Abstract
MicroRNAs (miRNAs) are short ~22-nucleotide noncoding RNA that have been found to influence the expression of many genes and cellular processes by either repressing translation or degrading messenger RNA transcripts. Platelet miRNA expression has been shown to be perturbed during ex vivo storage of platelets and in platelet-associated disorders. Although bioinformatics-based miRNA target predictions have been established, direct experimental validation of the role of miRNAs in platelet biology has been rather slow. Target prediction studies are, nonetheless, valuable in directing the design of appropriate experiments to test specific miRNA:messenger RNA interactions relevant to the underlying mechanisms of platelet function in general and in disease as well as in ex vivo storage-associated “storage lesions,” a collective term used to include physiologic, biochemical, and morphologic changes that occur in stored platelets. This brief review will focus on emerging human platelet miRNA studies to emphasize their potential role relevant to transfusion medicine field in terms of regulating platelet signaling pathways, markers of platelet associated disorders, and remote impactors of gene expression (intercellular biomodulators) as well as potential platelet quality markers of storage and pathogen reduction treatments.
Keywords: MicroRNA, Blood disorders, Platelets, Signaling pathways
MicroRNAs (miRNAs) are endogenously expressed small noncoding RNAs involved in posttranscriptional regulation of cellular gene (messenger RNA [mRNA]) expression [1–3]. It is estimated that more than 60% of human protein-coding genes are regulated by miRNAs [2]. Being small in size, miRNAs are relatively stable molecules and easily quantifiable. Recent technical developments in the field have allowed researchers to measure the absolute and relative levels of many cellular miRNAs that further led to the recognition of their prognostic value as biomarkers [4–7]. Subsequently, this concept has further evolved, and differential profiling of miRNAs as a tool to identify changes (ie, biomarkers) in a normal “biological entity” vs its testing cohort has emerged. The “biological entity” could be a human, an animal, or a cell in culture, etc. The testing cohort could be a genetic variant or a disease state, a drug or biologic treatment, an infection state, radiation, or ex vivo stored human cells for therapeutics. Example of ex vivo stored cells includes concentrated human platelets, which are the most important transfused therapeutic products used in trauma, surgery, and profusely bleeding patients [8]. For many years, investigators have studied the physiologic and biochemical changes that occur in ex vivo stored platelets, which are collectively known as storage lesions. Because human mature platelet is enucleated, in order for it to be alive and functional, especially in ex vivo storage, and to regulate all important physiological processes, the platelet has to use its posttranscriptional regulatory machinery available in the cytoplasm. One such mechanism is miRNA-based regulation of mRNA. Research on how miRNAs participate in the regulation of platelet mRNA expression is the key toward understanding the molecular mechanisms of stored platelets that could facilitate enhancements to the quality and shelf-life of ex vivo stored platelets.
This brief review will provide an overview on emerging trends in human platelet miRNA studies and the potential directions these studies will take in the near future to address a range of issues such as miRNA-based regulation of platelet signaling pathways, miRNA as markers of platelet-associated disorders, and miRNA as remote impactors of gene expression (intercellular biomodulators) as well as potential platelet markers of quality in storage and pathogen reduction treatments.
MicroRNAs as potential regulators of signaling pathways in platelets
Human platelets are generated from hematopoietic stem cells through a process known as hematopoiesis [9–11], and developmental differentiation of platelets from megakaryocytes is known as megakaryopoiesis. In these processes miRNAs have shown to play important roles as illustrated in Figure 1 [9–18]. Different members of the cytoplasmic miRNA processing complex that includes proteins such as RNase III Dicer, Ago2 (Argonaute protein, which is an essential component of the RNA-induced silencing complex aka RISC), and TRBP2 (TAR RNA binding protein 2) have been reported recently in platelets [19]. Although a functional cytoplasmic miRNA processing exists in platelets, the abundance of mature miRNAs over pre-miRNAs suggests that platelets inherit most of its mature miRNAs directly from the megakaryocytes. The observation of abundance of miRNAs in platelets relative to mRNAs suggests that the miRNA:mRNA ratio normally observed in nucleated cells of the hematopoietic system is significantly altered in platelets. In other words, a single mRNA can be targeted by multiple miRNAs. These observations support the idea that, in a platelet, mRNAs are perhaps decorated at multiple sites by Ago2-miRNA complexes resulting in rendering a stronger mRNA repression than in other cells [19]. In platelets, more than 75% of the expressed miRNAs were found to be predominantly from only 5 miRNA families that include let-7, miR-199, miR-103, miR-25, and miR-140 [20]. In addition, among these 5 families, the members of let-7 and miR-103 are also in the list of top 30 highly expressed platelet miRNAs [21], which clearly suggests their key role in various biological processes of platelets. Another miRNA, miR-326, was markedly increased in platelet storage and was shown to directly target and inhibit antiapoptotic Bcl-xL gene (ie, its mRNA) [22,23]. We have previously shown that 2 miRNAs, let-7b and miR-16, distinctly demonstrated an increasing trend, whereas miR-7 and miR-145 showed a decreasing profile during platelet storage [24]. Our previous observation of miR-16 increase during platelet storage and the increase of miR-326 in storage reported by another group [22], both of which target the genes involved in apoptosis, provides further circumstantial support that platelet apoptosis is regulated by miRNAs.
Fig 1.
MicroRNAs and their targets in megakaryopoiesis. MicroRNAs involved in positive and negative regulation of megakaryopoiesis and their target genes (superscript numbers in parentheses denote appropriate references listed in the references section).
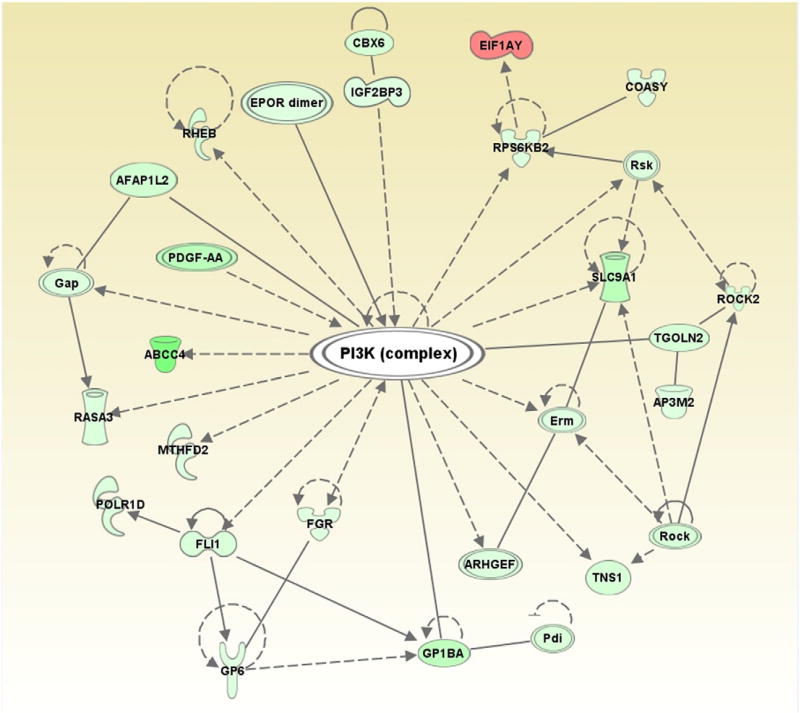
Recently, we have profiled both miRNAs and mRNAs in ex vivo stored platelets at different days of storage to examine storage-associated changes in mRNA levels and correlated them with differentially expressed miRNAs. The data analysis was performed using Ingenuity Pathway Analysis and identified top canonical pathways in which potential genes (mRNAs) that are targeted by miRNAs were uncovered (Table). Some of these pathways such as those involved in “actin cytoskeleton signaling” and “integrin signaling” pathways that we identified in our analysis here were also previously identified by others [25] as important players of platelet biology, which lends support to our observation. Furthermore, it is known that Ras-related protein 1 signaling pathway increases during platelet storage and inhibition of phosphatidylinositol-4,5-biphosphate 3-kinase was shown to reduce Ras-related protein 1 activity and concomitant improvement in platelet integrity and quality [26]. Through the network analysis of potential miRNA targets, we identified a set of 28 mRNAs that were implicated in phosphatidylinositol-4,5-biphosphate 3-kinase signaling pathway (Fig 2), which again argues in favor of the idea that miRNAs do play important roles in platelet biology.
Table.
Top canonical pathways identified by Ingenuity pathway analysis
| Top canonical pathways | P | Ratio | Genes |
|---|---|---|---|
| Day 5 | |||
| Amyloid processing | 9.21E-04 | 2/61 (0.033) | PRKACB, CSNK2A1 |
| Putrescine biosynthesis III | 1.77E-03 | 1/4 (0.25) | ODC1 |
| IGF-1 signaling | 3.28E-03 | 2/107 (0.019) | PRKACB, CSNK2A1 |
| NF-κB signaling | 1.01E-02 | 2/181 (0.011) | PRKACB, CSNK2A1 |
| RAR activation | 1.05E-02 | 2/195 (0.01) | PRKACB, CSNK2A1 |
| Day 9 | |||
| Actin cytoskeleton signaling | 1.02E-12 | 32/242 (0.132) | RAF1, MPRIP, PDGFA, ARPC5, EGF, SSH1, SLC9A1, PDGFC, ROCK2, PTK2, SHC1, DIAPH1, FGF18, MAP2K2, FLNA, EZR, MAP2K1, ARHGEF12, GRB2, CSK, CRKL, GNA12, ITGA2, RAC1, GSN, ROCK1, PIP5K1C, CYFIP1, MYH9, WASF2, ACTN4, PIP4K2A |
| Integrin signaling | 9.2E-11 | 28/208 (0.135) | RAF1, FYN, RAP2A, MPRIP, ARPC5, ILK, ITGB3, PTK2, NCK2, SHC1, AKT1, MAP2K2, MAP2K1, VASP, SRC, GRB2, ASAP1, CRKL, RALB, ITGA2, RAC1, TNK2, ROCK1, ARF1, ARF3, LIMS1, ACTN4, FNBP1 |
| Ephrin receptor signaling | 1.6E-10 | 26/210 (0.124) | RAF1, FYN, AXIN1, PDGFA, ARPC5, GNB5, EGF, PDGFC, GNB1, ROCK2, PTK2, NCK2, SHC1, AKT1, MAP2K2, ATF4, MAP2K1, SRC, GRB2, CRKL, GNA12, ITGA2, RAC1, GNAZ, ROCK1, ACP1 |
| Molecular mechanisms of cancer | 1.15E-09 | 36/388 (0.093) | RAF1, FYN, RAP2A, AXIN1, CTNNA1, PTK2, TGFBR2, CASP6, SHC1, AKT1, MAP2K2, ARHGEF3, MAP2K1, RALGDS, PRKCA, SRC, ARHGEF12, PRKCQ, CASP3, GRB2, GNA12, ADCY3, RALB, RAC1, GNAZ, RALBP1, FADD, BCL2L1, CBL, FOXO1, IRS1, CDKN1A, ARHGEF18, MAP2K3, DIABLO, FNBP1 |
| VEGF signaling | 1.18E-09 | 18/109 (0.165) | EIF1AY, SRC, RAF1, PTPN6, GRB2, PDGFC, ROCK2, PTK2, ROCK1, SHC1, BCL2L1, AKT1, FOXO1, MAP2K2, FOXO3, ACTN4, MAP2K1, PRKCA |
Fig 2.
Network analysis using Ingenuity Pathway Analysis. Network analysis of potential miRNA targets identified a set of 28 mRNAs or their products implicated in PI3K pathway. Solid and dashed lines represent direct and indirect interactions among different members of the PI3K complex, respectively.
MicroRNAs as Potential Markers of Platelet-Associated Disorders
The first human platelet miRNA profiling was reported as part of a study testing for differentially expressed miRNA in polycythemia vera patients and found that miR-26b was significantly higher in these patient platelets compared to healthy controls, which has opened the field toward identifying miRNAs as potential biomarkers of platelet disorders [27]. Platelet reactivity is another disorder in which miR-96 was shown to down-regulate vesicle-associated membrane protein 8 mRNA expression and the protein levels [28]. In atrial fibrillation (AF) associated with poor prognosis in heart failure patients, platelets contribute to the prothrombotic state, and in a recent study, miR-150 expression levels in platelets evaluated from systolic heart failure patients with AF were found to be significantly reduced and correlated to the cell-free circulating levels of this miRNA [29]. This suggests that perhaps for this disease, circulating miR-150 levels could serve as a surrogate marker. In sickle cell disease, increased platelet activation contributes to a state of hypercoagulability and thereby increasing the risk of thrombotic complications where 40 miRNAs were implicated in the pathogenesis of sickle cell disease in which 3 distinct miRNA families (hsa-miR-154, hsa-miR-329, and hsa-miR-376) were down-regulated when compared to healthy controls [30]. This is another example where miRNAs appears to be useful as markers of platelet-associated disease. In megakaryocytes and platelets, miR-223 is the most abundant miRNA, and 1 of the genes (mRNA) regulated by this miRNA is the ADP receptor P2Y12, a key target for current antiplatelet therapy. Human P2Y12 mRNA 3′ UTR has 1 miR-223 target site, a requirement for miRNA-based gene regulation. Many studies have demonstrated favorable trend toward a regulatory function of miR-223 in several clinical disorders, some of which include platelet-associated disorders such as high on-treatment platelet activity (HTPR). High on-treatment platelet activity is a strong predictor of major cardiovascular events in coronary heart disease patients receiving antiplatelet treatment, suggesting the value of presumably platelet-derived miR-223 as a diagnostic tool for HTPR in clinical settings [31].
Platelet Microparticle-Derived miRNAs as In Vivo Biomodulators
Earlier studies on circulating microparticles (MPs) suggest that most of these MPs are platelet-derived MPs (PMPs) [32] and PMPs contain various cytoplasmic components including miRNAs [33]. Platelet-derived MPs act as intercellular carriers to deliver and/or transfer genetic information from one cell to the other across the circulatory system and modulate cellular gene expression, vascular homeostasis, and inflammation [32–39].
One of them iRNAs identified in PMPs, the miR-126, is known to mediate vascular repair and progenitor cell recruitment to maintain vascular homeostasis [35,39]. Another miRNA, miR-223, although abundant in platelets and in PMPs, it is scarce in endothelial cells. When human umbilical vein endothelial cells were incubated with PMPs containing Ago2–miR-223 complex, it was observed that, in 6 hours, the expression of 2 endothelial genes (mRNAs), the FBXW7 and EFNA1, that contain miR-223 binding sites in their 3′ UTR, was significantly down-regulated and so was their protein expression in 18 hours and 96 hours, respectively [40,41]. These observations further generate a testable hypothesis that when stored platelets containing substantial amount of PMPs are transfused, a variety of miRNAs trapped in PMPs are also transferred to the recipient and modulate the gene (mRNA) expression in the recipients. Testing this hypothesis first in preclinical animal studies and subsequently through prospective clinical trials will unravel the long-term consequences, if any, of the stored platelet-derived PMP-miRNAs in transfusion recipients.
MicroRNAs as Potential Platelet Markers of Storage and Pathogen Reduction Treatments
Although ex vivo stored platelets at 22°C to 24°C (room temperature) with continuous gentle agitation do well in transfusion recipients, both the platelet storage solutions and the storage temperature are conducive to bacterial growth. As a result, often undetectable bacterial contamination (ie, residual) that occurs during collection from the donor and downstream processing has the potential to grow rapidly. This is an added risk for sepsis in patients, especially those with impaired immune systems. Several different techniques are available to reduce the bacterial burden in stored platelets. The 2 most common methods used for pathogen reduction are Mirasol Pathogen Reduction Technology (riboflavin + ultraviolet B light) [42] and Intercept Blood System (amotosalen + ultraviolet A light) [43].
We have previously reported that among 52 selected apoptosis-associated miRNAs, a few are differentially expressed during various time points of platelet storage, which suggested that miRNA profiles do respond to platelet treatments (stored vs fresh) and, therefore, selected differentially expressed platelet miRNAs have the potential to serve as storage biomarkers of quality [24]. In line with this concept, recently, Osman et al [44] have reported changes in platelet miRNAs associated with different pathogen reduction treatments relative to respective controls, which further suggests that miRNAs of platelets have the potential to be the markers for a given context.
Conclusions and Future Prospects
Within the last 5 years, identification and evaluation of platelet-associated miRNAs in different areas related to platelet function have expanded our understanding of molecular mechanisms underlying some of the platelet functions. The examples that we provided in this brief review clearly suggest that the platelet miRNAas a field has crossed the threshold of mere predictions through bioinformatics about what these tiny RNAs may or may not do functionally, to a stage where we can rely on miRNA with respect to unravelling many poorly understood events of platelets in storage, such as platelet storage lesions. Such studies should help facilitate improvements to platelet quality in storage for better patient outcomes after transfusion as well as enhancements to their storage shelf-life beyond current 5-day expiration.
Acknowledgments
CDA received funding for this study from the Center for Biologics Evaluation and Research, US Food and Drug Administration. ND and LV are recipients of a postdoctoral fellowship at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration. The collaborative work performed by the coauthors KGB, WHW, and YZ at the National Institute on Aging, NIH, was supported by their Intramural Research Program. We would like to thank Dr Valerie W. Hu from the Department of Biochemistry and Molecular Biology, The George Washington University School of Medicine and Health Sciences, for allowing us to use the Ingenuity Pathway Analysis software.
Footnotes
Disclaimers
None.
Conflict of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–53. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 5.De Santis G, Ferracin M, Biondani A, Caniatti L, Rosaria Tola M, Castellazzi M, et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol. 2010;226:165–71. doi: 10.1016/j.jneuroim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Shi Y, Yin Z, Xue X, Zhou B. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12:1–12. doi: 10.1186/1479-5876-12-159. [159] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckland J. Biomarkers: microRNAs under the spotlight in inflammatory arthritis. Nat Rev Rheumatol. 2010;6:1–20. doi: 10.1038/nrrheum.2010.112. [436] [DOI] [PubMed] [Google Scholar]
- 8.Holcomb JB, Tilley BC, Baraniuk S, Fox EE, Wade CE, Podbielski JM, et al. PROPPR Study Group. ransfusion of plasma, platelets, and red blood cells in a 1:1:1 vs a 1:1:2 ratio and mortality in patients with severe trauma: the PROPPR randomized clinical trial. JAMA. 2015;313:471–82. doi: 10.1001/jama.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romania P, Lulli V, Pelosi E, Biffoni M, Peschle C, Marziali G. MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors. Br J Haematol. 2008;143:570–80. doi: 10.1111/j.1365-2141.2008.07382.x. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. PNAS. 2009;106:238–43. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- 12.Lazare SS, Wojtowicz EE, Bystrykh LV, de Haan G. microRNAs in hematopoiesis. Exp Cell Res. 2014;329:234–8. doi: 10.1016/j.yexcr.2014.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Montagner S, Deho L, Monticelli S. MicroRNAs in hematopoietic development. BMC Immunol. 2014;15:14–25. doi: 10.1186/1471-2172-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Undi RB, Kandi R, Gutti RK. MicroRNAs as haematopoiesis regulators. Adv Hematol. 2013;2013:1–20. doi: 10.1155/2013/695754. [695754] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. PNAS. 2006;103:5078–83. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–53. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro F, Gutman D, Meire E, Cáceres M, Rigoutsos I, Bentwich Z, et al. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114:2181–92. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- 18.Yuan JY, Wang F, Yu J, Yang GH, Liu XL, Zhang JW. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J Cell Mol Med. 2009;13:4551–9. doi: 10.1111/j.1582-4934.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16:961–6. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ple H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7:1–14. doi: 10.1371/journal.pone.0050746. [e50746] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon LM, Edelstein LC, Nagalla S, Woodley AB, Chen ES, Kong X, et al. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood. 2014;123:e37–45. doi: 10.1182/blood-2013-12-544692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu S, Deng G, Qian D, Xie Z, Sun H, Huang D, et al. Detection of apoptosis-associated microRNA in human apheresis platelets during storage by quantitative real-time polymerase chain reaction analysis. Blood Transfus. 2014;12:541–7. doi: 10.2450/2014.0291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu S, Huang H, Deng G, Xie Z, Ye Y, Guo R, et al. miR-326 targets antiapoptotic Bcl-xL and mediates apoptosis in human platelets. PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0122784. [e0122784] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kannan M, Mohan KV, Kulkarni S, Atreya C. Membrane array-based differential profiling of platelets during storage for 52 miRNAs associated with apoptosis. Transfusion. 2009;49:1443–50. doi: 10.1111/j.1537-2995.2009.02140.x. [DOI] [PubMed] [Google Scholar]
- 25.Rosado JA, Meijer EM, Hamulyak K, Novakova I, Heemskerk JW, Sage SO. Fibrinogen binding to the integrin alpha(IIb)beta(3) modulates store-mediated calciumentry in human platelets. Blood. 2001;97:2648–56. doi: 10.1182/blood.v97.9.2648. [DOI] [PubMed] [Google Scholar]
- 26.Schubert P, Thon JN, Walsh GM, Chen CH, Moore ED, Devine DV, et al. A signaling pathway contributing to platelet storage lesion development: targeting PI3-kinasedependent Rap1 activation slows storage-induced platelet deterioration. Transfusion. 2009;49:1944–55. doi: 10.1111/j.1537-2995.2009.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruchova H, Merkerova M, Prchal JT. Aberrant expression of microRNA in polycythemia vera. Haematologica. 2008;93:1009–16. doi: 10.3324/haematol.12706. [DOI] [PubMed] [Google Scholar]
- 28.Kondkar AA, Bray MS, Leal SM, Nagalla S, Liu DJ, Jin Y, et al. VAMP8/endobrevin is overexpressed in hyperreactive human platelets: suggested role for platelet microRNA. J Thromb Haemost. 2010;8:369–78. doi: 10.1111/j.1538-7836.2009.03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goren Y, Meiri E, Hogan C, Mitchell H, Lebanony D, Salman N, et al. Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol. 2014;113:976–81. doi: 10.1016/j.amjcard.2013.11.060. [DOI] [PubMed] [Google Scholar]
- 30.Jain S, Kapetanaki MG, Raghavachari N, Woodhouse K, Yu G, Barge S, et al. Expression of regulatory platelet microRNAs in patients with sickle cell disease. PLoS One. 2013;8:1–13. doi: 10.1371/journal.pone.0060932. [e60932] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi R, Zhou X, Ji WJ, Zhang YY, Ma YQ, Zhang JQ, et al. The emerging role of miR-223 in platelet reactivity: implications in antiplatelet therapy. Biomed Res Int. 2015;2015:1–8. doi: 10.1155/2015/981841. [981841] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horstman LL, Ahn YS. Platelet microparticles: a wide-angle perspective. Crit Rev Oncol Hematol. 1999;30:111–42. doi: 10.1016/s1040-8428(98)00044-4. [DOI] [PubMed] [Google Scholar]
- 33.Mause SF, Weber C. Protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010;107:1047–57. doi: 10.1161/CIRCRESAHA.110.226456. [DOI] [PubMed] [Google Scholar]
- 34.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:1–13. doi: 10.1371/journal.pone.0003694. [e3694] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–60. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 36.Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N, et al. Microparticles:major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93:633–44. doi: 10.1093/cvr/cvs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gatsiou A, Boeckel JN, Randriamboavonjy V, Stellos K. MicroRNAs in platelet biogenesis and function: implications in vascular homeostasis and inflammation. Curr Vasc Pharmacol. 2012;10:524–31. doi: 10.2174/157016112801784611. [DOI] [PubMed] [Google Scholar]
- 38.Semple JW. Platelets deliver small packages of genetic function. Blood. 2013;122:155–6. doi: 10.1182/blood-2013-05-502609. [DOI] [PubMed] [Google Scholar]
- 39.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 40.Laffont B, Corduan A, Ple H, Duchez AC, Cloutier N, Boilard E, et al. Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood. 2013;122:253–61. doi: 10.1182/blood-2013-03-492801. [DOI] [PubMed] [Google Scholar]
- 41.Pan Y, Liang H, Liu H, Li D, Chen X, Li L, et al. Platelet secreted MicroRNA-223 promotes endothelial cell apoptosis induced by advanced glycation end products via targeting the insulin-like growth factor 1 receptor. J Immunol. 2014;192:437–46. doi: 10.4049/jimmunol.1301790. [DOI] [PubMed] [Google Scholar]
- 42.Castrillo A, Cardoso M, Rouse L. Treatment of buffy coat platelets in platelet additive solution with the mirasol((R)) pathogen reduction technology system. Transfus Med Hemother. 2013;40:44–8. doi: 10.1159/000345679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abedi MR, Doverud AC. Preparation and pathogen inactivation of double dose buffy coat platelet products using the INTERCEPT blood system. J Vis Exp. 2012;70:1–8. doi: 10.3791/4414. [e4414] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osman A, Hitzler WE, Meyer CU, Landry P, Corduan A, Laffont B, et al. Effects of pathogen reduction systems on platelet microRNAs, mRNAs, activation, and function. Platelets. 2015;26:154–63. doi: 10.3109/09537104.2014.898178. [DOI] [PMC free article] [PubMed] [Google Scholar]