Abstract
Rationale
The P2Y12 receptor inhibitor clopidogrel is widely used in patients with acute coronary syndrome, percutaneous coronary intervention or ischemic stroke. Platelet inhibition by clopidogrel shows wide inter-patient variability and high on-treatment platelet reactivity is a risk factor for atherothrombotic events, particularly in high-risk populations. CYP2C19 polymorphism plays an important role in this variability, but heritability estimates suggest that additional genetic variants remain unidentified. The aim of the International Clopidogrel Pharmacogenomics Consortium (ICPC) is to identify genetic determinants of clopidogrel pharmacodynamics and clinical response.
Study design
Based on the data published on www.clinicaltrials.gov, clopidogrel intervention studies containing genetic and platelet function data were identified for participation. Lead investigators were invited to share DNA samples, platelet function test results, patient characteristics and cardiovascular outcomes, to perform candidate gene and genome-wide association studies.
Results
In total, 17 study sites from 13 countries participate in the ICPC, contributing individual patient data from 8,829 patients. Available adenosine diphosphate (ADP) stimulated platelet function tests included Vasodilator-Stimulated Phosphoprotein (VASP) assay, Light Transmittance Aggregometry (LTA) and the VerifyNow P2Y12 assay. A proof of principle analysis based on genotype data provided by each group showed a strong and consistent association between CYP2C19*2 and platelet reactivity (p-value = 5.1×10−40).
Conclusion
The ICPC aims to identify new loci influencing clopidogrel efficacy by using state-of-the-art genetic techniques in a large cohort of clopidogrel-treated patients in order to better understand the genetic basis of on-treatment response variability.
Keywords: clopidogrel, anti-platelet therapy, genome-wide association, CYP2C19, genotyping, candidate genes, pharmacogenetics, platelet reactivity, percutaneous coronary intervention, acute coronary syndrome, coronary artery disease
Background
In patients with coronary artery disease in which percutaneous coronary intervention (PCI) is performed, dual antiplatelet therapy with the cyclooxygenase-1 (COX-1) inhibitor aspirin and a P2Y12 receptor inhibitor, commonly clopidogrel, is the recommended antiplatelet treatment to prevent recurrent atherothrombotic events, like stent thrombosis.1, 2 Although the incidence of stent thrombosis is declining in recent years due to advances in clinical care, such as utilization of new stent designs, it remains a serious complication with a high mortality rate.3 In addition, medical management with clopidogrel is effective in the prevention of recurrent cardiovascular events in patients with acute coronary syndrome (ACS), ischemic stroke and peripheral arterial disease.4–6 However, substantial variability in on-clopidogrel platelet reactivity is well-documented. This variability leads to an increased risk for adverse thrombotic and bleeding complications in patients with high or low platelet reactivity, respectively.7–9 Multiple factors influence variation in on-clopidogrel platelet reactivity, including genetic, anthropometric and clinical variables, as well as drug-drug interactions (e.g. calcium channel blockers, certain proton pump inhibitors such as omeprazole and esomeprazole, ketoconazole, morphine or St. John’s Wort).10–12 Smoking has been correlated to higher platelet inhibition by clopidogrel, but the data are controversial.13, 14
Conversion of clopidogrel into its active thiol metabolite by hepatic CYP P450 enzymes results in irreversible inhibition of platelet P2Y12 receptors.15 Polymorphisms in enzymes which are involved in the two conversion steps, in particular the CYP2C19*2 (rs4244285) and CYP2C19*3 (rs4986893) loss-of-function (LoF) alleles, result in lower clopidogrel active metabolite levels16, 17, higher levels of on-treatment platelet reactivity7, 17–23, and increased risk for on-treatment atherothrombotic events, in particular in the patients with the highest thrombotic risk.24–29 The allele frequency of the CYP2C19*2 polymorphism is ~15% in Caucasian populations and ~29–35% in Asian populations.30, 31 These findings led the FDA in 2010 to add a boxed warning to the clopidogrel label, warning physicians that an alternative antiplatelet drug should be considered in CYP2C19*2 homozygote (poor metabolizer) patients.32
In addition to the CYP2C19*2 and *3 polymorphisms, there are other genes in which polymorphism has been associated with impaired clopidogrel efficacy, such as ABCB1, CYP2B6 and PON1. However, their role as determinants of clopidogrel response is controversial as the findings could not be reproduced in more recent studies.33–37
In contrast to factors related to increased atherothrombotic risk, the putative gain-of-function allele CYP2C19*17 has been correlated with a higher clopidogrel active metabolite level and a reduction in on-treatment platelet reactivity.38, 39 However, most of these reports have not taken into account that this allele is genetically linked to the CYP2C19*2 allele, i.e., the allele containing the *17 polymorphism lacks the *2 polymorphism.40 Also a rare decrease of function variant in carboxyesterase 1 (CES1), the enzyme responsible for converting clopidogrel into biologically inactive metabolites, has been reported to be associated with increased clopidogrel responsivity.40 Recently, exome sequencing of patients with extreme pharmacodynamic responses to clopidogrel identified B4GALT2 as a determinant of low on-treatment platelet reactivity.41 Whether these variants result in better clopidogrel efficacy and/or increased bleeding risk remains to be determined.
Despite the relationship between CYP2C19*2 and impaired platelet inhibition, as assessed by ex vivo adenosine diphosphate (ADP)-stimulated platelet aggregation, it only accounts for approximately 4–12% of the observed inter-individual variation in antiplatelet effect.34, 42–44 Estimates suggest the heritability of the variability in response to clopidogrel to be as high as 70%, suggesting other genetic factors influencing clopidogrel efficacy.44
A previously conducted genome wide association study (GWAS) of clopidogrel response in 429 Amish subjects supported CYP2C19*2 as the single major genetic determinant of clopidogrel response.44 However, there were several additional common variants in or near other genes that showed nominal evidence of association with clopidogrel response in that investigation, but did not achieve genome-wide significance (p-value ~ 10−6). If some of these variants represent true positive signals, they may be uncovered by larger sample sizes. To this end, the International Clopidogrel Pharmacogenomics Consortium (ICPC) was established to better define the genetic architecture of variable clopidogrel response. Participating study sites (N=17) contributed DNA samples, results of ADP-induced platelet function testing and clinical outcome data of patients treated with clopidogrel. A high quality genomic discovery resource was assembled consisting of DNA samples, pharmacodynamic data, major adverse cardiovascular outcomes, and relevant clinical characteristics.
This article describes the characteristics of the ICPC and provides a framework for the organization and execution of large pharmacogenetics studies.
Methods
Study design and population
An organizing committee of international pharmacogenomics investigators was established to define the scientific goals of the ICPC. These goals were to identify genetic variants that influence platelet aggregation and secondary clinical events in clopidogrel-treated subjects with the long-term objective of supporting the use of genetic information that would allow clinicians to make more informed treatment decisions for their patients requiring antiplatelet therapy.
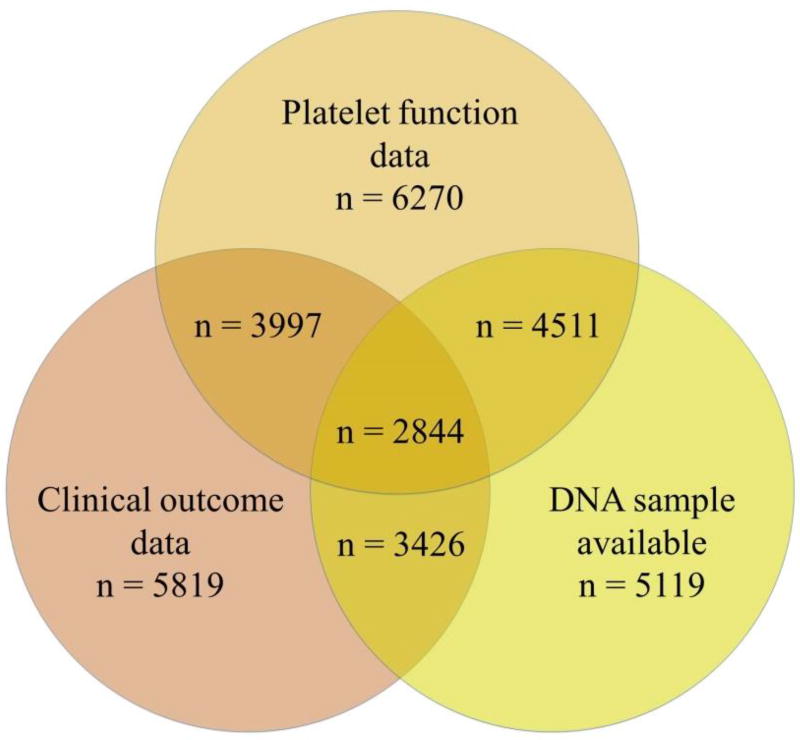
The organizing committee invited 150 lead investigators of clopidogrel-related clinical studies registered in www.clinicaltrials.gov as of June 2011 to participate. Criteria for participation included availability of on-clopidogrel platelet reactivity data, availability of DNA samples for genetic analysis, and for secondary analysis, availability of clinical outcomes; however, studies did not have to fulfill all of these criteria. Eligible studies had to have a minimum of 50 planned enrollees. The phenotypic variables must have been obtained using predefined protocols and methods, and all patients had to be consented for genetic analysis and data sharing. All participating investigators signed a memorandum of understanding (included in the Supplemental Data section). Investigators at Stanford University (PharmGKB) serve as the centralized data coordinating center. Platelet function test results, demographic and cardiovascular outcome data, and other clinical data with a potential influence on platelet reactivity, such as age, sex, diabetes, body mass index, renal function and medication usage (i.e. calcium channel blockers and proton-pump inhibitors) were obtained from each study’s investigators and curated. To date, 17 sites from 13 countries have joined the ICPC (Table I), contributing data representing 8,829 clopidogrel-exposed patients. The available sample size, including those with genotyping, and clinical outcome data, is described in Table II and Figure 1. DNA was available in 5119 ICPC subjects and sent to the University of Maryland where genetic analyses are coordinated. Cohort descriptions and the number of patients in which each platelet function test was performed are available in the online Supplemental Data section. The ICPC is supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number U01HL105198. The authors are solely responsible for the design and conduct of this study, all study analyses, the drafting and editing of the paper and its final contents.
Table I.
Participating Study Sites
| ICPC Primary Investigator | Country | No. patients |
|---|---|---|
| J.G. Shin | South Korea | 2280 |
| J.M. ten Berg | Netherlands | 1024 |
| D. Trenk | Germany | 797 |
| G.F. Gensini | Italy | 736 |
| J. Cleator, D. Roden | USA | 693 |
| A. Shuldiner | USA | 687 |
| P. Fontana, J. L. Reny | Switzerland, France | 538 |
| M. Schwab, T. Geisler | Germany | 442 |
| J.M. Siller-Matula | Austria | 416 |
| J. Déry | Canada | 325 |
| M. Valgimigli, G. Campo | Italy | 234 |
| D. Aradi | Hungary | 192 |
| M. Lee | Taiwan | 160 |
| L. Holmvang | Denmark | 106 |
| P. Gurbel | USA | 79 |
| I. Fernández-Cadenas | Spain | 70 |
| D. Alexopoulos | Greece | 50 |
| PharmGKB | ||
| T. Klein | USA | |
| L. Gong | USA | |
| P-STAR | ||
| M.D. Ritchie | USA |
Table II.
Baseline Characteristics of Study Participants
| Total cohort N=8829 |
Anticipated genotyped cohort* N=4511 |
|
|---|---|---|
| Self-reported race | ||
| White | 70.8 | 94.6 |
| Asian | 27.9 | 4.9 |
| Other | 1.3 | 0.5 |
| Gender (male) | 71.2 | 76.4 |
| Age (years) | 62.8 ± 13.0 | 63.5 ± 12.5 |
| Body Mass Index (kg/m2) | 26.9 ± 4.7 | 27.4 ± 4.4 |
| Diabetes | 26.0 | 23.3 |
| Current smoker | 19.5 | 22.3 |
| Hypercholesterolemia | 53.8 | 66.1 |
| LVEF <35% | 5.2 | 6.1 |
| Aspirin use | 94.7 | 93.2 |
| Statin use | 75.0 | 82.3 |
| CYP2C19 *2 carrier (site-reported) | 35.7 | 31.9 |
| CYP2C19 *17 carrier (site-reported) | 28.6 | 36.2 |
| Coronary artery disease (indication for clopidogrel use) | 81.4 | 94.3 |
| - acute coronary syndrome | 33.7 | 44.2 |
| - PCI performed | 92.5 | 87.6 |
| - for acute coronary syndrome | 36.4 | 56.6 |
| - for non-urgent PCI | 63.6 | 43.4 |
Data are expressed in % or Mean ± SD
All patients with both a DNA sample and platelet function test result available
Abbreviations: LVEF = left ventricular ejection fraction, PCI = percutaneous coronary intervention, SD = standard deviation
Figure 1.
Figure describing the subset of patients in which platelet function data, clinical outcome data and/or a DNA sample is available for analysis. Total number of patients is 8829.
Study endpoints
The primary ICPC study endpoint is platelet function. Both platelet function measurements in patients on clopidogrel maintenance dose or after adequate loading dose were used, which was defined as at least 2 hours between a 600mg clopidogrel loading dose and platelet function testing, 6 hours after 300mg clopidogrel loading dose and 5 days after start of 75mg maintenance dose without extra loading dose. As platelet function in each study was measured using different platelet function tests, i.e. Light Transmittance Aggregometry (LTA), Multiplate Analyzer (Multiple Electrode Aggregometry; MEA), Vasodilator-Stimulated Phosphoprotein (VASP), and VerifyNow P2Y12 assay, measurements were standardized across these different tests using a priority system laid out by the Phenotype Subcommittee of the ICPC: VASP assay > VerifyNow P2Y12 > ADP-induced LTA (higher ADP concentration > lower ADP concentration) > other tests. The platelet function test was then chosen from each site based on the highest ranking assay measured at that site that maximized the sample size. To standardize values across the different platelet function assays (all of which provide an assessment of ADP-stimulated platelet reactivity albeit expressed in different units), we standardized each measurement by subtracting out the mean platelet reactivity value and dividing by the standard deviation, thus expressing each measurement as a Z-score (i.e., the number of standard deviation units from the mean). These transformations were made within each site, thus allowing us to combine results by meta analysis despite the use of different platelet function assays. Standardized platelet function tests were available in 6270 ICPC subjects.
Secondary ICPC study endpoint included a composite clinical endpoint defined any of cardiovascular death, ischemic stroke, or spontaneous myocardial infarction. Stent thrombosis, defined by the Academic Research Consortium (ARC) criteria, was also ascertained but not considered as part of the composite clinical endpoint.45 This rare but potentially lethal complication of stent placement will be analyzed separately. Major bleeding complications are captured in the database, although various definitions for bleeding were used in different study cohorts. The secondary endpoints were adjudicated locally using site-specific criteria. The ICPC includes a total of 5,819 subjects in which all components of the combined clinical endpoint are available. In those patients, 290 major adverse cardiac events (MACE) were observed (event rate 5.0%) during a mean (standard deviation [SD]) follow-up duration of 13 ± 9 months. The event rate is lower compared to the clopidogrel treated cohorts in the PLATO and TRITON studies, which might be explained by the higher percentage of ACS patients in those cohorts.46, 47 In the subset of ICPC with DNA samples, 3426 patients have clinical data of whom 199 major adverse cardiac events were observed (event rate 5.8%) during a mean follow-up duration of 14 ± 11 months.
In addition to the main objectives of the ICPC, ICPC investigators as well as external investigators have the opportunity to access the combined data set for ancillary projects. Project proposals detailing research aims and analysis plans are reviewed and approved by the ICPC Steering Committee. Consortium members will not have access to personal identifiers. Consortium phenotype and genotype data will be deposited to public database (e.g. dbGAP) in accordance with NIH data sharing policies.
Genotyping
Genotyping will include both a candidate gene approach, in which a small number of genes and gene variants will be chosen based upon prior evidence for association with clopidogrel response, and an agnostic genome-wide approach using the Illumina Omni Express with Exome (OEE) chip, to identify novel loci associated with clopidogrel response. Data will be cleaned using the eMERGE QC pipeline.48 eMERGE is the electronic MEdical Record and GEnomics Network, a National Human Genome Research Institute (NHGRI) funded consortium which has been combining clinical cohorts with biobanks linked to electronic health records. The genotype data cleaning process includes evaluation of sample and marker call rate, gender mismatch, duplicate and HapMap concordance, batch effects, Hardy-Weinberg equilibrium, sample relatedness and population stratification. For GWAS, imputation to the 1000 Genomes reference dataset will be performed using the eMERGE Imputation Pipeline which includes SHAPEIT2 for phasing and IMPUTE2 for imputation.49 eMERGE has developed a robust imputation and quality control pipeline for the combining of multiple datasets, which will be appropriate for the ICPC.
Database validation
To validate the ICPC central database, the association between cross-study harmonized platelet reactivity and CYP2C19*2 was tested, as this association has been well-described in the literature.7, 17–22, 50 All Caucasian patients with site-reported CYP2C19*2 and harmonized ADP-induced platelet reactivity phenotype (n=5328) were selected. The analysis was adjusted for age and sex. All analyses were performed in SAS version 9.2 (SAS Institute Inc., Cary, North Carolina). Results confirmed that CYP2C19*2 was independently associated with higher on-treatment platelet reactivity (Beta = 0.37; p-value = 5.1×10−40).
Future directions
The ICPC is a collaborative platform for novel gene discovery related to clopidogrel effectiveness. It will serve as a well-powered resource to test new hypotheses and welcomes proposals from qualified investigators. ICPC will also serve as a replication cohort for genetic variants identified by other research groups worldwide. Proposals for new research questions can be send by email to the corresponding author of this paper.
Concluding remarks
With the introduction of the newer P2Y12 receptor inhibitors prasugrel and ticagrelor, multiple antiplatelet drugs are now available for clinicians to use. Although prasugrel and ticagrelor are more effective in reducing atherothrombotic events in ACS patients, they increase the risk of bleeding compared to clopidogrel, and are more expensive.46, 47 Given that alternative treatment options exist and pharmacodynamic and genetic testing options are available to predict or test the efficacy of clopidogrel, development of personalized antiplatelet strategies, to reduce atherothrombotic events and risk of bleeding, might significantly enhance patient care and potentially reduce costs; thus additional studies are warranted. Nevertheless, finding the optimal treatment strategy has proven to be difficult so far, as recent studies using different treatment modification strategies show contrasted results, owing in part to the different clinical settings and population vascular risk levels in the various studies.51–57 Identification of novel gene variants that predict clopidogrel efficacy may provide important information to improve predictive algorithms for genotype-directed therapy. These studies also promise to provide new insights into platelet biology and to identify novel targets for more effective and safe antiplatelet therapy.
Summary
The aim of the ICPC is to find novel genetic markers which influence clopidogrel efficacy, using GWAS and candidate gene approaches combined with pharmacodynamic and clinical outcome data.
Supplementary Material
Acknowledgments
The ICPC research reported in this publication was supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health (NIH) under Award Number U01HL105198 and a NIH National Institute of General Medical Sciences grant R24GM61374. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Genome wide SNP genotyping is supported by the Pharmacogenomics Research Network & CGM Global Alliance. P-STAR is funded by NIH grant HL065962. IFC thanks Dr. Christina-Gallego-Fabrega for compiling clinical data and performing genotyping. JLR recognizes Dr. Christophe Combescure’s contributions to the analysis plan. The authors thank Ankita Parihar, John Wallace and Shefali Setia for data QC and analyses performed. The authors gratefully acknowledge the editorial and data contributions of Prof. Bernd Jilma, principal investigator of the PEGASUS-PCI study.
The Taiwanese cohort research was supported by grants to Chao-Yung Wang from the National Health Research Institute (NHRI-EX100-9925SC), the National Science Council (98-2314-B-182A-082-MY3) and the Chang Gung Memorial Hospital (CMRPG391861), and to Ming-Shien Wen from NRPB (101TM1033) and the Chang Gung Memorial Hospital (CMRPG3A1071). The Korean cohort was supported by a grant of Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI15C1537)
Disclosures
Dr. Simon reports grants from AstraZeneca, Daiichi-Sankyo, Eli-Lilly, GSK, MSD, Novartis, and Sanofi, and personal fees from Board membership, consultancy, or lecture from AstraZeneca, Astellas, Bayer, BMS, Lilly, MSD, Novartis, and Sanofi, outside the submitted work. Dr. Hochholzer reports grants from German Heart Foundation and personal fees from AstraZeneca, The Medicines Company, Boehringer Ingelheim, and Daiichi Sankyo, outside the submitted work. Dr. Aradi reports personal fees from Roche Diagnostics, Verum Diagnostica, DSI/Lilly, AstraZeneca, Krka, from Pfizer, from Bayer AG, MSD Pharma, outside the submitted work. Drs. Schwab, Schaeffeler, Winter and Geisler are supported by the DFG Germany (grant number SCHW858/1–2) and in part by the EU Horizon 2020 UPGx grant (668353) and the Robert Bosch Stiftung Stuttgart, Germany. Dr. ten Berg reports receiving fees for board membership from AstraZeneca, consulting fees from AstraZeneca, Eli Lilly, and Merck, and lecture fees from Daiichi Sankyo and Eli Lilly, AstraZeneca, Sanofi and Accumetrics, outside the submitted work. Dr. Altman holds stock in Personalis Inc. and 23andMe, and is a paid advisor for Karius. Dr. Alexopoulos reports receiving advisory board fees from AstraZeneca, Bayer, Boehringer Ingelheim, and the Medicines Company, and lecture fees from AstraZeneca, outside the submitted work. Dr. Reny reports an unrestricted research grant from Daiichi-Sankyo and a travel grant from Bayer. Dr. Bergmeijer received a personal grant from the St. Antonius Research Fund. Dr. Shuldiner is an employee of Regeneron Pharmaceuticals, Inc.
References
- 1.Windecker S, Kolh P, Alfonso F, et al. 2014 ESC/EACTS guidelines on myocardial revascularization. EuroIntervention. 2015;10(9):1024–1094. doi: 10.4244/EIJY14M09_01. [DOI] [PubMed] [Google Scholar]
- 2.Levine GN, Bates ER, Blankenship JC, et al. 2011 ACCF/AHA/SCAI guideline for percutaneous coronary intervention: A report of the american college of cardiology Foundation/American heart association task force on practice guidelines and the society for cardiovascular angiography and interventions. Circulation. 2011;124(23):e574–651. doi: 10.1161/CIR.0b013e31823ba622. [DOI] [PubMed] [Google Scholar]
- 3.Grimfjard P, Erlinge D, Koul S, et al. Low real-world early stent thrombosis rates in ST-elevation myocardial infarction patients and the use of bivalirudin, heparin alone or glycoprotein IIb/IIIa inhibitor treatment: A nationwide swedish registry report. Am Heart J. 2016;176:78–82. doi: 10.1016/j.ahj.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 4.Yusuf S, Zhao F, Mehta SR, et al. Effects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. N Engl J Med. 2001;345(7):494–502. doi: 10.1056/NEJMoa010746. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Shen Q, Tang Y, et al. Efficacy and safety of adding clopidogrel to aspirin on stroke prevention among high vascular risk patients: A meta-analysis of randomized controlled trials. PLoS One. 2014;9(8):e104402. doi: 10.1371/journal.pone.0104402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CAPRIE Steering Committee. A randomised, blinded, trial of clopidogrel versus aspirin in patients at risk of ischaemic events (CAPRIE). CAPRIE steering committee. Lancet. 1996;348(9038):1329–1339. doi: 10.1016/s0140-6736(96)09457-3. [DOI] [PubMed] [Google Scholar]
- 7.Tantry US, Bonello L, Aradi D, et al. Consensus and update on the definition of on-treatment platelet reactivity to adenosine diphosphate associated with ischemia and bleeding. J Am Coll Cardiol. 2013;62(24):2261–2273. doi: 10.1016/j.jacc.2013.07.101. [DOI] [PubMed] [Google Scholar]
- 8.Brar SS, ten Berg J, Marcucci R, et al. Impact of platelet reactivity on clinical outcomes after percutaneous coronary intervention. A collaborative meta-analysis of individual participant data. J Am Coll Cardiol. 2011;58(19):1945–1954. doi: 10.1016/j.jacc.2011.06.059. [DOI] [PubMed] [Google Scholar]
- 9.Aradi D, Kirtane A, Bonello L, et al. Bleeding and stent thrombosis on P2Y12-inhibitors: Collaborative analysis on the role of platelet reactivity for risk stratification after percutaneous coronary intervention. Eur Heart J. 2015;36(27):1762–1771. doi: 10.1093/eurheartj/ehv104. [DOI] [PubMed] [Google Scholar]
- 10.Siller-Matula JM, Lang I, Christ G, Jilma B. Calcium-channel blockers reduce the antiplatelet effect of clopidogrel. J Am Coll Cardiol. 2008;52(19):1557–1563. doi: 10.1016/j.jacc.2008.07.055. [DOI] [PubMed] [Google Scholar]
- 11.Gilard M, Arnaud B, Cornily JC, et al. Influence of omeprazole on the antiplatelet action of clopidogrel associated with aspirin: The randomized, double-blind OCLA (omeprazole CLopidogrel aspirin) study. J Am Coll Cardiol. 2008;51(3):256–260. doi: 10.1016/j.jacc.2007.06.064. [DOI] [PubMed] [Google Scholar]
- 12.Siller-Matula JM, Trenk D, Krahenbuhl S, Michelson AD, Delle-Karth G. Clinical implications of drug-drug interactions with P2Y12 receptor inhibitors. J Thromb Haemost. 2014;12(1):2–13. doi: 10.1111/jth.12445. [DOI] [PubMed] [Google Scholar]
- 13.Hochholzer W, Trenk D, Mega JL, et al. Impact of smoking on antiplatelet effect of clopidogrel and prasugrel after loading dose and on maintenance therapy. Am Heart J. 2011;162(3):518–26. e5. doi: 10.1016/j.ahj.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Gurbel PA, Bliden KP, Logan DK, et al. The influence of smoking status on the pharmacokinetics and pharmacodynamics of clopidogrel and prasugrel: The PARADOX study. J Am Coll Cardiol. 2013;62(6):505–512. doi: 10.1016/j.jacc.2013.03.037. [DOI] [PubMed] [Google Scholar]
- 15.Kazui M, Nishiya Y, Ishizuka T, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010;38(1):92–99. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 16.Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 17.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost. 2008;6(8):1439–1441. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]
- 18.Hulot JS, Bura A, Villard E, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108(7):2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 19.Brandt JT, Close SL, Iturria SJ, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5(12):2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 20.Frere C, Cuisset T, Morange PE, et al. Effect of cytochrome p450 polymorphisms on platelet reactivity after treatment with clopidogrel in acute coronary syndrome. Am J Cardiol. 2008;101(8):1088–1093. doi: 10.1016/j.amjcard.2007.11.065. [DOI] [PubMed] [Google Scholar]
- 21.Geisler T, Schaeffeler E, Dippon J, et al. CYP2C19 and nongenetic factors predict poor responsiveness to clopidogrel loading dose after coronary stent implantation. Pharmacogenomics. 2008;9(9):1251–1259. doi: 10.2217/14622416.9.9.1251. [DOI] [PubMed] [Google Scholar]
- 22.Giusti B, Gori AM, Marcucci R, et al. Cytochrome P450 2C19 loss-of-function polymorphism, but not CYP3A4 IVS10 + 12G/A and P2Y12 T744C polymorphisms, is associated with response variability to dual antiplatelet treatment in high-risk vascular patients. Pharmacogenet Genomics. 2007;17(12):1057–1064. doi: 10.1097/FPC.0b013e3282f1b2be. [DOI] [PubMed] [Google Scholar]
- 23.Collet JP, Hulot JS, Anzaha G, et al. High doses of clopidogrel to overcome genetic resistance: The randomized crossover CLOVIS-2 (clopidogrel and response variability investigation study 2) JACC Cardiovasc Interv. 2011;4(4):392–402. doi: 10.1016/j.jcin.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: A cohort study. Lancet. 2009;373(9660):309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 25.Harmsze AM, van Werkum JW, Ten Berg JM, et al. CYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: A case-control study. Eur Heart J. 2010;31(24):3046–3053. doi: 10.1093/eurheartj/ehq321. [DOI] [PubMed] [Google Scholar]
- 26.Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: A meta-analysis. JAMA. 2010;304(16):1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rideg O, Komocsi A, Magyarlaki T, et al. Impact of genetic variants on post-clopidogrel platelet reactivity in patients after elective percutaneous coronary intervention. Pharmacogenomics. 2011;12(9):1269–1280. doi: 10.2217/pgs.11.73. [DOI] [PubMed] [Google Scholar]
- 28.Trenk D, Hochholzer W, Fromm MF, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008;51(20):1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 29.Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 30.Scott SA, Sangkuhl K, Gardner EE, et al. Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90(2):328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie HG, Kim RB, Wood AJ, Stein CM. Molecular basis of ethnic differences in drug disposition and response. Annu Rev Pharmacol Toxicol. 2001;41:815–850. doi: 10.1146/annurev.pharmtox.41.1.815. [DOI] [PubMed] [Google Scholar]
- 32.Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O'Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: Approaches to the FDA "boxed warning": A report of the american college of cardiology foundation task force on clinical expert consensus documents and the american heart association endorsed by the society for cardiovascular angiography and interventions and the society of thoracic surgeons. J Am Coll Cardiol. 2010;56(4):321–341. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Kassimis G, Davlouros P, Xanthopoulou I, Stavrou EF, Athanassiadou A, Alexopoulos D. CYP2C19*2 and other genetic variants affecting platelet response to clopidogrel in patients undergoing percutaneous coronary intervention. Thromb Res. 2012;129(4):441–446. doi: 10.1016/j.thromres.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Bouman HJ, Harmsze AM, van Werkum JW, et al. Variability in on-treatment platelet reactivity explained by CYP2C19*2 genotype is modest in clopidogrel pretreated patients undergoing coronary stenting. Heart. 2011;97(15):1239–1244. doi: 10.1136/hrt.2010.220509. [DOI] [PubMed] [Google Scholar]
- 35.Reny JL, Combescure C, Daali Y, Fontana P PON1 Meta-Analysis Group. Influence of the paraoxonase-1 Q192R genetic variant on clopidogrel responsiveness and recurrent cardiovascular events: A systematic review and meta-analysis. J Thromb Haemost. 2012;10(7):1242–1251. doi: 10.1111/j.1538-7836.2012.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mega JL, Close SL, Wiviott SD, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: A pharmacogenetic analysis. Lancet. 2010;376(9749):1312–1319. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 38.Harmsze AM, van Werkum JW, Hackeng CM, et al. The influence of CYP2C19*2 and *17 on ontreatment platelet reactivity and bleeding events in patients undergoing elective coronary stenting. Pharmacogenet Genomics. 2012;22(3):169–175. doi: 10.1097/FPC.0b013e32834ff6e3. [DOI] [PubMed] [Google Scholar]
- 39.Sibbing D, Koch W, Gebhard D, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121(4):512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 40.Lewis JP, Stephens SH, Horenstein RB, et al. The CYP2C19*17 variant is not independently associated with clopidogrel response. J Thromb Haemost. 2013;11(9):1640–1646. doi: 10.1111/jth.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott SA, Collet JP, Baber U, et al. Exome sequencing of extreme clopidogrel response phenotypes identifies B4GALT2 as a determinant of on-treatment platelet reactivity. Clin Pharmacol Ther. 2016 doi: 10.1002/cpt.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fontana P, James R, Barazer I, et al. Relationship between paraoxonase-1 activity, its Q192R genetic variant and clopidogrel responsiveness in the ADRIE study. J Thromb Haemost. 2011;9(8):1664–1666. doi: 10.1111/j.1538-7836.2011.04409.x. [DOI] [PubMed] [Google Scholar]
- 43.Hochholzer W, Trenk D, Fromm MF, et al. Impact of cytochrome P450 2C19 loss-of-function polymorphism and of major demographic characteristics on residual platelet function after loading and maintenance treatment with clopidogrel in patients undergoing elective coronary stent placement. J Am Coll Cardiol. 2010;55(22):2427–2434. doi: 10.1016/j.jacc.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 44.Shuldiner AR, O'Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302(8):849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cutlip DE, Windecker S, Mehran R, et al. Clinical end points in coronary stent trials: A case for standardized definitions. Circulation. 2007;115:2344–51. doi: 10.1161/CIRCULATIONAHA.106.685313. [DOI] [PubMed] [Google Scholar]
- 46.Wiviott SD, Braunwald E, McCabe CH, et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2007;357(20):2001–2015. doi: 10.1056/NEJMoa0706482. [DOI] [PubMed] [Google Scholar]
- 47.Wallentin L, Becker RC, Budaj A, et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2009;361(11):1045–1057. doi: 10.1056/NEJMoa0904327. [DOI] [PubMed] [Google Scholar]
- 48.Turner S, Armstrong LL, Bradford Y, et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet. 2011 doi: 10.1002/0471142905.hg0119s68. Chapter 1:Unit1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma SS, de Andrade M, Tromp G, et al. Imputation and quality control steps for combining multiple genome-wide datasets. Front Genet. 2014;5:370. doi: 10.3389/fgene.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siller-Matula JM, Lang IM, Neunteufl T, et al. Interplay between genetic and clinical variables affecting platelet reactivity and cardiac adverse events in patients undergoing percutaneous coronary intervention. PLoS One. 2014;9(7):e102701. doi: 10.1371/journal.pone.0102701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie X, Ma YT, Yang YN, et al. Personalized antiplatelet therapy according to CYP2C19 genotype after percutaneous coronary intervention: A randomized control trial. Int J Cardiol. 2013;168(4):3736–3740. doi: 10.1016/j.ijcard.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 52.Paarup Dridi N, Johansson PI, Lonborg JT, et al. Tailored antiplatelet therapy to improve prognosis in patients exhibiting clopidogrel low-response prior to percutaneous coronary intervention for stable angina or non-ST elevation acute coronary syndrome. Platelets. 2014:1–9. doi: 10.3109/09537104.2014.948837. [DOI] [PubMed] [Google Scholar]
- 53.Price MJ, Berger PB, Teirstein PS, et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: The GRAVITAS randomized trial. JAMA. 2011;305(11):1097–1105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]
- 54.Collet JP, Cuisset T, Range G, et al. Bedside monitoring to adjust antiplatelet therapy for coronary stenting. N Engl J Med. 2012;367(22):2100–2109. doi: 10.1056/NEJMoa1209979. [DOI] [PubMed] [Google Scholar]
- 55.Geisler T, Grass D, Bigalke B, et al. The residual platelet aggregation after deployment of intracoronary stent (PREDICT) score. J Thromb Haemost. 2008;6(1):54–61. doi: 10.1111/j.1538-7836.2007.02812.x. [DOI] [PubMed] [Google Scholar]
- 56.Reny JL, Fontana P, Hochholzer W, et al. Vascular risk levels affect the predictive value of platelet reactivity for the occurrence of MACE in patients on clopidogrel. systematic review and metaanalysis of individual patient data. Thromb Haemost. 2016;115(4):844–855. doi: 10.1160/TH15-09-0742. [DOI] [PubMed] [Google Scholar]
- 57.Gurbel PA, Jeong YH, Navarese EP, Tantry US. Platelet-mediated thrombosis: From bench to bedside. Circ Res. 2016;118(9):1380–1391. doi: 10.1161/CIRCRESAHA.115.307016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.