Abstract
Mutations in PITX2 have been implicated in several genetic disorders, particularly Axenfeld-Rieger syndrome. In order to determine the most reliable bioinformatics tools to assess the likely pathogenicity of PITX2 variants, the results of bioinformatics predictions were compared to the impact of variants on PITX2 structure and function. The MutPred, Provean, and PMUT bioinformatic tools were found to have the highest performance in predicting the pathogenicity effects of all 18 characterized missense variants in PITX2, all with sensitivity and specificity >93%. Applying these three programs to assess the likely pathogenicity of 13 previously uncharacterized PITX2 missense variants predicted 12/13 variants as deleterious, except A30V which was predicted as benign variant for all programs. Molecular modeling of the PITX2 homoedomain predicts that of the 31 known PITX2 variants, L54Q, F58L, V83F, V83L, W86C, W86S, and R91P alter PITX2’s structure. In contrast, the remaining 24 variants are not predicted to change PITX2’s structure. The results of molecular modeling, performed on all the PITX2 missense mutations located in the homeodomain, were compared with the findings of eight protein stability programs. CUPSAT was found to be the most reliable in predicting the effect of missense mutations on PITX2 stability. Our results showed that for PITX2, and likely other members of this homeodomain transcription factor family, MutPred, Provean, PMUT, molecular modeling, and CUPSAT can reliably be used to predict PITX2 missense variants pathogenicity.
Introduction
Paired-like homeodomain transcription factor 2 (PITX2, RefSeq NM 000325.5, MIM# 601542) is located at 4q25 and is expressed in the developing eye, brain, pituitary, lungs, heart, and gut [1]. Mutations in human PITX2 or the forkhead box transcription factor C1 (FOXC1; 6p25, RefSeq NM 001453.2, MIM# 601090) underlie the autosomal dominant disorder called Axenfeld-Rieger syndrome (ARS; MIM# 602482) [2–5]. ARS is a full penetrant, but clinically and genetically heterogeneous disorder characterized by developmental anomalies involving both ocular and non-ocular structures [6]. To date, 87 mutations within the PITX2 gene have been identified including deletions, insertions, splice-site mutations, and coding region frameshift, nonsense and missense mutations [7–13].
Identifying new disease-associated variants is becoming increasingly important for genetic testing and it is leading to a significant change in the scale and sensitivity of molecular genetic analysis [14]. One of the most frequent approaches for detecting novel variants in target genes is using direct gene sequencing. However, due to increasing number of newly identified missense variants, it is often difficult to interpret the pathogenicity of these variants as not all the mutations alter protein function, and the ones that do may also have different functional impacts in disease [15,16]. Thus, prior to detailed analyses, novel variants cannot be easily classified as either deleterious or neutral, because of their unknown functional and phenotypic consequences. Therefore, further research should be conducted to validate the genetic diagnosis when a novel missense variant is discovered. Preferably, in vitro characterization of novel variants should be undertaken; however, due to facility limitation, it is often not practicable to experimentally verify the impact of large number of mutations on protein function [17]. Another robust approach to substantiate the pathogenicity is using animal models by generating the homologous mutation that recapitulates the human phenotype; but, similar to in vitro studies, these are time-consuming, labor-intensive, difficult and expensive, making this approach unfeasible to experimentally determine the pathogenicity effects of all novel identified variants [18]. To circumvent the above mentioned limitations and to provide fast and efficient methods for predicting the functional effect of nonsynonymous variants on protein stability, structure, and function, several computational tools have been developed [19–21].
Protein stability and structure are key factors affecting function, activity, and regulation of proteins. Conformational changes are necessary for many proteins’ function and disease-causing variants can impair protein folding and stability. Missense variants are also capable of impairing protein structure, likely by affecting protein folding, protein-protein interaction, solubility or stability of protein molecules. The structural effect of mutational changes can be examined in silico on the basis of three-dimensional structure, multiple alignments of homologous sequences, and molecular dynamics [22–24]. Therefore, analysing sequence data in silico first and detecting a small number of predicted deleterious mutations for further experimental characterization is a key factor in today’s genetic and genomic studies.
In general, bioinformatics prediction methods obtain information on amino acid conservation through alignment with homologous and distantly related sequences. The most common criteria considered in many bioinformatics programs for predicting the functional effect of an amino acid substitution are amino acid sequence conservation across multiple species, physicochemical properties of the amino acids involved, database annotations, and potential protein structural changes [23,25,26]. As mentioned above, resources for in vitro and in vivo functional analysis of novel variants are constrained in most clinical laboratories. Therefore, identifying and reporting novel variants that are likely to be pathogenic often requires accurate prediction using computational tools.
In a previous study, we examined the effect of FOXC1 variants on protein structure and function by combining laboratory experiments and in silico techniques. Our results showed that integration of different algorithms with in vitro functional characterization serves as a reliable means of prioritizing, and then functional analyzing, candidate FOXC1 variants [27]. Unlike most previous studies that focused on using only PolyPhen and SIFT to predict the pathogenicity of missense mutations, here, we investigated the predictive value of SIFT, PolyPhen and nine other prediction tools by comparing their predictions to in vitro functional data for PITX2 variants. The bioinformatics programs found to be most reliable were then used to predict the likely consequences of 13 functionally-uncharacterized PITX2 variants. We also performed molecular modeling on all the PITX2 missense mutations located in the homeodomain and compared the results with the findings of protein stability algorithms to identify the most reliable tools in predicting the effect of missense mutations on PITX2 stability. To the best of our knowledge, this is the first study that incorporates the results of functional studies in conjunction with bioinformatics approaches for predicting the pathogenicity of mutations in PITX2 gene.
Materials and methods
Source of missense variants
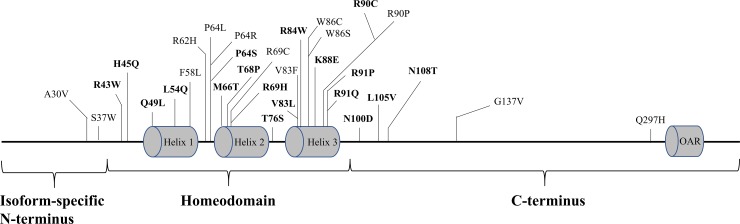
Lists of PITX2 missense variants were assembled from the previous literature and a search using the ClinVar [28], Human Gene Mutation Database (HGMD) [29], the Genome Aggregation Database (gnomAD), and the single nucleotide polymorphism database (dbSNP). This study found 47 PITX2 missense variants; 31 of which were described in the literature as being associated with ARS or coronary artery disease (CAD), while the remaining 16 variants, were considered as benign variants (Fig 1). Eighteen of the 31 variants were classified as pathogenic based on functional studies utilizing site-directed mutagenesis, expression studies, and other functional analysis (Table 1). Thirteen of 31 variants were described as associated with ARS and CAD in the absence of functional analyses on PITX2 structure or function. Sixteen SNPs, with population allele frequencies > 0.0005 were identified from the gnomAD and the ClinVar. Based upon the allele frequency (approximately 10-fold greater than the disease frequency of ARS) these have been considered benign polymorphisms. Nucleotide numbering of the mutations herein indicates cDNA numbering with +1 as the A of the ATG translation initiation codon in the NCBI reference sequence NM_000325.5, while the amino positions are based on the corresponding NCBI reference sequence NP_000316.2. This study is a retrospective case report that does not require ethics committee approval at our institution. All patients’ mutations and phenotypes were obtained from previously published studies.
Fig 1. Summary of all 31 known pathogenic missense variants in PITX2.
Characterized variants are shown in bold type.
Table 1. Position, effects on protein function and associated phenotype of previously characterised PITX2 missense variants.
| No | Variant | Exon | Domain | Phenotype | Effect on protein function | Reference |
|---|---|---|---|---|---|---|
| 1 | R43W | 5 | HD | ARS | Reduced DNA-binding and transactivational activity | Idress et al. 2006 [30] Footz et al., 2009 [31] |
| 2 | H45Q | 5 | HD | CHD | Reduced transactivational activity | Yuan et. 2013 [32] |
| 3 | Q49L | 5 | HD | TOF | Reduced transactivational activity | Sun et al. 2016 [33] |
| 4 | L54Q | 5 | HD | ARS | Reduced DNA-binding and transactivational activity | Semina et al. 1996 [34] Amendt et al. 1998 [35] |
| 5 | P64S | 5 | HD | AF | Reduced transactivational activity | Wang et al. 2014 [36] |
| 6 | M66T | 5 | HD | CHD | Reduced transactivational activity | Yuan et al. 2013 [32] |
| 7 | T68P | 5 | HD | ARS | Reduced DNA-binding and transactivational activity | Semina et al. 1996 [34] Amendt et al. 1998 [35] Amendt et al. 2000 [37] Kozlowski and Walter, 2000 [38] Espinoza et al. 2002 [39] Saadi et al. 2001 [40] |
| 8 | R69H | 5 | HD | ARS | Reduced DNA-binding activity | Kulak et al. 1998 [41] Amendt et al. 2000 [37] Strungaru et al. 2007 [42] Kozlowski and Walter, 2000 [38] |
| 9 | T76S | 5 | HD | CHD | Reduced transactivational activity | Wei et al. 2014 [43] |
| 10 | V83L | 5 | HD | ARS | Reduced DNA-binding activity, but increased transactivational activity | Priston et al. 2001 [44] |
| 11 | R84W | 5 | HD | ARS | Reduced DNA binding and transactivational activity | Alward et al. 1998 [45] Amendt et al. 2000 [37] Kozlowski and Walter, 2000 [38] Espinoza et al. 2002 [39] |
| 12 | K88E | 6 | HD | ARS | Reduced DNA binding and transactivational activity | Amendt et al. 2000 [37] Perveen et al. 2000 [46] Saadi et al. 2001 [40] |
| 13 | R90C | 6 | HD | ARS | Reduced DNA binding and transactivational activity | Perveen et al. 2000 [46] Footz et al. 2009 [31] |
| 14 | R91P | 6 | HD | ARS | Reduced DNA binding and transactivational activity | Semina et al. 1996 [34] Amendt et al. 1998 [35] Amendt et al. 2000 [37] Priston et al. 2001 [44] Kozlowski and Walter, 2000 [38] |
| 15 | R91Q | 6 | HD | CHD | Reduced transactivational activity | Wei et al. 2014 [43] |
| 16 | N100D | 6 | Downstream of HD | CHD | Reduced transactivational activity | Wang et al. 2013 [47] |
| 17 | L105V | 6 | Downstream of HD | ARS | Reduced DNA binding activity | Phillips, 2002 [48] Footz et al. 2009 [31] |
| 18 | N108T | 6 | Downstream of HD | ARS | Reduced DNA-binding activity, but increased transactivational activity | Phillips, 2002 [48] Footz et al. 2009 [31] |
AF; atrial fibrillation (AF), ARS; Axenfeld-Rieger syndrome (ARS), CHD; congenital heart disease, HD; homeodomain, TOF; tetralogy of Fallot
Predicting functional impact of missense mutation
PITX2 amino acid and DNA sequences were obtained from National Center for Biotechnology Information (NCBI) in FASTA format. The functional context of missense mutations was predicted using the default settings of eleven different in silico prediction algorithms, SIFT [49], PolyPhen-2 [50], PANTHER-PSEP [51], MutPred [52], MutationTaster [53], Provean [54], PMUT [55], FATHMM [56], nsSNPAnalyzer [57], Align GV-GD [58], and REVEL [59]. These programs were used to analyse 18 functionally characterised PITX2 missense variants plus 13 additional, functionally uncharacterized PITX2 missense variants.
SIFT program provides functional predictions for coding variants, based on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences, collected by PSI-BLAST algorithm [60]. The PolyPhen-2 (Polymorphism phenotyping-2) server predicts possible effect of an amino acid change on the structure and function of a protein using several sources of information such as straightforward physical and comparative considerations [61]. PANTHER-PSEP is a new application that analyses the length of time a given amino acid has been conserved in the lineage leading to the protein of interest. There is a direct association between the conservation time and the likelihood of functional impact [62]. MutPred is a free web-based application that utilizes a random forest algorithm with data based upon the probabilities of loss or gain of properties relating to many protein structures and dynamics, predicted functional properties, and amino acid sequence and evolutionary information [52]. MutationTaster is a tool that combines information derived from various biomedical databases and uses established analysis programs. Unlike SIFT or PolyPhen-2 which work on DNA level, MutationTaster processes substitutions of single amino acids and allows insertions and deletions [53]. Protein variation effect analyzer (PROVEAN) is a web server which uses an alignment-based score approach to generate predictions not only for single amino acid substitutions, but also for multiple amino acid substitutions, and in-frame insertions and deletions [54]. PMUT focuses on the annotation and prediction of pathological variants. PMUT is trained with a massive database of human disease-causing and neutral mutations. PMUT calculates mutational hot spots, which are provided by three different approaches, alanine scanning, genetically accessible mutations, and a very large database of mutation [55]. FATHMM, a web-server software, is able to predict not only the functional consequences coding variants, but also non-coding variants. To assess the large-scale cancer genomic datasets in a short time, FATHMM provides users with unlimited and near instant predictions for all possible amino acid substitutions within the human proteome [56]. sSNPAnalyzer uses the multiple sequence alignment and the 3D structure to evaluate the possible effect of nonsynonymous single nucleotide polymorphism (nsSNP) and also provides extra information about the SNP to aid the interpretation of results, including structural environment and multiple sequence alignment [57]. The Align-GVGD Web-based server uses the biophysical features of amino acids and protein multiple sequence alignments to predict the pathogenicity of missense variants. This tool is an extension of the original Grantham difference to multiple sequence alignments and true simultaneous multiple comparisons [58]. REVEL combines 13 individual prediction tools (MutPred, FATHMM, VEST, PolyPhen, SIFT, PROVEAN, MutationAssessor, MutationTaster, LRT, GERP++, SiPhy, phyloP, and phastCons) as features to predict the pathogenicity of missense variants. REVEL was trained with recently discovered pathogenic and rare neutral missense variants [59]. Please see Table 2 for more information on the prediction tools used in this study.
Table 2. Amino acid substitution (AAS) prediction methods used in this study.
| Program | Input | Algorithm | Output | URL | Reference |
|---|---|---|---|---|---|
| SIFT | PS and AAS, protein sequence alignment and AAS, dbSNP id, or protein id | Uses sequence homology, scores assessment is based on position-specific scoring matrices with Dirichlet priors | Score ranges from 0 to 1, where < = 0.05 is damaging and >0.05 is tolerated | http://sift.jcvi.org/www/SIFT_enst_submit.html | Ng and Henikoff, 2001 [63] |
| PolyPhen-2 | PS and AAS, dbSNP id, HGVbASE id, or protein id | Uses sequence conservation and structure to model location of amino acid substitution, Swiss-Prot and TrEMBL annotation | Score ranges from 0 to 1, where < = 0.05 is benign, and >0.05 is damaging | http://genetics.bwh.harvard.edu/pph2/ | Ramensky et al. 2002 [50] |
| PANTHER-PSEP | PS and AAS | Uses sequence homology; scores are based on PANTHER Hidden Markov Model families | Probably damaging: time > 450my possibly damaging: 450my > time > 200my probably benign: time < 200my) | http://www.pantherdb.org/tools/csnpScoreForm.jsp | Tang and Thomas, 2016 [64] |
| MutPred | Protein id, PS, or multiple sequence alignment | Prediction is based on one of two neural networks which uses internal databases, secondary structure prediction, and sequence conservation | Score ranges from 0 to 1, where 0 is polymorphism and high scores are predicted to be deleterious/disease-associated | http://mutpred.mutdb.org/ | Li et al. 2009 [65] |
| MutatioTaster | DNA sequence | Predictions are calculated by a naive Bayes classifier, which predicts the disease potential | Prediction is based one of four possible types: a) disease causing: probably deleterious b) disease causing automatic: known to be deleterious c) polymorphism: probably harmless d) polymorphism automatic: known to be harmless | http://www.mutationtaster.org/ | Schwarz et al. 2014 [53] |
| Provean | PS and AAS | Uses an alignment-based score approach to generate predictions not only for single amino acid substitutions, but also for multiple amino acid substitutions, and in-frame insertions and deletions | the default score threshold is currently set at -2.5, in which >-2.5 is neutral, and <-2.5 is deleterious | http://provean.jcvi.org/index.php | Choi and Chan, 2015 [54] |
| PMUT | PS and AAS, dbSNP, Uniprot or PDB ID of protein | Based on the application of neural networks which uses internal databases, secondary structure prediction, and sequence conservation | Score ranges from 0 to 1, where <0.50 is neutral and >0.50 is disease associated | http://mmb.pcb.ub.es/pmut2017/analyses/new/ | Ferrer-Costa et al. 2002 [55] |
| FATHMM | protein identifier and the amino acid substitution, dbSNP id | Uses sequence homology | The score threshold is set at -2.5, in which >-2.5 is neutral, and <-2.5 is deleterious | http://fathmm.biocompute.org.uk/index.html | Shihab et al. 2013 [56] |
| nsSNPAnalyzer | Protein sequence in FASTA format and a substitution file denoting the SNP identities to be analyzed | Uses information contained in the multiple sequence alignment and information contained in the three-dimensional protein structure to make predictions. | Normalized probability of the substitution calculated by the SIFT program | http://snpanalyzer.uthsc.edu/ | Bao et al. 2005 [57] |
| Align GV-GD | Protein sequence in FASTA format and a substitution file denoting the SNP identities to be analyzed | Uses biophysical features of amino acids and protein multiple sequence alignments | A value of C > 0 was considered deleterious; otherwise a variant was neutral | http://agvgd.hci.utah.edu/ | Tavtigian et al. 2006 [58] |
| REVEL | Precomputed REVEL scores are provided for all possible human missense variants | Prediction is based on a combination of scores from 13 individual tools | Score ranges from 0 to 1, where <0.50 is neutral and >0.50 is pathogenic | https://sites.google.com/site/revelgenomics/ | Ioannidis et al. 2016 [59] |
AAS; amino acid sequences, PS; protein sequence, PDB, protein data bank
Molecular modeling of the mutant protein structure
The NMR structure of the homeodomain of PITX2 complexed with a TAATCC DNA binding site (PDB: 2LKX) were analyzed by the SWISS-MODEL server (http://www.expasy.org/spdbv/; provided in the public domain by the Swiss Institute of Bioinformatics, Geneva, Switzerland). Model structures of wild-type and mutants were created in Swiss-Pdb Viewer and investigated using the ANOLEA server (http://melolab.org/anolea). For structure predictions of PITX2, sequence in FASTA format was obtained from NCBI database (NP_001191327.1).
Calculating changes in protein stability
Eight different protein stability programs (DUET, SDM, mCSM I-Mutant3.0, MUpro, iPTREE-STAB, CUPSAT, and iStable) were used to predict the effects of missense mutations on the stability of PITX2 protein. DUET is a web server that uses integrated computational approach to predict effect of missense mutations on protein stability [66]. DUET calculation is based on complementary data regarding the mutation including secondary structure [67] and a pharmacophore vector [68]. SDM, a computational method, has been demonstrated as the most appropriate method to use along with many other programs. SDM assesses the amino acid substitution occurring at specific structural environment that are tolerated within the family of homologous proteins of defined three dimensional structures and change them into substitution probability tables [69]. mCSM relies on graph-based signature concept and predicts not only the effect of single-point mutations on protein stability, but also protein–protein and protein–nucleic acid binding [70]. I-Mutant3.0 is a neural-network-based web server that predicts automatically protein stability changes upon single point protein mutations based on either protein sequence or protein structure. I-Mutant3.0 can predict the severity effect of a mutation on the stability of the folded protein [71]. MUpro is a set of machine learning programs that accurately calculates protein stability alterations based on primary sequence information particularly where the tertiary structure is unrevealed, overcoming a major restriction of previous methods which are based on the tertiary structure [72]. iPTREE-STAB is a web service and mainly provides two function modules of services including discriminating the stability of a protein upon single amino acid substitutions and predicting their numerical stability values [73]. CUPSAT uses protein environment specific mean force potentials (through solvent accessibility and secondary structure specificity) to analyse and predict protein stability changes upon point mutations [74]. iStable, a combined predictor, was designed by using sequence information and prediction data from various element predictors. iStable is available with two different input types: structural and sequential [75]. Please see Table 3 for more information on the stability predictors used in this study.
Table 3. Protein stability prediction methods used in this study.
| Program | Input | Algorithm | Output | URL | Reference |
|---|---|---|---|---|---|
| DUET | Protein structure | Uses SVM regression with a Radial Basis Function kernel, and RSA | Score ranges from negative to positive numbers, where negative number denote destabilizing, and positive number denote stabilizing | http://bleoberis.bioc.cam.ac.uk/duet/ | Pires et al. 2014 [66] |
| SDM | Protein structure | Uses conformationally constrained environment-specific substitution tables (ESSTs) | Score ranges from negative to positive numbers, where negative number denote destabilizing, and positive number denote stabilizing | http://131.111.43.103/prediction | Pandurangan et al. 2017 [69] |
| mCSM | Protein structure | Uses the concept of graph-based structural signatures | Score ranges from negative to positive numbers, where negative number denote destabilizing, and positive number denote stabilizing | http://biosig.unimelb.edu.au/mcsm/protein_protein | Pires et al. 2014 [70] |
| I-Mutant3.0 | Protein sequence alone or protein structure | Using SVM regression with a Radial Basis Function kernel, and RSA | Score ranges from negative to positive numbers, where negative number denote destabilizing, and positive number denote stabilizing | http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi | Capriotti et al. 2006 [71] |
| MUpro | Protein sequence | Uses feed-forward neural networks and SVMs | A score near 0 means unchanged stability. Score near -1 means high confidence in decreased stability. Score near +1 means high confidence in increased stability | http://www.ics.uci.edu/~baldig/mutation.html | Cheng et al. 2006 [72] |
| iPTREE-STAB | Protein sequence | Based on the neighboring residues of short window length | Score ranges from negative to positive numbers, where negative number denote destabilizing, and positive number denote stabilizing | http://210.60.98.19/IPTREEr/iptree.htm | Huang et al. 2007 [76] |
| CUPSAT | Existing PDB structures or custom protein structures | Uses structural environment specific atom potentials and torsion angle potentials | Score ranges from negative to positive numbers, where negative number denote destabilizing, and positive number denote stabilizing | http://cupsat.tu-bs.de/ | Parthiban et al. 2006 [74] |
| iStable | Protein sequence or PDB structure (PDB ID) | Uses SVM | Score ranges from negative to positive numbers, where negative number denote destabilizing, and positive number denote stabilizing | http://predictor.nchu.edu.tw/istable/indexSeq.php | Chen et al. 2013 [75] |
RSA; residue relative solvent accessibility, SVM; support vector machine
Variants classification
Previous analyses of missense variations in different human diseases predicted that the stability margin without any immediate effect on protein fitness is 1–3 kcal mol-1 [77–79]. Mutations that reduce the protein stability by >2 kcal mol-1 contribute to severe disease phenotypes [80,81]. Therefore, in this study, all variations were classified as predicted to be neutral (-1.5 < ΔΔG < 1.5), stabilizing (ΔΔG > 1.5) or destabilizing (ΔΔG < -1.5).
Results
Bioinformatics functional predictions
The protein sequence and/or protein structure with mutational position and amino acid residue of 18 previously functionally characterized pathogenic PITX2 missense variants, plus 16 SNPs with a population frequency of higher than 0.05% (thus considered benign polymorphisms), were used to test the predictive value of eleven common bioinformatics prediction programs; SIFT, PolyPhen-2, PANTHER-PSEP, MutPred, MutationTaster, Provean, PMUT, FATHMM, nsSNPAnalyzer, Align GV-GD, and REVEL (Table 4 and Table 5). To evaluate the performances of the programs, seven measures (sensitivity, specificity, accuracy, precision, positive predictive value (PPV), negative predictive value (NPV), and Matthews correlation coefficient (MCC)) were calculated by comparing the results of all programs with previously generated functional data.
Table 4. Functional characterization vs. bioinformatics programs.
Comparison of in silico program predictions of degrees of tolerance for 18 functionally characterized PITX2 missense mutation.
| No | Missense variants | SIFT Score | PolyPhen-2 Score | MutPred Score | MutationTaster Score | Provean Score | PANTHER-PSEP Score | PMUT Score | FATHMM Score | nsSNPAnalyzer Score | Align GV-GD Score | REVEL Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | R43W | 0 (√) | 0.003 (×) | 0.952 (√) | 101 (√) | -7.125 (√) | PD (√) | 0.91 (√) | -5.53 (√) | 0.00 (√) | C65 (√) | 0.599 (√) |
| 2 | H45Q | 0.21 (×) | 1.000 (√) | 0.672 (√) | 24 (√) | -7.176 (√) | PD (√) | 0.87 (√) | -3.86 (√) | 0.15 (×) | C15 (√) | 0.903 (√) |
| 3 | Q49L | 0.25 (×) | 0.995 (√) | 0.598 (√) | 113 (√) | -6.498 (√) | PD (√) | 0.76 (√) | -3.96 (√) | 0.84 (×) | C65 (√) | 0.555 (√) |
| 4 | L54Q | 0 (√) | 0.997 (√) | 0.959 (√) | 113 (√) | -5.598 (√) | PD (√) | 0.91 (√) | -6.23 (√) | 0.00 (√) | C65 (√) | 0.856 (√) |
| 5 | P64S | 0 (√) | 0.999 (√) | 0.867 (√) | 74 (√) | -7.547 (√) | PD (√) | 0.85 (√) | -5.05 (√) | 0.00 (√) | C65 (√) | 0.908 (√) |
| 6 | M66T | 0 (√) | 0.995 (√) | 0.566 (√) | 81 (√) | -5.555 (√) | PD (√) | 0.88 (√) | -3.64 (√) | 0.01 (√) | C65 (√) | 0.595 (√) |
| 7 | T68P | 0 (√) | 0.946 (√) | 0.854 (√) | 38 (√) | -5.094 (√) | PD (√) | 0.87 (√) | -3.81 (√) | 0.01 (√) | C35 (√) | 0.747 (√) |
| 8 | R69H | 0 (√) | 0.007 (×) | 0.985 (√) | 29 (√) | -4.733 (√) | PD (√) | 0.90 (√) | -4.42 (√) | 0.00 (√) | C25 (√) | 0.752 (√) |
| 9 | T76S | 0 (√) | 0.995 (√) | 0.655 (√) | 58 (√) | -3.652 (√) | PD (√) | 0.89 (√) | -4.01 (√) | 0.01 (√) | C55 (√) | 0.706 (√) |
| 10 | V83L | 0.01 (√) | 0.902 (√) | 0.944 (√) | 32 (√) | -2.758 (√) | PD (√) | 0.89 (√) | -4.91 (√) | 0.14 (×) | C25 (√) | 0.812 (√) |
| 11 | R84W | 0 (√) | 0.994 (√) | 0.841 (√) | 101 (√) | -7.350 (√) | PD (√) | 0.88 (√) | -4.01 (√) | 0.00 (√) | C65 (√) | 0.907 (√) |
| 12 | K88E | 0 (√) | 0.008 (×) | 0.828 (√) | 56 (√) | -3.800 (√) | PD (√) | 0.88 (√) | -3.92 (√) | 0.04 (√) | C55 (√) | 0.725 (√) |
| 13 | R90C | 0 (√) | 0.957 (√) | 0.975 (√) | 180 (√) | -7.599 (√) | PD (√) | 0.91 (√) | -4.45 (√) | 0.00 (√) | C65 (√) | 0.816 (√) |
| 14 | R91P | 0 (√) | 0.998 (√) | 0.959 (√) | 103 (√) | -6.649 (√) | PD (√) | 0.91 (√) | -5.73 (√) | 0.00 (√) | C65 (√) | 0.727 (√) |
| 15 | R91Q | 0 (√) | 0.997 (√) | 0.918 (√) | 43 (√) | -3.800 (√) | PD (√) | 0.91 (√) | -5.71 (√) | 0.00 (√) | C35 (√) | 0.726 (√) |
| 16 | N100D | 0.2 (×) | 0.863 (√) | 0.365 (×) | 23 (√) | -4.013 (√) | PD (√) | 0.81 (√) | -3.65 (√) | 0.20 (×) | C15 (√) | 0.861 (√) |
| 17 | L105V | 0.06 (×) | 0.974 (√) | 0.788 (√) | 32 (√) | -1.894 (×) | PD (√) | 0.80 (√) | -3.16 (√) | 0.27 (×) | C25 (√) | 0.861 (√) |
| 18 | N108T | 0.24 (×) | 0.990 (√) | 0.789 (√) | 65 (√) | -3.332 (√) | PD (√) | 0.68 (√) | -3.05 (√) | 0.35 (×) | C55 (√) | 0.443 (×) |
PD; probably damaging
√ correspond to functional characterization; ×, do not correspond to functional characterization.
Table 5. In silico analysis of the effect of 16 PITX2 benign variants.
| No | Missense variants | SIFT Score | PolyPhen-2 Score | MutPred Score | MutationTaster Score | Provean Score | PANTHER-PSEP Score | PMUT Score | FATHMM Score | nsSNPAnalyzer Score | Align GV-GD Score | REVEL Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P41S | 0.25 (√) | 0.002 (√) | 0.201 (√) | 74 (×) | 0.325 (√) | PD (×) | 0.16 (√) | -2.80 (×) | 0.38 (√) | C65 (×) | 0.237 (√) |
| 2 | Q75P | 0.33 (√) | 0.000 (√) | 0.371 (√) | 76 (×) | -0.065 (√) | PD (×) | 0.14 (√) | -2.77 (×) | 0.27 (√) | C65 (×) | 0.503 (×) |
| 3 | V81M | 0.11 (√) | 0.459 (×) | 0.063 (√) | 21 (√) | -0.023 (√) | PB (√) | 0.08 (√) | -2.74 (×) | 0.14 (√) | C15 (×) | 0.501 (×) |
| 4 | A188T | 0.68 (√) | 0.027 (√) | 0.329 (√) | 58 (×) | -1.018 (√) | PD (×) | 0.04 (√) | -2.86 (×) | 0.57 (√) | C55 (×) | 0.151 (√) |
| 5 | M207V | 0.62 (√) | 0.069 (√) | 0.386 (√) | 21 (×) | -1.461 (√) | PD (×) | 0.15 (√) | -2.61 (×) | 0.47 (√) | C15 (×) | 0.185 (√) |
| 6 | R203C | 0 (×) | 0.968 (×) | 0.150 (√) | 81 (×) | -0.455 (√) | PB (√) | 0.03 (√) | -2.51 (×) | 0.12 (√) | C65 (×) | 0.146 (√) |
| 7 | Q193T | 0.38 (√) | 0.270 (√) | 0.196 (√) | 84 (×) | 0.630 (√) | PD (×) | 0.02 (√) | -.2.60 (×) | 0.00 (×) | C35 (×) | 0.152 (√) |
| 8 | Y131D | 0 (×) | 0.812 (×) | 0.121 (√) | 35 (×) | -0.010 (√) | PD (×) | 0.03 (√) | -1.64 (√) | 0.01 (×) | C65 (×) | 0.251 (√) |
| 9 | G166D | 0 (×) | 0.557 (×) | 0.283 (√) | 76 (×) | -0.357 (√) | PB (√) | 0.31 (√) | -2.58 (×) | 0.01 (×) | C65 (×) | 0.224 (√) |
| 10 | H151Y | 0 (×) | 0.512 (×) | 0.045 (√) | 75 (×) | -2.400 (√) | PD (×) | 0.03 (√) | -2.62 (×) | 0.01 (×) | C65 (×) | 0.169 (√) |
| 11 | I138F | 0 (×) | 0.671 (×) | 0.056 (√) | 57 (×) | -0.390 (√) | PB (√) | 0.2 (√) | -2.69 (×) | 0.04 (×) | C0 (√) | 0.145 (√) |
| 12 | G205S | 0.68 (√) | 0.017 (√) | 0.193 (√) | 87 (×) | -0.372 (√) | PB (√) | 0.15 (√) | -2.62 (×) | 0.78 (√) | C55 (×) | 0.147 (√) |
| 13 | G186R | 0 (×) | 1.000 (×) | 0.212 (√) | 48 (×) | -1.092 (√) | PD (×) | 0.08 (√) | -2.86 (×) | 0.00 (×) | C65 (×) | 0.103 (√) |
| 14 | H57Q | 0 (×) | 0.736 (×) | 0.056 (√) | 63 (×) | -3.010 (×) | PB (√) | 0.03 (√) | -2.55 (×) | 0.00 (×) | C15 (×) | 0.112 (√) |
| 15 | A246D | 0 (×) | 1.000 (×) | 0.440 (√) | 44 (×) | -0.769 (√) | PD (×) | 0.18 (√) | -2.65 (×) | 0.01 (×) | C65 (×) | 0.181 (√) |
| 16 | G148W | 0 (×) | 0.844 (×) | 0.195 (√) | 65 (×) | -0.242 (√) | PB (√) | 0.31 (√) | -2.74 (×) | 0.04 (×) | C65 (×) | 0.109 (√) |
PB; probably benign, PD; probably damaging
√ correspond to functional characterization; ×, do not correspond to functional characterization.
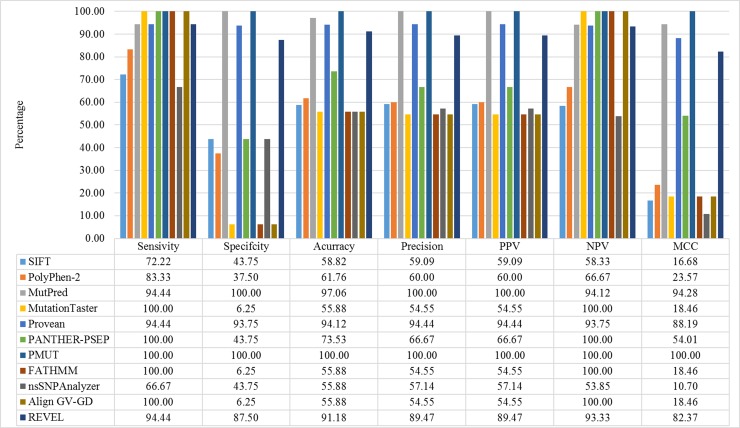
For PITX2, MutPred, Provean, and PMUT were the most reliable of the bioinformatics tools in predicting the pathogenicity effects of all 18 functionally characterized missense variants in PITX2, with sensitivity and specificity of > 93% (Fig 2). Then, REVEL tool showed high sensitivity and specificity, 94.44% and 87.50%, respectively. Analysis of the sensitivity and specificity SIFT showed that this program had good sensitivity (72.22%) but low specificity (43.75%). Although PolyPhen-2, MutationTaster, PANTHER-PSEP, FATHMM, and Align GV-GD exhibited over 83% sensitivity, they were unable to identify the benign polymorphisms, showing specificity of 37.50%, 6.25%, 43.75%, 6.25%, and 6.25%, respectively. The predictive value of nsSNPAnalayzer was similar to that of SIFT program, with sensitivity and specificity of 66.67% and 43.75%, respectively.
Fig 2. Reliability of eleven in silico programs used to analyze all 18 functionally characterized missense variants in PITX2.
True positives (TP) are missense variants correctly predicted to disrupt PITX2 protein function, and false negatives (FN) are those incorrectly predicted to be benign or tolerated. True negatives (TN) are neutral variants correctly predicted as benign or tolerated and false positives (FP) are neutral variants incorrectly predicted to disrupt PITX2 protein function. The total of variants for all methods was 34, 18 pathogenic variants and 16 benign variants. Values were converted to percentage. Values were converted to percentage. The statistics used were calculated as follows: Sensitivity = TP/(TP + FN); Specificity = TN/(TN + FP); Accuracy = (TP + TN)/(TP + TN + FP + FN); Precision = TP/(TP + FP); Negative predictive value (NPV) = TN/(TN + FN); Positive predictive value (PPV) = TP/(TP + FP); Matthews correlation coefficient (MCC) = (TP × TN − FP × FN)/-([TP + FP] × [TP + FN] × [TN + FP] × [TN + FN]R).
The most reliable programs found in this study’s analyses (MutPred, Provean, and PMUT) were then used to predict the likely pathogenicity of 13 PITX2 missense variants for which functional testing has not been performed (Table 6). Interestingly, the A30V variant unanimously was predicted as benign by all three programs. The remaining 12 PITX2 variants were predicted to be disease-associated mutations by all programs.
Table 6. Bioinformatics prediction of the degree of tolerance for 13 functionally uncharacterized PITX2 missense variants.
| No | Missense variants | References | Phenotype | Mutpred Score | Provean Score | PMUT Score |
|---|---|---|---|---|---|---|
| 1 | A30V | Zaidi et al. 2013 [82] | CHD | B, 0.152 | B, -0.948 | B, 0.10 |
| 2 | S37W | Yang et al. 2013 [83] | AF | PD, 0.503 | PD, -1.074 | PD, 0.81 |
| 3 | F58L | Vieira et al. 2006 [84] D'haene et al. 2011 [85] | ARS | PD, 0.947 | PD, -5.560 | PD, 0.90 |
| 4 | R62H | Amendt et al. 2000 [37] Xia et al. 2004 [86] | ARS | PD, 0.856 | PD, -4.686 | PD, 0.70 |
| 5 | P64L | Phillips JC, 2002 [48] Weisschuh et al. 2006 [87] Meyer-Marcotty et al. 2008 [88] Dressler et al. 2010 [89] | ARS | PD, 0.973 | PD, -9.421 | PD, 0.81 |
| 6 | P64R | Weisschuh et al. 2006 [87] | ARS | PD, 0.944 | PD, -8.496 | PD, 0.84 |
| 7 | R69C | Kimura et al. 2014 [90] | ARS | PD, 0.960 | PD, -7.575 | PD, 0.91 |
| 8 | V83F | Reis et al. 2012 [91] | ARS | PD, 0.912 | PD, -4.643 | PD, 0.91 |
| 9 | W86S | Dandan et al. 2008 [92] | ARS | PD, 0.868 | PD, -13.298 | PD, 0.91 |
| 10 | W86C | Reis et al. 2012 [91] | ARS | PD, 0.950 | PD, -12.282 | PD, 0.91 |
| 11 | R90P | Phillips JC, 2002 [48] | ARS | PD, 0.960 | PD, -6.649 | PD, 0.91 |
| 12 | G137V | Kniestedt et al. 2006 [93] | ARS | PD, 0.816 | B, -1.902 | PD, 0.61 |
| 13 | Q297H | Huang et al. 2015 [94] | ARS | PD, 0.682 | PD, -3.966 | PD, 0.91 |
AF; atrial fibrillation (AF), ARS; Axenfeld-Rieger syndrome (ARS), ASMD; Anterior segment mesenchymal dysgenesis, B; benign, CHD; congenital heart disease, PD; probably damaging
Molecular modeling of PITX2
Molecular models for the homeodomain of wild-type and variant-containing PITX2 proteins were designed using threading algorithms to assess impairment of PITX2 structure by missense variants.
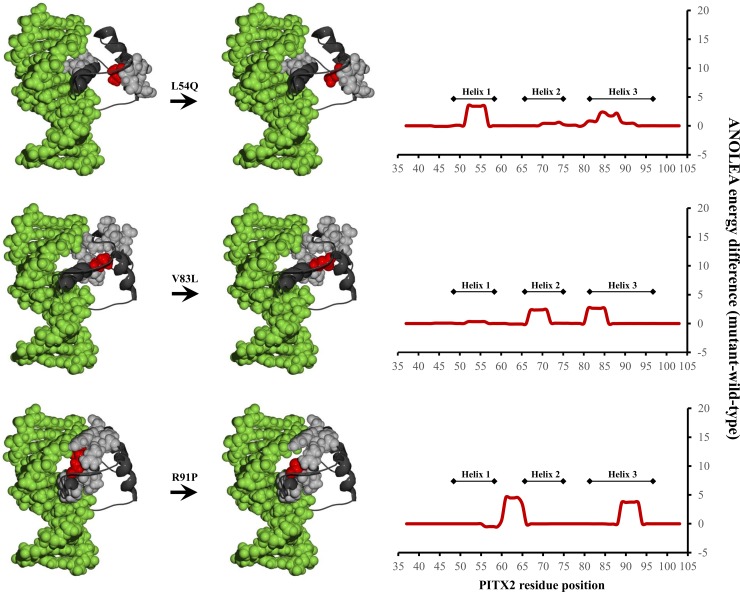
Three functionally characterised variants, N100D, L105V, and N108T, were excluded from these molecular modeling analyses since they are not located in the homeodomain, which is the only portion of PITX2 with a known structure. Wild-type amino acids were changed to variant residues to determine putative structural effects of the remaining 15 functionally analysed PITX2 variants through ANOLEA mean force potential calculations. The molecular modeling identified three mutations as high-risk (L54Q, V83L, and R91P) to change the structure of PITX2, particularly in the H1, H2, and H3 subdomains (Fig 3). The R91P variant was predicted to grossly disrupt the non-local amino acid side chain contacts. Similar, although less profound, effects were predicted when L54 and V83 were altered to glutamine and leucine, respectively. In contrast, the remaining twelve amino acid variants showed no predicted substantially altered pairwise interactions, indicating that these missense variants are predicted to have minor or no effects on PITX2’s structure (S1 Fig).
Fig 3. Homology models (left) and scatterplots (right) of in silico analyses of the L54Q, V83L, and R91P variants in the PITX2 gene.
The 3D model of PITX2 is presented with the protein backbone depicted in black ribbon, the co-crystallized DNA binding target in space-filled green model and the mutants positions in red. The wild-type and mutant-equivalent models were analyzed by the atomic nonlocal environment assessment (ANOLEA) server. Peaks on the scatterplots show the positions of amino acids that changed their pseudoenergy state, as a consequence of the mentioned variants.
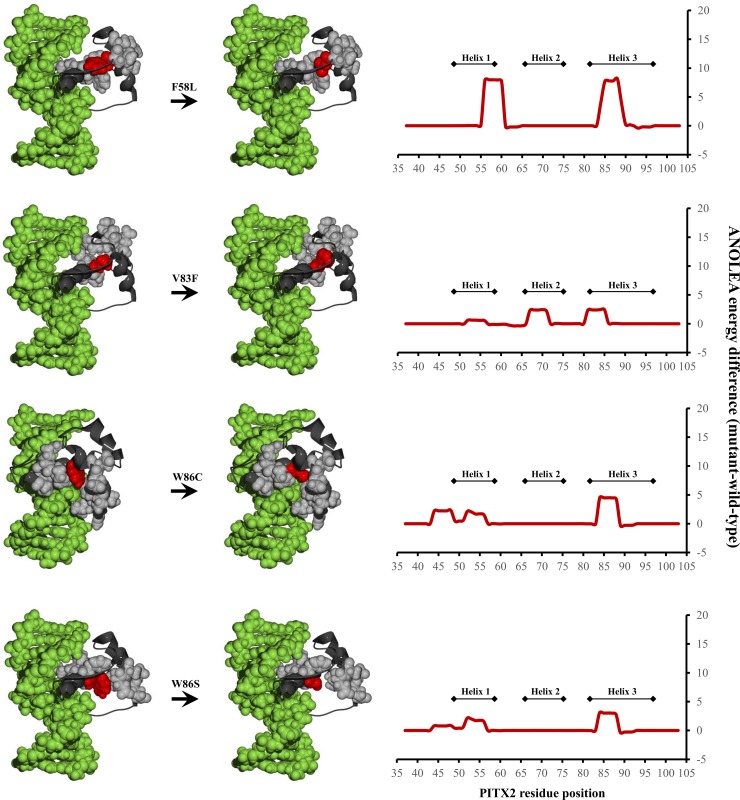
Molecular modeling was also performed on the nine functionally uncharacterised PITX2 missense mutations located in the homeodomain. Four mutations (F58L, V83F, W86C, W86S) were predicted to change the structure of PITX2 (Fig 4), while, the remaining five variants (R62H, P64L, P64R, R69C, and R90P) were predicted to have minor or no impact on PITX2’s structure (S2 Fig).
Fig 4. Homology models (left) and scatterplots (right) of in silico analyses of the F58L, V83F, W86C, and W86S variants in the PITX2 gene.
The 3D model of PITX2 is presented with the protein backbone depicted in black ribbon, the co-crystallized DNA binding target in space-filled green model and the mutants positions in red. The wild-type and mutant-equivalent models were analyzed by the atomic nonlocal environment assessment (ANOLEA) server. Peaks on the scatterplots show the positions of amino acids that changed their pseudoenergy state, as a consequence of the mentioned variants.
Evaluation of the different algorithms in predicting stability changes
To assess the performance of eight different stability predictor programs (DUET, SDM, mCSM, I-Mutant3.0, MUpro, iPTREE-STAB, CUPSAT, and iStable) in predicting the effect of missense mutations on PITX2 protein stability, the change in protein stability (ΔΔG) were computed for all 24 PITX2 homeodomain variants (15 functionally characterised and 9 functionally uncharacterised mutations) (Table 7).
Table 7. Evaluation of stability changes of 15 functionally characterized and 9 functionally uncharacterized PITX2 homeodomain missense variants using eight different protein stability prediction programs.
| No. | Variations | DUET | SDM | mCSM | I-Mutant3.0 SEQ | I-Mutant3.0 Structure | MUpro | iPTREE-STAB | CUPSAT | iStable |
|---|---|---|---|---|---|---|---|---|---|---|
| Characterised variants | ||||||||||
| 1 | R43W | -1.773 | 0.35 | -0.97 | 0.00 | -0.13 | -0.162 | 0.0337 | -75.87 | 0.0077 |
| 2 | H45Q | 0.158 | -0.15 | 0.027 | 0.07 | 0.17 | -0.112 | -2.9050 | 19.1 | 0.3529 |
| 3 | Q49L | 0.471 | 0.29 | 0.186 | 0.38 | 0.68 | 1 | 0.9422 | -5.82 | 0.6946 |
| 4 | L54Q* | -2.892 | -2.3 | -2.73 | -1.65 | -1.50 | -1 | -1.8488 | 3.85 | -0.9075 |
| 5 | P64S | -2.069 | -1.27 | -1.97 | -1.59 | -1.57 | -1 | -1.0233 | -45.88 | -0.9568 |
| 6 | M66T | 0.444 | -0.67 | 0.181 | -1.20 | -0.32 | -1 | 1.0943 | 10.62 | -0.1104 |
| 7 | T68P | -0.359 | -0.34 | -0.361 | -0.90 | -0.68 | 0.155 | -1.0594 | 0.38 | 0.2945 |
| 8 | R69H | -2.369 | -0.15 | -2.147 | -1.56 | -1.29 | -0.633 | -1.3667 | 8.49 | -0.7126 |
| 9 | T76S | -1.35 | -0.79 | -1.211 | -0.69 | -0.26 | -0.014 | 0.9377 | -16.05 | -0.0892 |
| 10 | V83L* | -0.305 | 0.09 | -0.44 | -0.91 | -0.72 | 0.224 | -1.3883 | -2.72 | -0.3060 |
| 11 | R84W | -1.056 | -0.06 | -1.125 | -0.52 | 0.41 | -0.966 | -2.9167 | -23.81 | -0.1240 |
| 12 | K88E | -1.759 | 0.87 | -1.777 | -0.32 | -0.24 | -0.024 | -0.9691 | 8.8 | -0.1765 |
| 13 | R90C | -2.014 | -0.49 | -2.019 | -0.86 | -0.89 | -0.567 | -0.6385 | -19.6 | -0.4268 |
| 14 | R91P* | -2.225 | -2.25 | -1.777 | -0.82 | -0.93 | -1 | -2.7464 | -75.47 | -0.6208 |
| 15 | R91Q | -1.308 | -0.08 | -1.3 | -0.95 | -1.04 | -1 | 0.3362 | 37.96 | -0.4777 |
| Uncharacterised variants | ||||||||||
| 1 | F58L* | -0.868 | 0.64 | -0.882 | -0.69 | -0.71 | 0.446 | -1.3492 | 8.37 | 0.4267 |
| 2 | R62H | -1.839 | 0.2 | -1.757 | -1.24 | -1.17 | -0.634 | -2.1794 | -1.72 | -0.6073 |
| 3 | P64L | -0.55 | 0.32 | -0.845 | -0.07 | -0.64 | -0.260 | -4.1000 | -5.02 | -0.1755 |
| 4 | P64R | -0.979 | -2.07 | -0.944 | -0.83 | -1.09 | -0.892 | -0.8385 | -13.21 | -0.5091 |
| 5 | R69C | -2.183 | 0.23 | -2.107 | -1.12 | -1.07 | -0.183 | 0.2429 | 0.51 | -0.5278 |
| 6 | V83F* | -1.437 | -1.32 | -1.265 | -1.16 | -1.12 | -0.496 | -1.3883 | -10.94 | -0.6159 |
| 7 | W86S* | -2.327 | -2.67 | -2.514 | -1.64 | -1.55 | -1 | -0.6167 | -31.26 | -1.0663 |
| 8 | W86C* | -0.931 | -1.57 | -1.018 | -1.52 | -1.40 | -0.971 | 0.6923 | -12.15 | -0.8733 |
| 9 | R90P | -1.623 | -2.25 | -1.319 | -0.71 | -0.74 | -0.346 | -2.8825 | -23.89 | -0.3739 |
*Predicted by molecular modeling to destabilize the structure and function of PITX2 protein.
Of these eight programs, CUPSAT was the most consistent with the results of our molecular modeling, by identifying 5 of 7 destabilizing mutations that were also predicted to be destabilizing by molecular modeling (V83L, V83F, W86S, W86C, and R91P).
SDM also showed high consistency with the results of our molecular modeling, by detecting 4 of 7 destabilizing mutations that were also predicted to be destabilizing by molecular modeling (L54Q, R91P, W86S, and W86C). DUET, mCSM, and I-Mutant3.0 identified 3 and iPTREE-STAB detected 2 of 7 destabilizing mutations detected by molecular modeling. MUpro and iStable were unable to identify any of the 7 destabilizing mutations predicted by molecular modeling.
Discussion
Although in silico programs are not a substitute for wet-lab experiments, they can provide a supportive role in the experimental validation of disease-associated alleles and can help further diagnostic strategies by prioritizing the most likely pathogenic novel variants.
While many tools are available for assessing the functional significance of variants, determining the reliability of prediction results is challenging. In this context, the current study investigated the combination of experimental findings, molecular modeling, in silico mutation prediction programs, and stability prediction software to assess the pathogenicity of PITX2 missense variants. In silico methods that correctly identify deleterious variants do not always inevitably work well for benign predictions. The methods determined by this study to be preferred for analyses of PITX2 variants were those best able to distinguish both pathogenic and benign variants, thus yielding the highest accuracy.
Our results showed that MutPred, Provean, and PMUT tools were the most accurate in predicting pathogenicity of PITX2 missense variants (Fig 2). The sensitivity and specificity of these three tools in recognizing PITX2 disease-causing variants were over 93%, indicating the strong performance of these programs in identifying as pathogenic only PITX2 variants with significant functional defects. After these three tools, REVEL showed highest sensitivity and specificity, 94.44% and 87.50%, respectively. SIFT showed good sensitivity (72.22%) but low specificity (43.75%). PolyPhen-2, MutationTaster and PANTHER-PSEP, FATHMM, and Align GV-GD demonstrated > 83% sensitivity, but, they were unable to identify the benign polymorphisms, showing the specificity of 37.50%, 6.25%, 43.75%, 6.25%, and 6.25%, respectively. The predictive value of nsSNPAnalayzer was similar to that of SIFT program, with sensitivity and specificity of 66.67% and 43.75%, respectively. Our results showed, therefore, that MutPred, Provean, and PMUT can be utilized with high confidence to test whether or not a PITX2 missense variant is likely to be deleterious. Interestingly, MutPred was the only in silico program that ranked in the top three programs in identifying both pathogenic and benign PITX2 and FOXC1 variants [27]. A likely explanation for MutPred’s high ranking is that it evaluates the most factors in making assessments. However, since the number of variants available for testing in this study were small, a larger dataset would confirm that our results are reproducible and generally applicable.
The three programs that were found to be the most reliable (MutPred, Provean, and PMUT) were then used to assess the likely pathogenicity of thirteen PITX2 missense variants for which functional analyses have not been performed, but which have been associated with ARS or CAD (Table 6). Our results showed that MutPred, Provean, and PMUT predicted as pathogenetic 12/13 of the variants. The A30V variant was scored as non-pathogenetic/benign by all three programs. While it is possible that A30V is an example of a false negative for all three programs, it is likely that this variant is instead benign. Functional testing of the A30V variant is needed to determine which of these possibilities is accurate.
Various intramolecular interactions are involve in stabilizing and folded state of protein, including hydrophobic, electrostatic, and hydrogen-bonding [95–98]. The stability state of a protein is key factor in its proper functionality. In fact, up to 80% of Mendelian disease-causing mutations in protein coding regions are predicted to be caused by altering protein stability [99]. In recent years, due to the availability of high-throughput array-based genotyping methods [100] and next generation sequencing platforms [101,102], a large number of SNPs has been reported. However, the association of missense variants with protein stability has often been difficult to predict. Fortunately, recent advances in computational prediction of protein stability offers potential insight into this question. We used two parallel prediction methods to investigate the possible effects on PITX2 protein structure and stability of missense variants.
Knowledge of a protein's 3D structure can be used to predict the functionality of protein and the possible impact of variants on protein conformation and structure. We thus first used molecular modelling analyses to assess and compared the total energy difference between native and mutated modeled structure of PITX2 proteins. The results predicted that while most PITX2 variants did not dramatically affect the protein tertiary structure, seven variants (L54Q, F58L, V83F, V83L, W86C, W86S, and R91P) altered the total energy level in comparison with the native structure, suggesting that these amino acid substitutions changed the structure of the PITX2 protein. Molecular modeling of the PITX2 homeodomain predicted that these variants impair the required energy to maintain the proper folding of helix 1–3 and cause global destabilization of the structure of PITX2. These seven amino acids are either invariant (e.g., W86) or highly conserved in the approximately 300 homeobox proteins analyzed, consistent with a pivotal role of these residues in the homeodomain [103–105]. These seven amino acids are tightly packed hydrophobic amino acids responsible for holding helices of the PITX2 homeodomain together, supporting our molecular modeling predicting that mutations of these amino acids disrupt PITX2 structure. For F58L, V83F, and V83L, the native wild-type residues and the introduced mutant residues differ in size, probably causing loss of hydrophobic interactions in the core of the protein, particularly involving helix 1–3. For L54Q, W86C, W86S, and R91P, the wild-type residues and the mutant residues are different in both size and charge, likely disturb the local structure of protein thereby altering protein structure and function.
Residues V83, W86, and R91 are located within the third helix which is specifically responsible for binding with the major groove of the DNA [106]. Thus, the prediction that these mutations impair the capacity of this helix to interact with DNA is consistent with this knowledge and with previous functional characterizations that showed reduced DNA-binding capacities of the V83L and R91P mutant PITX2 proteins [5,107]. Consistent with bioinformatics predictions of deleterious affects of mutation of W86, mutations of the neighboring amino acids (R84W and K88E) have been shown to decrease the ability of the mutant proteins to interact with DNA [39,108].
Residues L54 and F58 are located in helix 1 of the homeodomain, responsible for contacting with the minor groove of the DNA. Molecular modeling of L54Q is consistent with the suggestion that mutations in these highly-conserved residues in helix 1 of the homeodomain might disturb the DNA-protein binding affinity. Our prediction is supported by the fact that changing the leucine to a glutamine (L54Q) disrupts DNA–protein complex, indicating the necessity of leucine at position 54 for PITX2 binding ability [109]. Thus, consistent with our recent studies on FOXC1 protein [110], the results of molecular modeling of PITX2 are strongly consistent with the functional characterization of PITX2 missense variants.
The results from our molecular modeling analysis were also compared to the predictions of eight stability predictor methods (DUET, SDM, mCSM, I-mutant3.0, MUpro, iPTREE-STAB, CUPSAT, and iStable). Based on our analyses, it appears that CUPSAT performs the best of the seven methods evaluated here in predicting the effect of missense mutations on PITX2 protein stability, with SDM, DUET, mCSM, and I-Mutant3.0, performing weaker, consistent with the results of previous studies [111,112]. Our results indicate that further studies are required to improve ΔΔG predictions, especially for buried amino acids.
In this study, for the first time, we evaluated the impact of missense variants on PITX2 stability, structure and function by integrating stability prediction algorithms, bioinformatics mutation prediction tools, and molecular modeling. Our results showed that MutPred, Provean, PMUT, molecular modeling, and CUPSAT are reliable methods to assess PITX family missense variants in the absence of laboratory experiments. However, for our analyses, it must be noted that we used sixteen SNPs as non-pathogenetic control variants to investigate the performance of prediction programs. Although we considered SNPs with a population frequency of >0.05% as benign, we cannot formally exclude that these SNPs might have un-documented pathogenic effects on PITX2. In addition, while the prediction methods used in this study are not gene-specific, generalization of the performance of these programs to other human genes may be inappropriate without additional study. When assessing the pathogenicity of missense variants, it is necessary to be cautious on depending merely on in silico programs without wet-lab experiments. According to standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology, in silico predictions only serve as one supporting factor, whereas functional tests are frequently needed to assess the pathogenicity of missense variants. In particular, as per clinical guidelines for the interpretation of single substitution variants, the output of computational tools should be interpreted in the light of functional studies results, population frequency data and segregation in affected families.
Supporting information
The 3D model of PITX2 is presented with the protein backbone depicted in black ribbon, the co-crystallized DNA binding target in space-filled green model and the mutants positions in red. The wild-type and mutant-equivalent models were analyzed by the atomic nonlocal environment assessment (ANOLEA) server. Peaks on the scatterplots show the positions of amino acids that changed their pseudoenergy state, as a consequence of the mentioned variants.
(TIF)
The 3D model of PITX2 is presented with the protein backbone depicted in black ribbon, the co-crystallized DNA binding target in space-filled green model and the mutants positions in red. The wild-type and mutant-equivalent models were analyzed by the atomic nonlocal environment assessment (ANOLEA) server. Peaks on the scatterplots show the positions of amino acids that changed their pseudoenergy state, as consequence of the mentioned variants.
(TIF)
Acknowledgments
The authors would like to thank the members of the Walter Laboratory for critical reading of the manuscript and for helpful comments.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Canadian Institute for Health Research (M.A. Walter, [grant number G118160216], http://www.cihr-irsc.gc.ca/e/193.html) and Alberta Innovates Health Solutions graduate studentship (M. Seifi, [award number 201500029], http://www.aihealthsolutions.ca/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vaux C, Sheffield L, Keith CG, Voullaire L. Evidence that Rieger syndrome maps to 4q25 or 4q27. J Med Genet. 1992;29: 256–8. doi: 10.1136/jmg.29.4.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tümer Z, Bach-Holm D. Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. European journal of human genetics: EJHG. Nature Publishing Group; 2009;17: 1527–39. doi: 10.1038/ejhg.2009.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strungaru MH, Dinu I, Walter MA. Genotype-Phenotype Correlations in Axenfeld-Rieger Malformation and Glaucoma Patients with FOXC1 and PITX2 Mutations. Investigative Opthalmology & Visual Science. 2007;48: 228 doi: 10.1167/iovs.06-0472 [DOI] [PubMed] [Google Scholar]
- 4.Lines MA, Kozlowski K, Walter MA. Molecular genetics of Axenfeld-Rieger malformations. Human molecular genetics. 2002;11: 1177–84. doi: 10.1093/hmg/11.10.1177 [DOI] [PubMed] [Google Scholar]
- 5.Kozlowski K, Walter MA. Variation in residual PITX2 activity underlies the phenotypic spectrum of anterior segment developmental disorders. Human molecular genetics. 2000;9: 2131–9. [DOI] [PubMed] [Google Scholar]
- 6.Shields MB. Axenfeld-Rieger syndrome: a theory of mechanism and distinctions from the iridocorneal endothelial syndrome. Transactions of the American Ophthalmological Society. 1983;81: 736–84. [PMC free article] [PubMed] [Google Scholar]
- 7.Xia K, Wu L, Liu X, Xi X, Liang D, Zheng D, et al. Mutation in PITX2 is associated with ring dermoid of the cornea. Journal of medical genetics. 2004;41: e129 doi: 10.1136/jmg.2004.022434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seifi M, Footz T, Taylor SAM, Elhady GM, Abdalla EM, Walter MA. Novel PITX2 gene mutations in patients with Axenfeld-Rieger syndrome. Acta Ophthalmologica. 2016;94: e571–e579. doi: 10.1111/aos.13030 [DOI] [PubMed] [Google Scholar]
- 9.Saadi I, Toro R, Kuburas A, Semina E, Murray JC, Russo AF. An unusual class of PITX2 mutations in Axenfeld-Rieger syndrome. Birth defects research Part A, Clinical and molecular teratology. 2006;76: 175–81. doi: 10.1002/bdra.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’haene B, Meire F, Claerhout I, Kroes HY, Plomp A, Arens YH, et al. Expanding the spectrum of FOXC1 and PITX2 mutations and copy number changes in patients with anterior segment malformations. Investigative ophthalmology & visual science. 2011;52: 324–33. doi: 10.1167/iovs.10-5309 [DOI] [PubMed] [Google Scholar]
- 11.Brooks BP, Moroi SE, Downs CA, Wiltse S, Othman MI, Semina E V, et al. A novel mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome. Ophthalmic genetics. 2004;25: 57–62. Available: http://www.ncbi.nlm.nih.gov/pubmed/15255117 [DOI] [PubMed] [Google Scholar]
- 12.Weisschuh N, Dressler P, Schuettauf F, Wolf C, Wissinger B, Gramer E. Novel Mutations of FOXC1 and PITX2 in Patients with Axenfeld-Rieger Malformations. Investigative Opthalmology & Visual Science. 2006;47: 3846 doi: 10.1167/iovs.06-0343 [DOI] [PubMed] [Google Scholar]
- 13.Perveen R, Lloyd IC, Clayton-Smith J, Churchill A, van Heyningen V, Hanson I, et al. Phenotypic variability and asymmetry of Rieger syndrome associated with PITX2 mutations. Investigative ophthalmology & visual science. 2000;41: 2456–60. Available: http://www.ncbi.nlm.nih.gov/pubmed/10937553 [PubMed] [Google Scholar]
- 14.Judkins T, Hendrickson BC, Deffenbaugh AM, Scholl T. Single nucleotide polymorphisms in clinical genetic testing: the characterization of the clinical significance of genetic variants and their application in clinical research for BRCA1. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2005;573: 168–179. doi: 10.1016/j.mrfmmm.2004.07.024 [DOI] [PubMed] [Google Scholar]
- 15.Young DL, Fields S. The role of functional data in interpreting the effects of genetic variation. Molecular biology of the cell. American Society for Cell Biology; 2015;26: 3904–8. doi: 10.1091/mbc.E15-03-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campuzano O, Allegue C, Fernandez A, Iglesias A, Brugada R. Determining the Pathogenicity of Genetic Variants Associated with Cardiac Channelopathies. Scientific Reports. Nature Publishing Group; 2015;5: 7953 doi: 10.1038/srep07953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia L, Yarlagadda R, Reed CC, Haeberlein M, Chen H, Ma S. Structure Based Thermostability Prediction Models for Protein Single Point Mutations with Machine Learning Tools. Zhang Y, editor. PLOS ONE. Plenum Press; 2015;10: e0138022 doi: 10.1371/journal.pone.0138022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan SE, Patch A-M, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genetic testing and molecular biomarkers. 2010;14: 533–537. doi: 10.1089/gtmb.2010.0036 [DOI] [PubMed] [Google Scholar]
- 19.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Current Opinion in Structural Biology. 2009;19: 596–604. doi: 10.1016/j.sbi.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 20.Gromiha MM. Prediction of protein stability upon point mutations. Biochemical Society Transactions. 2007;35: 1569–1573. doi: 10.1042/BST0351569 [DOI] [PubMed] [Google Scholar]
- 21.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the Functional Effect of Amino Acid Substitutions and Indels. de Brevern AG, editor. PLoS ONE. Public Library of Science; 2012;7: e46688 doi: 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefl S, Nishi H, Petukh M, Panchenko AR, Alexov E. Molecular mechanisms of disease-causing missense mutations. Journal of molecular biology. NIH Public Access; 2013;425: 3919–36. doi: 10.1016/j.jmb.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laskowski RA, Tyagi N, Johnson D, Joss S, Kinning E, McWilliam C, et al. Integrating population variation and protein structural analysis to improve clinical interpretation of missense variation: application to the WD40 domain. Human molecular genetics. Oxford University Press; 2016;25: 927–35. doi: 10.1093/hmg/ddv625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao L, Cui Y. Prediction of the phenotypic effects of non-synonymous single nucleotide polymorphisms using structural and evolutionary information. Bioinformatics. 2005;21: 2185–2190. doi: 10.1093/bioinformatics/bti365 [DOI] [PubMed] [Google Scholar]
- 25.Wallqvist A, Fukunishi Y, Murphy LR, Fadel A, Levy RM. Iterative sequence/secondary structure search for protein homologs: comparison with amino acid sequence alignments and application to fold recognition in genome databases. Bioinformatics (Oxford, England). 2000;16: 988–1002. [DOI] [PubMed] [Google Scholar]
- 26.Geourjon C, Combet C, Blanchet C, Deléage G. Identification of related proteins with weak sequence identity using secondary structure information. Protein science: a publication of the Protein Society. Wiley-Blackwell; 2001;10: 788–97. doi: 10.1110/ps.30001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seifi M, Footz T, Taylor SAM, Walter MA. Comparison of Bioinformatics Prediction, Molecular Modeling, and Functional Analyses of FOXC1 Mutations in Patients with Axenfeld-Rieger Syndrome. Human Mutation. 2017;38: 169–179. doi: 10.1002/humu.23141 [DOI] [PubMed] [Google Scholar]
- 28.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Research. 2014;42: D980–D985. doi: 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenson PD, Mort M, Ball E V., Shaw K, Phillips AD, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Human Genetics. 2014;133: 1–9. doi: 10.1007/s00439-013-1358-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Idrees F, Bloch-Zupan A, Free SL, Vaideanu D, Thompson PJ, Ashley P, et al. A novel homeobox mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome associated with brain, ocular, and dental phenotypes. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2006;141B: 184–91. doi: 10.1002/ajmg.b.30237 [DOI] [PubMed] [Google Scholar]
- 31.Footz T, Idrees F, Acharya M, Kozlowski K, Walter MA. Analysis of mutations of the PITX2 transcription factor found in patients with Axenfeld-Rieger syndrome. Investigative ophthalmology & visual science. 2009;50: 2599–606. doi: 10.1167/iovs.08-3251 [DOI] [PubMed] [Google Scholar]
- 32.Yuan F, Zhao L, Wang J, Zhang W, Li X, Qiu X-B, et al. PITX2c loss-of-function mutations responsible for congenital atrial septal defects. International journal of medical sciences. 2013;10: 1422–9. doi: 10.7150/ijms.6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun YM, Wang J, Qiu XB, Yuan F, Xu YJ, Li RG, et al. PITX2 loss-of-function mutation contributes to tetralogy of Fallot. Gene. 2016;577: 258–264. doi: 10.1016/j.gene.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 34.Semina E V, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nature genetics. 1996;14: 392–9. doi: 10.1038/ng1296-392 [DOI] [PubMed] [Google Scholar]
- 35.Amendt BA, Sutherland LB, Semina E V, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. The Journal of biological chemistry. 1998;273: 20066–72. Available: http://www.ncbi.nlm.nih.gov/pubmed/9685346 [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Zhang D-F, Sun Y-M, Yang Y-Q. A novel PITX2c loss-of-function mutation associated with familial atrial fibrillation. European journal of medical genetics. 2014;57: 25–31. doi: 10.1016/j.ejmg.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 37.Amendt BA, Semina E V, Alward WL. Rieger syndrome: a clinical, molecular, and biochemical analysis. Cellular and molecular life sciences: CMLS. 2000;57: 1652–66. Available: http://www.ncbi.nlm.nih.gov/pubmed/11092457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozlowski K, Walter MA. Variation in residual PITX2 activity underlies the phenotypic spectrum of anterior segment developmental disorders. Human molecular genetics. 2000;9: 2131–9. Available: http://www.ncbi.nlm.nih.gov/pubmed/10958652 [DOI] [PubMed] [Google Scholar]
- 39.Espinoza HM, Cox CJ, Semina E V, Amendt BA. A molecular basis for differential developmental anomalies in Axenfeld-Rieger syndrome. Human molecular genetics. 2002;11: 743–53. [DOI] [PubMed] [Google Scholar]
- 40.Saadi I, Semina E V, Amendt BA, Harris DJ, Murphy KP, Murray JC, et al. Identification of a dominant negative homeodomain mutation in Rieger syndrome. The Journal of biological chemistry. 2001;276: 23034–41. doi: 10.1074/jbc.M008592200 [DOI] [PubMed] [Google Scholar]
- 41.Kulak SC, Kozlowski K, Semina E V., Pearce WG, Walter MA. Mutation in the RIEG1 gene in patients with iridogoniodysgenesis syndrome. Human Molecular Genetics. 1998;7: 1113–1117. doi: 10.1093/hmg/7.7.1113 [DOI] [PubMed] [Google Scholar]
- 42.Strungaru MH, Dinu I, Walter MA. Genotype-Phenotype Correlations in Axenfeld-Rieger Malformation and Glaucoma Patients with FOXC1 and PITX2 Mutations. Investigative Opthalmology & Visual Science. 2007;48: 228 doi: 10.1167/iovs.06-0472 [DOI] [PubMed] [Google Scholar]
- 43.Wei D, Gong X-H, Qiu G, Wang J, Yang Y-Q. Novel PITX2c loss-of-function mutations associated with complex congenital heart disease. International Journal of Molecular Medicine. 2014;33: 1201–8. doi: 10.3892/ijmm.2014.1689 [DOI] [PubMed] [Google Scholar]
- 44.Priston M, Kozlowski K, Gill D, Letwin K, Buys Y, Levin A V, et al. Functional analyses of two newly identified PITX2 mutants reveal a novel molecular mechanism for Axenfeld-Rieger syndrome. Human molecular genetics. 2001;10: 1631–8. Available: http://www.ncbi.nlm.nih.gov/pubmed/11487566 [DOI] [PubMed] [Google Scholar]
- 45.Alward WL, Semina E V, Kalenak JW, Héon E, Sheth BP, Stone EM, et al. Autosomal dominant iris hypoplasia is caused by a mutation in the Rieger syndrome (RIEG/PITX2) gene. American journal of ophthalmology. 1998;125: 98–100. Available: http://www.ncbi.nlm.nih.gov/pubmed/9437321 [DOI] [PubMed] [Google Scholar]
- 46.Perveen R, Lloyd IC, Clayton-Smith J, Churchill A, van Heyningen V, Hanson I, et al. Phenotypic variability and asymmetry of Rieger syndrome associated with PITX2 mutations. Investigative ophthalmology & visual science. 2000;41: 2456–60. Available: http://www.ncbi.nlm.nih.gov/pubmed/10937553 [PubMed] [Google Scholar]
- 47.Wang J, Xin Y-F, Xu W-J, Liu Z-M, Qiu X-B, Qu X-K, et al. Prevalence and spectrum of PITX2c mutations associated with congenital heart disease. DNA and cell biology. Mary Ann Liebert, Inc.; 2013;32: 708–16. doi: 10.1089/dna.2013.2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips JC. Four novel mutations in the PITX2 gene in patients with Axenfeld-Rieger syndrome. Ophthalmic research. 34: 324–6. doi: 10.1159/000065602 [DOI] [PubMed] [Google Scholar]
- 49.Ng PC, Henikoff S. Predicting Deleterious Amino Acid Substitutions. Genome Research. 2001;11: 863–874. doi: 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic acids research. 2002;30: 3894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang H, Thomas PD. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32: 2230–2232. doi: 10.1093/bioinformatics/btw222 [DOI] [PubMed] [Google Scholar]
- 52.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics (Oxford, England). 2009;25: 2744–50. doi: 10.1093/bioinformatics/btp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014;11: 361–2. doi: 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- 54.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics (Oxford, England). 2015;31: 2745–7. doi: 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferrer-Costa C, Orozco M, de la Cruz X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. Journal of Molecular Biology. 2002;315: 771–786. doi: 10.1006/jmbi.2001.5255 [DOI] [PubMed] [Google Scholar]
- 56.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GLA, Edwards KJ, et al. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Human Mutation. 2013;34: 57–65. doi: 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bao L, Zhou M, Cui Y. nsSNPAnalyzer: Identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Research. 2005;33 doi: 10.1093/nar/gki372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tavtigian S V, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. Journal of medical genetics. 2006;43: 295–305. doi: 10.1136/jmg.2005.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. American Journal of Human Genetics. 2016;99: 877–885. doi: 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4: 1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 61.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. Journal of Molecular Biology. 2001;305: 567–580. doi: 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 62.Mi H, Thomas P. PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools. 2009. pp. 123–140. doi: 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic acids research. 2003;31: 3812–4. Available: http://www.ncbi.nlm.nih.gov/pubmed/12824425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang H, Thomas PD. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32: 2230–2232. doi: 10.1093/bioinformatics/btw222 [DOI] [PubMed] [Google Scholar]
- 65.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics (Oxford, England). 2009;25: 2744–50. doi: 10.1093/bioinformatics/btp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pires DE V, Ascher DB, Blundell TL. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic acids research. 2014;42: W314–9. doi: 10.1093/nar/gku411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pires DE V., Ascher DB, Blundell TL. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30: 335–342. doi: 10.1093/bioinformatics/btt691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Worth CL, Preissner R, Blundell TL. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Research. 2011;39: W215–W222. doi: 10.1093/nar/gkr363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandurangan AP, Ochoa-Montaño B, Ascher DB, Blundell TL. SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Research. 2017;45: W229–W235. doi: 10.1093/nar/gkx439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pires DE V, Ascher DB, Blundell TL. MCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30: 335–342. doi: 10.1093/bioinformatics/btt691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics (Oxford, England). 2006;22: 2729–34. doi: 10.1093/bioinformatics/btl423 [DOI] [PubMed] [Google Scholar]
- 72.Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62: 1125–32. doi: 10.1002/prot.20810 [DOI] [PubMed] [Google Scholar]
- 73.Huang L-T, Gromiha MM, Ho S-Y. iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics (Oxford, England). 2007;23: 1292–3. doi: 10.1093/bioinformatics/btm100 [DOI] [PubMed] [Google Scholar]
- 74.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Research. 2006;34 doi: 10.1093/nar/gkl190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen CW, Lin J, Chu YW. iStable: Off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinformatics. 2013;14 doi: 10.1186/1471-2105-14-S2-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang L-T, Gromiha MM, Ho S-Y. iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics (Oxford, England). 2007;23: 1292–3. doi: 10.1093/bioinformatics/btm100 [DOI] [PubMed] [Google Scholar]
- 77.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Current Opinion in Structural Biology. 2009;19: 596–604. doi: 10.1016/j.sbi.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 78.Mayer S, Rüdiger S, Ang HC, Joerger AC, Fersht AR. Correlation of levels of folded recombinant p53 in escherichia coli with thermodynamic stability in vitro. Journal of molecular biology. 2007;372: 268–76. doi: 10.1016/j.jmb.2007.06.044 [DOI] [PubMed] [Google Scholar]
- 79.Calloni G, Zoffoli S, Stefani M, Dobson CM, Chiti F. Investigating the Effects of Mutations on Protein Aggregation in the Cell. Journal of Biological Chemistry. 2005;280: 10607–10613. doi: 10.1074/jbc.M412951200 [DOI] [PubMed] [Google Scholar]
- 80.Randles LG, Lappalainen I, Fowler SB, Moore B, Hamill SJ, Clarke J. Using model proteins to quantify the effects of pathogenic mutations in Ig-like proteins. The Journal of biological chemistry. 2006;281: 24216–26. doi: 10.1074/jbc.M603593200 [DOI] [PubMed] [Google Scholar]
- 81.Lindberg MJ, Byström R, Boknäs N, Andersen PM, Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102: 9754–9. doi: 10.1073/pnas.0501957102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498: 220–3. doi: 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Y-Q, Xu Y-J, Li R-G, Qu X-K, Fang W-Y, Liu X. Prevalence and spectrum of PITX2c mutations associated with familial atrial fibrillation. International journal of cardiology. 2013;168: 2873–6. doi: 10.1016/j.ijcard.2013.03.141 [DOI] [PubMed] [Google Scholar]
- 84.Vieira V, David G, Roche O, de la Houssaye G, Boutboul S, Arbogast L, et al. Identification of four new PITX2 gene mutations in patients with Axenfeld-Rieger syndrome. Molecular vision. 2006;12: 1448–60. Available: http://www.ncbi.nlm.nih.gov/pubmed/17167399 [PubMed] [Google Scholar]
- 85.D’haene B, Meire F, Claerhout I, Kroes HY, Plomp A, Arens YH, et al. Expanding the spectrum of FOXC1 and PITX2 mutations and copy number changes in patients with anterior segment malformations. Investigative ophthalmology & visual science. 2011;52: 324–33. doi: 10.1167/iovs.10-5309 [DOI] [PubMed] [Google Scholar]
- 86.Xia K, Wu L, Liu X, Xi X, Liang D, Zheng D, et al. Mutation in PITX2 is associated with ring dermoid of the cornea. Journal of medical genetics. 2004;41: e129 doi: 10.1136/jmg.2004.022434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weisschuh N, Dressler P, Schuettauf F, Wolf C, Wissinger B, Gramer E. Novel Mutations of FOXC1 and PITX2 in Patients with Axenfeld-Rieger Malformations. Investigative Opthalmology & Visual Science. 2006;47: 3846 doi: 10.1167/iovs.06-0343 [DOI] [PubMed] [Google Scholar]
- 88.Meyer-Marcotty P, Weisschuh N, Dressler P, Hartmann J, Stellzig-Eisenhauer A. Morphology of the sella turcica in Axenfeld-Rieger syndrome with PITX2 mutation. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2008;37: 504–10. doi: 10.1111/j.1600-0714.2008.00650.x [DOI] [PubMed] [Google Scholar]
- 89.Dressler S, Meyer-Marcotty P, Weisschuh N, Jablonski-Momeni A, Pieper K, Gramer G, et al. Dental and Craniofacial Anomalies Associated with Axenfeld-Rieger Syndrome with PITX2 Mutation. Case reports in medicine. 2010;2010: 621984 doi: 10.1155/2010/621984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kimura M, Tokita Y, Machida J, Shibata A, Tatematsu T, Tsurusaki Y, et al. A novel PITX2 mutation causing iris hypoplasia. Human genome variation. 2014;1: 14005 doi: 10.1038/hgv.2014.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reis LM, Tyler RC, Volkmann Kloss BA, Schilter KF, Levin A V, Lowry RB, et al. PITX2 and FOXC1 spectrum of mutations in ocular syndromes. European Journal of Human Genetics. 2012;20: 1224–1233. doi: 10.1038/ejhg.2012.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li D, Zhu Q, Lin H, Zhou N, Qi Y. A novel PITX2 mutation in a Chinese family with Axenfeld-Rieger syndrome. Molecular vision. Emory University; 2008;14: 2205–10. Available: http://www.ncbi.nlm.nih.gov/pubmed/19052653 [PMC free article] [PubMed] [Google Scholar]
- 93.Kniestedt C, Taralczak M, Thiel MA, Stuermer J, Baumer A, Gloor BP. A novel PITX2 mutation and a polymorphism in a 5-generation family with Axenfeld-Rieger anomaly and coexisting Fuchs’ endothelial dystrophy. Ophthalmology. 2006;113: 1791e1-8. doi: 10.1016/j.ophtha.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 94.Huang X, Xiao X, Jia X, Li S, Li M, Guo X, et al. Mutation analysis of the genes associated with anterior segment dysgenesis, microcornea and microphthalmia in 257 patients with glaucoma. International journal of molecular medicine. 2015;36: 1111–7. doi: 10.3892/ijmm.2015.2325 [DOI] [PubMed] [Google Scholar]
- 95.Aydin H, Al-Khooly D, Lee JE. Influence of hydrophobic and electrostatic residues on SARS-coronavirus S2 protein stability: Insights into mechanisms of general viral fusion and inhibitor design. Protein Science. 2014;23: 603–617. doi: 10.1002/pro.2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aydin H, Al-Khooly D, Lee JE. Influence of hydrophobic and electrostatic residues on SARS-coronavirus S2 protein stability: insights into mechanisms of general viral fusion and inhibitor design. Protein science: a publication of the Protein Society. Wiley-Blackwell; 2014;23: 603–17. doi: 10.1002/pro.2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pace CN, Fu H, Lee Fryar K, Landua J, Trevino SR, Schell D, et al. Contribution of hydrogen bonds to protein stability. Protein science: a publication of the Protein Society. Wiley-Blackwell; 2014;23: 652–61. doi: 10.1002/pro.2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vagenende V, Yap MGS, Trout BL. Mechanisms of Protein Stabilization and Prevention of Protein Aggregation by Glycerol. Biochemistry. 2009;48: 11084–11096. doi: 10.1021/bi900649t [DOI] [PubMed] [Google Scholar]
- 99.Wang Z, Moult J. SNPs, protein structure, and disease. Human Mutation. 2001;17: 263–270. doi: 10.1002/humu.22 [DOI] [PubMed] [Google Scholar]
- 100.Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, et al. Whole‐Genome Genotyping. Methods in enzymology. 2006. pp. 359–376. doi: 10.1016/S0076-6879(06)10017-8 [DOI] [PubMed] [Google Scholar]
- 101.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452: 872–876. doi: 10.1038/nature06884 [DOI] [PubMed] [Google Scholar]
- 102.Metzker ML. Sequencing technologies—the next generation. Nature Reviews Genetics. 2010;11: 31–46. doi: 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- 103.Qian YQ, Billeter M, Otting G, Müller M, Gehring WJ, Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989;59: 573–80. [DOI] [PubMed] [Google Scholar]
- 104.Kornberg TB. Understanding the homeodomain. The Journal of biological chemistry. 1993;268: 26813–6. [PubMed] [Google Scholar]
- 105.Gehring WJ, Affolter M, Burglin T. Homeodomain Proteins. Annual Review of Biochemistry. 1994;63: 487–526. doi: 10.1146/annurev.bi.63.070194.002415 [DOI] [PubMed] [Google Scholar]
- 106.Chaney BA, Clark-Baldwin K, Dave V, Ma J, Rance M. Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome. Biochemistry. 2005;44: 7497–511. doi: 10.1021/bi0473253 [DOI] [PubMed] [Google Scholar]
- 107.Priston M, Kozlowski K, Gill D, Letwin K, Buys Y, Levin A V, et al. Functional analyses of two newly identified PITX2 mutants reveal a novel molecular mechanism for Axenfeld-Rieger syndrome. Human molecular genetics. 2001;10: 1631–8. [DOI] [PubMed] [Google Scholar]
- 108.Saadi I, Semina E V, Amendt BA, Harris DJ, Murphy KP, Murray JC, et al. Identification of a dominant negative homeodomain mutation in Rieger syndrome. The Journal of biological chemistry. 2001;276: 23034–41. doi: 10.1074/jbc.M008592200 [DOI] [PubMed] [Google Scholar]
- 109.Amendt BA, Sutherland LB, Semina E V, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. The Journal of biological chemistry. 1998;273: 20066–72. [DOI] [PubMed] [Google Scholar]
- 110.Seifi M, Footz T, Taylor SAM, Walter MA. Comparison of Bioinformatics Prediction, Molecular Modeling, and Functional Analyses of FOXC1 Mutations in Patients with Axenfeld-Rieger Syndrome. Human Mutation. 2016; doi: 10.1002/humu.23141 [DOI] [PubMed] [Google Scholar]
- 111.Doss CGP, Rajith B, Garwasis N, Mathew PR, Raju AS, Apoorva K, et al. Screening of mutations affecting protein stability and dynamics of FGFR1—A simulation analysis. Applied & Translational Genomics. 2012;1: 37–43. doi: 10.1016/j.atg.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khan S, Vihinen M. Performance of protein stability predictors. Human Mutation. 2010;31: 675–684. doi: 10.1002/humu.21242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 3D model of PITX2 is presented with the protein backbone depicted in black ribbon, the co-crystallized DNA binding target in space-filled green model and the mutants positions in red. The wild-type and mutant-equivalent models were analyzed by the atomic nonlocal environment assessment (ANOLEA) server. Peaks on the scatterplots show the positions of amino acids that changed their pseudoenergy state, as a consequence of the mentioned variants.
(TIF)
The 3D model of PITX2 is presented with the protein backbone depicted in black ribbon, the co-crystallized DNA binding target in space-filled green model and the mutants positions in red. The wild-type and mutant-equivalent models were analyzed by the atomic nonlocal environment assessment (ANOLEA) server. Peaks on the scatterplots show the positions of amino acids that changed their pseudoenergy state, as consequence of the mentioned variants.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.