Abstract
Hepcidin is a crucial peptide for regulating cellular iron efflux. Because iron is essential for cell survival, especially for highly active cells, such as tumor cells, it is imperative to understand how tumor cells manipulate hepcidin expression for their own metabolic needs. Studies suggest that hepcidin expression and regulation in tumor cells show important differences in comparison with those in non-tumorous cells. These differences should be investigated to develop new strategies to fight cancer cells. Manipulating hepcidin expression to starve cancer cells for iron may prove to be a new therapy in the anticancer arsenal.
Introduction
Cancer cells are highly active cells in terms of their metabolic needs. Their survival and proliferation depend on the supply of important micro- and macronutrients. One of these crucial micronutrients is iron. Iron is important for DNA synthesis, which is one of the reasons why iron depletion causes cell cycle arrest.1 When cancer cells are fed chronic subtoxic levels of iron, they transform into a more aggressive phenotype that is prone to metastasis.2 On the other hand, starving cancer cells with the use of iron-sequestering drugs inhibits tumor growth.2 This increased need for iron forces cancer cells to adapt to their metabolic needs by changing the expression of proteins involved in iron supply and iron export from cells.3 In terms of regulation of iron transport, hepcidin manipulation by cancer cells is of great interest to scientists since a hepcidin disturbance can significantly affect iron export and cause iron sequestration in tumor cells.4 Therefore, understanding the differences in hepcidin regulation between non-cancerous and cancerous cells is important for our knowledge of tumor cell survival and proliferation and can help us find new strategies to fight cancer.
Hepcidin regulation and action in non-cancerous cells
Hepcidin is a small peptide produced by the liver.4 Although it is classified as an antimicrobial peptide, it has been a focus of studies because of its crucial role in iron metabolism. Hepcidin importance in iron metabolism stems from its ability to control iron export from cells. This action of hepcidin is realized through the actions of ferroportin (FPN), which is known to be the main iron export protein.4 Hepcidin induces FPN degradation; therefore, it blocks iron export from cells such as macrophages and enterocytes.4
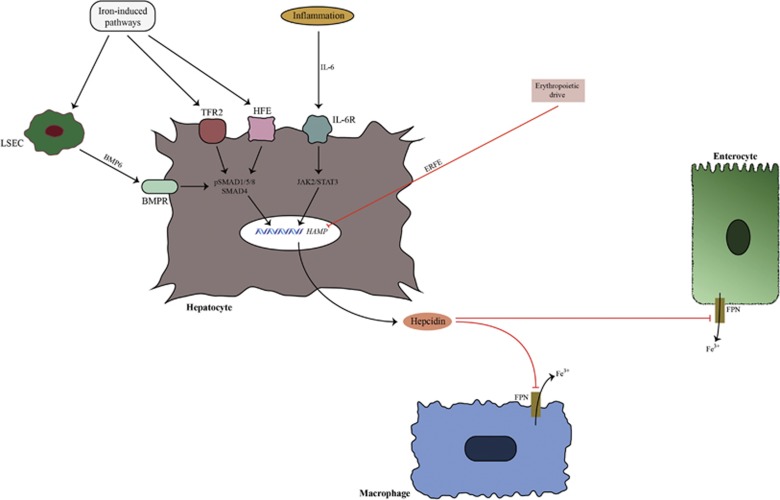
The levels of hepcidin are strictly controlled by different stimuli. Iron status is the prime controller of hepcidin expression under basal conditions.5 It is believed that iron status induces production of bone morphogenetic protein 6 (BMP6) from liver sinusoidal endothelial cells (LSEC) through an as yet unknown mechanism.6 BMP6 acts in a paracrine manner through the BMP receptor (BMPR) in hepatocytes. In this way, BMP6 activates the intracellular S-mothers against decapentaplegic (SMAD) pathway, which then induces the HAMP promoter to produce hepcidin.4 BMPR action is controlled by its coreceptor hemojuvelin (HJV).4 Loss of HJV causes significant iron overload because of impaired hepcidin expression.7 HJV is the substrate for matriptase 2 (MT2), which cleaves HJV.8 It is believed that the actions of MT2, HJV and BMPR are stabilized by neogenin, which serves as the ‘backbone’ structure for assembly of the BMPR, HJV and MT2 complex.9, 10 These observations demonstrate how hepcidin expression is controlled in a balanced manner by different regulatory mechanisms.
Transferrin receptor 2 and hemochromatosis protein (HFE) are other iron-sensing proteins in hepatocytes.4 They induce hepcidin through as yet unresolved pathways, with transferrin receptor 2 being more potent than HFE in this respect.11 Inflammatory stimuli are also important upregulators of hepcidin expression. Cytokines produced during inflammation, such as interleukin-6 (IL-6), activate the janus kinase 2-signal transducer and activator of transcription 3 pathway, which increases hepcidin expression (Figure 1).12
Figure 1.
Hepcidin regulation and action in non-cancerous tissue. The major regulators of hepcidin expression in hepatocytes are iron status, inflammatory signals and erythropoietic drive. Iron status is sensed by bone morphogenetic protein 6 (BMP6), transferrin receptor 2 (TFR2) and hemochromatosis protein (HFE). BMP6 is produced by non-parenchymal liver cells (LSEC). BMP6 binds with BMP receptor (BMPR), which activates the SMAD pathway. Phosphorylated SMAD molecules induce HAMP (hepcidin antimicrobial peptide) expression. Inflammation induces hepcidin expression by activating the janus kinase 2-signal transducer and activator of transcription 3 (JAK2/STAT3) pathway. Erythropoietic drive is a negative hepcidin regulator that acts by suppressing hepcidin expression through newly discovered erythroferrone (ERFE). Hepcidin mode of action is realized through induction of ferroportin (FPN) degradation in target cells, which causes sequestration of iron in cells.
Hepcidin is also negatively regulated by erythropoietic signaling. Precursors of erythrocytes produce erythroferrone (ERFE) in response to erythropoietin. ERFE then mediates hepcidin downregulation in hepatocytes through as yet unknown pathways.13 Other factors can also control liver hepcidin expression, such as hormones, growth factors and heparins.14, 15, 16 Although hepcidin is mainly produced by the liver, other organs are capable of expressing local hepcidin. Data suggest that local hepcidins are produced by the heart, prostate, lungs, kidneys and other organs.17 Still, local hepcidins have not been studied as extensively as liver hepcidin; therefore, their exact regulation and role in organ homeostasis are still not clear.
Hepcidin regulation in cancer
Different BMPs control hepcidin expression in tumor tissues
BMPs are the main and most powerful stimulators of hepcidin expression. Although BMP6 stands out as the prime BMP molecule that controls hepcidin expression, other BMP molecules have the ability to induce hepcidin expression, often more potently than inflammatory stimuli.18 This ability of different BMPs to control hepcidin expression explains reports from studies of different tumors in which BMP6 does not seem to be the only BMP molecule to control hepcidin expression. For example, in prostate cancer, local BMP4 and especially BMP7 are linked with prostatic hepcidin expression, although the same cannot be said about local BMP6.19 Still, local BMP6 is an important factor in high-grade prostate cancer and is linked to tumor survival during androgen depletion.20, 21 Local BMP6 in prostate cancer can be produced through signaling from bone stromal cells.22
High BMP7 levels are linked with hepcidin overexpression in prostate cancer, especially in metastatic cancer, irrespective of androgen suppression therapy, implicating prostatic hepcidin as a factor that promotes cancer cell survival.19 The role of local BMP7 in metastatic prostate cancer has been documented; therefore, its control of hepcidin expression is an important additional strategy of prostate neoplastic cells to promote survival and tumor progression.23, 24 However, many questions remain to be answered, such as the reason why local BMP7 is able to induce hepcidin expression, while local BMP6 is not involved in hepcidin regulation in prostate cancer. Future studies should resolve this mystery.
BMPs seem to be important inducers of hepcidin expression in lung cancer as well, as the serum hepcidin levels in these patients are related to the serum BMP2 levels.25 BMP2 has been shown to induce hepcidin expression.26 Furthermore, studies have shown that BMP2 is upregulated in lung cancer.27 BMP2 is a more potent stimulator of hepcidin than IL-6, making it an important controller of hepcidin expression.18
We still do not know the exact mechanisms of BMP2 regulation in lung cancer and its relationship with other BMPs, such as BMP7, which is also linked to progression of lung cancer.28 Another important issue that arises is the source of BMP2 in lung cancer since, in addition to lung tumor cells, pulmonary endothelial cells and LSEC also express BMP2.18, 29, 30 Moreover, blocking local BMP2 can reduce tumor growth in lung cancer.30 Unfortunately, local BMP2 expression in tumor cells has not been evaluated in studies that have reported locally elevated levels of hepcidin in lung cancer.25
In breast cancer, BMPs are also linked with increased expression of hepcidin. In early breast cancer there is no correlation of hepcidin with IL-6, erythropoietin and ERFE.31 BMP7 may be involved in hepcidin overexpression in early breast cancer. BMP7 is overexpressed in breast cancer tissue and is linked with cancer metastasis, while BMP7 administration has shown that it can recover hepcidin expression in BMP6 knockout mice.32, 33 In a recent study on breast cancer patients, the levels of BMP6 were lower in cancer tissue compared with noncancerous tissue, making local BMP6 an unlikely inducer of hepcidin expression.34 Since BMP6 can show antiproliferative and antimetastatic potential in breast cancer, it is not a surprise that BMP6 inhibition favors cancer cell survival.34 A similar protective role of BMP6 has also been suggested in prostate cancer.35
However, hepcidin production in breast cancer does not only originate from cancer tissue. The liver is an important source of increased hepcidin levels in breast cancer, and this increase is related to BMP6 expression.36 It seems that the iron load is responsible for increasing BMP6 levels in the liver of breast cancer patients.36 Further studies are needed to examine the correlation between different BMPs and hepcidin in breast cancer.
One of the main negative controllers of hepcidin expression, MT2, is downregulated in breast cancer, which can additionally cause hepcidin overexpression in breast cancer.37 However, we still do not know the role of local MT2 in breast tissue, especially with respect to hepcidin expression.
In multiple myeloma (MM), hepcidin expression is related to BMP2, but the cellular origin of BMP2 in MM is unclear.38 FPN downregulation in MM patients is significantly associated with poor prognosis, indicating FPN downregulation as a factor in promoting the aggressive behavior of tumor cells.39 The use of cellular iron chelators in MM suppresses tumor growth, which shows the importance of iron metabolism for tumor survival.39 However, the increase in serum hepcidin levels in MM is associated with low hepcidin expression in myeloma cells, which suggests that the rise in hepcidin levels in MM originates from the liver.40 Additionally, low hepcidin in myeloma cells shows that in MM compared with other cancers, local FPN downregulation is not dependent on local hepcidin action. Consequently, manipulating liver hepcidin or local FPN rather than local hepcidin could be the best option to suppress tumor growth in MM.
BMPs are also suspected to be inducers of hepcidin expression in non-Hodgkin lymphoma (NHL) because of the strong correlation between hepcidin and ferritin observed in this cancer.41 Studies have shown that ferritin can increase the levels of BMP6 and hepcidin in the liver.42 This would make BMP6 a potential cause of high hepcidin levels in NHL, although other BMPs should be taken into account. Interestingly, the results from the molecular profiling of tumor samples in patients with NHL have shown increased expression of BMP6 in cancer tissue.43 Unfortunately, studies in this cancer have not evaluated the relation of local BMPs with systemic and tumor hepcidin expression.
In colon cancer, local hepcidin expression is generally low, but iron accumulation is more prominent in cancer cells than normal cells.44, 45 Although iron accumulation in colorectal cancer is not uniform, when present, it is associated with higher levels of hepcidin.44 Higher local hepcidin expression in this cancer does not seem to be dependent on IL-6.44 On the other hand, it is not known which BMPs are responsible for hepcidin expression in this cancer. BMP7 has been linked with the metastatic capacity of colon cancer, while BMP2 has been linked with tumor suppression; therefore, BMP2 does not seem to be a part of tumor proliferation strategy.46, 47, 48 It is interesting to note that SMAD4 is downregulated in colon cancer, while the BMP7 ability to promote the invasive behavior of colon cancer cells is independent of SMAD4 molecules.49, 50, 51 BMP7 is not the only BMP to be linked with tumor proliferation in colon cancer. BMP4 has also been linked with metastatic forms of colon cancer.52 Furthermore, the role of BMP4 in inducing the invasive properties of colon cancer is related to reduced SMAD4 signaling.53 Future studies should resolve the roles of BMP4 and BMP7 in inducing local hepcidin expression in colon cancer.
Hepcidin is increased in renal carcinoma as well and is associated with the metastatic potential of this cancer.54 Although local expression of hepcidin in renal cancer is low compared with adjacent normal cells, the levels of hepcidin are higher in metastatic cancer and are related to poor survival.54 Unfortunately, these studies have not been complemented with an examination of the role of BMPs in inducing hepcidin expression.
It is interesting to note that hepcidin overexpression by BMPs fits the overall strategy of cancer cells in preserving survival and expansion. Overexpression of hepcidin by certain BMPs appears to be part of the metastatic invasion strategy of cancer cells. BMPs can promote metastasis through lymphatic and blood vessels, while blocking BMP signaling reduces the metastatic capacity of cancer cells.55, 56
Role of inflammatory stimuli in controlling hepcidin expression in cancer
The levels of IL-6 are frequently increased in cancer and are related to the hepcidin levels, especially in established disease.57 However, in the early stages of breast cancer, there is no correlation between inflammatory stimuli and hepcidin increase.31 This is in line with studies that correlate the high levels of IL-6 in cancer with aggressive tumor behavior, unfavorable histologic grade and metastatic potential.58 These observations are further confirmed by studies of breast cancer patients with metastasis, where serum IL-6 is significantly related to serum hepcidin.59 Similar observations have been found in renal carcinoma,54 while in prostatic cancer, significant serum hepcidin elevations have been observed in a subset of patients with bone metastasis, which are probably caused by increased levels of IL-6.60 IL-6 can directly stimulate cancer cells to produce local hepcidin. This action has been observed in prostate cancer and can be reversed with the use of anti-IL-6 antibodies.19, 61
IL-6 is not consistently elevated in cancer and can be caused by different tumor histologies as well as by differences in inflammatory activities between early and established cancer; inflammatory cells are significantly increased in established cancer compared with early stages of disease.62 In MM, IL-6 has been clearly linked with tumor progression and survival of myeloma cells.63 The significant rise of IL-6 in MM is seen in later stages of the disease, which is in line with observations that have shown increased levels of hepcidin in the later stages of MM.40, 63 Even in MM, hepcidin expression appears to be related to BMP molecules, such as BMP2.38 Although data suggest that the higher hepcidin levels in MM are due to increased IL-6 and BMP2, BMP2 seems to be a more potent inducer of hepcidin than IL-6.38, 40 In NHL serum, hepcidin is also associated with IL-6, but this association is weak, which means that IL-6 alone cannot account for increased hepcidin production.41 Although MM studies have not evaluated local hepcidin expression, in another type of lymphoma named Waldenstrom’s macroglobulinemia, lymphoplasmacytic tumor cells have been shown to be a source of hepcidin expression.64
Hepcidin expression in patients with acute leukemia is dependent on the activity of the disease; during increased production of blast cells, the levels of serum hepcidin are elevated and related to IL-6 and only decrease during remission, probably due to erythropoietic signaling.65 Similar observations have been found in different types of myelodysplastic syndromes, with lower levels of hepcidin observed in myelodysplastic syndrome patients without excess blasts compared to those with excess blasts.66 Blast cells in acute leukemia are an important source of cytokines, such as IL-6, which would make the increased numbers of blast cells the ‘culprits’ behind the increased hepcidin levels.67
In lung cancer, IL-6 is negatively correlated with hepcidin, which makes IL-6 an unimportant inducer of serum hepcidin elevation in this cancer.25
Other factors that control hepcidin expression in cancer
Erythropoietic signaling through ERFE is not important for hepcidin expression in breast cancer, at least in the early stages of the disease.31 In acute leukemia remission and myelodysplastic syndromes without excess blasts, erythropoietic signals inhibit hepcidin expression due to the increased need for iron of erythrocyte precursors.65, 66 The increased need for iron due to the erythropoietic drive in myelodysplastic syndromes can cause an insufficient hepcidin increase in response to the cellular iron load, which is reflected by the low hepcidin/ferritin ratio observed in these patients.68 This means that anemia and hypoxia could be important regulators of hepcidin expression in these patients. How these changes in hepcidin levels are related to tumor progression is unclear, although iron deprivation has been shown to induce antiproliferative effects in leukemia cells.69
SOSTDC1, an important new actor in inducing local hepcidin expression
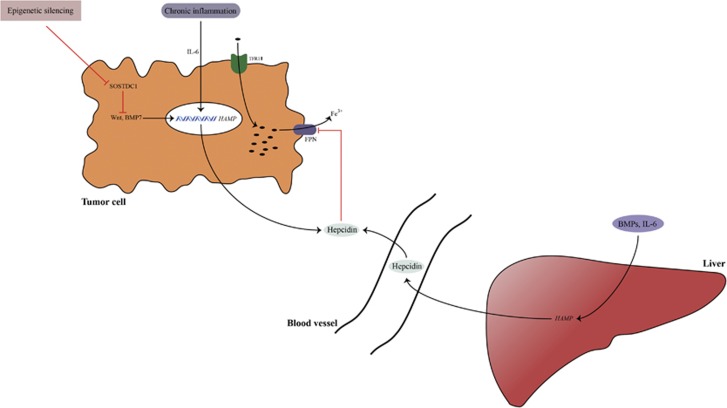
In prostate cancer, hepcidin is not induced by identical stimuli compared with the liver or systemic hepcidin. Although local IL-6 has similar effects on hepcidin expression in prostatic tissue compared with liver tissue, the local BMPs responsible for hepcidin expression are different.19 Additionally, studies suggest that the Wnt pathway is a novel regulator of hepcidin expression in prostatic tissue, which has not been observed in the liver.19 This is important because Wnt pathway dysregulation is involved in prostatic tumorigenesis. Increased cellular iron sequestration has been linked with increased Wnt signaling, which promotes tumorigenesis.19 Blocking the Wnt pathway and BMP pathway with sclerostin domain-containing protein 1 (SOSTDC1) markedly reverses hepcidin expression (Figure 2).19
Figure 2.
Hepcidin regulation in cancer tissue. Hepcidin in cancer tissue is produced by local tumor cells and liver. Hepcidin overexpression in tumor tissue is related to different bone morphogenetic protein (BMP) molecules (such as BMP7) as well as to inflammatory stimuli, such as interleukin-6 (IL-6). Studies suggest the existence of new regulators of hepcidin in cancer tissue such as sclerostin domain-containing protein 1 (SOSTDC1) (which is downregulated in cancer through epigenetic silencing) and the Wnt pathway (which is upregulated in cancer). Increased hepcidin in the tumor milieu induces iron sequestration in tumor cells through its actions on ferroportin (FPN). Hepcidin increase is accompanied by tumor transferrin receptor 1 (TFR1) upregulation, which increases iron supply into tumor cells. Increased iron depots in tumor cells help them survive and proliferate.
SOSTDC1 is expressed in different organs, especially the kidneys.70 SOSTDC1 is downregulated in the prostate, breast, non-small cell lung carcinoma and renal cancer.19, 71, 72, 73 This downregulation can cause local overexpression of hepcidin, which can result in iron sequestration in cells through hepcidin effects on FPN. On the other hand, the role of SOSTDC1 as a tumor suppressor has been observed in in vivo studies with SOSTDC1 overexpression.72
Because SOSTDC1 can affect local hepcidin production, it would be interesting to examine whether SOSTDC1 inhibition affects iron depots in cells and whether this is the reason for excessive Wnt signaling in cancer.
In renal carcinoma, SOSTDC1 is downregulated and has antitumor properties.73 Another interesting observation from renal tissue shows that SOSTDC1 is localized closely to BMP7. The actions of SOSTDC1 include suppression of BMP7 and the Wnt pathway, which means that SOSTDC1 inhibition in renal carcinoma might elevate hepcidin levels in a similar manner to prostate cancer.19, 70 SOSTDC1 appears to be downregulated in less prevalent tumors, such as thyroid tumors.74 Further studies should examine the intriguing idea that SOSTDC1 inhibition could be a universal pathway through which different cancers regulate hepcidin expression.
Hepcidin disturbance in cancer: a role for local and systemic (liver) isoforms
Subtle regulatory differences in tumors with increased levels of hepcidin
An increase in hepcidin levels has been reported in lung cancer, breast cancer, renal carcinoma, prostate cancer and other cancers. Although in many cancers local FPN downregulation is related to high local hepcidin expression, there are subtle differences between different cancers. In colorectal cancer, hepcidin expression is significantly increased compared with the nearly non-existent expression of local hepcidin in normal tissue.44 Local hepcidin in colon cancer can induce FPN downregulation, but the protein levels of FPN in this cancer are increased.44, 75 FPN in colon cancer is located in the cytoplasm and not in the basolateral membrane, which means that FPN cannot induce iron export in colon cancer cells, rendering it ineffective as an iron exporter.75 The reduced iron export in colon cancer might be due to disturbances of another protein called hephaestin. Hephaestin interacts with FPN to promote iron export from cells, but its levels are downregulated in colon cancer.75, 76 Loss of hephaestin is known to cause iron overload in enterocytes and reduced iron delivery to plasma.77 All of these observations show that iron export from tumor cells is reduced in colon cancer as well, but the culprit behind this disturbance is not entirely related to the action hepcidin.
In lung cancer, the existence of a correlation between the serum hepcidin levels and metastatic potential of tumors is accompanied by a lack of correlation between mRNA of local hepcidin and the metastatic potential of the tumor.25 This means that the aggressive behavior of lung cancer is dependent on the systemic levels of hepcidin. This association of tumor behavior with serum hepcidin levels might be a result of the beneficial effect of the addition of liver hepcidin in a tumor environment for survival and proliferation of neoplastic cells.25
Increased levels of hepcidin: part of the tumor strategy to increase cellular iron
The pattern of increased serum hepcidin levels in cancer is accompanied with increased local mRNA expression of hepcidin. Increased local hepcidin causes FPN degradation, which prevents iron export from cells and increases the iron labile pool in tumor cells, helping them to survive and proliferate.19 Local hepcidin production is not only observed in tumor epithelial cells but has also been observed in activated macrophages and leukocytes of the tumor milieu.76 It is interesting to note the increased FPN expression observed in these cells.78 Increased FPN expression might secure a larger iron supply for tumor cells, in which FPN is frequently downregulated. Increased FPN expression in adjacent normal cells can also serve tumor cells in a similar manner, by which strategy tumor cells would ‘hack’ the iron metabolism of neighboring cells for their own survival. The increased iron accumulation in cancer cells is partly dependent on hepcidin activity through its effect on FPN. Increased iron supply through TFR1 is another crucial strategy for cancer cells, which is why TFR1 is often overexpressed in cancer cells.79 The increase in local breast TFR1 mRNA correlates with the increase of iron depots and metastatic potential of tumors.80 However, inhibition of TFR1 might be hindered by other potential pathways that are supposed to serve as alternative routes of iron entry into cells.81, 82
In the liver, carcinoma hepcidin levels are low compared with other cancers
In hepatocellular carcinoma (HCC), hepcidin levels are lower compared with controls, which differs from the paradigm of high levels of serum hepcidin found in other cancers (Table 1).83 This downregulation is observed even in the early stages of HCC.83 IL-6 is known to be overactivated in HCC; therefore, low hepcidin levels in HCC are not related to an IL-6 disturbance.84 On the other hand, studies suggest that the low levels of BMP6 in HCC are due to DNA hypermethylation.85 Because BMP6 is the main regulator of hepcidin expression, this might affect the hepcidin levels in HCC. One of the main regulators of hepcidin expression is HJV, and its activity is downregulated in HCC as well, which also accounts for the lower expression of hepcidin observed in HCC.86 One of the most mutated genes in HCC is tumor suppressor gene p53.87 p53 can activate the HAMP promoter, and its loss is associated with lower hepcidin expression.88 However, there are observations that suggest a modest regulatory role of p53 in inducing hepcidin expression, which does not significantly affect its serum levels.89
Table 1. Major regulators of hepcidin in different cancers.
| Type of cancer | Cancer tissue expression of hepcidin regulators | Serum levels of hepcidin regulators in cancer | Factors that regulate hepcidin expression in cancer | Local tumor hepcidin expression | Systemic (liver) hepcidin expression | References |
|---|---|---|---|---|---|---|
| Breast cancer | BMP7 ↑, BMP6 ↓, MT2 ↓ | IL-6 ↑ | BMP6a, IL-6a | ↑ | ↑ | 31, 32, 34, 36, 37, 59 |
| Lung cancer | BMP2 ↑, BMP7 ↑ | BMP2 ↑ | BMP2a | ↑ | ↑ | 25, 27, 28, 30 |
| Prostate cancer | BMP4/6/7 ↑, IL-6 ↑ | IL-6 ↑ | BMP4/7b, IL-6b, IL-6a | ↑ | ↑ | 19, 20, 21, 60 |
| Multiple myeloma | ? | BMP2 ↑, IL-6 ↑ | BMP2a, IL-6a | ↓ | ↑ | 38, 40 |
| Non-Hodgkin lymphoma | BMP6 ↑ | IL-6 ↑ | IL-6a | ↑ | ↑ | 41, 43 |
| Liver carcinoma | BMP6 ↓, HJV ↓, IL-6 ↑ | IL-6 ↑ | BMP6c, HJVc | ↓ | ↓ | 83, 84, 85, 86 |
| Acute leukemia | IL-6 ↑ | IL-6 ↑ | IL-6a | ? | ↑ | 65, 67 |
Abbreviations: BMP, bone morphogenetic protein; HJV, hemojuvelin, IL-6, interleukin-6; MT2, matriptase 2.
Regulators of liver hepcidin expression.
Regulators of local hepcidin expression.
BMP6 and HJV are major regulators of liver hepcidin expression, but studies have yet to directly prove that their downregulation in hepatocellular carcinoma is the reason behind low levels of hepcidin observed in this cancer.
Many factors that increase the risk of cancer, such as hepatitis virus C and alcohol, cause low levels of hepcidin.90, 91 Studies that have evaluated the expression of iron regulatory genes in HCC reveal an interesting model of iron disturbance. The iron load in HCC is observed in adjacent non-cancerous cells, while iron depletion is observed in cancerous cells.92 Expression of iron regulatory genes in cancer cells shows a pattern of low hepcidin, but high TFR1 expression, while adjacent non-cancerous cells show increased levels of hepcidin and FPN.92, 93 Increased levels of hepcidin may serve to counteract the increased iron supply from enterocytes and macrophages, while increased FPN activity serves to protect normal cells from iron overload. The protein levels of FPN in HCC are not correlated with increased mRNA of FPN1 and are similar to the FPN levels in normal cells.92 This might be caused by reduced expression of ceruloplasmin in cancerous cells.92, 93 Ceruloplasmin post-translationally affects FPN similarly to hepcidin, although not as potently.94
The questions then arises, why are low liver hepcidin levels found in HCC compared with increased hepcidin levels in other cancers? Increasing local hepcidin sequesters iron in tumor cells and increases the proliferative properties of the tumor. In the liver, hepcidin produced by hepatocytes is also the main reservoir of systemic hepcidin; therefore, if liver tumor cells are able to secure the much-needed iron supply as do other tumor cells, they would have to manipulate liver hepcidin expression. By lowering liver hepcidin levels, liver tumor cells are able to secure abundant iron from the main suppliers of iron, such as enterocytes and macrophages. This iron would preferentially be used by cancerous cells because of the higher TFR1 levels observed in tumor cells. If liver hepcidin increases, then iron is sequestered in macrophages and enterocytes and liver tumor cells are iron starved. Additionally, the retained hepcidin expression from non-cancerous liver cells secures additional iron sequestration by inducing FPN degradation and increases the iron labile pool in cancer cells. Recent data also suggest that hepcidin has other protective properties in liver tissue outside of iron metabolism. Hepcidin signaling can secure the quiescent state of hepatic stellate cells.95 Since activated hepatic stellate cells are the hallmark of liver cirrhosis, this action would make hepcidin an important peptide that protects the liver from cirrhosis and possibly liver cancer. In any case, it is still too early to associate hepcidin activity outside its iron metabolism-related actions. More studies are needed to confirm this potential of hepcidin.
Complex regulation of hepcidin in brain cancers
Characteristic regulation of hepcidin has also been observed in brain tumor cells. The levels of local hepcidin are low in some tumors, while in others, he hepcidin levels are comparable to those of normal brain tissue.96 It is interesting to note that HJV is not expressed in brain tissue, while neogenin is present but downregulated in many brain tumors.96 Iron metabolism in the brain is under differential regulation by specific cell types. For example, in glioblastoma, tumor stem cells extract iron from the tumor environment more effectively than other tumor cells.97 The important role of glial cells and pericytes in hepcidin regulation in brain tissue might explain the specific hepcidin regulation in brain tumors.98 In any case, hepcidin in brain tissue can downregulate FPN, similar to other tissues.98, 99, 100 Furthermore, this effect of hepcidin in neurons and vascular endothelial cells has been shown to be related to the downregulation of TFR1 and reduction of transferrin-bound iron from the periphery into the brain.99, 100 The interesting downregulation of TFR1 by hepcidin has been observed in astrocytes as well.101 This is important because TFR1 upregulation helps brain cancer cells survive. It is still not clear how these actions of hepcidin might help in manipulating iron metabolism in the brain since there are conflicting studies in terms of hepcidin’s role in reducing the iron load in brain cells.100, 102 The iron accumulation in neurons and microglia observed by Urrutia et al.102 is likely caused by specific actions induced by inflammatory signaling, but not directly by hepcidin. The action of hepcidin on iron metabolism of brain tumors is important because research has shown that cutting the iron supply to brain tumor cells can have important antineoplastic effects.103, 104
CLINICAL RELEVANCE OF HEPCIDIN MANIPULATION IN CANCER
We have to be careful when examining hepcidin levels in cancer patients. The absolute levels of serum hepcidin might not be as important as the dynamics between local and systemic hepcidin levels. In cancer tissues, most models indicate that there is higher hepcidin and lower FPN expression compared with normal adjacent tissue.19, 36, 38, 39 A high hepcidin/low FPN model accompanied by high TFR1 expression36, 79, 97 further strengthens the idea that in cancer tissue, increased iron supply is the norm, promoting cellular proliferation in cancer tissues. Suppressing liver hepcidin inhibits cancer cell proliferation, and the same result can be realized through knockout of tumor hepcidin.36 A similar result can be achieved by blocking hepcidin expression through anti-HJV antibodies.105
A local anti-hepcidin strategy could involve blocking the action of local hepcidin with anti-hepcidin antibodies. In prostate cancer, the use of anti-hepcidin antibodies restores FPN expression and prevents cancer proliferation.19 Another strategy could involve blocking the action of local hepcidin on FPN in cancer cells. This could be done by mutating the FPN gene in cancer cells, which would produce an FPN mutant that is resistant to the action of hepcidin.
The use of hepcidin as a therapeutic target has already been accompanied by the production of liver hepcidin agonists and antagonists. One method to antagonize hepcidin is to use molecules that cause RNA interference, such as small interfering RNA.106, 107 Small interfering RNA have been shown to block liver expression of hepcidin, but a specific hepcidin small interfering RNA with proven beneficial properties in human trials has yet to be developed.106, 107 Another method is to use molecules with specific affinity to hepcidin. These experimental drugs have already been developed, such as NOX-H94 (also known as an anti-hepcidin spiegelmer), which can significantly increase serum iron and hemoglobin levels in human patients with anemia due to inflammation and anemia due to cancer, but not in erythropoiesis-stimulating agent-hyporesponsive anemia in dialysis patients.106, 107, 108 On the other hand, specific hepcidin antibodies, such as LY2787106, have already been incorporated in human clinical trials to treat anemia due to cancer. Although the first results suggested good patient tolerance, the observed efficacy of this drug was unfortunately short-lived, probably due to homeostatic compensatory production of hepcidin, which should be taken into account when blocking liver hepcidin with specific antibodies.109 Hepcidin antagonism also includes interfering with the action of regulators of hepcidin expression, such as BMP molecules, including BMPR, HJV and MT2.107
In cases in which liver hepcidin expression has to be restored (like in liver cancer), the strategy could include the use of hepcidin agonists that mimic hepcidin action and are chemically remodeled to improve their pharmacokinetic properties. These so-called mini-hepcidins have already shown beneficial results in reducing the iron load in animal models with β-thalasemia, polycythaemia vera and hemochromatosis.107 Furthermore, the experimental drug LJPC-401, a hepcidin agonist, is being studied in human clinical trials.107 The results from a phase I study show that this drug significantly reduces serum iron in acute and long-term settings without toxicities.110 The pharmaceutical company that developed this drug intends to use it in clinical trials with β-thalasemia patients in mid-2017.110
Until now, the only established indication for the use of hepcidin therapeutics in cancer has been anemia of cancer.106, 107 This condition is characterized by high levels of serum hepcidin, which cause low levels of serum iron and hemoglobin. High levels of hepcidin cause downregulation of FPN and resultant cellular iron sequestration in enterocytes and macrophages, which are the major iron suppliers of plasma.38, 40, 41, 64, 105, 109 It remains to be seen whether local or systemic hepcidin therapy can affect cancer cell proliferation as well.
Manipulation of hepcidin can also be performed with pharmacotherapeutic agents. The anticancer properties of drugs, such as sorafenib, were shown to be enhanced by iron-chelation therapy.111 It is interesting to note that sorafenib can induce hepcidin expression through inhibition of the Ras/MAPK pathway, but whether this means that the prohepcidin action of sorafenib adds to its anticancer properties in HCC is still unclear.112
Heparins are strong suppressors of hepcidin expression through their actions on the BMP/SMAD pathway,16, 106. More importantly, heparins can bind different BMPs, which is why heparins can suppress hepcidin, even in BMP6 knockout models.16, 106, 113 Chemically modified heparins with non-anticoagulant properties, such as SST0001, have already shown promise in MM and sarcoma models by reducing tumor growth alone or in combination with standard chemotherapy.114, 115 However, the mechanisms behind the antitumor properties of SST001 are extensive, and it is still not clear if they are related to hepcidin suppression.
Conclusion
Hepcidin is an important peptide for cellular iron homeostasis. This is important since the level of iron load affects the cellular redox status.3 Oxidative damage is a pathogenic factor that is related to different conditions.107 Therefore, it is not much of a surprise that hepcidin disbalance has already been linked to conditions such as diabetes, liver cirrhosis and heart dysfunction.116, 117, 118
Although the role of hepcidin in cancer has been studied less extensively, accumulating data suggest that a hepcidin disbalance is prevalent in cancer as well. In most cancers, the hepcidin levels are increased, except in HCC and some brain tumors, in which the hepcidin levels are low. A hepcidin disbalance in cancer is important because it provides tumors with the necessary iron for their survival. The strategy that cancer cells use to achieve this is by increasing cellular iron import through upregulation of TFR1 and by downregulating cellular iron export, which is realized through the actions of hepcidin on FPN.107
This review discussed the regulation of hepcidin in cancer by different BMP molecules and inflammatory stimuli, such as IL-6. Data suggest that BMP molecules are more potent regulators of hepcidin expression than IL-6 in cancer. BMP6 does not seem to be the main inducer of hepcidin expression in neoplastic tissue. This role in cancer is attributed to BMP2, BMP4 and BMP7. Cancer cells use noncanonical pathways to induce hepcidin expression such as the Wnt pathway. This is important because Wnt pathway disturbances are frequently dysregulated in cancer. Wnt and BMPs are both regulated by SOSTDC1. SOSTDC1 is downregulated in cancer cells, which causes Wnt and BMP overexpression. Studies suggest that SOTDC1 inhibition might be a common route through which cancer cells increase hepcidin levels, although this hypothesis remains to be confirmed by future studies.
Although some of the actors involved in hepcidin expression in cancer have been revealed, important questions remain to be answered. First, we still do not know the exact mechanism of regulating hepcidin expression in cancer cells by BMPs. This can be determined by studies that examine associations between local hepcidin expression and suspected regulators of hepcidin. This has been done in studies with prostatic cancer, while data from other cancers are partial or lacking. On the other hand, we still do not know the exact cellular origin of BMPs in all cancers. BMPs can be produced by endothelial cells, stromal cells and tumor cells.
We also need to develop a better understanding of the dynamic between local hepcidin expression and liver expression of hepcidin as well as its relation to FPN expression in different stages of cancer. This will help us explain the exact role of local and systemic (liver) hepcidin in cancer. Data suggest that this interaction is complex and characterized by specific subtleties. For example, the levels of cancer hepcidin production are up to eight times higher in prostate cancer cell lines than in normal cell lines,19 whereas serum hepcidin levels rise only in patients with bone metastasis.60 In lung cancer, serum and local hepcidin expression are both high, with results from cell lines suggesting that local elevation of hepcidin is not as prominent as in prostate cancer.25 Interestingly, the serum levels of hepcidin in lung cancer are consistently higher than in prostate cancer and seem to contribute to the total levels of hepcidin in the tumor microenvironment.25 In breast cancer, the serum hepcidin levels are high, with tumor hepcidin expression only marginally increased.36 On the other hand, suppression of liver hepcidin and knockdown of tumor hepcidin by in vitro and in vivo experiments inhibits tumor proliferation in breast cancer.36 This might indicate that both sources of hepcidin, liver and local, help in promoting tumor progression. In MM, the local levels of hepcidin are low, while the serum levels of hepcidin are high and correlate with IL-6 and BMP2. Low levels of hepcidin and the inability of IL-6 to induce hepcidin expression in MM cells indicates that most hepcidin in MM comes from the liver.40
In addition, data from lung cancer studies show that the hepcidin serum levels are related to the metastatic potential and clinical stage of the tumor.25 Local expression of hepcidin in prostate cancer has also been linked with the metastatic potential of the cancer.19 Similarly, serum hepcidin as well as local hepcidin expression in renal cancer are related to the metastatic potential of the cancer.54 Furthermore, local tumor expression of hepcidin in renal carcinoma is an independent prognostic factor related to shorter overall survival.54 In NHL, the serum hepcidin levels are related to the disease activity and international prognostic index score, which is a clinical parameter that predicts long-term survival in NHL.41
In any case, more studies (especially in vivo studies) need to address the specific contribution of liver and local hepcidin to the total hepcidin levels in the tumor milieu of different cancers.
Studies focused on hepcidin disturbances in cancer will have to address the issue of the exact interactions between activated leukocytes and tumor cells in terms of the iron metabolism dynamics in the cancer environment. Finally, studies should also reveal the exact ‘iron-hacking’ strategies of cancerous cells towards non-cancerous cells through manipulation of hepcidin expression and other iron proteins.
Blocking local hepcidin action in certain cancers can inhibit tumor progression. In other cancers, such as breast cancer, blocking liver hepcidin (in addition to local hepcidin suppression) has also been shown to suppress tumor proliferation. Blocking hepcidin in MM with anti-IL-6 antibodies has shown mixed results, indicating that hepcidin suppression in MM would be more successful by direct actions on hepcidin. Although these data are rather encouraging, similar studies from other cancers will further enhance our knowledge of the role of hepcidin therapy in cancer. We must be careful when blocking liver hepcidin because this action might cause side effects that should be taken into account.
In other cancers, such as HCC, the hepcidin levels are low; therefore, the aim of the antitumor strategy is to restore the hepcidin levels. In brain cancer, the situation is more complicated; the hepcidin levels in many tumors of the brain are low, although this observation does not extend to all brain tumors. Additionally, different brain cell types show differential regulation of hepcidin expression. The low levels of hepcidin observed in brain tumors seem to be in agreement with observations that suggest a protective role of hepcidin therapy in brain tissue.99, 100, 119 Still, the question remains whether hepcidin manipulation can help inhibit brain tumor growth.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare no conflict of interest.
References
- Terada N, Or R, Szepesi A, Lucas JJ, Gelfand EW. Definition of the roles for iron and essential fatty acids in cell cycle progression of normal human T lymphocytes. Exp Cell Res 1993; 204: 260–267. [DOI] [PubMed] [Google Scholar]
- Chanvorachote P, Luanpitpong S. Iron induces cancer stem cells and aggressive phenotypes in human lung cancer cells. Am J Physiol 2016; 310: C728–C739. [DOI] [PubMed] [Google Scholar]
- Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer 2013; 13: 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T, Nemeth E. Hepcidin and iron homeostasis. Biochim Biophys Acta 2012; 1823: 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriopoulos B Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet 2009; 41: 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canali S, Zumbrennen-Bullough KB, Core AB, Wang C-Y, Nairz M, Bouley R et al. Endothelial cells produce bone morphogenetic protein 6 required for iron homeostasis in mice. Blood 2017; 129: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederkofler V, Salie R, Arber S, Enns C, Gerhard G. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest 2005; 115: 2180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Nizzi CP, Anderson SA, Wang J, Ueno A, Tsukamoto H et al. Low intracellular iron increases the stability of matriptase-2. J Biol Chem 2015; 290: 4432–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Maxson JE, Zhang RH, Wahedi M, Enns CA, Zhang A-S. Neogenin facilitates the induction of hepcidin expression by hemojuvelin in the liver. J Biol Chem 2016; 291: 12322–12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns CA, Ahmed R, Zhang A-S. Neogenin interacts with matriptase-2 to facilitate hemojuvelin cleavage. J Biol Chem 2012; 287: 35104–35117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DF, Summerville L, Crampton EM, Frazer DM, Anderson GJ, Subramaniam VN. Combined deletion of Hfe and transferrin receptor 2 in mice leads to marked dysregulation of hepcidin and iron overload. Hepatology 2009; 50: 1992–2000. [DOI] [PubMed] [Google Scholar]
- Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood 2006; 108: 3204–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautz L, Jung G, Valore EV, Rivella S, Nemeth E, Ganz T. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet 2014; 46: 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtihet M, Bonde Y, Beckman L, Berinder K, Hoybye C, Rudling M et al. Circulating hepcidin-25 is reduced by endogenous estrogen in humans. PLoS ONE 2016; 11: e0148802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnough JB, Ramos E, Nemeth E, Ganz T. Inhibition of hepcidin transcription by growth factors. Hepatology 2012; 56: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli M, Girelli D, Campostrini N, Maccarinelli F, Finazzi D, Luscieti S et al. Heparin: a potent inhibitor of hepcidin expression in vitro and in vivo. Blood 2011; 117: 997–1004. [DOI] [PubMed] [Google Scholar]
- Krause A, Neitz S, Mägert HJ, Schulz A, Forssmann WG, Schulz-Knappe P et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 2000; 480: 147–150. [DOI] [PubMed] [Google Scholar]
- Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA 2006; 103: 10289–10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfay L, Clausen KA, Kim JW, Hegde P, Wang X, Miller LD et al. Hepcidin regulation in prostate and its disruption in prostate cancer. Cancer Res 2015; 75: 2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen H-F, Chan Y-P, Cheung W-L, Wong Y-C, Wang X, Chan K-W. The prognostic significance of BMP-6 signaling in prostate cancer. Mod Pathol 2008; 21: 1436–1443. [DOI] [PubMed] [Google Scholar]
- Barnes J, Anthony CT, Wall N, Steiner MS. Bone morphogenetic protein-6 expression in normal and malignant prostate. World J Urol 1995; 13: 337–343. [DOI] [PubMed] [Google Scholar]
- Lee GT, Kang DI, Ha Y-S, Jung YS, Chung J, Min K et al. Prostate cancer bone metastases acquire resistance to androgen deprivation via WNT5A-mediated BMP-6 induction. Br J Cancer 2014; 110: 1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey C, Brown LG, Pitts TEM, Vessella RL, Corey E. Bone morphogenetic protein 7 is expressed in prostate cancer metastases and its effects on prostate tumor cells depend on cell phenotype and the tumor microenvironment. Neoplasia 2010; 12: 192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Fukabori Y, Nakano K, Takezawa Y, CSuzuki T, Yamanaka H. Increased expression of bone morphogenetic protein-7 in bone metastatic prostate cancer. Prostate 2003; 54: 268–274. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wang L, Ma Y, Wu X, Jin L, Yu F. Increased hepcidin expression in non-small cell lung cancer tissue and serum is associated with clinical stage. Thorac Cancer 2014; 5: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Choesang T, Li H, Sun S, Pham P, Bao W et al. Increased hepcidin in transferrin-treated thalassemic mice correlates with increased liver BMP2 expression and decreased hepatocyte ERK activation. Haematologica 2016; 101: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenfeld EM, Bojnowski J, Perone J, Langenfeld J. Expression of bone morphogenetic proteins in human lung carcinomas. Ann Thorac Surg 2005; 80: 1028–1032. [DOI] [PubMed] [Google Scholar]
- Chen J, Ye L, Xie F, Yang Y, Zhang L, Jiang WG. Expression of bone morphogenetic protein 7 in lung cancer and its biological impact on lung cancer cells. Anticancer Res 2010; 30: 1113–1120. [PubMed] [Google Scholar]
- Bieniasz M, Oszajca K, Eusebio M, Kordiak J, Bartkowiak J, Szemraj J. The positive correlation between gene expression of the two angiogenic factors: VEGF and BMP-2 in lung cancer patients. Lung Cancer 2009; 66: 319–326. [DOI] [PubMed] [Google Scholar]
- Langenfeld EM, Calvano SE, Abou-Nukta F, Lowry SF, Amenta P, Langenfeld J. The mature bone morphogenetic protein-2 is aberrantly expressed in non-small cell lung carcinomas and stimulates tumor growth of A549 cells. Carcinogenesis 2003; 24: 1445–1454. [DOI] [PubMed] [Google Scholar]
- Ciniselli CM, De Bortoli M, Taverna E, Varinelli L, Pizzamiglio S, Veneroni S et al. Plasma hepcidin in early-stage breast cancer patients: no relationship with interleukin-6, erythropoietin and erythroferrone. Expert Rev Proteomics 2015; 12: 695–701. [DOI] [PubMed] [Google Scholar]
- Buijs JT, Henriquez NV, van Overveld PGM, van der Horst G, Que I, Schwaninger R et al. Bone morphogenetic protein 7 in the development and treatment of bone metastases from breast cancer. Cancer Res 2007; 67: 8742–8751. [DOI] [PubMed] [Google Scholar]
- Pauk M, Grgurevic L, Brkljacic J, Kufner V, Bordukalo-Niksic T, Grabusic K et al. Exogenous BMP7 corrects plasma iron overload and bone loss in Bmp6−/− mice. Int Orthop 2015; 39: 161–172. [DOI] [PubMed] [Google Scholar]
- Hu F, Meng X, Tong Q, Liang L, Xiang R, Zhu T et al. BMP-6 inhibits cell proliferation by targeting microRNA-192 in breast cancer. Biochim Biophys Acta 2013; 1832: 2379–2390. [DOI] [PubMed] [Google Scholar]
- Haudenschild DR, Palmer SM, Moseley TA, You Z, Reddi AH. Bone morphogenetic protein (BMP)-6 signaling and BMP antagonist noggin in prostate cancer. Cancer Res 2004; 64: 8276–8284. [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen Y, Guo W, Yuan L, Zhang D, Xu Y et al. Disordered hepcidin-ferroportin signaling promotes breast cancer growth. Cell Signal 2014; 26: 2539–2550. [DOI] [PubMed] [Google Scholar]
- Tuhkanen H, Hartikainen JM, Soini Y, Velasco G, Sironen R, Nykopp TK et al. Matriptase-2 gene (TMPRSS6) variants associate with breast cancer survival, and reduced expression is related to triple-negative breast cancer. Int J Cancer 2013; 133: 2334–2340. [DOI] [PubMed] [Google Scholar]
- Maes K, Nemeth E, Roodman GD, Huston A, Esteve F, Freytes C et al. In anemia of multiple myeloma, hepcidin is induced by increased bone morphogenetic protein 2. Blood 2010; 116: 3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Wang H, Xia J, Yang Y, Jin Z, Xu H et al. Decreased ferroportin promotes myeloma cell growth and osteoclast differentiation. Cancer Res 2015; 75: 2211–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Nemeth E, Chen Y-H, Goodnough J, Huston A, Roodman GD et al. Involvement of hepcidin in the anemia of multiple myeloma. Clin Cancer Res 2008; 14: 3262–3267. [DOI] [PubMed] [Google Scholar]
- Tisi MC, Bozzoli V, Giachelia M, Massini G, Ricerca BM, Maiolo E et al. Anemia in diffuse large B-cell non-Hodgkin lymphoma: the role of interleukin-6, hepcidin and erythropoietin. Leuk Lymphoma 2014; 55: 270–275. [DOI] [PubMed] [Google Scholar]
- Feng Q, Migas MC, Waheed A, Britton RS, Fleming RE. Ferritin upregulates hepatic expression of bone morphogenetic protein 6 and hepcidin in mice. AJP Gastrointest Liver Physiol 2012; 302: G1397–G1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002; 346: 1937–1947. [DOI] [PubMed] [Google Scholar]
- Pusatcioglu CK, Nemeth E, Fantuzzi G, Llor X, Freels S, Tussing-Humphreys L et al. Systemic and tumor level iron regulation in men with colorectal cancer: a case control study. Nutr Metab 2014; 11: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DG, Roberts K, Brookes MJ, Joy H, Martin A, Ismail T et al. Increased hepcidin expression in colorectal carcinogenesis. World J Gastroenterol 2008; 14: 1339–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Fu J, Li Y, Wang Y, Zhang L, Liu Y. Bone morphogenetic protein 7 is associated with the nodal invasion of colon cancer. Oncol Lett 2016; 11: 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama K, Tanaka F, Kosaka Y, Mimori K, Uetake H, Inoue H et al. Clinical significance of BMP7 in human colorectal cancer. Ann Surg Oncol 2008; 15: 1530–1537. [DOI] [PubMed] [Google Scholar]
- Vishnubalaji R, Yue S, Alfayez M, Kassem M, Liu F-F, Aldahmash A et al. Bone morphogenetic protein 2 (BMP2) induces growth suppression and enhances chemosensitivity of human colon cancer cells. Cancer Cell Int 2016; 16: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grijelmo C, Rodrigue C, Svrcek M, Bruyneel E, Hendrix A, Dewever O et al. Proinvasive activity of BMP-7 through SMAD4/src-independent and ERK/ Rac/JNK-dependent signaling pathways in colon cancer cells. Cell Signal 2007; 19: 1722–1732. [DOI] [PubMed] [Google Scholar]
- Voorneveld PW, Kodach LL, Jacobs RJ, Liv N, Zonnevylle AC, Hoogenboom JP et al. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology 2014; 147: 196–208.e13. [DOI] [PubMed] [Google Scholar]
- Yao L-Q, Zhong Y-S, Ren Z, Li Q-L, Zhou P-H, Xu M-D et al. Analysis of the role of the BMP7-Smad4-Id2 signaling pathway in SW480 colorectal carcinoma cells. Mol Med Rep 2011; 4: 627–631. [DOI] [PubMed] [Google Scholar]
- Deng H, Makizumi R, Ravikumar TS, Dong H, Yang W, Yang W-L. Bone morphogenetic protein-4 is overexpressed in colonic adenocarcinomas and promotes migration and invasion of HCT116 cells. Exp Cell Res 2007; 313: 1033–1044. [DOI] [PubMed] [Google Scholar]
- Deng H, Ravikumar TS, Yang W-L. Overexpression of bone morphogenetic protein 4 enhances the invasiveness of Smad4-deficient human colorectal cancer cells. Cancer Lett 2009; 281: 220–231. [DOI] [PubMed] [Google Scholar]
- Kamai T, Tomosugi N, Abe H, Arai K, Yoshida K-I. Increased serum hepcidin-25 level and increased tumor expression of hepcidin mRNA are associated with metastasis of renal cell carcinoma. BMC Cancer 2009; 9: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens P, Pickup MW, Novitskiy SV, Giltnane JM, Gorska AE, Hopkins CR et al. Inhibition of BMP signaling suppresses metastasis in mammary cancer. Oncogene 2015; 34: 2437–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DM, Jin S-W. Bone morphogenetic protein functions as a context-dependent angiogenic cue in vertebrates. Semin Cell Dev Biol 2011; 22: 1012–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egler RA, Burlingame SM, Nuchtern JG, Russell HV. Interleukin-6 and soluble interleukin-6 receptor levels as markers of disease extent and prognosis in neuroblastoma. Clin Cancer Res 2008; 14: 7028–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara T, DeClerck YA. Interleukin-6 in bone metastasis and cancer progression. Eur J Cancer 2010; 46: 1223–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Cao F, Tao M. The clinical value of hepcidin in breast cancer and its bone metastasis. Ann Clin Lab Sci 2017; 47: 120–128. [PubMed] [Google Scholar]
- Tanno T, Rabel A, Alleyne M, Lee YT, Dahut WL, Gulley JL et al. Hepcidin, anaemia, and prostate cancer. BJU Int 2011; 107: 678–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Smith MA, Doshi P, Sasser K, Fulp W, Altiok S et al. Antitumor efficacy of the anti-interleukin-6 (IL-6) antibody siltuximab in mouse xenograft models of lung cancer. J Thorac Oncol 2014; 9: 974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachbaur DM, Herold M, Maneschg A, Huber H. Serum levels of interleukin-6 in multiple myeloma and other hematological disorders: correlation with disease activity and other prognostic parameters. Ann Hematol 1991; 62: 54–58. [DOI] [PubMed] [Google Scholar]
- Ciccarelli BT, Patterson CJ, Hunter ZR, Hanzis C, Ioakimidis L, Manning R et al. Hepcidin is produced by lymphoplasmacytic cells and is associated with anemia in Waldenström’s macroglobulinemia. Clin Lymphoma Myeloma Leuk 2011; 11: 160–163. [DOI] [PubMed] [Google Scholar]
- Cheng P-P, Sun Z-Z, Jiang F, Tang Y-T, Jiao X-Y. Hepcidin expression in patients with acute leukaemia. Eur J Clin Invest 2012; 42: 517–525. [DOI] [PubMed] [Google Scholar]
- Santini V, Girelli D, Sanna A, Martinelli N, Duca L, Campostrini N et al. Hepcidin levels and their determinants in different types of myelodysplastic syndromes. PLoS ONE 2011; 6: e23109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H, Inoue K, Ogawa H, Yamagami T, Soma T, Miyake S et al. The expression of IL-6 and its related genes in acute leukemia. Leuk Lymphoma 1996; 21: 49–52. [DOI] [PubMed] [Google Scholar]
- Cui R, Gale RP, Zhu G, Xu Z, Qin T, Zhang Y et al. Serum iron metabolism and erythropoiesis in patients with myelodysplastic syndrome not receiving RBC transfusions. Leuk Res 2014; 38: 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callens C, Coulon S, Naudin J, Radford-Weiss I, Boissel N, Raffoux E et al. Targeting iron homeostasis induces cellular differentiation and synergizes with differentiating agents in acute myeloid leukemia. J Exp Med 2010; 207: 731–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagita M, Oka M, Watabe T, Iguchi H, Niida A, Takahashi S et al. USAG-1: a bone morphogenetic protein antagonist abundantly expressed in the kidney. Biochem Biophys Res Commun 2004; 316: 490–500. [DOI] [PubMed] [Google Scholar]
- Clausen KA, Blish KR, Birse CE, Triplette MA, Kute TE, Russell GB et al. SOSTDC1 differentially modulates Smad and beta-catenin activation and is down-regulated in breast cancer. Breast Cancer Res Treat 2011; 129: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wu S, Yang Y, Cai J, Zhu X, Wu J et al. SOSTDC1 is down-regulated in non-small cell lung cancer and contributes to cancer cell proliferation. Cell Biosci 2016; 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blish KR, Wang W, Willingham MC, Du W, Birse CE, Krishnan SR et al. A human bone morphogenetic protein antagonist is down-regulated in renal cancer. Mol Biol Cell 2008; 19: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chen J, Feng J, Xu Y, Zheng W, Wang J. SOSTDC1 inhibits follicular thyroid cancer cell proliferation, migration, and EMT via suppressing PI3K/Akt and MAPK/Erk signaling pathways. Mol Cell Biochem 2017; 435: 87–95. [DOI] [PubMed] [Google Scholar]
- Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut 2006; 55: 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K, Yeh M, Glass J. Interactions between ferroportin and hephaestin in rat enterocytes are reduced after iron ingestion. Gastroenterology 2011; 141: 292–299.e1. [DOI] [PubMed] [Google Scholar]
- Fuqua BK, Lu Y, Darshan D, Frazer DM, Wilkins SJ, Wolkow N et al. The multicopper ferroxidase hephaestin enhances intestinal iron absorption in mice. PLoS ONE 2014; 9: e98792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques O, Porto G, Rêma A, Faria F, Cruz Paula A, Gomez-Lazaro M et al. Local iron homeostasis in the breast ductal carcinoma microenvironment. BMC Cancer 2016; 16: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels TR, Bernabeu E, Rodríguez JA, Patel S, Kozman M, Chiappetta DA et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta 2012; 1820: 291–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Wang F, Elliott RL, Head JF. Expression of transferrin receptor and ferritin H-chain mRNA are associated with clinical and histopathological prognostic indicators in breast cancer. Anticancer Res 2001; 21: 541–549. [PubMed] [Google Scholar]
- Xu G, Ahn J, Chang S, Eguchi M, Ogier A, Han S et al. Lipocalin-2 induces cardiomyocyte apoptosis by increasing intracellular iron accumulation. J Biol Chem 2012; 287: 4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of mammalian iron metabolism. Cell 2010; 142: 24–38. [DOI] [PubMed] [Google Scholar]
- Kessler SM, Laggai S, Kiemer AK, Barghash A, Helms V. Hepatic hepcidin expression is decreased in cirrhosis and HCC. J Hepatol 2015; 62: 977–979. [DOI] [PubMed] [Google Scholar]
- Schmidt-Arras D, Rose-John S. IL-6 pathway in the liver: from physiopathology to therapy. J Hepatol 2016; 64: 1403–1415. [DOI] [PubMed] [Google Scholar]
- He Y, Cui Y, Xu B, Gu J, Wang W, Luo X. Hypermethylation leads to bone morphogenetic protein 6 downregulation in hepatocellular carcinoma. PLoS ONE 2014; 9: e87994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegdefrau U, Arndt S, Kivorski G, Hellerbrand C, Bosserhoff A-K. Downregulation of hemojuvelin prevents inhibitory effects of bone morphogenetic proteins on iron metabolism in hepatocellular carcinoma. Lab Invest 2011; 91: 1615–1623. [DOI] [PubMed] [Google Scholar]
- Hsu H, Peng S, Lai P, Chiu J, Lee P. Mutations of p53 gene in hepatocellular-carcinoma (hcc) correlate with tumor progression and patient prognosis—a study of 138 patients with unifocal hcc. Int J Oncol 1994; 4: 1341–1347. [DOI] [PubMed] [Google Scholar]
- Weizer-Stern O, Adamsky K, Margalit O, Ashur-Fabian O, Givol D, Amariglio N et al. Hepcidin, a key regulator of iron metabolism, is transcriptionally activated by p53. Br J Haematol 2007; 138: 253–262. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang W, Tsuji Y, Torti SV, Torti FM. Post-transcriptional modulation of iron homeostasis during p53-dependent growth arrest. J Biol Chem 2008; 283: 33911–33918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girelli D, Pasino M, Goodnough JB, Nemeth E, Guido M, Castagna A et al. Reduced serum hepcidin levels in patients with chronic hepatitis C. J Hepatol 2009; 51: 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridle KR, Cheung T-K, Murphy TL, Walters MM, Anderson GJ, Crawford DHG et al. Hepcidin is down-regulated in alcoholic liver injury: implications for the pathogenesis of alcoholic liver disease. Alcohol Clin Exp Res 2006; 30: 106–112. [DOI] [PubMed] [Google Scholar]
- Tan MGK, Kumarasinghe MP, Wang SM, Ooi LLPJ, Aw SE, Hui KM. Modulation of iron-regulatory genes in human hepatocellular carcinoma and its physiological consequences. Exp Biol Med 2009; 234: 693–702. [DOI] [PubMed] [Google Scholar]
- Tseng H-H, Chang J-G, Hwang Y-H, Yeh K-T, Chen Y-L, Yu H-S. Expression of hepcidin and other iron-regulatory genes in human hepatocellular carcinoma and its clinical implications. J Cancer Res Clin Oncol 2009; 135: 1413–1420. [DOI] [PubMed] [Google Scholar]
- Musci G, Polticelli F, Bonaccorsi di Patti MC. Ceruloplasmin-ferroportin system of iron traffic in vertebrates. World J Biol Chem 2014; 5: 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CY, Koo JH, Kim SH, Gardenghi S, Rivella S, Strnad P et al. Hepcidin inhibits Smad3 phosphorylation in hepatic stellate cells by impeding ferroportin-mediated regulation of Akt. Nat Commun 2016; 7: 13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänninen MM, Haapasalo J, Haapasalo H, Fleming RE, Britton RS, Bacon BR et al. Expression of iron-related genes in human brain and brain tumors. BMC Neurosci 2009; 10: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberg DL, Miller TE, Wu Q, Flavahan WA, Das NK, Hale JS et al. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell 2015; 28: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RC, Kosman DJ. Glial cell ceruloplasmin and hepcidin differentially regulate iron efflux from brain microvascular endothelial cells. PLoS ONE 2014; 9: e89003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Qian Z-M, Luo Q, Yung W-H, Ke Y. Hepcidin suppresses brain iron accumulation by downregulating iron transport proteins in iron-overloaded rats. Mol Neurobiol 2015; 52: 101–114. [DOI] [PubMed] [Google Scholar]
- Zhou Y-F, Zhang C, Yang G, Qian Z-M, Zhang M-W, Ma J et al. Hepcidin protects neuron from hemin-mediated injury by reducing iron. Front Physiol 2017; 8: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Qian C, Ming Qian Z, Wu X-M, Xie H, Yung W-H et al. Hepcidin directly inhibits transferrin receptor 1 expression in astrocytes via a cyclic AMP-protein kinase a pathway. Glia 2011; 59: 936–945. [DOI] [PubMed] [Google Scholar]
- Urrutia P, Aguirre P, Esparza A, Tapia V, Mena NP, Arredondo M et al. Inflammation alters the expression of DMT1, FPN1 and hepcidin, and it causes iron accumulation in central nervous system cells. J Neurochem 2013; 126: 541–549. [DOI] [PubMed] [Google Scholar]
- Dayani PN, Bishop MC, Black K, Zeltzer PM. Desferoxamine (DFO)-mediated iron chelation: rationale for a novel approach to therapy for brain cancer. J Neurooncol 2004; 67: 367–377. [DOI] [PubMed] [Google Scholar]
- Valdés PA, Samkoe K, O’Hara JA, Roberts DW, Paulsen KD, Pogue BW. Deferoxamine iron chelation increases δ-aminolevulinic acid induced protoporphyrin IX in xenograft glioma model. Photochem Photobiol 2010; 86: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac S, Böser P, Cui Y, Ferring-Appel D, Casarrubea D, Huang L et al. Anti-hemojuvelin antibody corrects anemia caused by inappropriately high hepcidin levels. Haematologica 2016; 101: e173–e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli M, Asperti M, Ruzzenenti P, Regoni M, Arosio P. Hepcidin antagonists for potential treatments of disorders with hepcidin excess. Front Pharmacol 2014; 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crielaard BJ, Lammers T, Rivella S. Targeting iron metabolism in drug discovery and delivery. Nat Rev Drug Discov 2017; 16: 400–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOX-H94. Noxxon Pharma. Available at: http://www.noxxon.com/index.php?option=com_content&view=article&id=22&Itemid=479.
- Vadhan-Raj S, Abonour R, Goldman JW, Smith DA, Slapak CA, Ilaria RL et al. A first-in-human phase 1 study of a hepcidin monoclonal antibody, LY2787106, in cancer-associated anemia. J Hematol Oncol 2017; 10: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LJPC-401. LaJolla Pharmaceutical. Available at: http://lajollapharmaceutical.com/product-pipeline/ljpc-401/.
- Urano S, Ohara T, Noma K, Katsube R, Ninomiya T, Tomono Y et al. Iron depletion enhances the effect of sorafenib in hepatocarcinoma. Cancer Biol Ther 2016; 17: 648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mleczko-Sanecka K, Roche F, Rita da Silva A, Call D, D’Alessio F, Ragab A et al. Unbiased RNAi screen for hepcidin regulators links hepcidin suppression to proliferative Ras/RAF and nutrient-dependent mTOR signaling. Blood 2014; 123: 1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli M, Asperti M, Naggi A, Campostrini N, Girelli D, Corbella M et al. Glycol-split nonanticoagulant heparins are inhibitors of hepcidin expression in vitro and in vivo. Blood 2014; 123: 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Magen H, Einsele H, Chatterjee M, Grasso M, Specchia G et al. Roneparstat (SST0001), an innovative heparanase (HPSE) inhibitor for multiple myeloma (MM) therapy: first in man study. Blood 2015; 126: 3246. [Google Scholar]
- Cassinelli G, Lanzi C, Tortoreto M, Cominetti D, Petrangolini G, Favini E et al. Antitumor efficacy of the heparanase inhibitor SST0001 alone and in combination with antiangiogenic agents in the treatment of human pediatric sarcoma models. Biochem Pharmacol 2013; 85: 1424–1432. [DOI] [PubMed] [Google Scholar]
- Pechlaner R, Weiss G, Bansal S, Mayr M, Santer P, Pallhuber B et al. Inadequate hepcidin serum concentrations predict incident type 2 diabetes mellitus. Diabetes Metab Res Rev 2016; 32: 187–192. [DOI] [PubMed] [Google Scholar]
- Tan TCH, Crawford DHG, Franklin ME, Jaskowski LA, Macdonald GA, Jonsson JR et al. The serum hepcidin:ferritin ratio is a potential biomarker for cirrhosis. Liver Int 2012; 32: 1391–1399. [DOI] [PubMed] [Google Scholar]
- Lakhal-Littleton S, Wolna M, Chung YJ, Christian HC, Heather LC, Brescia M et al. An essential cell-autonomous role for hepcidin in cardiac iron homeostasis. Elife 2016; 5: e19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Du F, Qian ZM, Luo QQ, Sheng Y, Yung W et al. Pre-treatment of rats with ad-hepcidin prevents iron-induced oxidative stress in the brain. Free Radic Biol Med 2016; 90: 126–132. [DOI] [PubMed] [Google Scholar]