Abstract
BACKGROUND
The association between X-ray repair cross-complementing group 1 (XRCC1) Arg399Gln gene polymorphism and hepatocellular carcinoma (HCC) has been investigated in several populations. However, the findings are controversial. The aim of this study was to address the association between XRCC1 Arg399Gln polymorphism and HCC in an Iranian population.
METHODS
We have evaluated the association between XRCC1 Arg399Gln gene polymorphism and HCC in 151 Iranian individuals (50 patients with HCC and 101 healthy matched controls) using polymerase chain reaction-restriction fragment length polymorphisms (PCR-RFLP) method.
RESULTS
Significant association was found for the XRCC1 A allele and HCC [OR = 1.93, 95% CI (1.16 - 3.25), P = 0.0099]. Also, genotype analysis by SNPStats online software showed a significant association between XRCC1 gene polymorphisms and HCC under co-dominant, dominant, and recessive genetic models.
CONCLUSION
Our study provides evidence that the XRCC1 Arg399Gln polymorphism may be associated with the risk of HCC development in Iranian population.
Keywords: X-ray repair cross-complementing group 1, XRCC1 Arg399Gln polymorphism, Hepatocellular carcinoma
INTRODUCTION
Liver cancer is the sixth most common cancer and the second leading cause of cancer-related deaths worldwide. Hepatocellular carcinoma (HCC) is the most common histological type of liver cancer.1 In Middle Eastern countries, such as Iran, the prevalence of HCC is lower compared with sub-Saharan Africa and some Far East countries.2 Many environmental factors, such as alcohol consumption, insulin resistance, and viral hepatitis, including chronic hepatitis B (HBV) or hepatitis C (HCV) have been evaluated for their effects on HCC susceptibility.3 The low incidence of HCC in Iran seems more likely to be related to low incidence of alcoholic cirrhosis and the relatively low prevalence of HBV and HCV infections.4 However, genetic factors might play the key role in the pathogenesis of HCC.5 Many studies show that the incidence of HCC is increasing in Iran.6 Therefore, the plan for the control and identification of this cancer is crucial.
The presence of specific single nucleotide polymorphisms (SNPs) inhuman genome has been associated with susceptibility to carcinogenesis.7,8 Therefore, the identification of genetic factors influencing the risk of developing HCC is crucial.Previous studies have shown that the human X-ray repair cross-complementing group 1 gene (XRCC1) is an important candidate gene influencing HCC susceptibility.9,10 This gene is a major DNA repair gene involved in base excision repair (BER) pathway.11 The XRCC1 Arg399Gln gene polymorphism is a common functional SNP in codon 399 of exon 10, a non-synonymous G/A.
polymorphism changing arginine to glutamine.12 DNA repair ability could be impaired by amino acid substitution, and this can increase the probability of cancer.13 Therefore, XRCC1 + 399G/A (rs25487) polymorphism may increase the risk of susceptibility to HCC. Previous studies showed that XRCC1 rs25487 could result in increased risk of tumorigenesis in HCC.12,14-16, However, some studies have indicated that the rs25487 polymorphism has no significant association with HCC.17 The aim of this study was to assess whether XRCC1 (Arg399Gln) gene polymorphism affects the susceptibility of HCC in an Iranian population.
MATERIALS AND METHODS
Study subjects
The sample sizes were calculated as 50 for the patients and 101 for the control group. The patients with the HCC were recruited from the hospitals of Mazandaran, Tehran, Shiraz, Isfahan, Sistan and Baluchestan, and Gorgan provinces. Control individuals, were randomly recruited from Mazandaran province and were matched with the patients with HCC for age, sex, and ethnical origin. The patients with HCC were diagnosed by expert physicians based on pathological and histopathological criteria.18 Written informed consent was obtained from all the subject.
DNA extraction
DNA was extracted from paraffin-embedded tissues of the patients, using a CinnaPure DNA kit (sinnaclon, Iran) according to the manufacturer’s instructions, and from blood cells of the controls, using the boiling procedure. DNA concentration was determined by Picodrop UV/Vis Spectrophotometer (Picodrop Ltd, UK).
XRCC1 genotyping
Genotyping of XRCC1 single-nucleotide polymorphism rs25487 was performed by restriction fragment length polymorphisms (RFLP) method, as previously described.19 The primers used were as follows: 5´TTGTGCTTTCTCTGTGTCCA3´ (Forward) and 5´TCCTCCAGCCTTTTCTGATA3´ (Reverse). Briefly, the region was amplified by polymerase chain reaction (PCR). The PCR was performed using the following conditions: an initial denaturation step at 95°C for 5 min followed by 35 cycles of 30 s at 95°C; 30 s at 61°C; 30 s at 72°C, and a final extension step of at 72°C for 5 min. The PCR products were digested through 15 min with MspI at 37°C, separated by electrophoresis on a 3% agarose gel in Tris base EDTA (TBE) buffer stained with ethidium bromide and visualized by Gel Doc Imaging System (E-Box VILBER, Japanese). MspI digestion produced 240 and 375 bp fragments for G allele and 615 bp fragment for A allele. The length of the genotype bands has been shown in figure 1.
Fig.1.
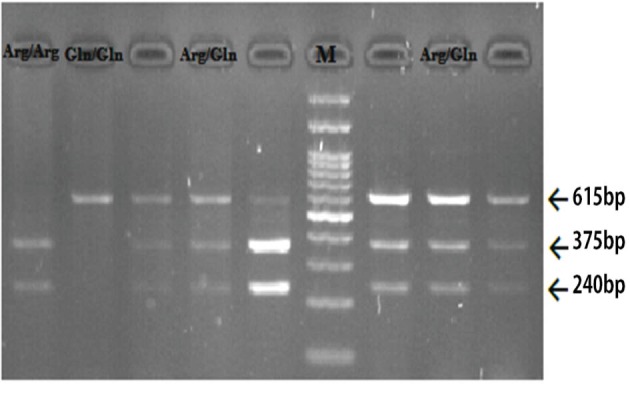
Electrophoresis digested products with MspI restricted enzyme. The marker (M) that used was 100 base pairs
Statistical analysis
The association between XRCC1 genepolymorphism (rs25487) and HCC risk and the Hardy-Weinberg equilibrium exact test were performed using the online SNPstats software.20 The SNPstats software analyses the associations by linear or logistic regression according to the response variable.21 This web-based software uses the multiple inheritance models (co-dominant, dominant, recessive, overdominant, and log-additive) for SNP association analysis.21 Inheritance models compare Gln/Gln genotypes to Arg/Arg-Arg/Gln genotypes in recessive model and Arg/Gln-Gln/Gln to Arg/Arg genotypes in dominant model. These inheritance models, also, compare Arg/Gln genotypes to Arg/Arg-Gln/Gln genotypes in overdominant model and Gln/Gln, Arg/Arg to Arg/Arg genotypes in co-dominant model.
RESULTS
Our study included 50 patients with HCC and 101 controls aged over 57.5 years. Genotype and allelic frequencies of XRCC1 polymorphism (rs 25487) are shown in table 1. The genotype distribution was in Hardy–Weinberg equilibrium in both groups (p > 0.05). Allele frequency for A allele in the patients with HCC was significantly higher than the healthy controls [OR = 1.93, 95% CI (1.16 - 3.25), p = 0.0099] (table 2). A significant association was observed between the genotype frequency of the patients with HCC and healthy controls under co-dominant (p = 0.0015), recessive (p = 0.0004), and overdominant (p = 0.019) models (table 2). In contrast, there was no significant association between the genotype frequency of the patients with HCC and healthy controls under dominant (p = 0.42) model (table 2).
Table 1. Genotype and allelic frequencies of XRCC1 Arg399Gln gene polymorphism (rs25487) in cases and controls .
| Variables | All subjects | Case | Control | ||||
| Count | Proportion | Count | Proportion | Count | Proportion | ||
| Allele | G | 160 | 0.53 | 42 | 0.42 | 118 | 0.58 |
| A | 142 | 0.47 | 58 | 0.58 | 84 | 0.42 | |
| Genotype | A/A | 34 | 0.23 | 20 | 0.4 | 14 | 0.14 |
| G/A | 74 | 0.49 | 18 | 0.36 | 56 | 0.55 | |
| G/G | 43 | 0.28 | 12 | 0.24 | 31 | 0.31 | |
Table 2. Association between XRCC1 Arg399Gln genepolymorphism (rs25487) and HCC(n = 151, adjusted by sex) .
| Model | Genotype | Case(%) | Control(%) | OR (95% CI) | P value |
| Co-dominant | G/G(Arg/Arg) | 12 (24%) | 31 (30.7%) | 1.00 | 0.0015 |
| A/G(Arg/Gln) | 18 (36%) | 56 (55.5%) | 1.25 (0.53-2.95) | ||
| A/A(Gln/Gln) | 20 (40%) | 14 (13.9%) | 0.27 (0.10-0.70) | ||
| Dominant | G/G | 12 (24%) | 31 (30.7%) | 1.00 | 0.42 |
| A/G-A/A | 38 (76%) | 70 (69.3%) | 0.73 (0.33-1.58) | ||
| Recessive | G/G-A/G | 30 (60%) | 87 (86.1%) | 1.00 | 0.0004 |
| A/A | 20 (40%) | 14 (13.9%) | 0.24 (0.11-0.53) | ||
| Overdominant | G/G-A/A | 32 (64%) | 45 (44.5%) | 1.00 | 0.019 |
| A/G | 18 (36%) | 56 (55.5%) | 2.30 (1.13-4.65) | ||
| Log-additive | --- | --- | --- | 0.51 (0.31-0.85) | 0.0074 |
| Allele | G | 42 (42%) | 118 (58%) | 1.00 | 0.0099 |
| A | 58 (58%) | 84 (42%) | 1.93 (1.16-3.25) |
DISCUSSION
HCC is the sixth most common cancer and the second cause of cancer-related death worldwide.1 XRCC1 is a crucial component of BER, which is the predominant DNA damage repair pathway for processing of small base lesions. So any alteration in the activity of these genes due to polymorphism can lead to the increased risk of the carcinoma development. Different studies suggested that G to A substitution at codon 399 in exon 10 of XRCC1 gene could injure the DNA repair capabilities and increase the risk of various cancers.22 Many research studies have evaluated the association between XRCC1 Arg399Gln gene polymorphism and HCC risk, but the results are controversial.12,14,16,17
In the present study, we observed that XRCC1 + 399G/A (rs25487) polymorphism presented as a risk factor for HCC in the studied population. Our data showed that the frequency of XRCC1 + 399 A allele in the patients with HCC was significantly higher than the healthy controls (p = 0.0099) (table 2). It seems that XRCC1 + 399 A allele and Gln/Gln genotype is a risk allele for the patients with HCC. These genotyping data are opposite to some studies 17 and in line with others.12,14,16 The results, are identical to the findings of Li and colleagues who showed that XRCC1 Arg399Gln polymorphism was significantly associated with HCC susceptibility in Asian population.23 But X.Y.Zeng shows that XRCC1 Arg399Gln was not significantly correlated to susceptibility to HCC.17 Also, our study showed that the Gln/Gln genotype may increase the risk of HCC among the patients. However, Yu and co-workers have reported that the Gln/Gln genotype could have a protective effect for patients.24 This suggests that the increased risk of HCC might be dependent on the population and sample sizes. Different populations also have different allele frequencies, such as African, European, and Asian populations.
Genotypic analysis, also, showed that there was a significant association between the frequency of Arg/Gln and Gln/Gln genotypes among the patients with HCC and healthy controls under co-dominant and recessive models, but not under a dominant model (table 2). Our genetic data provide suitable information about XRCC1 + 399 polymorphism and susceptibility to HCC in Iranian population, but it is necessary to replicate the study with larger sample size.
CONCLUSIONS
Our study showed that XRCC1 Arg399Gln polymorphism may be associated with the risk of HCC development and also, the Gln/Gln genotype may increase the risk of HCC in Iranian population. However, further studies with large sample size should be conducted to clarify the association betweenXRCC1 Arg399Gln gene polymorphism and HCC in Iranian population.
Please cite this paper as:
Bazgir A, Agha Gholizadeh M, Khosravi A, Mansour Samaei N. The X-ray Repair Cross-Complementing Group 1 Arg399Gln Genetic Polymorphism and Risk of Hepatocellular Carcinoma in an Iranian Population. Middle East J Dig Dis 2018;10:40-44. doi: 10.15171/mejdd.2017.89.
Footnotes
ETHICAL APPROVAL There is nothing to be declared.
CONFLICT OF INTEREST The authors declare no conflict of interest related to this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Poustchi H, Sepanlou S, Esmaili S, Mehrabi N, Ansarymoghadam A. Hepatocellular carcinoma in the world and the middle East. Middle East J Dig Dis. 2010;2:31–41. [PMC free article] [PubMed] [Google Scholar]
- 3.Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14:4300–8. doi: 10.3748/wjg.14.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moghaddam SD, Haghdoost AA, Hoseini SH, Ramazani R, Rezazadehkermani M. Incidence of hepatocellular carcinoma in southeast iran. Hepat Mon. 2010;10:270–4. [PMC free article] [PubMed] [Google Scholar]
- 5.Ho DW-H, Lo RC-L, Chan L-K, Ng IO-L. Molecular pathogenesis of hepatocellular carcinoma. Liver cancer. 2016;5:290–302. doi: 10.1159/000449340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirzaei M, Ghoncheh M, Pournamdar Z, Soheilipour F, Salehiniya H. Incidence and Trend of Liver Cancer in Iran. J Coll Physicians Surg Pak. 2016;26:306–9. [PubMed] [Google Scholar]
- 7.Gholizadeh M, Khosravi A, Torabian P, Gholipoor N, Samaei NM. Association of the epidermal growth factor gene+ 61A> G polymorphism with hepatocellular carcinoma in an Iranian population. Gastroenterol Hepatol Bed Bench. 2017;10:284–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Khosravi A, Javan B, Tabatabaiefar MA, Ebadi H, Fathi D, Shahbazi M. Association of interleukin-1 gene cluster polymorphisms and haplotypes with multiple sclerosis in an Iranian population. J Neuroimmunol. 2015;288:114–9. doi: 10.1016/j.jneuroim.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Bi J, Zhong C, Li K, Chu H, Wang B. Association study of single nucleotide polymorphisms in XRCC1 gene with risk of hepatocellular carcinoma in Chinese Han population. Biomed Res Int. 2013;2013:138785. doi: 10.1155/2013/138785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi Y, Cui L, Song Y, Li N. XRCC1 Arg399Gln genetic polymorphism and the risk of hepatocellular carcinoma: a meta-analysis. Mol Biol Rep. 2014;41:879–87. doi: 10.1007/s11033-013-2929-0. [DOI] [PubMed] [Google Scholar]
- 11. Izumi T, Mellon I. Base Excision Repair and Nucleotide Excision Repair. Genome Stability: Elsevier 2016. p. 275-302.
- 12.Li Qw, Lu C-r, Ye M, Xiao W-h, Liang J. Evaluation of DNA repair gene XRCC1 polymorphism in prediction and prognosis of hepatocellular carcinoma risk. Asian Pac J Cancer Prev. 2012;13:191–4. doi: 10.7314/APJCP.2012.13.1.191. [DOI] [PubMed] [Google Scholar]
- 13. Sterpone S, Cozzi R. Influence of XRCC1 genetic polymorphisms on ionizing radiation-induced DNA damage and repair. J Nucleic Acids 2010;2010. pii: 780369. [DOI] [PMC free article] [PubMed]
- 14.Duan WH, Zhu ZY, Liu JG, Dong M-S, Chen J-Z, Liu Q-D. et al. XRCC1 Arg399Gln genepolymorphism and hepatocellular carcinoma risk in the Chinese Han population: a meta-analysis. Asian Pac J Cancer Prev. 2012;13:3601–4. doi: 10.7314/APJCP.2012.13.8.3601. [DOI] [PubMed] [Google Scholar]
- 15.Hu Z, Ma H, Chen F, Wei Q, Shen H. XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiol Biomarkers Prev. 2005;14:1810–8. doi: 10.1158/1055-9965. [DOI] [PubMed] [Google Scholar]
- 16.Pan Y, Zhao L, Chen X-M, Gu Y, Shen J-G, Liu L-M. The XRCC1 Arg399Gln genetic polymorphism contributes to hepatocellular carcinoma susceptibility: an updated meta-analysis. Asian Pac J Cancer Prev. 2013;14:5761–7. doi: 10.7314/APJCP.2013.14.10.5761. [DOI] [PubMed] [Google Scholar]
- 17.Zeng X, Huang J, Xu J, Xu Y, Yu H, Ji L. et al. Meta-analysis demonstrates lack of a relationship between XRCC1-399 gene polymorphisms and susceptibility to hepatocellular carcinoma. Genet Mol Res. 2013;12:1916–23. doi: 10.4238/2013. [DOI] [PubMed] [Google Scholar]
- 18.Kojiro M, Wanless IR, Alves V, Badve S, Balabaud C, Bedosa P. et al. Pathologic diagnosis of early hepatocellular carcinoma: a report of the international consensus group for hepatocellular neoplasia. Hepatology. 2009;49:658–64. doi: 10.1002/hep.22709. [DOI] [PubMed] [Google Scholar]
- 19.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–9. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 20. SNPstats software. Available : http://bioinfo.iconcologia.net/snpstats/start.htm. Accessed 2016.
- 21.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Peng Q, Chen Y, You J, Chen Z, Deng Y. et al. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk: a meta-analysis. PloS One. 2013;8:e73448. doi: 10.1371/journal.pone.0073448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Li Z, Feng L, Guo W, Zhang S. Polymorphisms of DNA repair gene XRCC1 and hepatocellular carcinoma risk among East Asians: a meta-analysis. Tumour Biol. 2013;34:261–9. doi: 10.1007/s13277-012-0546-5. [DOI] [PubMed] [Google Scholar]
- 24.Yu MW, Yang SY, Pan IJ, Lin CL, Liu CJ, Liaw YF. et al. Polymorphisms in XRCC1 and Glutathione S-Transferase Genes and Hepatitis B–Related Hepatocellular Carcinoma. J Natl Cancer Ins. 2003;95:1485–8. doi: 10.1093/jnci/djg051. [DOI] [PubMed] [Google Scholar]


