Abstract
In a previous study, two genes responsible for white flower phenotypes in carnation were identified. These genes encoded enzymes involved in anthocyanin synthesis, namely, flavanone 3-hydroxylase (F3H) and dihydroflavonol 4-reductase (DFR), and showed reduced expression in the white flower phenotypes. Here, we identify another candidate gene for white phenotype in carnation flowers using an RNA-seq analysis followed by RT-PCR. This candidate gene encodes a transcriptional regulatory factor of the basic helix-loop-helix (bHLH) type. In the cultivar examined here, both F3H and DFR genes produced active enzyme proteins; however, expression of DFR and of genes for enzymes involved in the downstream anthocyanin synthetic pathway from DFR was repressed in the absence of bHLH expression. Occasionally, flowers of the white flowered cultivar used here have red speckles and stripes on the white petals. We found that expression of bHLH occurred in these red petal segments and induced expression of DFR and the following downstream enzymes. Our results indicate that a member of the bHLH superfamily is another gene involved in anthocyanin synthesis in addition to structural genes encoding enzymes.
Keywords: anthocyanin, basic helix-loop-helix, carnation, transcriptional regulatory factor, variegation
Introduction
White flowers of several plant species are popular worldwide for use at ceremonial events as a symbol of innocence and purity. Additionally, white flowers are also of value for inclusion in floral arrangements together with colored flowers. Commercial growers therefore have a ready market for white flowers, especially in Japan where white carnations and chrysanthemums are required all year round for ceremonial events.
The pink, red and crimson colors of carnation flowers are derived from anthocyanins, and those of yellow and cream flowers are produced by chalcononaringenin and flavonols, respectively (Iwashina et al. 2010, Onozaki et al. 1999). These flower color variants have been generated by conventional breeding techniques by identification of spontaneous mutants, and then selection and cross-hybridization. More recently, advances in biotechnological techniques have allowed the generation of new carnation flower color varieties, especially those with blue hues, which are not found in natural mutants (Fukui et al. 2003, Okamura et al. 2013). Genetic studies over the last half century identified the genes responsible for flower color phenotypes; for example, defects in the A and I genes of carnations responsible for white and yellow flower color phenotypes, respectively (Geissman and Mehlquist 1947, Mehlquist 1939), and these variants have been used to establish new carnation flower color varieties. Molecular biological analyses revealed that the A and I genes encoded enzymes involved in the anthocyanin synthesis pathway, namely, dihydroflavonol 4-reductase (DFR) and chalcone isomerase, respectively (Itoh et al. 2002, Stich et al. 1992). Defective expression of flavanone 3-hydroxylase (F3H) has also been found to produce a white flower phenotype; this enzyme has a role in the catalytic step from naringenine to dihydroflavonol just before the DFR step (Mato et al. 2000). Thus, mutations in two structural genes for anthocyanin synthesis, DFR and F3H, are able to generate white flower phenotypes in carnations.
In commercial varieties of white flowered carnations, the occasional appearance of red spots, speckles and stripes on the white petals may occur (Fig. 1C). In some cases, fully red petals emerge in bud sports and are established as new cultivars (Mato et al. 2000, 2001, Yoshida et al. 2004). Our group showed that the white sport varieties ‘Kaly’ and ‘White Mind’ have defective expression of F3H and DFR, respectively, which results in their white flower phenotype (Mato et al. 2000). Here, we analyzed another white flower cultivar, ‘2-241-1 MB’, and found that the phenotype may be caused by the down-regulation of expression of the gene for a transcriptional regulatory factor that is a member of the basic helix-loop-helix (bHLH) protein superfamily.
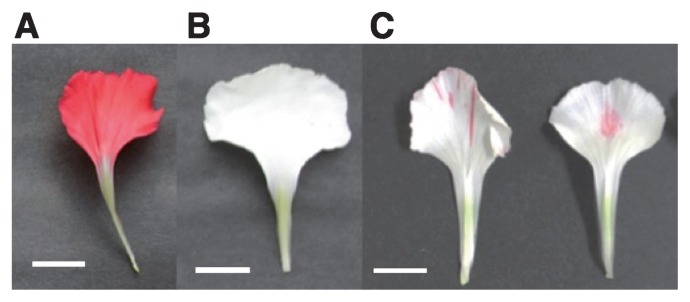
Fig. 1.
The phenotypes of carnation cultivars. (A) 4-94-1 MR, (B) 2-241-1 MB, (C) 2-241-1 MB variegated. White bar indicates 1 cm.
Materials and Methods
Plant materials and chemicals
The carnation (Dianthus caryophyllus) varieties used here, red-flowered ‘4-94-1 MR’ and white-flowered ‘2-241-1 MB’ (Fig. 1A, 1B), were a gift from Japan Agribio Company. Additionally, we used ‘2-241-1 MB variegated’ plants (Fig. 1C) that occurred spontaneously among ‘2-241-1 MB’ plants and had red spots and speckles on white petals. The definition of developmental stages of petals was as follows; stage 1, closed flower buds and petals in buds did not accumulate anthocyanin: stage 2, petals in closed buds of ‘4-94-1 MR’ began synthesis of anthocyanin to be red: stage 3, just opening flower buds in which top of petals made appearance: stage 4, petals just before full open: stage 5, petals of full open flowers (Mato et al. 2000, Matsuba et al. 2010). Harvested petals were immediately frozen in liquid nitrogen and stored at −80°C until use.
RNA-seq and the data analysis
Total RNAs were extracted from petals of ‘4-94-1 MR’ and ‘2-241-1 MB’ plants at individual stages 1–4. Total RNA integrity was assessed using a Fragment Analyzer with High Sensitivity RNA Analysis Kit (Advances Analytical Technologies, Inc., IA, USA). To construct RNA-seq libraries, poly(A)+ RNA was purified from 5 μg total RNA using the NEBNext Poly(A)mRNA Magnetic Isolation Module (New England BioLabs, MA, USA) according to the manufacturer’s instructions. Poly(A)+ RNA integrity was assessed using the same Fragment Analyzer kit as above. Libraries for Illumina paired-end sequencing were prepared using NEXTflex Rapid Directional mRNA-Seq Kit with NEXTFlex Adapters (Bioo Scientific, TX, USA). The size of libraries was confirmed using Fragment Analyzer with High Sensitivity NGS Fragment Analysis Kit (Advances Analytical Technologies, Inc.). Quantification of each library was determined using the StepOnePlus Real-time PCR system (Applied Biosystems, CA, USA) with Illumina Library Quantification Kit Universal qPCR Mix (Kapa Biosystems, Inc., MA, USA). Sequencing was performed on the NextSeq 500 system (150 bp, paired-end) (Illumina Inc., CA, USA). The conversion of base call (BCL) files to FASTQ files was performed using bcl2fastq2 Conversion v2.16.0.10 (Illumina). Processing of raw reads was performed according to the following three steps: first, low-quality sequences of the last 20 bases in each read were trimmed; second, short length reads (<100 bases) were removed; and third, paired-end reads were selected by filtering (parameters were set at q20, p80) from the trimmed 100–130 base reads. In total, 109,760,610 high quality reads (81.2% of the raw data) were obtained. These sequence data were registered as DDBJ DRA accession number DRA005974.
Nucleotide sequences from petal stages 1–4 of ‘4-94-1 MR’ and ‘2-241-1 MB’ were trimmed of adaptor and low-quality sequence regions. All sequences were mapped to the predicted transcriptome sequences and annotation data of carnation ‘Francesco’ genome (DCA_r1.0_cds.fna, downloaded from Carnation DB, http://carnation.kazusa.or.jp/) by bowtie2 version 2.2.9. Fragments per kilobase of exon per million fragments mapped (FPKM) were calculated using eXpress version 1.5.1. The FPKM values of the following genes for anthocyanin biosynthesis were compared: DFR (Carnation DB gene ID: Dca4324), anthocyanidin synthase (ANS) (Dca23371), UDP-glucose: anthocyanin 3-glucosyltransferase (3GT) (Dic3GT1: accession no. AB191245, 3GT1: Dca17573 and 3GT2: Dca47835) (Ogata et al. 2004), glutathione S-transferase (GST) (DcGSTF2: AB688111: Dca57804) (Sasaki et al. 2013) and bHLH (Dca31716).
Phylogenetic analysis
The amino acid sequences of the bHLH gene products were obtained from Carnation DB and accession numbers are as described by Yagi et al. (2014). A phylogenetic tree was produced using the ClustalW alignment and the Neighbor-joining algorithm of MEGA version 7.0.21. Bootstrap values were obtained from 1,000 replications.
Quantitative RT-PCR analysis
Stage 5 petals from ‘2-241-1 MB’ were used for a quantitative RT-PCR analysis. For ‘2-241-1 MB variegated’ plants, the red blotchy regions of the petals were used to prepare RNA. The first strand cDNA reaction mixture was diluted three-fold with water, and then 1 μL of the diluted mixture was used as the PCR template in a 10 μL PCR reaction mixture. DFR, ANS, GST, and bHLH primers were designed based on Carnation DB, and the Actin primers were described in our previous report (Matsuba et al. 2010) (Supplemental Table 1). Quantitative RT-PCR was performed using Thunderbird SYBR qPCR Mix (TOYOBO Co. Ltd., Osaka, Japan) and a DNA Engine Opticone 2 (Bio-Rad Laboratories, CA, USA). The PCR conditions were 1 min denaturation at 95°C followed by 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. Actin was used as the reference gene for comparison of expression level of each gene. The assay was repeated three times.
Results and Discussion
Genes for flavonoid biosynthesis enzymes can be separated into an “early gene” group including chalcone synthase, chalcone isomerase, F3H, flavonoid 3′-hydroxylase and 3GT, and a “late gene” group including DFR, ANS and GST (Yonekura-Sakakibara et al. 2007). Four kaempferol glycosides are accumulated in the petals of white carnations (Iwashina et al. 2010). This suggests that ‘2-241-1 MB’ might produce kaempferol but not anthocyanin, and therefore that expression of genes encoding enzymes involved in the early genes of the upstream flavonoid biosynthetic pathway (up to F3H) should be active, but those downstream of DFR might be repressed. In the RNA-seq analysis, the nucleotide sequence of transcripts for late genes were essentially identical to those encoded in the wild type cultivar ‘Francesco’ (Yagi et al. 2014). Detailed nucleotide sequences for F3H and DFR, which are defective in the white flower phenotype cultivars ‘Kaly’ and ‘White Mind’, respectively (Mato et al. 2000), were determined. The deduced amino acid sequence of F3H was identical between white-flowered ‘2-241-1 MB’ and red-flowered ‘4-94-1 MR’ (Supplemental Fig. 1A). Compared to the DcDFR-R1 cDNA being the wild-type nucleotide sequence, the DcDFR-R2 and DcDFR-B1 cDNAs had a nine base insertion at the proximal region of the N-terminal end, which was identified as the footprint sequence of the dTdic1 excision event (Itoh et al. 2002). This nine base insertion resulted in an additional three amino acids without interruption of the coding frame (Supplemental Fig. 1B, 1C). The DcDFR-B2 cDNA from ‘2-241-1 MB’ had an eight base insertion to become frameshift and generate a stop codon (Supplemental Fig. 1B, 1C). Recombinant proteins, produced using Escherichia coli cells transformed with DcDFR-R2 or DcDFR-B1 cDNA in the expression vector, showed DFR enzyme activity in vitro (data not shown). These results along with the RNA-seq analysis data indicated that the late genes, DFR, ANS, and GST in ‘2-241-1 MB’ and ‘2-241-1 MB variegated’ were able to produce enzymes with the capacity to synthesize anthocyanin.
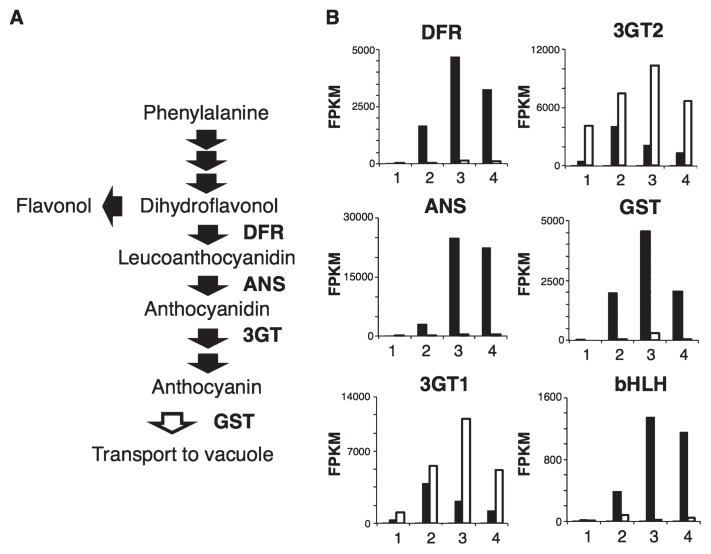
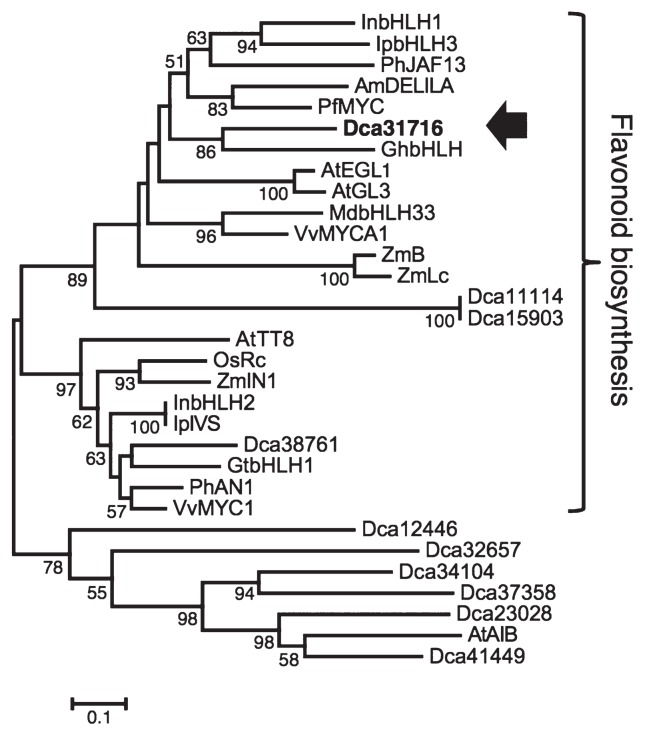
Expression of the late genes is regulated by a trio of transcription factors, namely, bHLH, MYB, and WD40 in flowers, leaves, stems, and fruits (Hichri et al. 2011). Although any genes for transcriptional regulatory factors belonged to MYB and WD40 superfamilies could not be found to show remarkable spatiotemporal expression profiles coordinated with those of the structural genes for anthocyanin synthetic pathway indicated by FPKM values in RNA-seq analysis, one bHLH gene (Dca31716), which belonged to the bHLH superfamily (Yagi et al. 2014), had a reduced FPKM value, as did the late genes, in white petals of ‘2-241-1 MB’. However, the FPKM values for bHLH (Dca31716) and the late genes, DFR, ANS and GST, were both high in red petals of ‘4-94-1 MR’ (Fig. 2B) during flower developmental stages. Because 3GT involved in the downstream anthocyanin synthetic pathway from ANS was included in the early gene but not the late genes, 3GT1 (Dca17573) and 3GT2 (Dca47835) showed higher FPKM value in ‘2-241-1 MB’ at stage 3 than both in ‘4-94-1 MR’ and the peak of expression of 3GT1 and 3GT2 in ‘4-94-1 MR’ was observed at stage 2 earlier than that of DFR, ANS and GST, included in the late gene coupled with bHLH at stage 3 (Fig. 2B), suggesting that 3GT expression might not be correlated with bHLH expression. A phylogenetic tree analysis showed that bHLH (Dca31716) was located in the same clade as bHLH amino acid sequences of other plant species that are known to regulate flavonoid biosynthesis. The other three bHLHs, Dca11114, Dca15903, and Dca38761, were also located in the same clade as Dca31716 (Fig. 3). Although they belonged to the flavonoid biosynthesis clade, the FPKM values of the additional three bHLHs were very low in petals of ‘4-94-1 MR’ (data not shown).
Fig. 2.
Schematic representation of flavonoid biosynthesis and gene expression profiles. (A) Summary of the anthocyanin/flavonoid biosynthetic pathway. Dihydroflavonol 4-hydroxylase (DFR), anthocyanidin synthase (ANS) and UDP-glucose: anthocyanin 3-glucosyltransferase (3GT) are anthocyanin biosynthesis enzymes. Glutathione S-transferase (GST) plays a role in anthocyanin transportation into vacuoles. (B) The expression profiles of DFR (Dca4324), ANS (Dca23371), 3GT1 (Dca47835), 3GT2 (Dca17573), GST (Dca57804) and bHLH (Dca31716) in red petals of ‘4-94-1 MR’ (black bar) and in white petals of ‘2-241-1 MB’ (white bar) at petal stages 1 to 4.
Fig. 3.
Phylogenetic tree of bHLH-type transcription factors. The amino acid sequences analyzed were as follow: Arabidopsis thaliana (AtTT8, NM_117050; AtEGL1, NM_001198373; AtGL3, NM_148067, AtAIB, NM_130216), Antirrhinum majus (AmDELILA, M84913), Oryza sativa (OsRc, AB247503), Petunia hybrida (PhAn1, AF260919; PhJAF13, AF020545), Vitis vinifera (VvMYC1, EU447172; VvMYCA1, EF193002), Zea mays (ZmB, X57276; ZmLc, M26227; ZmlN1, U57899), Malus × domestica (MdbHLH33, DQ266451), Gerbera hybrida (GhbHLH, AJ007709), Perilla frutescens (PfMYC, AB024050), Ipomoea purpurea (IpIVS, AB154369) and Gentiana triflora (GtbHLH1, AB459661). These bHLH-type transcription factors were as described by Yagi et al. (2014). Bar = 0.1 amino acid substitutions/site. The bootstrap values only show upper 50%.
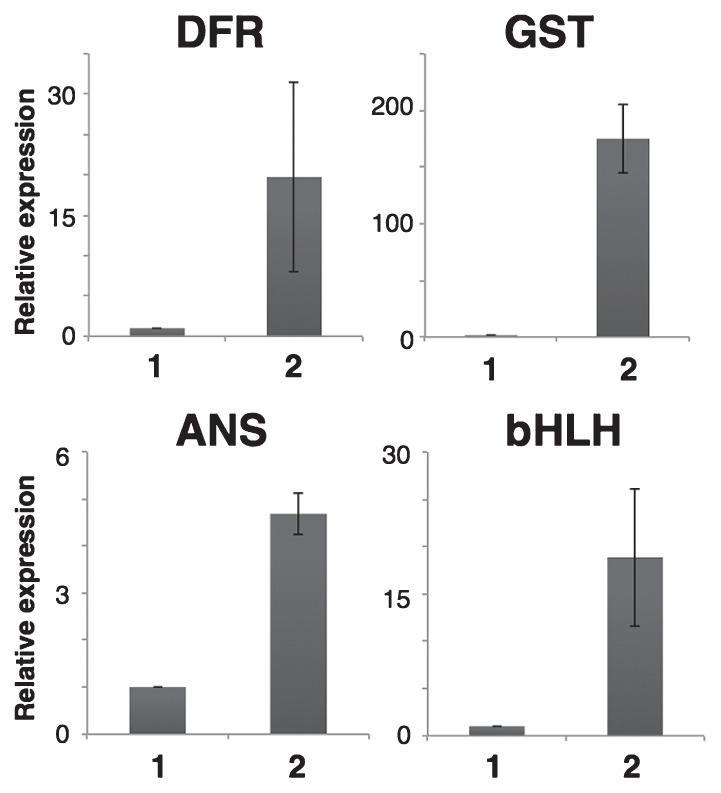
To confirm the expression level of the genes, quantitative RT-PCR analysis of cDNAs from ‘2-241-1 MB’ and ‘2-241-1 MB variegated’ was performed (Fig. 4). Expression of bHLH and late genes was reduced in ‘2-241-1 MB’ but was up-regulated in ‘2-241-1 MB variegated’; all of the late genes showed more than four-fold greater expression in ‘2-241-1 MB variegated’ than in the white petals of ‘2-241-1 MB’ (Fig. 4).
Fig. 4.
Quantitative RT-PCR of late genes for flavonoid biosynthesis. 1: ‘2-241-1 MB’, 2: ‘2-241-1 MB variegated’. Error bars indicate ± SD for three biological replicates.
It has been suggested that ‘2-241-1 MB’ might have a defective bHLH (Dca31716) that up-regulated expression of the late genes. The results here, and presented by our group previously, indicate that the white flower phenotypes in carnations might be due to three possible mechanisms: disruption of the A gene encoding DFR activity (Itoh et al. 2002, Stich et al., 1992); deficient F3H gene expression (Mato et al. 2000); repression of bHLH resulting in reduced expression of the late genes involved in anthocyanin synthetic pathway.
Currently, we are investigating the effects of reduced bHLH gene expression using transgenic techniques for gene knock-downs and also performing complementation assays. If alteration of the nucleotide sequence of bHLH reduces its expression, then gene specific DNA marker(s) for bHLH could be developed and used to isolate and establish carnation cultivars with a stable and perfect white phenotype, without the possibility of red variegation.
Supplementary Information
Acknowledgements
This work was supported by “Breeding of floricultural plants adapted for high practical needs and development of low cost cultivation technique” of Ministry of Agriculture, Forestry and Fisheries, project number 15653424.
Literature Cited
- Fukui, Y., Tanaka, Y., Kusumi, T., Iwashita, T. and Nomoto, K. (2003) A rationale for the shift in colour towards blue in transgenic carnation flowers expressing the flavonoid 3′,5′-hydroxylase gene. Phytochemistry 63: 15–23. [DOI] [PubMed] [Google Scholar]
- Geissman, T.A. and Mehlquist, G.A.L. (1947) Inheritance in the carnation, Dianthus caryophyllus. IV. The chemistry of flower color variation, I. Genetics 32: 410–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S. and Lauvergeat, V. (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62: 2465–2483. [DOI] [PubMed] [Google Scholar]
- Itoh, Y., Higeta, D., Suzuki, A., Yoshida, H. and Ozeki, Y. (2002) Excision of transposable elements from the chalcone isomerase and dihydroflavonol 4-reductase genes may contribute to the variegation of the yellow-flowered carnation (Dianthus caryophyllus). Plant Cell Physiol. 43: 578–585. [DOI] [PubMed] [Google Scholar]
- Iwashina, T., Yamaguchi, M., Nakayama, M., Onozaki, T., Yoshida, H., Kawanobu, S., Ono, H. and Okamura, M. (2010) Kaempferol glycosides in the flowers of carnation and their contribution to the creamy white flower color. Nat. Prod. Commun. 5: 1903–1906. [PubMed] [Google Scholar]
- Mato, M., Onozaki, T., Ozeki, Y., Higeta, D., Itoh, Y., Yoshimoto, Y., Ikeda, H., Yoshida, H. and Shibata, M. (2000) Flavonoid biosynthesis in white-flowered Sim carnations (Dianthus caryophyllus). Sci. Hortic. 84: 333–347. [Google Scholar]
- Mato, M., Onozaki, T., Ozeki, Y., Higeta, D., Itoh, Y., Hisamatsu, T., Yoshida, H. and Shibata, M. (2001) Flavonoid biosynthesis in pink-flowered cultivars derived from ‘William Sim’ carnation (Dianthus caryophyllus). J. Japan Soc. Hort. Sci. 70: 315–319. [Google Scholar]
- Matsuba, Y., Sasaki, N., Tera, M., Okamura, M., Abe, Y., Okamoto, E., Nakamura, H., Funabashi, H., Takatsu, M., Saito, M. et al. (2010) A novel glucosylation reaction on anthocyanins catalyzed by acylglucose-dependent glucosyltransferase in the petals of carnation and delphinium. Plant Cell 22: 3374–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlquist, G.A.L. (1939) Inheritance in the carnation, Dianthus caryophyllus. I. Inheritance of flower color. Proc. Am. Soc. Hortic. Sci. 37: 1019–1021. [Google Scholar]
- Ogata, J., Itoh, Y., Ishida, M., Yoshida, H. and Ozeki, Y. (2004) Cloning and heterologous expression of cDNAs encoding flavonoid glucosyltransferases from Dianthus caryophyllus. Plant Biotechnol. 21: 367–375. [Google Scholar]
- Okamura, M., Nakayama, M., Umemoto, N., Cano, E.A., Hase, Y., Nishizaki, Y., Sasaki, N. and Ozeki, Y. (2013) Crossbreeding of a metallic color carnation and diversification of the peculiar coloration by ion-beam irradiation. Euphytica 191: 45–56. [Google Scholar]
- Onozaki, T., Mato, M., Shibata, M. and Ikeda, H. (1999) Differences in flower color and pigment composition among white carnation (Dianthus caryophyllus L.) cultivars. Sci. Hortic. 82: 103–111. [Google Scholar]
- Sasaki, N., Nishizaki, Y., Uchida, Y., Wakamatsu, E., Umemoto, N., Momose, M., Okamura, M., Yoshida, H., Yamaguchi, M., Nakayama, M. et al. (2013) Identification of the glutathione S-transferase gene responsible for flower color intensity in carnations. Plant Biotechnol. 29: 233–227. [Google Scholar]
- Stich, K., Eidenberger, T., Wurst, F. and Forkmann, G. (1992) Enzymatic conversion of dihydroflavonols to flavan-3,4-diols using flower extracts of Dianthus caryophyllus L. (carnation). Planta 187: 103–108. [DOI] [PubMed] [Google Scholar]
- Yagi, M., Kosugi, S., Hirakawa, H., Ohmiya, A., Tanase, K., Harada, T., Kishimoto, K., Nakayama, M., Ichimura, K., Onozaki, T. et al. (2014) Sequence analysis of the genome of carnation (Dianthus caryophyllus L.). DNA Res. 21: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Tohge, T., Niida, R. and Saito, K. (2007) Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282: 14932–14941. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Akimoto, H., Yamaguchi, M., Shibata, M., Habu, Y., Iida, S. and Ozeki, Y. (2004) Alteration of methylation profiles in distinct cell lineages of the layers during vegetative propagation in carnation (Dianthus caryophyllus). Euphytica 135: 247–253. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.