nat Neurosci 11: 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. 2010. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt M, Fanselow MS. 1999. The neuroanatomical and neurochemical basis of conditioned fear. Neurosci Biobehav Rev 23: 743–760. [DOI] [PubMed] [Google Scholar]
- Fernandez RI, Lyons LC, Levenson J, Khabour O, Eskin A. 2003. Circadian modulation of long-term sensitization in Aplysia. Proc Natl Acad Sci 100: 14415–14420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiro MG, Rea MS. 2010. The effects of red and blue lights on circadian variations in cortisol, alpha amylase, and melatonin. Int J Endocrinol 2010: 829351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang W-Y, Mao Y-W, Gräff J, Guan J-S, Pan L, Mak G, Kim D, Su SC, Tsai L-H. 2010. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466: 1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner JR, Yin JC. 2010. Circadian rhythms and memory formation. Nat Rev Neurosci 11: 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner JR, Smith GG, Lenz O, Perron IJ, Buono RJ, Ferraro TN. 2014. BMAL1 controls the diurnal rhythm and set point for electrical seizure threshold in mice. Front Syst Neurosci 8: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciarena M, Depino AM, Pitossi FJ. 2010. Prenatal inflammation impairs adult neurogenesis and memory related behavior through persistent hippocampal TGFβ1 downregulation. Brain Behav Immun 24: 1301–1309. [DOI] [PubMed] [Google Scholar]
- Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. 2003. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol 90: 1152–1159. [DOI] [PubMed] [Google Scholar]
- Guzman-Marin R, Ying Z, Suntsova N, Methippara M, Bashir T, Szymusiak R, Gomez-Pinilla F, McGinty D. 2006. Suppression of hippocampal plasticity-related gene expression by sleep deprivation in rats. J Physiol 575(Pt 3): 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. 2010. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One 5: e15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Karelina K, Sakamoto K, Wayman GA, Impey S, Obrietan K. 2013. miRNA-132: a dynamic regulator of cognitive capacity. Brain Struct Funct 218: 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Aten S, Snider KH, Loeser J, Hesse AM, Page CE, Pelz C, Arthur JS, Impey S, Obrietan K. 2016. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn Mem 23: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Lucas RJ, Mrosovsky N, Thompson S, Douglas RH, Hankins MW, Lem J, Biel M, Hofmann F, Foster RG. 2003. Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature 424: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Rapp J, Smith PY, Filali M, Goupil C, Planel E, Magill ST, Goodman RH, Hébert SS. 2015. Memory formation and retention are affected in adult miR-132/212 knockout mice. Behav Brain Res 287: 15–26. [DOI] [PubMed] [Google Scholar]
- Holloway FA. 1978. State-dependent retrieval based on time of day. Drug discrimination and state-dependent learning, pp. 319–343, Academic, New York. [Google Scholar]
- Holloway FA, Sturgis RD. 1976. Periodic decrements in retrieval of the memory of nonreinforcement as reflected in resistance to extinction. J Exp Psychol Anim Behav Process 2: 335–341. [Google Scholar]
- Holloway FA, Wansley R. 1973a. Multiphasic retention deficits at periodic intervals after passive-avoidance learning. Science 180: 208–210. [DOI] [PubMed] [Google Scholar]
- Holloway FA, Wansley R. 1973b. Multiple retention deficits at periodic intervals after active and passive avoidance learning. Behav Biol 9: 1–14. [DOI] [PubMed] [Google Scholar]
- Hoogerwerf WA, Hellmich HL, Cornélissen G, Halberg F, Shahinian VB, Bostwick J, Savidge TC, Cassone VM. 2007. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology 133: 1250–1260. [DOI] [PubMed] [Google Scholar]
- Impey S, Davare M, Lesiak A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. 2010. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci 43: 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C, Lahens NF, Hogenesch JB, Sehgal A. 2015. Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res 25: 1836–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasińska M, Miłek J, Cymerman IA, Łęski S, Kaczmarek L, Dziembowska M. 2016. miR-132 Regulates Dendritic Spine Structure by Direct Targeting of Matrix Metalloproteinase 9 mRNA. Mol Neurobiol 53: 4701–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. 2007. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci 10: 1513–1514. [DOI] [PubMed] [Google Scholar]
- Lambert TJ, Storm DR, Sullivan JM. 2010. MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PLoS One 5: e15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart BW, Bensen AL, Washburn EK, Perederiy JV, Su KG, Li Y, Kernie SG, Parada LF, Westbrook GL. 2011. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One 6: e19077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons LC. 2011. Critical role of the circadian clock in memory formation: lessons from Aplysia. Front Mol Neurosci 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. 2010. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci 107: 20382–20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Paz A, Sanchez-Mut JV, Samitier-Martí M, Petazzi P, Sáez M, Szczesna K, Huertas D, Esteller M, Ausió J. 2015. Circadian cycle-dependent MeCP2 and brain chromatin changes. PLoS One 10: e0123693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. 1996. Control of memory formation through regulated expression of a CaMKII transgene. Science 274: 1678–1683. [DOI] [PubMed] [Google Scholar]
- McGowan D. 2006. MeCP2 and memory mechanisms. Nat Rev Neurosci 7: 87. [Google Scholar]
- McGowan NM, Coogan AN. 2013. Circadian and behavioural responses to shift work-like schedules of light/dark in the mouse. J Mol Psychiatry 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, Edbauer D, Sur M. 2011. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci 14: 1240–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michán S, Li Y, Chou MM, Parrella E, Ge H, Long JM, Allard JS, Lewis K, Miller M, Xu W. 2010. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci 30: 9695–9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk TH, Buysse DJ, Reynolds CF 3rd, Berga SL, Jarrett DB, Begley AE, Kupfer DJ. 1997. Circadian rhythms in human performance and mood under constant conditions. J Sleep Res 6: 9–18. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. 2006. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci 26: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N, Zinovyev A, Nonne N, Pritchard LL, Gorban AN, Harel-Bellan A. 2012. Kinetic signatures of microRNA modes of action. RNA 18: 1635–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser EI, Kropff E, Moser MB. 2008. Place cells, grid cells, and the brain's spatial representation system. Annu Rev Neurosci 31: 69–89. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. 1992. NeuN, a neuronal specific nuclear protein in vertebrates. Development 116: 201–211. [DOI] [PubMed] [Google Scholar]
- Na ES, Nelson ED, Adachi M, Autry AE, Mahgoub MA, Kavalali ET, Monteggia LM. 2012. A mouse model for MeCP2 duplication syndrome: MeCP2 overexpression impairs learning and memory and synaptic transmission. J Neurosci 32: 3109–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na ES, Nelson ED, Kavalali ET, Monteggia LM. 2013. The impact of MeCP2 loss- or gain-of-function on synaptic plasticity. Neuropsychopharmacology 38: 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. 2010. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20: 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Smith D, Athos J, Storm DR. 1999. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem 274: 17748–17756. [DOI] [PubMed] [Google Scholar]
- O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. 2008. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320: 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka GJ, Watson CM, Radziewic T, Hayward M, Lahooti H, Christodoulou J, Tam PP. 2006. Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain 129(Pt 4): 887–898. [DOI] [PubMed] [Google Scholar]
- Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. 2014. Common features of microRNA target prediction tools. Front Genet 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TX, Chan GC, Sindreu CB, Eckel-Mahan KL, Storm DR. 2011. The diurnal oscillation of MAP (mitogen-activated protein) kinase and adenylyl cyclase activities in the hippocampus depends on the suprachiasmatic nucleus. J Neurosci 31: 10640–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro A, Hayer K, Lahens NF, Hogenesch JB. 2013. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res 41(Database issue): D1009–D1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Ko CH, Antoniadis EA, Seco P, Irani F, Presta C, McDonald RJ. 2002. The significance of circadian phase for performance on a reward-based learning task in hamsters. Behav Brain Res 136: 179–184. [DOI] [PubMed] [Google Scholar]
- Rawashdeh O, Jilg A, Jedlicka P, Slawska J, Thomas L, Saade A, Schwarzacher SW, Stehle JH. 2014. PERIOD1 coordinates hippocampal rhythms and memory processing with daytime. Hippocampus 24: 712–723. [DOI] [PubMed] [Google Scholar]
- Remenyi J, Hunter CJ, Cole C, Ando H, Impey S, Monk CE, Martin KJ, Barton GJ, Hutvagner G, Arthur JS. 2010. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem J 428: 281–291. [DOI] [PubMed] [Google Scholar]
- Remenyi J, van den Bosch MW, Palygin O, Mistry RB, McKenzie C, Macdonald A, Hutvagner G, Arthur JS, Frenguelli BG, Pankratov Y. 2013. miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One 8: e62509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby NF, Hwang CE, Wessells C, Fernandez F, Zhang P, Sapolsky R, Heller HC. 2008. Hippocampal-dependent learning requires a functional circadian system. Proc Natl Acad Sci 105: 15593–15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Collette F, Cajochen C, Peigneux P. 2007. A time to think: circadian rhythms in human cognition. Cogn Neuropsychol 24: 755–789. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Kobayashi Y, Nakatsuji E, Yamazaki M, Shimba S, Sakimura K, Fukada Y. 2016. SCOP/PHLPP1β mediates circadian regulation of long-term recognition memory. Nat Commun 7: 12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smarr BL, Jennings KJ, Driscoll JR, Kriegsfeld LJ. 2014. A time to remember: the role of circadian clocks in learning and memory. Behav Neurosci 128: 283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider KH, Dziema H, Aten S, Loeser J, Norona FE, Hoyt K, Obrietan K. 2016. Modulation of learning and memory by the targeted deletion of the circadian clock gene Bmal1 in forebrain circuits. Behav Brain Res 308: 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan FK, Kovacevic NS. 1978. Multiple retention deficit in passive avoidance in rats is eliminated by suprachiasmatic lesions. Behav Biol 22: 456–462. [DOI] [PubMed] [Google Scholar]
- Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. 2009. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol 23: 1876–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Sawa K, Okada T. 2013. The diurnal variation of performance of the novel location recognition task in male rats. Behav Brain Res 256: 488–493. [DOI] [PubMed] [Google Scholar]
- Tapp WN, Holloway FA. 1981. Phase shifting circadian rhythms produces retrograde amnesia. Science 211: 1056–1058. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Kolker DE, Vitaterna MH, Ferrari E, Takahashi JS, Turek FW. 2001. Effect of circadian phase on context and cued fear conditioning in C57BL/6J mice. Animal Learning and Behavior 29: 133–142. [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. 2005. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci 102: 16426–16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansley RA, Holloway FA. 1975. Multiple retention deficits following one-trial appetitive training. Behav Biol 14: 135–149. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH. 2008. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci 105: 9093–9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chen J, Cheng AS, Ko BC. 2014. Depletion of sirtuin 1 (SIRT1) leads to epigenetic modifications of telomerase (TERT) gene in hepatocellular carcinoma cells. PLoS One 9: e84931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang HJ, Wang DR, Qu WM, Huang ZL. 2017. Red light at intensities above 10 lx alters sleep–wake behavior in mice. Light: Science & Applications 6: e16231. [DOI] [PMC free article] [PubMed] [Google Scholar]