Abstract
Bacterial infectious diseases produced by Vibrio are the main cause of economic losses in aquaculture. During recent years it has been shown that the expression of virulence genes in some Vibrio species is controlled by a population-density dependent gene-expression mechanism known as quorum sensing (QS), which is mediated by the diffusion of signal molecules such as N-acylhomoserine lactones (AHLs). QS disruption, especially the enzymatic degradation of signalling molecules, known as quorum quenching (QQ), is one of the novel therapeutic strategies for the treatment of bacterial infections. In this study, we present the detection of AHLs in 34 marine Vibrionaceae strains. Three aquaculture-related pathogenic Vibrio strains, V. mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193 were selected for further studies based on their virulence and high production of AHLs. This is the first report where the signal molecules have been characterized in these emerging marine pathogens and correlated to the expression of virulence factors. Moreover, the results of AHL inactivation in the three selected strains have been confirmed in vivo against brine shrimps (Artemia salina) and Manila clams (Venerupis philippinarum). This research contributes to the development of future therapies based on AHL disruption, the most promising alternatives for fighting infectious diseases in aquaculture.
Introduction
Outbreaks of bacterial disease are considered as a significant constraint to the development of aquaculture in the world, causing serious economic losses in this sector [1]. Although many of the causative pathogens are still unknown, most of them belong to the family Vibrionaceae [2]. Vibriosis is one of the most common diseases in aquaculture, affecting a large number of fishes, mollusks, crustaceans and corals [3,4]. Vibrio species, such as V. harveyi, V. vulnificus, V. anguillarum, V. ordalii and V. parahaemolyticus, are the major causes of vibriosis and are ubiquitous in global marine environments [5]. To date, other emerging species, such as V. neptunius, V. mediterranei and V. coralliilyticus are recently being related to mortality outbreaks worldwide [6,7].
During the last few years virulence of different pathogenic bacteria has been associated to quorum sensing (QS), a sophisticated mechanism which coordinates gene expression by means of small signal molecules known as autoinducers [8,9]. Amongst others, autoinducers include N-acylhomoserine lactones (AHLs) produced by the Proteobacteria, oligopeptides produced by the Firmicutes and furanosylborate diester (AI-2) which are produced by both Proteobacteria and Firmicutes and are used for interspecies communication [10,11]. In general terms, the basic mechanism of QS systems begins with a basal expression of a signal molecule (autoinducer). The concentration of the autoinducer increases with cell density, and upon reaching a threshold level, it binds and activates a LuxR-type regulator. The autoinducer LuxR-complex then causes the expression of the autoinducer synthase, thereby establishing a positive feedback loop. The result of this induction is the regulation (either positive or negative) of different target genes, some of which involve the expression of virulence factors and exoenzymes, conjugal DNA transfer, control of plasmid-copy number, production of and susceptibility to antibiotics, biofilm formation and exopolysaccharide production [10,11].
To date, QS systems have been studied in different marine pathogenic species belonging to the genera Vibrio, Edwardsiella, Aeromonas, Pseudomonas and Yersinia. In most cases, the expression of virulence genes such as exoenzymes or pigment production has been reported to be regulated by QS [12–15]. The majority of these studies have focused on the genus Vibrio, the main etiological agent of aquatic-related diseases [15–18] and responsible for mass mortalities in a great number of bivalve mollusks [6,19–21]. For instance, AHL-type autoinducers control the production of metalloprotease, siderophores and chitinase A amongst other virulence factors in Vibrio campbellii [22–24]; as well as biofilm formation and metalloprotease and siderophore production in V. anguillarum [13,25].
Besides the ability to produce and use AHL-based communication systems, the ability to interfere with QS systems or to degrade AHLs has been reported for many marine bacteria [26–30], suggesting that these organisms have developed various mechanisms to interfere with or disrupt these cellular communication systems [31–34]. One of the most promising mechanisms that strongly perturbs or even abolishes QS-regulated functions is related to the production of enzymes capable of degrading the AHL-signal molecules, known as quorum quenching (QQ) [33]. To date, different groups of enzymes have been identified according to the enzymatic mechanism involved: the AHL lactonases (lactone hydrolysis), the AHL acylases (amide hydrolysis) and the AHL oxidases/oxidoreductases (oxidoreduction) [35–37].
Research on QS-dependent pathogenic bacteria is becoming a very important tool for understanding and controlling the diseases they cause, and their inhibition by QQ and other strategies has already proven to be one of the most promising alternatives for fighting infectious diseases in aquaculture [38–40]. To increase the information about QS systems and their role in emerging aquaculture-related pathogenic bacteria, we have analyzed the production of AHL signal molecules in 34 strains isolated from healthy and diseased corals and seawater by a well-diffusion agar-plate assay using the biosensor strains Agrobacterium tumefaciens NTL4 (pZLR4) and Chromobacterium violaceum CV026. The fingerprints and the molecular structure of the AHLs produced by three selected pathogenic strains were then analyzed and compared by TLC and characterized by HPLC/FT-HRMS. In order to investigate the role of QS in the virulence of the selected Vibrio strains, AHL lactonases were expressed in each strain, and different phenotypes were investigated in the AHL-defective transformants in comparison with the wild type strains. Additionally, and considering the economic importance of clam and crustacean production in aquaculture and their susceptibility to vibriosis [41,42], Manila clams (Venerupis philippinarum) and brine shrimp (Artemia salina) were used as infection models to conduct preliminary in vivo assays to evaluate the impact of AHL interference upon the virulence of the three selected Vibrio strains.
The information generated in this research contributes to the development of new therapies based on AHL disruption, one of the most innovative and promising alternatives for combating infectious diseases in the aquaculture sector, and suggests that QQ is an appropriate strategy to prevent infections caused by these emerging pathogens.
Material and methods
2.1. Bacterial strains, media and growth conditions
The 34 marine strains tested in this study were previously isolated from Oculina patagonica and Cladocora caespitosa healthy and diseased corals and from seawater and they were identified according to their 16S rRNA gene (~1,500 bp) (Table 1) [43]. They were routinely cultured at 25°C in marine broth (MB), as well as Vibrio anguillarum ATCC 19264T [25]. Agrobacterium tumefaciens NTL4 (pZLR4) [44] was cultured at 28°C in Luria Bertani (LB) medium or AB medium supplemented with 80 μg/mL of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) when needed [45]. Chromobacterium violaceum CV026 [46] was grown at 28°C in LB medium. In the laboratory, stocks of the bacterial strains were maintained at -80°C with a cryopreservative.
Table 1. Detection of AHLs produced by marine Vibrionaceae strains detected by well-diffusion agar-plate assay.
| Strain | Phylogenetically most related species |
Isolation source | NTL4 | CV026 |
|---|---|---|---|---|
| Vcl-20 | Vibrio comitans | Cladocora caespitosa (diseased) | + | - |
| VibC-Oc-021 | V. splendidus | Oculina patagonica (healthy) | - | - |
| VibC-Oc-024 | V. gigantis | O. patagonica (healthy) | - | - |
| VibC-Oc-027 | V. gigantis | O. patagonica (healthy) | + | - |
| VibC-Oc-044 | V. comitans | O. patagonica (healthy) | + | - |
| VibC-Oc-062 | V. breoganii | O. patagonica (healthy) | - | - |
| VibC-Oc-063 | V. hepatarius | O. patagonica (healthy) | + | - |
| VibC-Oc-068 | V. coralliilyticus | O. patagonica (diseased) | + | - |
| VibC-Oc-071 | V. hepatarius | O. patagonica (healthy) | + | - |
| VibC-Oc-079 | V. fortis | O. patagonica (healthy) | + | - |
| Vcl-080 | V. tubiashii | C. caespitosa (diseased) | + | - |
| VibC-Oc-082 | V. maritimus | O. patagonica (healthy) | ++ | ++ |
| VibC-Oc-086 | Photobacterium lutimaris | Seawater | - | - |
| VibC-Oc-089 | P. phosphoreum | Seawater | + | - |
| Voc-097 | V. maritimus | O. patagonica (diseased) | +++ | - |
| VibC-Oc-097 | V. mediterranei | O. patagonica (diseased) | +++ | + |
| VibC-Oc-106 | V. owensii | Seawater | ++ | ++ |
| VibC-Oc-109 | V. fortis | O. patagonica (healthy) | - | - |
| VibC-Oc-110 | V. rotiferianus | Seawater | + | - |
| VibC-Oc-112 | V. alginolyticus | O. patagonica (healthy) | + | - |
| Vcl-121 | V. campbellii | C. caespitosa (diseased) | + | - |
| Voc-130 | P. rosenbergii | O. patagonica (healthy) | + | - |
| Voc-170 | Listonella pelagia | O. patagonica (diseased) | - | + |
| Vcl-178 | V. ponticus | C. caespitosa (diseased) | + | - |
| VibC-Oc-193 | V. coralliilyticus | O. patagonica (diseased) | ++ | - |
| Vcl-234 | V. breoganii | C. caespitosa (diseased) | + | - |
| Vcl-245 | V. comitans | C. caespitosa (diseased) | + | - |
| Vcl-258 | V. coralliilyticus | C. caespitosa (diseased) | + | - |
| Voc-264 | V. natriegens | O. patagonica (healthy) | + | - |
| Voc-271 | V. natriegens | O. patagonica (healthy) | + | - |
| Voc-273 | V. parahaemolyticus | O. patagonica (healthy) | + | - |
| Voc-277 | V. harveyi | O. patagonica (healthy) | + | - |
| Voc-279 | Vibrio sp. BWDY-66 | O. patagonica (healthy) | + | - |
| Voc-1146 | V. mediterraneii | O. patagonica (diseased) | - | - |
++ strong activation, + medium activation and - no activation of the biosensors Chromobacterium violaceum CV026 and Agrobacterium tumefaciens NTL4 (pZLR4).
When required, antibiotics were used at the following final concentrations: kanamycin (Km) 50 μg/mL, gentamicin (Gm) 50 μg/mL and tetracycline (Tc) 10 and 75 μg/mL.
2.2. Ethical statement
This study was carried out following the fundamental ethical principles with regard to the use of Artemia salina and Venerupis philippinarum. According to the European normative that regulates minimum fishing size of (Council Regulation No. 1967/2006), only clams of commercial size with minimum length of 25 mm were bought at the local market for this study. Artemia salina and Venerupis philippinarum are not considered endangered or protected species in any European or international species catalogue, including the CITES list (www.cites.org).
2.3. Screening for AHL activity by a well-diffusion agar-plate assay
The detection of AHLs was done as previously described in our laboratory [29,30]. The 34 bacterial strains were grown in 5 mL of MB at 25°C until the early stationary phase (OD600 2.8). An overnight culture of one of the biosensor strains [C. violaceum CV026 or A. tumefaciens NTL4 (pZLR4)] was diluted to 1:10 in 5 mL of LB 0.7% (w/v) agar and poured respectively onto AB medium supplemented with X-gal and LB agar plates. Then 6-mm-diameter wells were hollowed in the medium with the back of a sterile Pasteur pipette and 100 μL-aliquots of each culture were loaded in the wells. The plates were incubated at 28°C for 24 h to check for the appearance of a coloured halo around the wells. V. anguillarum ATCC 19264T was used as positive control and MB medium as a negative control.
2.4. AHL extraction and thin-layer chromatography analysis
To extract AHL molecules, 30 mL cultures of the three selected bacteria (Vibrio mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193) were grown until the early stationary phase (OD600 2.8). Then, the entire cultures were extracted twice with equal volumes of dichloromethane [47,48]. The extracts were dried and suspended in 100 μL of 70% (v/v) methanol.
To characterize the AHLs produced by each strain, the samples were analyzed by thin-layer chromatography (TLC). 25 μL of each AHL extract and standards were spotted onto a TLC plate (Partisil KC18 Whatman 20 × 20 cm) and developed with 60% (v/v) methanol. After migration, the plates were air-dried and overlaid with top agar containing the appropriate biosensor and incubated at 28°C for 24 h [47,48].
2.5. AHL identification by high performance liquid chromatography-Fourier transform-high resolution mass spectrometry
To identify the AHLs produced by the selected Vibrio strains by HPLC/FT-HRMS, a litre of whole culture of each strain was grown in MB medium to stationary phase (OD600 2.8) and extracted twice with an equal volume of dichloromethane, evaporated and resuspended in CH3CN [47,48].
For the analysis, each extract was purified by solid phase extraction [49] and analyzed by high-performance liquid chromatography coupled with Fourier-transform and high resolution mass spectrometry (HPLC/FT-HRMS) [50,51] using full scan (FS) and selected ion monitoring (SIM) modes in a Thermo Scientific Accela LC system coupled to a LTQ Orbitrap. The software Xcalibur 3.0 was used for data processing in both cases. For the HPLC/FT-HRMS FS mode analysis, the extract was passed through a 0.22 μm PTFE syringe filter before injection and 10 μl of each filtered solution was analyzed (mass range from m/z 50 to 500). Analyses were carried out using an Atlantis C18 column (100 mm × 4.6 mm, 5 μm); and a gradient mixture of CH3CN (solvent A) and H2O (solvent B) containing 0.1% (v/v) formic acid at flow rate of 350 μL/min (S1 Table). In the case of the analysis by HPLC/FT-HRMS SIM mode, each extract was passed through an Oasis HLB cartridge, which had previously been conditioned and equilibrated with 500 μL of CH3CN and 500 μL of H2O. Then, two fractions were eluted with 500 μL of CH3CN and H2O mixtures (1:1 and 1:0), and 10 μL of the fraction eluted with a mixture of CH3CN/H2O (1:0) was analyzed. In this case the analysis was conducted in a Scharlau KromaPhase C18 column (150 mm × 4.6 mm, 5 μm) using again a gradient mixture of CH3CN (solvent A) and H2O (solvent B), containing 0.1% (v/v) formic acid at flow rate of 80 μL/min (S1 Table).
Precision and accuracy were calculated from the relative standard deviation of the replicates (<15%) and by direct comparison of mean measured levels of spiked analytes with expected concentrations for unextracted standards, respectively. The external standard method was used for quantification. LC-MS/MS peak areas were calculated and used to build calibration curves of peak area ratio against analyte concentration using unweighted linear regression analysis (S1 Fig and S2 Table). The lower limit of quantification was defined as the concentration at which a signal/noise ratio of 10:1 was achieved.
2.6. Expression of AHL-lactonase genes in Vibrio spp
The selected Vibrio strains (V. mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193) were transformed by electroporation [52] with the recombinant plasmids pME6010::attM, pME6010::aiiA, pME6010::hqiA (coding each for an AHL-lactonase gene) [53–55] and with the empty plasmid pME6010 [56] (TcR). Competent cells were obtained after some modifications of the classical protocol [57]. Briefly, 25 mL of overnight cultures of each strain were centrifuged and cells were washed twice and resuspended in sucrose 300 mM (50 mL, 25 mL and 500 μL respectively) at 4°C. Then, 40 ng of each plasmid was transformed into 40 μL of each Vibrio competent cells by electroporation [52].
The transformants were confirmed by PCR amplification of the corresponding AHL-lactonase gene. The plasmid DNA of each transformant was extracted by the alkaline lysis protocol [58] and used as the template for the PCR amplification. A specific primer set for each AHL-lactonase gene was used [53–55]: aiiA forward 5’-ATGAAGAAAATGAACCATGGA-3’ and aiiaA reverse 5’-CTATACAAGCAGTCCAAAAGCC-3’, attM forward 5’-GACGCAATGAAACAGAGCCG-3’ and attM reverse 5’- AAGAGCGACCTGAACGAAGC-3’ and hqiA-T forward 5′-ATGAGTGAAATCACGTTGGC-3′ and hqiA-T reverse 5′-CTTTACCCGAAGGATCGTAA-3′.
To determine AHL production in each transformant, AHL molecules were extracted [47,48] from 5 mL-cultures of each Vibrio strain. Then, 10 μL of the crude extracts were added to 5-mm sterile disks placed onto AB medium supplemented with 80 μg/mL X-gal with an overlay of A. tumefaciens NTL4 (pZLR4), made as previously explained. The plates were incubated overnight at 28°C to allow the biosensor to grow and surround the paper disks with blue haloes.
2.7. Phenotypic analyses
10 μL of the wild type Vibrio and its transformant cultures (harbouring a plasmid with or without an AHL lactonase) (DO600 2.2) were spotted on different media. Proteolytic activity was detected in casein medium [59]. Hemolytic activity was determined in Columbia agar supplemented with 5% (w/v) sheep blood [59]. Chitinase activity was assayed in marine broth supplemented with 1% (w/v) colloidal chitin [60]. DNase activity was tested in DNase agar medium [61]. Lipase activity was determined in marine agar (MA) supplemented with 1% (v/v) Tween 20 or Tween 80 [62]. Amylase activity was assayed in MA added with 1% (w/v) starch [59]. Gelatinase activity was measured in MA supplemented with 1% (v/v) gelatin [63]. The hydrolysis of esculin was tested in MA added with 1% (w/v) esculin and 0.05% (w/v) ferric chloride[64]. The swimming motility test was conducted in MB supplemented with 0.3% (w/v) agar [65], while swarming motility was assayed in MB supplemented with 0.5% (w/v) agar [66]. Acid phosphatase activity was detected in PVK medium [67] and alkaline phosphatase was evaluated in MA supplemented with phenolphthalein phosphate 0.01% (w/v) [68]. In all these media the results were acquired by measuring haloes around the spotted area after a 7-day incubation, excluding the motility tests where growth due to the migration of cells away from the inoculation site was measured after 20 h.
2.8. In vivo assays
The virulence of the wild type Vibrio strains and their transformants (harbouring pME6010 with or without attM, aiiA and hqiA) were preliminarily tested in vivo in Artemia salina (brine shrimp) nauplii. Additionally, the virulence of Vibrio coralliilyticus VibC-Oc-193 was evaluated in Venerupis philippinarum (Manila clams) obtained from the local market.
In the case of the brine shrimp evaluation test, the hatching cysts of Artemia salina (JBL Artemio Pur) wereobtained following the manufacturer instructions using filtered and autoclaved seawater (SFSW) (salinity 36 g/L, 20°C, pH 7.3). The virulence test was performed as described in the protocol by Ha et al. [69], but with minor modifications. After hatching, 20 brine shrimp nauplii were transferred into a Petri dish (35 × 10 mm) containing 20 mL of SFSW. Then, the bacterial cells were washed twice in SFSW and added in a final concentration of 106 CFU/mL (bacterial concentration was determined by the plate counting method on TCBS) in order to infect the brine shrimps in each condition, and incubated at 25°C for 3 days. As a control, the same volume of SFSW was added to the shrimp larvae and incubated in equal conditions. The survival of the shrimps (detected by the loss of their motility) was scored daily after the addition of bacteria.
The evaluation of virulence against Manila clams was conducted following the methodology described by Drummond et al. [70]. Cultures of Vibrio spp. were centrifuged and pellets were washed twice and resuspended in the same volume of SFSW. Eight 25-mm of healthy adult Manila clams were placed in separate empty aquariums, each forming a single non-overlapping layer. 100 mL of SFSW were added to each tank to cover all clams. The bacterial suspension was added to each aquarium, giving a final concentration of 106 CFU/mL, and mixed to ensure even distribution of bacteria amongst the clams. The clams were left undisturbed for 1 h, while they were monitored in order to verify that their shells were open and that they were actively filter-feeding, thereby facilitating the uptake of bacteria. The water was then removed and the clams remained out of water for30 min in order to encourage closure of the valves and the subsequent incorporation of the bacterium within the pallial cavity. In separate control aquariums, control clams were maintained and treated as above, but without the addition of the bacterial suspension. After the infection, the clams were transferred to aerated tanks each containing 4 L of SFSW. Mortalities (detected by the irreversible relax of the adductor muscle and wide opening of the shell valves) were counted during 7 days after inoculation.
For plate counts of the number of vibrios in the aquariums in each in vivo experiment, 10-fold serial dilutions of seawater from the tanks were prepared in SFSW, plated on TCBS agar and incubated for 24 h at 25°C.
Each Manila clam assay was done in duplicate and brine shrimp tests were done in triplicate. Differences between mortalities in each experiment were statistically analysed using an ANOVA test (P<0.05) and a Tukey test using R.
Results
3.1. Production of AHLs in the Vibrionaceae strains
The synthesis of AHL-type QS signal molecules has been investigated in 34 marine strains (mainly Vibrio spp.) isolated from Oculina patagonica and Cladocora caespitosa corals and from seawater by using a well-diffusion agar-plate assay. The biosensor strains used, Agrobacterium tumefaciens NTL4 (pZLR4) and Chromobacterium violaceum CV026, developed a blue and purple colour respectively in the presence of exogenous AHLs. These experiments were carried out in triplicate and the same results were obtained. It was found that 27 strains activated A. tumefaciens NTL4 (pZLR4) and four bacteria induced the purple pigment formation of C. violaceum CV026 (Table 1).
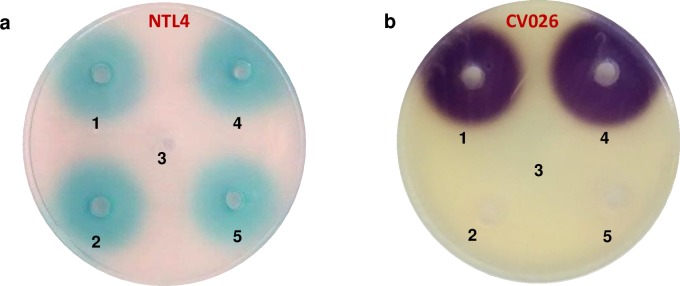
Three strains (Vibrio mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193) were chosen for further analyses based on the fact that they are considered to be emerging marine pathogenic species and also due to their high virulence and AHL production (Fig 1).
Fig 1. Well-diffusion agar-plate assay to detect AHL production by Vibrio spp.
100 μL aliquots of each culture were placed in each well: Vibrio owensii VibC-Oc-106 (1), V. coralliilyticus VibC-Oc-193 (2), V. mediterranei VibC-Oc-097 (5), V. anguillarum ATCC 19264T (positive control) (4) and MB (negative control) (3). The biosensors strains used were Agrobacterium tumefaciens NTL4 (pZLR4) (a) and Chromobacterium violaceum CV026 (b).
3.2. Analysis by TLC of the AHLs produced by the three selected Vibrio strains
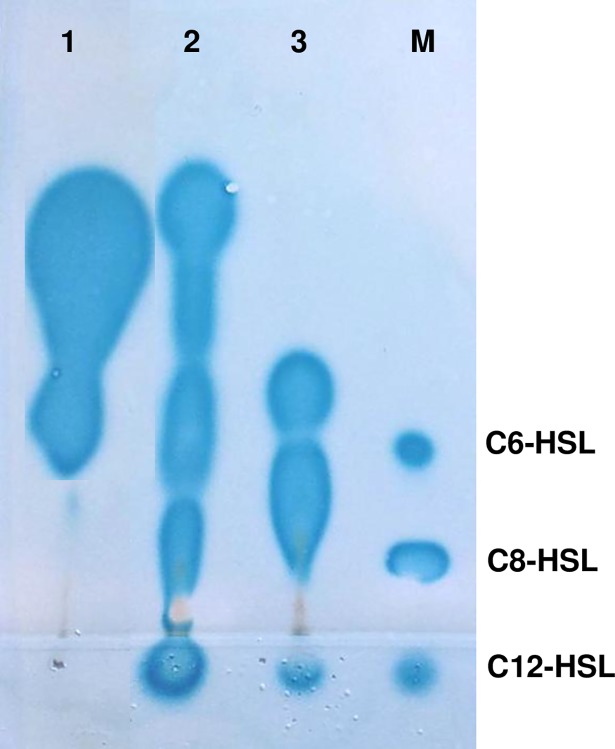
Culture extracts of the three selected strains were first analyzed by TLC in combination with an overlay of the A. tumefaciens NTL4 (pZLR4) biosensor. The AHL profiles were different amongst the tested strains, with the detection of at least two active compounds in each strain (Fig 2). In many cases, the signals migrated as tailed spots, indicating that they might be oxo-substituted AHLs. V. mediterranei VibC-Oc-097 showed the highest diversity of AHLs, with four compounds being produced. Three of them migrated similarly to those of the standards used (C6-HSL, C8-HSL and C12-HSL) and one of them migrated above the C6-HSL control. V. coralliilyticus VibC-Oc-193 produced two spots with motilities above and similar to that of C6-HSL. V. owensii VibC-Oc-106 synthesized at least three detectable signals, one of them chromatographed with a mobility similar to that of C12-HSL and the other two tailed spots migrated above the C6-HSL and C8-HSL standards.
Fig 2. TLC analysis of the AHLs produced by Vibrio spp. using the biosensor strain Agrobacterium tumefaciens NTL4 (pZLR4).
Lane 1, Vibrio coralliilyticus VibC-Oc-193; lane 2, V. mediterranei VibC-Oc-097; lane 3, V. owensii VibC-Oc-106 and lane M, synthetic AHL standards used as markers: C6-HSL (804 pmol), C8-HSL (31.6 pmol) and C12-HSL (4.8 nmol). Lanes 1 to 3 contain 25 μL of bacterial culture extract.
3.3. Identification of AHLs by HPLC/FT-HRMS analysis
Signal molecules produced by V. mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193 were identified and quantified from a large quantity of an early stationary phase culture of each strain.
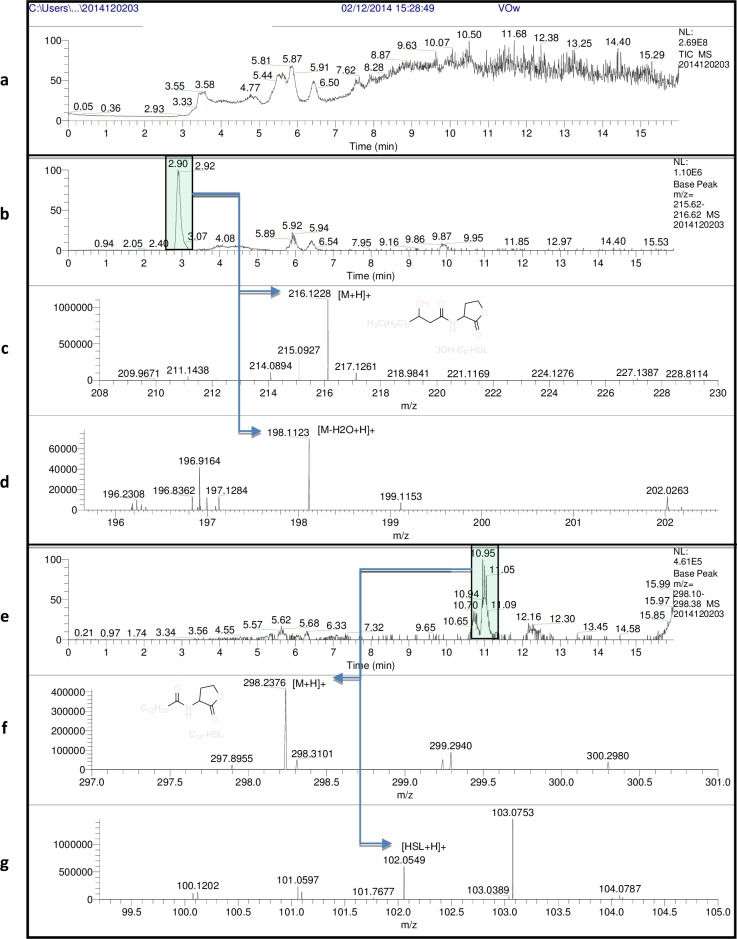
The identification of AHLs in each strain was initially made by HPLC/FT-HRMS SIM mode analysis using a mixture of twelve AHL standards (Table 2). Detection of further autoinducer molecules was performed by HPLC/FT-HRMS FS mode analysis, which showed the [M+H]+ ion peaks of additional AHLs (C9-HSL, C13-HSL, 3-OH-C5-HSL, 3-OH-C6-HSL and 3-OH-C7-HSL) (Table 3). However, the existence of these AHLs could not be confirmed due to the lack of the appropriate standards. Nevertheless, their presence was corroborated by the occurrence in their (+)-high-resolution electrospray ionisation mass spectrometry analysis [(+)-HRESIMS] of the characteristic [102 + H]+ ion peak corresponding to the homoserine lactone fragment and the [M+H-18]+ and [M+H-101]+ ion peaks which correspond to the loss of H2O and the HSL moiety (Table 3, S2 Fig). For example, the presence of 3-OH-C6-HSL and C13-HSL in V. owensii VibC-Oc-106 was corroborated by HPLC/FT-HRMS FS mode analysis through the detection of the corresponding [M-18+H]+ ion peak at m/z 198.1123 (calc. for C10H16NO3, 198.1125) for the first HSL and the detection of the [HSL+H]+ ion peak at m/z 102.0549 (calc. for C4H8NO2, 102.0550), for the second HSL (Table 3, Fig 3 and S3 Fig). In a similar way, presence of 3-OH-C5-HSL in V. mediterranei VibC-Oc-097 was confirmed by HPLC/FT-HRMS FS mode analysis through the detection of the corresponding [M-18+H]+ ion peak at m/z 184.0968 (calc. for C9H14NO3+, 184.0968) (Table 3, S4 Fig).
Table 2. AHLs identified by HPLC/FT-HRMS SIM mode analysis in Vibrio mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193.
| AHL identified | Concentration of AHLs (nmol/L)a | ||||
|---|---|---|---|---|---|
| Ion quantification (m/z) | Retention timeb (min) |
V. mediterranei VibC-Oc-097 | V. owensii VibC-Oc-106 | V. coralliilyticus VibC-Oc-193 | |
| C4-HSL | 172.0965 | 3.82 | 4.00 | nq | 3.30 |
| C6-HSL | 200.1281 | 3.53 | 1.33 | 0.08 | nq |
| C8-HSL | 228.1594 | 3.94 | 0.15 | 0.11 | nq |
| C12-HSL | 284.2220 | 4.87 | 0.67 | 4.12 | nq |
| C16-HSL | 304.2846 | 4.88 | 0.64 | 0.14 | 0.34 |
| 3-O-C10-HSL | 270.1700 | 5.63 | nq | 0.03 | nq |
| 3-O-C12-HSL | 298.2013 | 6.71 | 0.02 | nq | nq |
| 3-O-C13-HSL | 312.2169 | 7.27 | 1.76 | nq | nq |
| 3-O-C14-HSL | 326.2326 | 7.81 | nq | nq | nq |
| 3-OH-C10-HSL | 272.1856 | 5.24 | 0.36 | 0.25 | 0.69 |
| 3-OH-C12-HSL | 300.2169 | 6.36 | 8.08 | 2.77 | 0.12 |
| 3-OH-C14-HSL | 328.2482 | 7.53 | nq | nq | nq |
aCalculations were made from the adjusted peak areas from each strain. Data relate to those AHLs which were positively identified against a standard and those that produce a significant quantification ion signal (R>30,000; mass tolerance < 1 ppm).
bRetention time of the standards used in the quantification process. nq not quantifiable (limit of detection of AHLs < 0.10 ng/mL).
Table 3. AHLs detected by HPLC/FT-HRMS FS mode analysis in Vibrio mediterranei VibC-Oc-097 and Vibrio owensii VibC-Oc-106.
| Microorganism | AHL detected |
Retention time (min) |
Principal product ions (m/z) | Identity of principal product ions |
|---|---|---|---|---|
|
V. mediterranei VibC-Oc-097 |
3-OH-C5-HSL | 6.37 | 202.1072 184.0968 |
[M+H]+, [M-H2O+H]+ a |
| V. owensii VibC-Oc-106 | 3-OH-C6-HSL | 2.90 | 216.1228, 198.1123 |
[M+H]+, [M- H2O+H]+ |
| 3-OH-C7-HSL | 3.82 | 230.1381 | [M+H]+ | |
| C13-HSL | 11.00 | 298.2376 102.0549 |
[M+H]+, [HSL+H]+ b |
a[M-H2O +H]+ is the result of a loss of water
b[HSL+H]+ corresponds to the HSL moiety
Fig 3. HPLC/FT-HRESIMS (FS mode) experiments for the detection of N-acylhomoserine lactones from Vibrio owensii VibC-Oc-106.
Total ion chromatogram (a). Extracted mass chromatogram (m/z 215.62–216.62) showing the peak at tR = 2.90 min (b). Expanded regions of the (+)-HRESIMS of the peak at tR = 2.90 min identified as 3-OH-C6-HSL showing the ion peaks [M+H]+ at m/z 216.1228 (calc. for C10H18NO4 216.1230) (c) and [M-H2O+H]+ at m/z 198.1123 (calc. for C10H16NO3 198.1125) (d). Extracted mass chromatogram (m/z 298.10–29838) showing the peak at tR = 11.00 min (e). Expanded regions of the (+)-HRESIMS of the peak at tR = 11.00 min identified as C13-HSL showing the ion peaks [M+H]+ at m/z 298.2376 (calc. for C17H32NO3 298.2377) (f) and [HSL+H]+ at m/z 102.0549 (calc. for C4H8NO2 102.0550) (g).
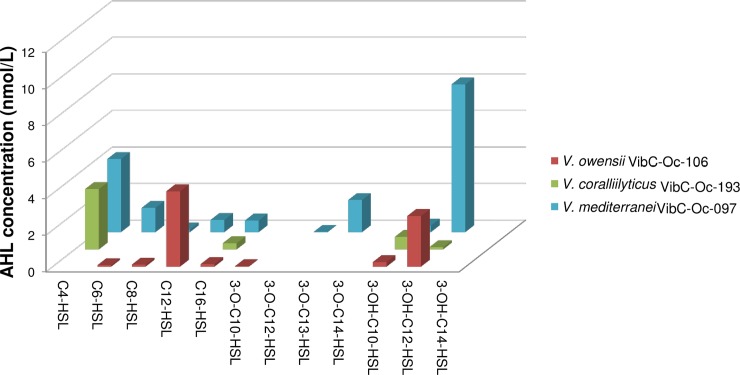
As can be seen in the results obtained, the AHL-profile for each bacterium strain is different (Tables 2 and 3). If the percentage in which the molecules are present in the strains tested is considered, 3-OH-C12-HSL is the most predominant in V. mediterranei VibC-Oc-097, C4-HSL is the most abundant in V. coralliilyticus VibC-Oc-193 and C12-HSL is the most commonly produced by V. owensii VibC-Oc-106. Furthermore, the highest concentration of all the identified AHLs corresponds to 3-OH-C12-HSL in V. mediterranei VibC-Oc-097 (Table 2, Fig 4).
Fig 4. Concentration of the major AHLs quantified in Vibrio mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193 using HPLC/FT-HRESIMS (SIM mode).
AHL concentration values are shown in nmol/L.
3.4. Selection of AHL-producing defective Vibrio strains
In order to evaluate the cellular functions controlled by AHLs, two strategies were used to reduce the AHLs produced by the selected Vibrio strains.
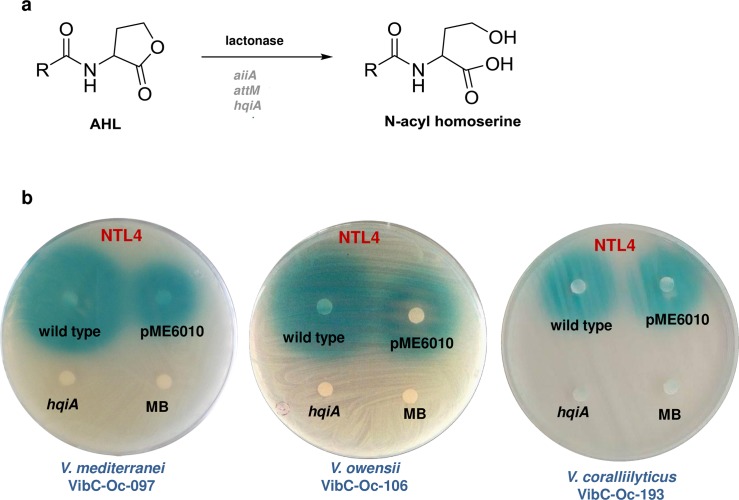
As a first strategy, AHL-lactonase genes were expressed in each Vibrio strain. Competent cells of the three pathogenic Vibrio strains were transformed with recombinant plasmids expressing different AHL lactonase enzymes such as the AiiA, AttM and HqiA, which hydrolyze the lactone ring of AHL signals (Fig 5A). The empty plasmid pME6010 was also transformed into Vibrio strains as a negative control. Positive transformants were firstly confirmed by the amplification of the corresponding AHL-lactonase gene (~750–800 bp). Secondly, the transformants containing each of the lactonase genes that had lost the ability to activate the biosensor A. tumefaciens NTL4 (pZLR4) were selected. In each case, Vibrio containing the plasmid pME6010, wild type strains and MB were used as negative controls (Fig 5B).
Fig 5. AHL degradation by lactonase-type quorum quenching enzymes.
AHL degradation mechanism of the lactonase enzymes AiiA, AttM and HqiA (a). Well-diffusion agar-plate assay to detect AHL production of each wild type Vibrio sp. and its transformants (expressing hqiA gene) using the biosensor Agrobacterium tumefaciens NTL4 (pZLR4). 10 μL-aliquots of each bacterial culture extracts were placed in each sterile disk (b).
Additionally, co-cultures of each of the three pathogenic strains V. mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193 with the AHL-degrading strains PQQ-42 and PQQ-44 [67] were performed in parallel (data not shown). In all the cases, the addition of these QQ strains resulted in a significant reduction of AHLs. None of these strains had a negative effect on the growth of any of the Vibrio strains tested (data not shown).
3.5. AHL degradation reduces some virulence factors produced by the Vibrio spp
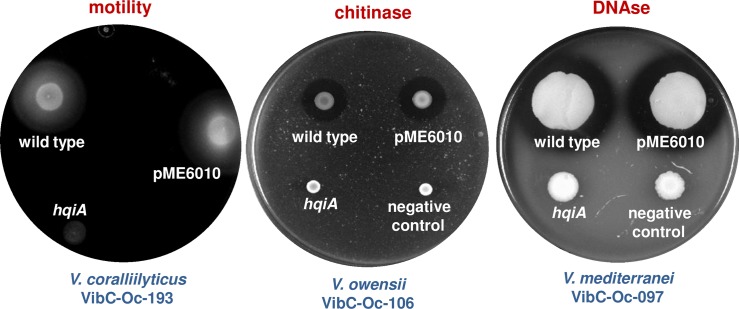
Different phenotypes were analyzed in the AHL defective Vibrio strains (expressing AHL-lactonase genes). The results obtained are the same forthe three lactonases tested, but we only present the outcome after the expression of hqiA in Table 4. As shown, the reduction in the accumulation of AHLs in the Vibrio strains diminished different phenotypes. For instance, swimming motility was reduced and caseinase and amylase activity was eliminated in V. coralliilyticus VibC-Oc-193; chitinase and DNAse activities were diminished in V. owensii VibC-Oc-106 and V. mediterranei VibC-Oc-097, respectively (Fig 6, Table 4).
Table 4. Effect of AHL-degradation upon some of the virulence factors produced by Vibrio spp. after the expression of the hqiA QQ gene.
| Phenotype |
V. owensii VibC-Oc-106 |
V. mediterranei VibC-Oc-097 |
V. coralliilyticus VibC-Oc-193 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| wild type | pME6010 | hqiA | wild type | pME6010 | hqiA | wild type | pME6010 | hqiA | |
| AHLs | ++ | ++ | - | ++ | ++ | - | ++ | ++ | - |
| Swimming motility | ++ | ++ | + | ++ | ++ | + | ++ | ++ | + |
| Swarming motility | ++ | + | + | ++ | ++ | ++ | ++ | ++ | + |
| Hemolysis | ++ | ++ | + | ++ | ++ | ++ | ++ | ++ | + |
| Caseinase | ++ | ++ | ++ | + | + | + | +++ | ++ | - |
| Amylase | ++ | ++ | + | - | - | - | ++ | ++ | - |
| DNAse | ++ | ++ | + | ++ | ++ | + | ++ | ++ | ++ |
| Chitinase | ++ | ++ | + | ++ | ++ | + | ++ | ++ | +++ |
| Lipase (Tween 20) | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Lipase (Tween 80) | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Acid phosphatase | + | + | + | + | + | + | - | - | - |
| Alkaline phosphatase | ++ | ++ | + | ++ | ++ | ++ | ++ | ++ | + |
| Esculin | + | + | + | + | + | + | + | + | + |
| Gelatinase | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
++ strong activity, + medium activity and - no activity after expressing the pME6010 plasmid with and without the hqiA QQ gene.
Fig 6. Examples of phenotypes affected by AHL reduction in the three Vibrio strains tested.
3.6. AHL degradation reduces virulence of Vibrio spp. on Artemia salina and Venerupis philippinarum
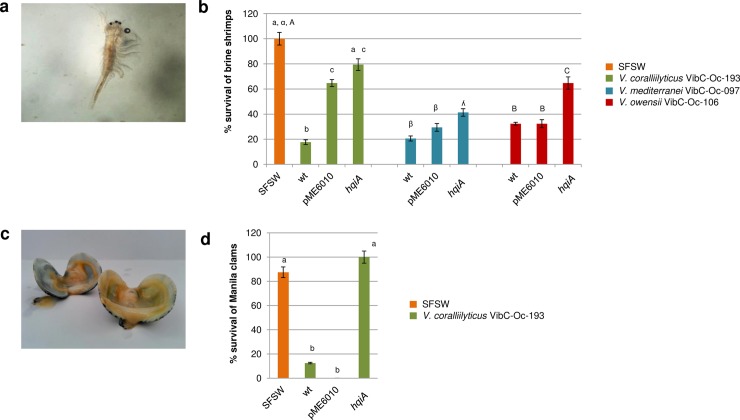
To test whether the interference of AHL production in V. mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193 had an impact on virulence, in vivo preliminary assays were conducted on Artemia salina (brine shrimp) nauplii. Since the expression of some phenotypes and virulence factors were diminished or eliminated after the expression of AHL-lactonase genes, the AHL defective strains harbouring the hqiA gene were chosen for these experiments. The loss of motility of the nauplii was observed with a magnifying glass during three days and the results showed that the expression of the hqiA AHL-lactonase gene in the tested Vibrio spp. significantly reduced the mortality upon Artemia nauplii in comparison with the wild type and the pME6010-expressing Vibrio strains (Fig 7). Based on the results obtained with the different Vibrio species tested, it was noted that V. owensii VibC-Oc-106 was the most virulent species since it produced a higher mortality on the brine shrimp nauplii.
Fig 7. Artemia salina and Venerupis philippinarum in vivo assays.
Artemia salina nauplii observed under a magnifying glass (a). Percentage of survival of brine shrimp after inoculation with V. coralliilyticus VibC-Oc-193, V. mediterranei VibC-Oc-097, V. owensii VibC-Oc-106, and their transformants (b). Dead Manila clams showing open valves and decontracted muscle (c). Percentage of survival of Manila clams after the inoculation of Vibrio coralliilyticus VibC-Oc-193 and its transformants (d). Error bars represent standard deviations. Different letters above the bars indicate that the values are significantly different (P<0.05).
Additionally, the virulence of V. coralliilyticus VibC-Oc-193 was also preliminarily tested on Venerupis philippinarum (Manila clams), since the other two strains produced a very slow mortality in this invertebrate model and the assay could not be maintained in our laboratory facilities. For each aquarium, death symptoms on the clams (wide open valves and decontracted adductor muscle) were observed during seven days. The results indicated that V. coralliilyticus VibC-Oc-193 expressing the AHL lactonase gene significantly reduced mortality upon V. philippinarum in comparison with the controls (Fig 7).
Regarding the number of CFU of Vibrio spp. in water in all the experiments conducted, in each case the concentration of bacteria showed similar values throughout the experiment (data not shown).
Discussion
Marine pathogenic bacteria can completely kill mollusks, fish and coral populations that are cultivated in aquaculture, causing drastic economic losses [71,72]. Even though antibiotics have been used for many years to control infections, the emergence of antibiotic-resistant strains [73,74] and sometimes the inefficacy of the treatments [75] make the situation more and more alarming. Global efforts are needed to search for novel strategies to control pathogens in aquaculture and to promote a responsible use of antibiotics to make the industry more sustainable as well as to maintain a healthy environment [76]. In the case of pathogens which depend on QS to regulate virulence, such as some species of Vibrio, the degradation of the AHL signal molecules through QQ could become a good alternative for fighting pathogenicity as pathogen numbers are reduced rather than directly being killed [39,77–79].
To obtain a better understanding of the production of communication signals in aquaculture-related bacteria we have investigated the QS systems in 34 marine strains isolated from healthy and diseased corals and seawater belonging to the genera Vibrio, Photobacterium and Listonella [43]. Although many of these strains have been associated to coral diseases such as bleaching [80–82], most of these diseases have also been related to mollusks, crustaceans and fish. For example, V. coralliilyticus is now extending its host range to bivalve species such as the hard clam (Mercenaria mercenaria), the flat oyster (Ostrea edulis), the bay scallop (Argopecten irradians), the green-lipped mussel (Perna canaliculus) and the naval shipworm (Teredo navalis), constituting a serious threat for the bivalve industry, being one of the most important emerging pathogens responsible for larval mortality detected in bivalve hatcheries in France, New Zealand and the United States [6,83,84]. Other species, such as V. owensii and V. mediterranei have also been associated to other organisms apart from coral: crustaceans such as the ornate spiny lobster (Panulirus ornatus) [85,86] and mollusks such as the blue mussel (Mytilus edulis) or the Manila clam (Venerupis philippinarum) [87,88]. Nevertheless, the misidentification of several V. coralliilyticus such as V. tubiashii and V. neptunius, and the reclassification of V. shiloi as V. mediterranei and of V. communis as V. owensii [83,89–91] has complicated the role of these pathogens in fish and mollusk mortality and as the potential etiological agents involved in aquaculture-related diseases.
Nowadays, several methods have been described to easily detect the production of AHLs, such as the "cross-streak" method [92] or the double-layer microplate high-throughput assay that has been performed to test a high number of isolates [16]. In this study we have chosen a well-diffusion agar-plate assay and two biosensor strains based on previous experience in our laboratory [29,30,47]. The biosensors used were Chromobacterium violaceum CV026, which responds to exogenously added short-chain AHLs (C4-HSL and C6-HSL), and Agrobacterium tumefaciens NTL4 (pZLR4), a sensitive, broad-spectrum AHL-responsive reporter. Our results indicate that four strains induce the biosensor CV026 and 27 out of the 34 bacteria synthesized signal molecules to activate NTL4. In a previous study, Yang et al.[15] have also found that a high number of bacteria (21 out of 25 strains) belonging to the family Vibrionaceae induced the biosensor A. tumefaciens. To date, the use of biosensors that are able to detect the production of AHLs has been very useful for the identification of a large number of AHL-based QS systems [16,93,94].
Additionally, the use of biosensor microorganisms in combination with TLC provides a simple and rapid way of determining the number and nature of the AHLs produced by a particular strain [44]. Based on these techniques some authors have shown the production of AHLs in different strains of the family Vibrionaceae [15,94–96].
In this study, we have carried out an in-depth characterization of the AHL signal molecules of three Vibrio strains selected by the analysis of their AHL profiles and the virulence capacity of the species to which they are related: V. mediterranei VibC-Oc-097, V. owensii VibC-Oc-106 and V. coralliilyticus VibC-Oc-193. By using a TLC analysis, our results indicate the existence of a high AHL diversity amongst the three species, as other authors have previously reported for other species [15,16].
Since the TLC technique only provides limited information concerning the nature of signal compounds present in the extracts, HPLC/FT-HRMS analyses was essential for determining the structures of these potential signal molecules [50,51]. In this analysis, the whole culture of each strain was used to prepare the extract instead of only using the cell-free supernatant. For this reason, we were able to enhance the extraction of long-chain AHLs, because otherwise their low permeability through the cell membrane limits their extraction and detection [97]. Additionally, no differences were found in the AHL extraction when the supernatant was acidified (data not shown), as suggested by other authors [98]. With respect to the organic solvent needed for the AHL extraction, dichloromethane was used, although no differences were detected when ethyl acetate was used (data not shown). As explained, TLC and HPLC/FT-HRMS analysis showed a high diversity for the production of AHLs molecules by the tested strains, the long-acyl chains AHLs being more predominant in three of the Vibrio species analyzed.
The different AHL molecules produced by the three Vibrio spp. tested may be the result of the presence of several LuxI synthases acting in a multiple signalling system, as occurs in Pseudomonas aeruginosa [99], or the same LuxI synthesizing multiple AHLs based on the intracellular availability of acyl-carrier proteins, as described in Halomonas anticariensis [100].
To date, it has been shown that virulence is regulated by QS in many Vibrio spp. affecting fish and shellfish. Despite the extensive knowledge about the molecules involved and the biological processes controlled by intercellular communication in some Vibrio strains, less is known about the diversity and role of AHLs produced by a wide range of emerging pathogenic species, such as V. coralliilyticus or V. mediterranei. The in-depth study of QS is important for a better understanding of their pathogenicity and for the development of future treatments for the diseases, they cause. In a previous study, we demonstrated that AHL-degradation protects Oculina patagonica coral from bleaching caused by V. mediterranei VibC-Oc-097. The addition of the AHL-degrading strain PQQ-42 completely eliminated the AHLs produced by V. mediterranei VibC-Oc-097, reducing its swimming motility and protease activity, which therefore seem to be regulated by QS systems [30]. AHL degradation in other Vibrio species has also helped to understand the role of QS in the regulation of virulence factors, such as biofilm or hemolysin production [16,101]. In this study, the results obtained indicate that the expression of AHL-lactonase genes (aiiA, attM or hqiA) in the pathogenic selected Vibrio strains resulted in a significant decrease of AHL accumulation and a drastic reduction of some potential virulence factors, such as the production of protease and chitinase, swimming motility and hemolysis [39,102,103]. Additionally, the AHL defective strains of each selected Vibrio produced a significant reduction of the virulence in vivo in brine shrimp (Artemia salina). In the case of V. coralliilyticus VibC-Oc-193, the AHL defective strain was significantly less virulent against Manila clams (Venerupis philippinarum) when compared with the control, originating a 100% survival, which is the second most important bivalve in aquaculture worldwide with a very important commercial value [40]. Although these results are preliminary, they indicate that the expression of the QQ enzyme HqiA [55] plays an important role in attenuating the virulence in the invertebrate models used, reinforcing the idea that QS is the mechanism involved in the regulation of the virulence of these Vibrio strains.
To the best of our knowledge, this is the first report where the QS signal molecules have been characterized and correlated to the expression of virulence factors in the emerging pathogenic species V. owensii, V. mediterranei and V. coralliilyticus. Therefore, this research paves the way to the future development of new therapies based on quorum quenching, an AHL-disruption approach that has already been successfully undertaken for the control of infection in many marine animals.
Supporting information
800 ng/mL (a) and 1,000 ng/mL (b and c). External calibration curves by adding C4-HSL, C16-HSL, 3-O-C10-HSL, 3-O-C12-HSL, 3-O-C13-HSL 3-OH-C10-HSL, 3-OH-C12-HSL, 3-OH-C13-HSL and 3-OH-C14-HSL.
(DOCX)
General AHL structure (a). Mass spectrometry fragmentation pathway of AHLs (b).
(DOCX)
Total ion chromatogram (a). Extracted mass chromatogram (m/z 230.13–230.14) showing the peak at tR = 3.82 min (b). Expanded regions of the (+)-HRESIMS of the peak at tR = 3.82 min identified as 3-OH-C7-HSL showing the [M+H]+ ion peak at m/z 230.1381 (calc. for C11H20NO4 230.1387) (c).
(DOCX)
Total ion chromatogram (a). Extracted mass chromatogram (m/z 202.00–202.22) showing the peak at tR = 6.37 min (b). Expanded region of the (+)-HRESIMS of the peak at tR = 6.37 min identified as 3-OH-C5-HSL showing the ion peaks [M+H]+ at m/z 202.1073 (calc. for C9H16NO4 202.1074) (c). Expanded region of the (+)-HRESIMS of the peak at tR = 6.37 min identified as 3-OH-C5-HSL showing the [M-H2O+H]+ ion peak at m/z 184.0968 (calc. for C9H14NO3 184.0968) (d).
(DOCX)
(PDF)
Numbers are counts in the mass spectra.
(PDF)
Acknowledgments
The authors thank Angela Tate for her critical reading of this manuscript, as well as Josefa Antón and Esther Rubio-Portillo for providing the Vibrio strains.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported in part by grants from the Ministerio de Economía y Competitividad (Spain) (http://www.mineco.gob.es/portal/site/mineco/idi) (AGL2012-39274-C02-02; AGL2015-68806-R; AGL2015-63740-C2-2-R). José Carlos Reina is supported by a FPU fellowship from the Ministerio de Educación, Cultura y Deporte (Spain) (https://www.mecd.gob.es/portada-mecd/) (FPU15-01717) and a Initiation to Research fellowship for Master Students (2016) from the University of Granada (Spain) (http://www.ugr.es). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Food and Agriculture Organization of the United Nations. The State of world fisheries and aquaculture. 2016.
- 2.Rosenberg E, Kushmaro A. Microbial diseases of corals: pathology and ecology. Coral reefs: an ecosystem in transition. Dordrecht: Springer Netherlands; 2011. pp. 451–464. doi: 10.1007/978-94-007-0114-4_26 [Google Scholar]
- 3.Austin B, Zhang XH. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol. 2006;43: 119–124. doi: 10.1111/j.1472-765X.2006.01989.x [DOI] [PubMed] [Google Scholar]
- 4.Austin B, Austin D. Bacterial fish pathogens: disease of farmed and wild fish 3rd edition. Verlag K, editor. Berlin, Germany: Springer; 1999. [Google Scholar]
- 5.Igbinosa EO, Okoh AI. Emerging Vibrio species: an unending threat to public health in developing countries. Res Microbiol. 2008;159: 495–506. doi: 10.1016/j.resmic.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 6.Dubert J, Barja JL, Romalde JL. New insights into pathogenic vibrios affecting bivalves in hatcheries: present and future prospects. Front Microbiol. 2017;8: 1–16. doi: 10.3389/fmicb.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romalde JL, Dieguez AL, Lasa A, Balboa S. New Vibrio species associated to molluscan microbiota: a review. Front Microbiol. 2014;4: 413 doi: 10.3389/fmicb.2013.00413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng. 2008;10: 145–167. doi: 10.1146/annurev.bioeng.10.061807.160536 [DOI] [PubMed] [Google Scholar]
- 9.Ng W, Bassler B. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43: 197–222. doi: 10.1146/annurev-genet-102108-134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González JE, Marketon MM. Quorum sensing in nitrogen-fixing Rhizobia. Microbiol Mol Biol Rev. 2003;67: 574–592. doi: 10.1128/MMBR.67.4.574-592.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead NA, Barnard AML, Slater H, Simpson NJL, Salmond GPC. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25: 365–404. [DOI] [PubMed] [Google Scholar]
- 12.Bruhn JB, Dalsgaard I, Nielsen KF, Buchholtz C, Larsen JL, Gram L. Quorum sensing signal molecules (acylated homoserine lactones) in Gram-negative fish pathogenic bacteria. Dis Aquat Organ. 2005;65: 43–52. doi: 10.3354/dao065043 [DOI] [PubMed] [Google Scholar]
- 13.Croxatto A, Pride J, Hardman A, Williams P, Cámara M, Milton D. A distinctive dual-channel quorum sensing system operates in Vibrio anguillarum. Mol Microbiol. 2004;52: 1677–1689. doi: 10.1111/j.1365-2958.2004.04083.x [DOI] [PubMed] [Google Scholar]
- 14.Morohoshi T, Inaba T, Kato N, Kanai K, Ikeda T. Identification of quorum-sensing signal molecules and the LuxRI homologs in fish pathogen Edwardsiella tarda. J Biosci Bioeng. 2004;98: 274–281. doi: 10.1016/S1389-1723(04)00281-6 [DOI] [PubMed] [Google Scholar]
- 15.Yang Q, Han Y, Zhang XH. Detection of quorum sensing signal molecules in the family Vibrionaceae. J Appl Microbiol. 2011;110: 1438–1448. doi: 10.1111/j.1365-2672.2011.04998.x [DOI] [PubMed] [Google Scholar]
- 16.García-Aljaro C, Vargas-Cespedes GJ, Blanch AR. Detection of acylated homoserine lactones produced by Vibrio spp. and related species isolated from water and aquatic organisms. J Appl Microbiol. 2011;112: 383–389. doi: 10.1111/j.1365-2672.2011.05199.x [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen BB, Nielsen KF, MacHado H, Melchiorsen J, Gram L, Sonnenschein EC. Global and phylogenetic distribution of quorum sensing signals, acyl homoserine lactones, in the family of Vibrionaceae. Mar Drugs. 2014;12: 5527–5546. doi: 10.3390/md12115527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Kuang W, Long L, Zhang S. Production of quorum-sensing signals by bacteria in the coral mucus layer. Coral Reefs. 2017;36: 1235–1241. doi: 10.1007/s00338-017-1616-3 [Google Scholar]
- 19.Prado S, Dubert J, Barja JL. Characterization of pathogenic vibrios isolated from bivalve hatcheries in Galicia, NW Atlantic coast of Spain. Description of Vibrio tubiashii subsp. europaensis subsp. nov. Syst Appl Microbiol. 2015;38: 26–29. doi: 10.1016/j.syapm.2014.11.005 [DOI] [PubMed] [Google Scholar]
- 20.Gómez-León J, Villamil L, Lemos ML, Novoa B, Figueras A. Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl Environ Microbiol. 2005;71: 98–104. doi: 10.1128/AEM.71.1.98-104.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojas R, Miranda CD, Santander J, Romero J. First report of Vibrio tubiashii associated with a massive larval mortality event in a commercial hatchery of scallop Argopecten purpuratus in Chile. Front Microbiol. 2016;7: 1–13. doi: 10.3389/fmicb.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Defoirdt T, Verstraete W, Bossier P. Luminescence, virulence and quorum sensing signal production by pathogenic Vibrio campbellii and Vibrio harveyi isolates. J Appl Microbiol. 2008;104: 1480–1487. doi: 10.1111/j.1365-2672.2007.03672.x [DOI] [PubMed] [Google Scholar]
- 23.Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36: 940–954. Available: http://www.ncbi.nlm.nih.gov/pubmed/10844680 [DOI] [PubMed] [Google Scholar]
- 24.Ruwandeepika HAD, Defoirdt T, Bhowmick PP, Karunasagar I, Karunasagar I, Bossier P. In vitro and in vivo expression of virulence genes in Vibrio isolates belonging to the Harveyi clade in relation to their virulence towards gnotobiotic brine shrimp (Artemia franciscana). Environ Microbiol. 2011;13: 506–517. doi: 10.1111/j.1462-2920.2010.02354.x [DOI] [PubMed] [Google Scholar]
- 25.Milton DL, Hardman A, Camara M, Chhabra SR, Bycroft BW, Stewart GSAB, et al. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-L-homoserine lactone. J Bacteriol. 1997;179: 3004–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saurav K, Costantino V, Venturi V, Steindler L. Quorum sensing inhibitors from the sea discovered using bacterial N-acyl-homoserine lactone-based biosensors. Mar Drugs. 2017;15: 53 doi: 10.3390/md15030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manefield M, de Nys R, Naresh K, Roger R, Givskov M, Peter S, et al. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology. 1999;145: 283–291. doi: 10.1099/13500872-145-2-283 [DOI] [PubMed] [Google Scholar]
- 28.Romero M, Martin-Cuadrado AB, Roca-Rivada A, Cabello AM, Otero A. Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol Ecol. 2011;75: 205–217. doi: 10.1111/j.1574-6941.2010.01011.x [DOI] [PubMed] [Google Scholar]
- 29.Torres M, Romero M, Prado S, Dubert J, Tahrioui A, Otero A, et al. N-acylhomoserine lactone-degrading bacteria isolated from hatchery bivalve larval cultures. Microbiol Res. 2013;168: 547–554. doi: 10.1016/j.micres.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 30.Torres M, Rubio-Portillo E, Antón J, Ramos-Esplá AA, Quesada E, Llamas I. Selection of the N-acylhomoserine lactone-degrading bacterium Alteromonas stellipolaris PQQ-42 and of its potential for biocontrol in aquaculture. Front Microbiol. 2016;7: 646 doi: 10.3389/fmicb.2016.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uroz S, D’Angelo-Picard C, Carlier A, Elasri M, Sicot C, Petit A, et al. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology. 2003;149: 1981–1989. doi: 10.1099/mic.0.26375-0 [DOI] [PubMed] [Google Scholar]
- 32.Uroz S, Heinonsalo J. Degradation of N-acyl homoserine lactone quorum sensing signal molecules by forest root-associated fungi. FEMS Microbiol Ecol. 2008;65: 271–278. doi: 10.1111/j.1574-6941.2008.00477.x [DOI] [PubMed] [Google Scholar]
- 33.Uroz S, Dessaux Y, Oger P. Quorum sensing and quorum quenching: the yin and yang of bacterial communication. ChemBioChem. 2009;10: 205–216. doi: 10.1002/cbic.200800521 [DOI] [PubMed] [Google Scholar]
- 34.Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol. 2011;37: 121–140. doi: 10.3109/1040841X.2010.532479 [DOI] [PubMed] [Google Scholar]
- 35.Dong Y, Zhang L. Quorum sensing and quorum-quenching enzymes. J Microbiol. 2005;43: 101–109. doi:2133 [pii] [PubMed] [Google Scholar]
- 36.Uroz S, Chhabra SR, Cámara M, Williams P, Oger P, Dessaux Y. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151: 3313–3322. doi: 10.1099/mic.0.27961-0 [DOI] [PubMed] [Google Scholar]
- 37.Park S, Kang H, Jang H, Koo B, Yum D, Park S, et al. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl Environ Microbiol. 2005;71: 2632–2641. doi: 10.1128/AEM.71.5.2632-2641.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tinh NTN, Dierckens K, Sorgeloos P, Bossier P. A review of the functionality of probiotics in the larviculture food chain. Mar Biotechnol. 2008;10: 1–12. doi: 10.1007/s10126-007-9054-9 [DOI] [PubMed] [Google Scholar]
- 39.Natrah F, Defoirdt T, Sorgeloos P, Bossier P. Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture. Mar Biotechnol. 2011;13: 109–126. doi: 10.1007/s10126-010-9346-3 [DOI] [PubMed] [Google Scholar]
- 40.Chu W, Zhou S, Zhu W, Zhuang X. Quorum quenching bacteria Bacillus sp. QSI-1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Sci Rep. 2014;4: 5446 doi: 10.1038/srep05446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordero D, Delgado M, Liu B, Ruesink J, Saavedra C. Population genetics of the Manila clam (Ruditapes philippinarum) introduced in North America and Europe. Sci Rep. 2017;7: 1–13. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorgeloos P, Persoone G. Technological improvements for the cultivation of invertebrates as food for fishes and crustaceans. Hatching and culturing of the brine shrimp, Artemia salina. Aquaculture. 1975;6: 303–317. doi: 10.1016/0044-8486(75)90110-6 [Google Scholar]
- 43.Rubio-Portillo E, Yarza P, Peñalver C, Ramos-Esplá A a, Antón J. New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J. 2014;2003: 1–14. doi: 10.1038/ismej.2014.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Rinehart KL, et al. Detecting and characterizing N-acylhomoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci. 1997;94: 6036–6041. doi: 10.1073/pnas.94.12.6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci. 1974;71: 3672–3676. doi: 10.1073/pnas.71.9.3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, et al. Quorum sensing and Chromobacteriurn violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology. 1997;143: 3703–3711. doi: 10.1099/00221287-143-12-3703 [DOI] [PubMed] [Google Scholar]
- 47.Llamas I, Quesada E, Martínez-Cánovas MJ, Gronquist M, Eberhard A, González JE. Quorum sensing in halophilic bacteria: detection of N-acyl-homoserine lactones in the exopolysaccharide-producing species of Halomonas. Extremophiles. 2005;9: 333–341. doi: 10.1007/s00792-005-0448-1 [DOI] [PubMed] [Google Scholar]
- 48.Marketon MM, Gonzalez JE. Identication of two quorum-sensing systems in Sinorhizobium meliloti. Society. 2002;184: 3466–3475. doi: 10.1128/JB.184.13.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espada A, Anta C, Bragado A, Rodríguez J, Jiménez C. An approach to speed up the isolation of hydrophilic metabolites from natural sources at semipreparative level by using a hydrophilic–lipophilic balance/mixed-mode strong cation exchange–high-performance liquid chromatography/mass spectrometry system. J Chromatogr. 2011;1218: 1790–1794. doi: 10.1016/j.chroma.2011.01.072 [DOI] [PubMed] [Google Scholar]
- 50.Osorio CR, Rivas AJ, Balado M, Fuentes-Monteverde JC, Rodríguez J, Jiménez C, et al. A transmissible plasmid-borne pathogenicity island confers piscibactin biosynthesis in the fish pathogen Photobacterium damselae subsp. piscicida. Appl Environ Microbiol. 2015;81: 5867–5879. doi: 10.1128/AEM.01580-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balado M, Souto A, Vences A, Careaga VP, Valderrama K, Segade Y, et al. Two catechol siderophores, acinetobactin and amonabactin, are simultaneously produced by Aeromonas salmonicida subsp. salmonicida sharing part of the biosynthetic pathway. ACS Chem Biol. 2015;10: 2850–2860. doi: 10.1021/acschembio.5b00624 [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Russell D. Molecular cloning: a laboratory manual New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 53.Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci. 2000;97: 3526–3531. doi: 10.1073/pnas.060023897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang XH, Wang L, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci. 2002;99: 4638–4643. doi: 10.1073/pnas.022056699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres M, Uroz S, Salto R, Fauchery L, Quesada E, Llamas I. HqiA, a novel quorum-quenching enzyme which expands the AHL lactonase family. Sci Rep. 2017;7: 943 doi: 10.1038/s41598-017-01176-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact. 2000;13: 232–237. doi: 10.1094/MPMI.2000.13.2.232 [DOI] [PubMed] [Google Scholar]
- 57.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166: 557–580. [DOI] [PubMed] [Google Scholar]
- 58.Morelle G. A plasmid extraction procedure on a miniprep scale. BRL Focus. 1989;11: 7–8. [Google Scholar]
- 59.Barrow G.I. and Feltham RKA. Cowan and Steel’s Manual for the identification of medical bacteria Barrow GI, Feltham RKA, editors. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- 60.Wu Y, Cheng C, Li Y. Cloning and expression of chitinase A from Serratia marcescens for large-scale preparation of N,N-Diacetyl chitobiose. J Chinese Chem Soc. 2009;56: 688–695. doi: 10.1002/jccs.200900103 [Google Scholar]
- 61.Jeffries CD, Holtmian DF, Guse DG. Rapid method for determining the activity of microorganisms on nucleic acids. J Bacteriol. 1957;73: 590–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mourey A, Kilbertus G. Simple media containing stabilized tributyrin for demonstrating lipolytic bacteria in foods and soils. J Appl Bacteriol. 1976;40: 47–51. doi: 10.1111/j.1365-2672.1976.tb00589.x [DOI] [PubMed] [Google Scholar]
- 63.Pickett MJ, Greenwood JR, Harvey SM. Tests for detecting degradation of gelatin: comparison of five methods. J Clin Microbiol. 1991;29: 2322–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choopun N, Louis V, Huq A, Colwell RR. Simple procedure for rapid identification of Vibrio cholerae from the aquatic environment. Society. 2002;68: 995–998. doi: 10.1128/AEM.68.2.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rui H, Liu Q, Ma Y, Wang Q, Zhang Y. Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus. FEMS Microbiol Lett. 2008;285: 155–162. doi: 10.1111/j.1574-6968.2008.01185.x [DOI] [PubMed] [Google Scholar]
- 66.O’May C, Tufenkji N. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl Environ Microbiol. 2011;77: 3061–3067. doi: 10.1128/AEM.02677-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pikovskaya RI. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Mikrobiologiya. 1948;17: 362–370. [Google Scholar]
- 68.Baird-Parker A. A classification of micrococci and staphylococci based on physiological and biochemical tests. J Gen Microbiol. 1963;30: 409–427. doi: 10.1099/00221287-30-3-409 [DOI] [PubMed] [Google Scholar]
- 69.Ha C, Kim SK, Lee MN, Lee JH. Quorum sensing-dependent metalloprotease VvpE is important in the virulence of Vibrio vulnificus to invertebrates. Microb Pathog. 2014;71–72: 8–14. doi: 10.1016/j.micpath.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 70.Drummond LC, Balboa S, Beaz R, Mulcahy MF, Barja JL, Culloty SC, et al. The susceptibility of Irish-grown and Galician-grown Manila clams, Ruditapes philippinarum, to Vibrio tapetis and brown ring disease. J Invertebr Pathol. 2007;95: 1–8. doi: 10.1016/j.jip.2006.11.010 [DOI] [PubMed] [Google Scholar]
- 71.Lafferty K, Harvell C, Conrad J, Friedman C, Kent M, Kuris A, et al. Infectious diseases affect marine fisheries and aquaculture economics. Ann Rev Mar Sci. 2015;7: 471–496. doi: 10.1146/annurev-marine-010814-015646 [DOI] [PubMed] [Google Scholar]
- 72.Paillard C, Le Roux F, Borrego JJ. Bacterial disease in marine bivalves, a review of recent studies: Trends and evolution. Aquat Living Resour. 2004;17: 477–498. doi: 10.1051/alr:2004054 [Google Scholar]
- 73.Smith P. Antimicrobial resistance in aquaculture. Rev Sci Tech. 2008;27: 243–264. [PubMed] [Google Scholar]
- 74.Akinbowale OL, Peng H, Barton MD. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J Appl Microbiol. 2006;100: 1103–1113. doi: 10.1111/j.1365-2672.2006.02812.x [DOI] [PubMed] [Google Scholar]
- 75.Dubert J, Aranda-Burgos JA, Ojea J, Barja JL, Prado S. Mortality event involving larvae of the carpet shell clam Ruditapes decussatus in a hatchery: isolation of the pathogen Vibrio tubiashii subsp. europaeus. J Fish Dis. 2017; 1185–1193. doi: 10.1111/jfd.12593 [DOI] [PubMed] [Google Scholar]
- 76.Defoirdt T, Sorgeloos P, Bossier P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol. 2011;14: 251–258. doi: 10.1016/j.mib.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 77.Defoirdt T, Boon N, Bossier P, Verstraete W. Disruption of bacterial quorum sensing: an unexplored strategy to fight infections in aquaculture. Aquaculture. 2004;240: 69–88. doi: 10.1016/j.aquaculture.2004.06.031 [Google Scholar]
- 78.Bjarnsholt T, van Gennip M, Jakobsen TH, Christensen LD, Jensen PØ, Givskov M. In vitro screens for quorum sensing inhibitors and in vivo confirmation of their effect. Nat Protoc. 2010;5: 282–293. doi: 10.1038/nprot.2009.205 [DOI] [PubMed] [Google Scholar]
- 79.Zhao J, Chen M, Quan C, Fan S. Mechanisms of quorum sensing and strategies for quorum sensing disruption in aquaculture pathogens. J Fish Dis. 2015;38: 771–786. doi: 10.1111/jfd.12299 [DOI] [PubMed] [Google Scholar]
- 80.Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E. Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol. 2001;51: 1383–1388. doi: 10.1099/00207713-51-4-1383 [DOI] [PubMed] [Google Scholar]
- 81.Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, et al. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol. 2003;53: 309–315. doi: 10.1099/ijs.0.02402-0 [DOI] [PubMed] [Google Scholar]
- 82.Ushijima B, Smith A, Aeby GS, Callahan SM. Vibrio owensii induces the tissue loss disease Montipora white syndrome in the Hawaiian reef coral Montipora capitata. PLoS One. 2012;7: e46717 doi: 10.1371/journal.pone.0046717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson L. Two pathogens of GreenshellTM mussel larvae, Perna canaliculus: Vibrio splendidus and a V. coralliilyticus/neptunius -like isolate. J Fish Dis. 2009;32: 499–507. doi: 10.1111/j.1365-2761.2009.01006.x [DOI] [PubMed] [Google Scholar]
- 84.Richards GP, Watson MA, Needleman DS, Church KM, Häse CC. Mortalities of Eastern and Pacific oyster larvae caused by the pathogens Vibrio coralliilyticus and Vibrio tubiashii. Appl Environ Microbiol. 2015;81: 292–297. doi: 10.1128/AEM.02930-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cano-Gomez A, Goulden EF, Owens L, Højj L. Vibrio owensii sp. nov., isolated from cultured crustaceans in Australia. FEMS Microbiol Lett. 2010;302: 175–181. doi: 10.1111/j.1574-6968.2009.01850.x [DOI] [PubMed] [Google Scholar]
- 86.Goulden EF, Hall MR, Bourne DG, Pereg LL, Høj L. Pathogenicity and infection cycle of Vibrio owensii in larviculture of the ornate spiny lobster (Panulirus ornatus). Appl Environ Microbiol. 2012;78: 2841–2849. doi: 10.1128/AEM.07274-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beaz-Hidalgo R, Diéguez AL, Cleenwerck I, Balboa S, Doce A, de Vos P, et al. Vibrio celticus sp. nov., a new Vibrio species belonging to the Splendidus clade with pathogenic potential for clams. Syst Appl Microbiol. 2010;33: 311–315. doi: 10.1016/j.syapm.2010.06.007 [DOI] [PubMed] [Google Scholar]
- 88.Tarazona E, Lucena T, Arahal DR, Macián MC, Ruvira MA, Pujalte MJ. Multilocus sequence analysis of putative Vibrio mediterranei strains and description of Vibrio thalassae sp. nov. Syst Appl Microbiol. 2014;37: 320–328. doi: 10.1016/j.syapm.2014.05.005 [DOI] [PubMed] [Google Scholar]
- 89.Thompson FL, Hoste B, Thompson CC, Huys G, Swings J. The coral bleaching Vibrio shiloi Kushmaro et al. 2001 is a later synonym of Vibrio mediterranei Pujalte and Garay 1986. Syst Appl Microbiol. 2001;24: 516–519. doi: 10.1078/0723-2020-00065 [DOI] [PubMed] [Google Scholar]
- 90.Cano-Gomez A, Høj L, Owens L, Andreakis N. Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst Appl Microbiol. 2011;34: 561–565. doi: 10.1016/j.syapm.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 91.Richards GP, Needleman DS, Watson MA, Bono JL. Complete genome sequence of the larval shellfish pathogen Vibrio tubiashii type strain ATCC 19109. Genome Announc. 2014;2: e01252-14–e01252-14. doi: 10.1128/genomeA.01252-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steindler L, Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett. 2007;266: 1–9. doi: 10.1111/j.1574-6968.2006.00501.x [DOI] [PubMed] [Google Scholar]
- 93.Golberg K, Eltzov E, Shnit-Orland M, Marks RS, Kushmaro A. Characterization of quorum sensing signals in coral-associated bacteria. Microb Ecol. 2011;61: 783–792. doi: 10.1007/s00248-011-9848-1 [DOI] [PubMed] [Google Scholar]
- 94.Tait K, Hutchison Z, Thompson FL, Munn CB. Quorum sensing signal production and inhibition by coral-associated vibrios. Environ Microbiol Rep. 2010;2: 145–150. doi: 10.1111/j.1758-2229.2009.00122.x [DOI] [PubMed] [Google Scholar]
- 95.Valiente E, Bruhn JB, Nielsen KF, Larsen JL, Roig FJ, Gram L, et al. Vibrio vulnificus produces quorum sensing signals of the AHL-class: FEMS Microbiol Ecol. 2009;69: 16–26. doi: 10.1111/j.1574-6941.2009.00691.x [DOI] [PubMed] [Google Scholar]
- 96.Buchholtz C, Nielsen KF, Milton DL, Larsen JL, Gram L. Profiling of acylated homoserine lactones of Vibrio anguillarum in vitro and in vivo: influence of growth conditions and serotype. Syst Appl Microbiol. 2006;29: 433–445. doi: 10.1016/j.syapm.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 97.Llamas I, Keshavan N, Gonza JE. Use of Sinorhizobium meliloti as an indicator for specific detection of long-chain N-acyl homoserin lactones. Society. 2004;70: 3715–3723. doi: 10.1128/AEM.70.6.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Purohit AA, Johansen JA, Hansen H, Leiros HKS, Kashulin A, Karlsen C, et al. Presence of acyl-homoserine lactones in 57 members of the Vibrionaceae family. J Appl Microbiol. 2013;115: 835–847. doi: 10.1111/jam.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179: 3127–3132. doi:10 3127-3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tahrioui A, Schwab M, Quesada E, Llamas I. Quorum sensing in some representative species of Halomonadaceae. Life. 2013;3: 260–275. doi: 10.3390/life3010260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, Wang H, Liang W, Hay AJ, Zhong Z, Kan B, et al. Quorum sensing regulatory cascades control Vibrio fluvialis pathogenesis. J Bacteriol. 2013;195: 3583–3589. doi: 10.1128/JB.00508-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci. 2002;99: 3129–3134. doi: 10.1073/pnas.052694299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meron D, Efrony R, Johnson WR, Schaefer AL, Morris PJ, Rosenberg E, et al. Role of flagella in virulence of the coral pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2009;75: 5704–5707. doi: 10.1128/AEM.00198-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
800 ng/mL (a) and 1,000 ng/mL (b and c). External calibration curves by adding C4-HSL, C16-HSL, 3-O-C10-HSL, 3-O-C12-HSL, 3-O-C13-HSL 3-OH-C10-HSL, 3-OH-C12-HSL, 3-OH-C13-HSL and 3-OH-C14-HSL.
(DOCX)
General AHL structure (a). Mass spectrometry fragmentation pathway of AHLs (b).
(DOCX)
Total ion chromatogram (a). Extracted mass chromatogram (m/z 230.13–230.14) showing the peak at tR = 3.82 min (b). Expanded regions of the (+)-HRESIMS of the peak at tR = 3.82 min identified as 3-OH-C7-HSL showing the [M+H]+ ion peak at m/z 230.1381 (calc. for C11H20NO4 230.1387) (c).
(DOCX)
Total ion chromatogram (a). Extracted mass chromatogram (m/z 202.00–202.22) showing the peak at tR = 6.37 min (b). Expanded region of the (+)-HRESIMS of the peak at tR = 6.37 min identified as 3-OH-C5-HSL showing the ion peaks [M+H]+ at m/z 202.1073 (calc. for C9H16NO4 202.1074) (c). Expanded region of the (+)-HRESIMS of the peak at tR = 6.37 min identified as 3-OH-C5-HSL showing the [M-H2O+H]+ ion peak at m/z 184.0968 (calc. for C9H14NO3 184.0968) (d).
(DOCX)
(PDF)
Numbers are counts in the mass spectra.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.