Abstract
The Japanese morning glory (Ipomoea nil) and petunia (Petunia hybrida), locally called “Asagao” and “Tsukubane-asagao”, respectively, are popular garden plants. They have been utilized as model plants for studying the genetic basis of floricultural traits, especially anthocyanin pigmentation in flower petals. In their long history of genetic studies, many mutations affecting flower pigmentation have been characterized, and both structural and regulatory genes for the anthocyanin biosynthesis pathway have been identified. In this review, we will summarize recent advances in the understanding of flower pigmentation in the two species with respect to flower hue and color patterning. Regarding flower hue, we will describe a novel enhancer of flavonoid production that controls the intensity of flower pigmentation, new aspects related to a flavonoid glucosyltransferase that has been known for a long time, and the regulatory mechanisms of vacuolar pH being a key determinant of red and blue coloration. On color patterning, we describe particular flower patterns regulated by epigenetic and RNA-silencing mechanisms. As high-quality whole genome sequences of the Japanese morning glory and petunia wild parents (P. axillaris and P. inflata, respectively) were published in 2016, further study on flower pigmentation will be accelerated.
Keywords: anthocyanin, epigenetics, floral pigmentation pattern, Ipomoea, petunia, RNA silencing, vacuolar pH
Introduction
The Japanese morning glory (Ipomoea nil) has been a floricultural plant in Japan since the 17th century Edo period. While its wild-type plants produce blue flowers, a number of spontaneous mutants displaying various flower colors and pigmentation patterns have been isolated and maintained to the present. Recent molecular studies on the mutants showed that Tpn1 family transposons are the major mutagen in I. nil cultivars (Chopra et al. 2006, Iida et al. 2004). Most of the I. nil mutations are insertions of transposons or footprints caused by transposon excisions. Tpn1 family transposons are class II DNA elements that can transpose into new locations via a cut-and-paste mechanism. Even now, they can cause new spontaneous mutations exhibiting novel flower colors and patterns.
The petunia (Petunia hybrida) originated from interspecific hybridization between two petunia wild parents, P. axillaris and P. integrioforia (or its related species, P. inflata) (Stehmann et al. 2009, Vandenbussche et al. 2016). P. axillaris has large white flowers, while P. integrioforia and P. inflata display small purple flowers. The first hybrids were produced by European breeders in 19th century, which is considered to have been produced from multiple crossings between different accessions of the wild parent species (Sink 1984). Recent varieties of flower traits in commercial petunias have resulted from intense breeding since the first interspecific hybridizations. Like in the case of I. nil, class II DNA transposons have contributed to the phenotypic diversity of petunia flowers (Vandenbussche et al. 2016). The transposons in I. nil and petunia are useful tools for isolating genes responsible for flower pigmentation.
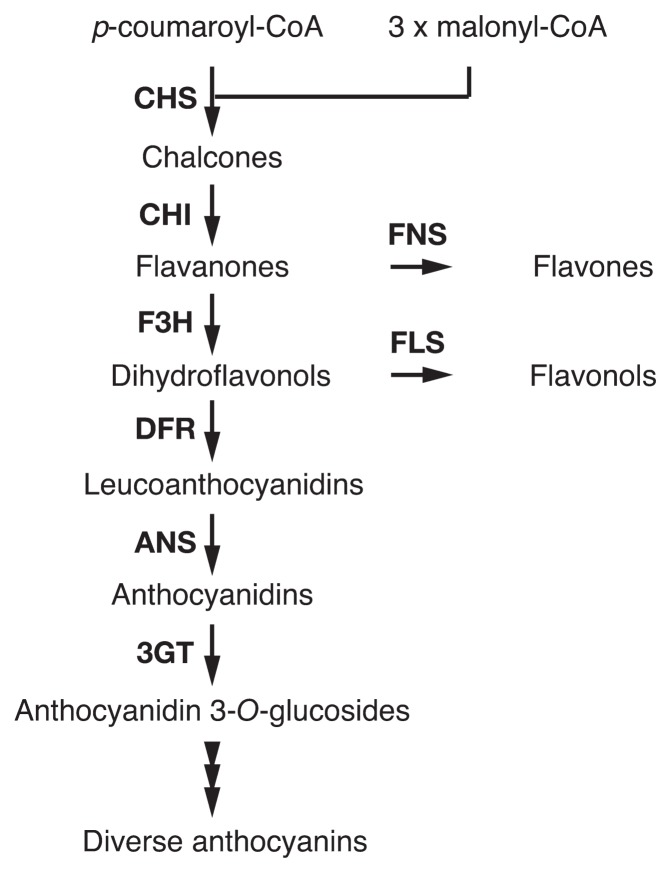
Flavonoids are major flower pigments that accumulate in vacuoles (Tanaka et al. 2008, Winkel-Shirley 2001). Anthocyanins are members of flavonoids responsible for a wide range of flower colors—red, orange, blue, and purple pigmentations. In the anthocyanin biosynthesis pathway (Fig. 1), chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H) catalyze “early” steps required for the synthesis of all flavonoids. Dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose:flavonoid 3-O-glucosyltransferase (3GT) mediate “late” steps that lead to anthocyanin and proanthocyanidin biosynthesis. Enzymes from CHS to ANS produce anthocyanidins that are anthocyanin aglycones and the central chromophores of anthocyanins. Anthocyanidins are generally glucosylated at the 3 position by 3GT to give anthocyanidin 3-glucosides that are the first stable anthocyanin. Further modifications, such as additional glycosylation and acylation to anthocyanidin 3-glucosides, occur in a species-specific manner. Transcriptional regulators for structural genes encoding enzymes in the pathway are known to include proteins containing an R2R3-MYB domain, an R3-MYB domain, a basic-helix-loop-helix (bHLH) domain, and WD40 repeats (WDR). The proteins constitute complexes that control the transcription of structural genes (Albert et al. 2014, Quattrocchio et al. 2006a).
Fig. 1.
Simplified flavonoid biosynthesis pathway. The enzymes in the pathway are: CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; 3GT, UDP-glucose:flavonoid 3-O-glucosyltransferase; FNS, flavone synthase; FLS, flavonol synthase. The arrowheads indicate modification steps of anthocyanins, which are mediated by glycosyltransferases, acyltransferases, and methyltransferases. Hypothesis 1 of EFP function: EFP enhances CHS activity. Hypothesis 2 of EFP function: EFP interacts with biosynthesis enzymes and forms a metabolon complex.
The diversity of anthocyanin structures is an important factor in the wide range flower color. In addition, the pH inside the vacuole, the vacuolar lumen, largely affects flower color (Yoshida et al. 2009b). Anthocyanins show reversible structural transformation in their chromophores dependent on the pH shift of the solvent, and the structural transformation dramatically affects their absorption spectra. The vacuolar lumen is generally acidified by H+-ATPase (V-ATPase) and H+-pyrophosphatase (PPase) on the vacuolar membrane, tonoplast (Eisenach et al. 2014, Gaxiola et al. 2007). Anthocyanins normally show reddish to purplish colors in the acidic vacuoles, and change to bluish colors due to increased pH in the vacuole. Other than vacuolar pH, co-pigments and epidermal cell shapes also affect flower color.
Thanks to a number of flower color mutations, I. nil and petunia have been utilized as ideal model plants to study flower pigmentation (Iida et al. 2004, Quattrocchio et al. 2006a, Tornielli et al. 2009). These plants have contributed to the advancement of studies on flower pigmentation, e.g., discovery of the blue gene (Holton et al. 1993). In this review, we describe recent advances in flower colors of the two species, especially with regard to a novel enhancer of flower pigmentation, new aspects on 3GT, color patterning regulated by epigenetics and RNA silencing, and vacuolar pH regulation.
A novel color intensifier, EFP (Enhancer of Flavonoid Production)
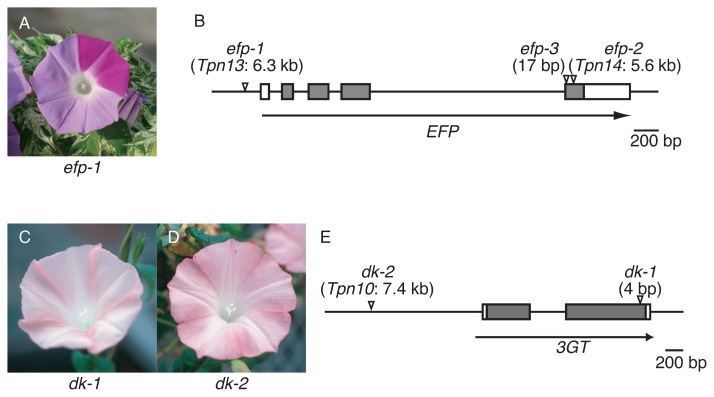
Classical genetic studies of I. nil reported several mutations resulting in alterations of flower color intensity that were probably due to changes in anthocyanin quantity (Imai 1931). To elucidate mechanisms underlying the regulation of anthocyanin quantity in flower petals, we employed a newly isolated mutation that confers pale-colored flowers (Fig. 2A). In the mutant, the total amount of anthocyanin was less than approximately 22% of that in the parental wild-type plant, which displays normally colored flowers. In addition, the colorless flavonoid content, flavonol (Fig. 1), was also decreased by 27% in the mutant. Flavonoid analysis indicated that the responsible gene plays a key role in flavonoid biosynthesis, increasing flavonoid total content by approximately 3–4 times. We named the gene Enhancer of Flavonoid Production (EFP), and isolated it by transposon tagging (Morita et al. 2014). The EFP gene encodes a CHI-like protein that is classified as a type IV CHI protein of unknown function (Ralston et al. 2005). EFP is not a functional CHI enzyme, because the chi mutant of I. nil accumulates chalcone derivatives rather than anthocyanins (Hoshino et al. 2001, Saito et al. 2011). The mutation named efp-1 was an insertion of Tpn13 into the EFP promoter sequence, resulting in the complete suppression of EFP expression (Fig. 2B). EFP and the flavonoid biosynthesis genes showed similar spatiotemporal expression patterns and were coordinately activated by the regulatory genes, InMYB1 and InWDR1 (Morita et al. 2006, 2014). We also isolated cDNA for the EFP in the petunia and torenia (Torenia hybrida) and obtained RNAi knockdown mutants of the EFP genes to examine whether EFP proteins in other ornamental flowers also increase flavonoid production efficiency (Morita et al. 2014). As expected, the transgenic RNAi knockdown petunia and torenia plants conferred pale-colored flowers. In the knockdown petunia, anthocyanins and other flavonoid (flavonols, Fig. 1) contents of the flower decreased to approximately 28% and 12%, respectively, of those in the control plants. Moreover, in the knockdown torenia, anthocyanins and other flavonoid (flavones, Fig. 1) contents of the flower also decreased to approximately 49% and 31%, respectively, of those in the control plants. These results clearly show that EFPs of I. nil, petunia, and torenia enhance flower pigmentation by increasing flavonoid production ability. Recently, we also reported the remarkable expression of EFP homologs in the flower petals of the carnation (Dianthus caryophyllus), chrysanthemum (Chrysanthemum morifolium), and snapdragon (Antirrhinum majus) (Morita et al. 2015b). The EFP gene may contribute to flower color intensity in these floricultural crops.
Fig. 2.
Flower phenotypes of flower hue mutants and the genomic structure of the mutated loci. The gray and white boxes indicate the coding and the untranslated regions, respectively. Flower phenotype of efp-1 mutant (A) and the structure of the EFP gene (B). The efp-1 mutant shows pale-colored flowers with normal pigmented spots and sectors. The arrowheads indicate insertion sites of Tpn13, Tpn14, and a 17-bp sequence of the efp-1, efp-2, and efp-3 mutations, respectively. Flower phenotypes of dk-1 (C) and dk-2 (D) mutants and the structure of the 3GT gene (E). The dk-1 and dk-2 mutants display pale and dull-colored flowers. The flower color in the dk-2 mutant is slightly darker than that in the dk-1 mutant. The arrowheads show a 4-bp sequence and Tpn10 insertion sites of the dk-1 and dk-2 mutations, respectively.
The molecular mechanisms of how EFP enhances flavonoid production remain to be elucidated. The reduction of flavone or flavonol contents in mutants of I. nil, petunia, and torenia suggests that EFPs activate at least the early steps of flavonoid biosynthesis (Fig. 1). In Arabidopsis (Arabidopsis thaliana), the EFP homologue AtCHIL and the genes for the enzymes that mediate the early steps are coordinately regulated (Yonekura-Sakakibara et al. 2008, 2012). In the moss Physcomitrella patens, the EFP homologue is coordinately expressed with CHS genes under a flavonoid-producing condition promoted by UV irradiation (Wolf et al. 2010). These observations are consistent with the idea that EFP activates the early steps of flavonoid biosynthesis.
We hypothesize two possibilities to explain the activation of the early steps by EFP: 1) EFP enhances CHS activity, 2) EFP is one component of the flavonoid biosynthesis enzyme complex (Fig. 1). In I. purpurea, an I. nil-related morning glory, dosage-dependent expression of the CHS gene led to the incomplete dominance of flower coloration (Johzuka-Hisatomi et al. 2011), suggesting that the reduction of CHS activity confers pale-colored flowers in I. nil. Incomplete dominance has not been reported in other genes in the early steps. In the efp-1 mutant flower, the levels of flavonoid precursor-related compounds, chlorogenic acid and caffeoyl glucoside were increased remarkably. The null chs mutants also accumulate these compounds (Hoshino et al. 2009, Saito et al. 1994). These observations support the first hypothesis. With respect to the second hypothesis, metabolon is an enzyme complex consisting of sequential enzymes of a metabolic pathway; it enables the effective synthesis of specific products and the avoidance of metabolic interference (Jorgensen et al. 2005). In Arabidopsis, CHS, CHI, F3H, and DFR are thought to form a complex ensuring efficient flavonoid biosynthesis (Burbulis and Winkel-Shirley 1999, Winkel-Shirley 1999, 2001). As the secondary structure of EFP is closely similar to that of CHI (Morita et al. 2014, Ngaki et al. 2012), EFP probably constitutes a metabolon with flavonoid biosynthesis enzymes that involve the early steps and enhance flavonoid production through the metabolon. Elucidating the mechanism of the activation of the early steps by EFP will provide insight into how flavonoid synthesis is controlled as well as the evolution of the flavonoid biosynthesis pathway.
Mutations of the 3GT gene conferring pale and brownish or grayish flowers in morning glories
In the anthocyanin biosynthesis pathway, anthocyanidin 3-glucosides are generally the first stable anthocyanins, and 3GT catalyzes glucosylation at the 3-position of anthocyanidins. 3GT belongs to the family 1 glycosyltransferases (UGTs) that uses UDP-sugars as the sugar donor. Although the gene for 3GT was first isolated from maize (Zea mays) more than 30 years ago (Fedoroff et al. 1984), no 3GT affecting flower petal color had been reported. Mutations in other flavonoid UGT genes for anthocyanidin 3-glucoside glucosyltransferase (3GGT) and anthocyanidin 3-glucoside rhamnosyltranferase (RT) were identified in I. nil and petunia, respectively (Kroon et al. 1994, Morita et al. 2005). They result in brownish or grayish flowers due to the accumulation of anthocyanins with fewer modifications of glycosylation and acylation, which are necessary for species-specific anthocyanin production and the bright coloration of flowers. The Dusky (Dy) locus encodes 3GGT in I. nil.
The duskish (dk) mutants of I. nil display brownish or grayish flowers (Fig. 2C, 2D). The flower colors of dk and dy mutants are similar; however, dk mutants show paler pigmentation than do dy mutants. Recently, we found that the Dk gene encodes 3GT, and we identified two allelic mutations, dk-1 and dk-2 (Fig. 2E, Morita et al. 2015a). The dk-1 and dk-2 mutations are a 4-bp insertion and an insertion mutation of Tpn1 family transposon, Tpn10, respectively. Pale brownish or grayish flowers are also found in cultivars of I. purpurea. We found a single base deletion in the 3GT gene in the cultivars, and we named the allele ip3gt-1 (Morita et al. 2015a). Among these mutations, dk-1 and ip3gt-1 seem to be null mutations, while dk-2 is a leaky mutation (see below).
Anthocyanin analysis of the dk-1 and ip3gt-1 mutants suggested that the absence of 3GT leads to the reduction of anthocyanin accumulation and the inhibition of glucosylation and acylation found in the anthocyanins in wild-type plants (Saito et al. 1998, Toki et al. 2001). The mutants accumulate small amounts of anthocyanidin 3-glucoside and its derivatives. Since the I. nil genome carries the unique 3GT gene, glucosylation at the 3 position of anthocyanidins is assumed to be mediated by glucosyltransferases other than Dk protein, the bona fide 3GT. UGT78D2 is the unique 3GT gene in Arabidopsis. Because its mutant accumulates small amounts of anthocyanidin 3-glucosides, the presence of secondary 3GT activities has also been suggested (Tohge et al. 2005). The formation of 3-glucosylated anthocyanins without bona fide 3GT may be conserved among divers plant species. In wild-type I. nil and I. purpurea, anthocyanidins are glucosylated at its 5 position, and their glucose moiety at the 3 position is decorated with glucose and caffeoyl moieties. Such glucosylation and acylation are not found in the major anthocyanins in dk-1 and ip3gt-1 mutants. This indicates that bona fide 3GT is necessary for the appropriate glucosylation and acylation of anthocyanidin 3-O-glucoside in the species. It can be speculated that glucosyltransferases and acyltransferases for anthocyanidin 3-O-glucoside form metabolon with 3GT in the cytosol to ensure effective and precise anthocyanin production. Therefore, the absence of 3GT could inhibit the appropriate modification of anthocyanidin 3-O-glucoside through the inhibition of metabolon formation.
A particular flower variegation involving epigenetic control of 3GT gene expression
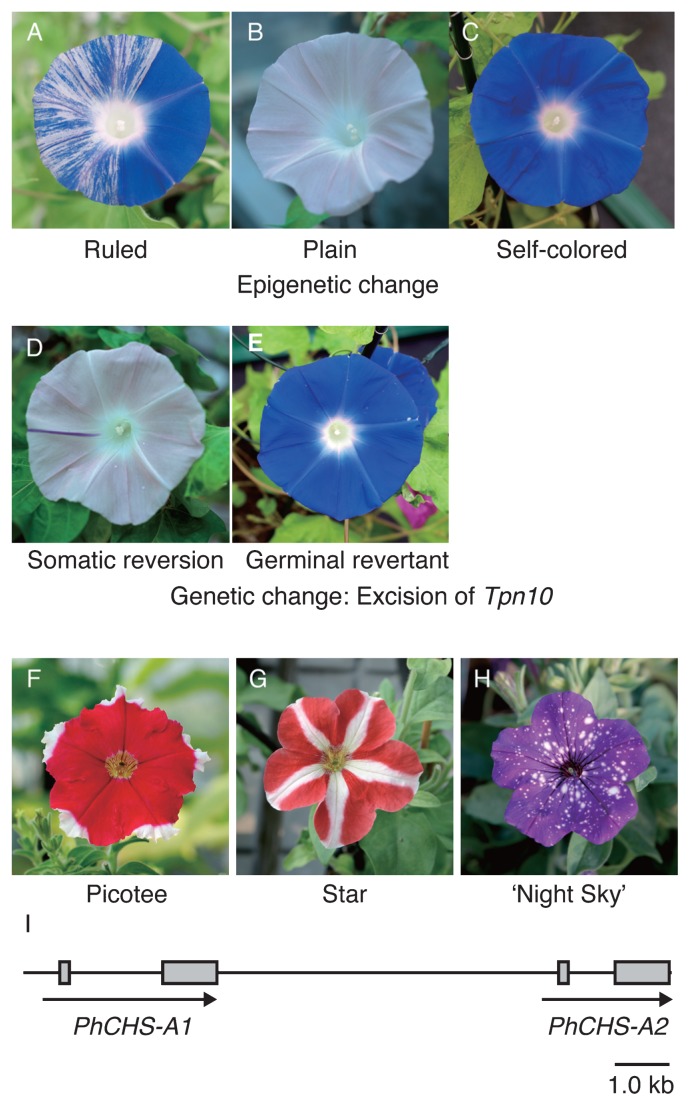
Among dk-2 mutants, the Q531 line shows genetically and epigenetically unstable expression of the 3GT gene (Fig. 3A–3E, Morita et al. 2015a). The line’s phenotype is variable, and plants with variegated flowers (Fig. 3A), pale grayish-purple flowers (Fig. 3B), and fully pigmented flowers (Fig. 3C) are segregated. Usually, flower variegation is due to recurrent somatic mutations caused by the excision of transposons and shows clonal chimera spots and sectors. Tpn1 family transposons inserted into genes for anthocyanin pigmentation can cause such flower variegation (Fukada-Tanaka et al. 2000, Hoshino et al. 2001, Inagaki et al. 1994, Morita et al. 2014). The Q531 line shows both clonal and non-clonal spots and sectors. Imai (1931, 1935) called the flower variegation phenotype with non-clonal and apparently non-chimeric spots and sectors in other dk mutants in particular “ruled”. In ruled flowers, fully pigmented areas appear within the pale grayish-purple areas and vice versa (Fig. 3A). The non-clonal variegation seems to be generated by reversible expression changes of the 3GT gene between an active state and an inactive state. Such a reversible change of the expression state is hard to explain by recurrent somatic mutations caused by transposons. The dk-2 mutation is an insertion mutation of the Tpn1 family transposon, Tpn10, 1.3 kb upstream of the 3GT start codon, leading to the gene’s leaky expression (Fig. 2E, Morita et al. 2015a). No footprint sequences generated by excisions of Tpn10 could amplify in the ruled flowers, even in fully colored sectors (Fig. 3A, Morita et al. unpublished). This indicates that reversible changes of the 3GT expression state resulting in the ruled phenotype occur independently from somatic excisions of Tpn10. It is speculated that epigenetic changes in DNA methylation and/or histone modification promote reversible changes in the ruled flowers (Morita et al. 2015a).
Fig. 3.
Flower color patterns of the Japanese morning glory and petunia. A–E: Flower phenotypes of the Q531 line of the dk2-mutant of the Japanese morning glory. (A) Ruled plants confer non-clonal spots and sectors, hakeme-shibori (brush marks variegation). Apparently clonal spots and sectors are also occasionally observed in ruled plants. (B) Plain plants display pale pigmentation petals. (C) Self-colored plants display fully pigmented flowers. These plants carry an identical dk-2 mutation, and Tpn10 seems to be able to transpose only in plain plants. (D) Somatic reversions caused by Tpn10 excisions are occasionally observed in plain plants. (E) Germinal revertant from a plain plant. (F–I) Naturally occurring bicolor RNAi mutants of the petunia. Flower phenotypes of Star (F), Picotee (G), and the bicolor cultivar ‘Night Sky’ (Ball Seed Co.) (I). (H) Genome structure of CHS siRNA-producing locus of the bicolor petunia. Two copies of CHS genes are tandemly located.
Imai called the segregated plants with pale grayish-purple flowers and fully pigmented flowers “plain” (Fig. 3B) and “self-colored” (Fig. 3C), respectively. Somatic changes of the 3GT expression state observed in ruled flowers are suppressed in these plants. Interestingly, the transposition of Tpn10 has been observed only in plain plants; clonal and chimera sectors and spots, as well as germinal revertants, appeared in the plants (Fig. 3D, 3E, Imai 1931, 1935, Morita et al. 2015a). In self-colored plants, the reversion of 3GT gene expression is also unrelated to the somatic excisions of Tpn10. We hypothesize three epigenetic states on the dk-2 locus to explain the phenotypes of the dk-2 mutants: 1) an epigenetic state in the ruled plants can induce variable expression of the 3GT gene, resulting in non-clonal variegation, and suppresses Tpn10 transposition; 2) an epigenetic state in plain plants can suppress the expression of the 3GT gene but not Tpn10 transposition; and 3) an epigenetic state in the self-colored plants can ensure the stable expression of the 3GT gene and might suppress Tpn10 transposition.
In other ornamental plants, transposons sometimes generate variations in floral phenotypes. Transposons integrated within the promoter sequence of flower pigmentation genes might be involved in flower variegation, as in the dk-2 mutants.
Naturally occurring RNAi and flower coloration patterns
The petunia has contributed to the discovery of the RNAi phenomenon. The first molecular observations associated with RNAi occurred after the appearance of irregular bicolored or white flowers in transgenic petunias that harbored exogenous flower pigmentation genes (Jorgensen 1995, Napoli et al. 1990, van der Krol et al. 1990). Picotee and Star, naturally occurring bicolor petunia cultivars, have pigmented petals with white margins and stars, respectively (Fig. 3F, 3G). These popular bicolor traits are believed to have arisen during the early impregnation events of interspecific hybridization breeding in the 19th century (Ewart 1984, Jorgensen 1995). Analyses of Star (Koes et al. 1987) and Picotee (Saito et al. 2006, 2007) petunias have indicated that the main cause of bicolor floral traits is the spatially regulated repression of CHS genes in the white tissue of petals (Fig. 1). Transcripts for both CHS copies, CHS-A and CHS-J, which are responsible for flower pigmentation, were down-regulated in the white tissue. Phenotypic similarities between transgenic and naturally occurring bicolor petunias suggested the involvement of an RNAi phenomenon in the expression of the Picotee and Star bicolor traits. Koseki et al. (2005) showed that while the CHS-A gene is transcribed into precursor mRNA in both white and pigmented tissues of Star petunias, mature CHS-A mRNA is accumulated only in the pigmented tissue. As the 21-nt CHS-A siRNA can be detected only in the white tissue, it was concluded that spatially expressed CHS-A siRNA guides the degradation of the mature CHS-A mRNA, resulting in the display of the Star phenotype. The spatially regulated production of CHS-A siRNAs was considered to be the main cause of bicolor floral traits; however, details of the molecular mechanisms, even in the genomic structure of CHS-A siRNA-producing locus, remained to be elucidated.
Morita et al. (2012) revealed the structure of the CHS-A siRNA-producing locus of Picotee and Star petunias. Interestingly, Picotee and Star petunias carry the same CHS-A siRNA-producing locus, consisting of two intact CHS-A copies in a tandem head-to-tail orientation (Fig. 3I). The CHS-A siRNAs were found to originate from the exon 2 region of both CHS-A copies, and those that restrict accumulations in the white tissues of Picotee and Star flowers are closely correlated with the spatial disappearance of the CHS mature mRNAs. These results suggest the existence of trans-acting factors, which regulate the spatiotemporal production of CHS-A siRNAs, originating from tandemly oriented CHS-A genes. Recent studies of naturally occurring seed pigmentation mutants of soybeans (Glycine max) reported that the CHS siRNA production from inversely located CHS gene clusters is controlled by AGO5, which is one of ARUGONAUTE (AGO) proteins (Cho et al. 2017). AGO proteins directly bind small RNAs and constitute a component of the RNA-induced silencing complex (RISC). Mutations in the gene for AGO5 lead to the altered spatial distribution of CHS siRNAs in the seed coat. Genetic diversities in the genes for RISC components may also lead to the spatiotemporal production of CHS-A siRNAs in bicolor petunias.
Morita et al. (2012) also predicted that tandemly oriented CHS-A genes were introduced into the petunia by interspecific hybridization breeding. As each copy of the tandemly oriented CHS-A genes is distributed independently in the genomes of wild Petunia species, the tandem arrangement was formed by a chromosomal rearrangement of the CHS-A locus. Dedicated screening was performed using many accessions of wild Petunia species collected from South America; however, the same tandem structure in the CHS-A locus was not to detectable (Morita et al. 2012). Surprisingly, P. inflata accession S6, which was employed for whole genome sequencing (Bombarely et al. 2016), was found to carry a tandemly oriented CHS-A genes almost identical with those of Picotee and Star petunias (data not shown). It was suggested that the CHS-A siRNA-producing locus was introduced from a wild population of P. inflata (or P. integrifolia) into the petunia genome, whereas other loci for hypothetical trans-acting factors regulating the spatiotemporal production of CHS-A siRNAs were from P. axillaris. We consider that recent bicolor cultivar, ‘Night Sky’ (Fig. 3H), which displayed irregular white spots in the pigmented petal, also carries the tandemly oriented CHS-A genes as a CHS-A siRNA-producing locus. The identification of the regulating locus in bicolor petunia cultivars, Picotee, Star, and ‘Night Sky’, might hold the key to understanding mechanisms for spatiotemporal siRNA production.
Vacuolar pH control—the proton transporters
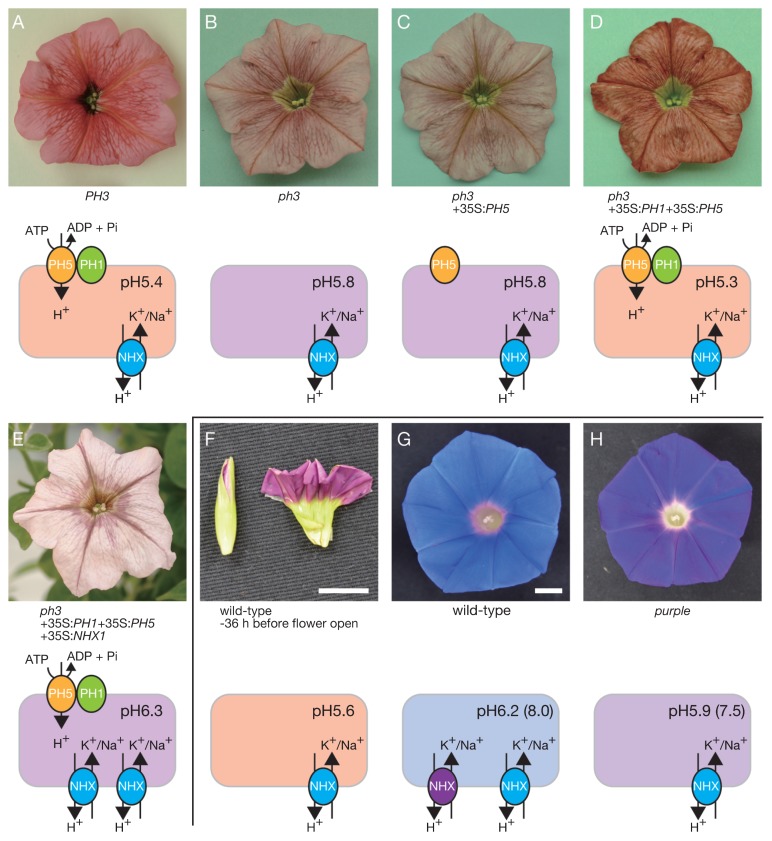
Petunias (Fig. 4A–4E) and the morning glories of I. nil (Fig. 4F–4H) and I. tricolor are ideal model plants to investigate vacuolar pH affecting flower color. In the petunia, purplish flower buds change to reddish open flowers due to decreased pH in the vacuole. In contrast to the petunia, the vacuolar pH of morning glories increases during flower opening, and reddish buds turn to blue open flowers (Fig. 4F, 4G). The pH values of petunia flowers were estimated from petal homogenates, and those of purplish buds and red flowers are around 6.3 and 5.3, respectively (Quattrocchio et al. 2006b). The pH values in the epidermal cells of I. tricolor were measured using microelectrodes, and those of reddish buds and blue flowers were 6.6 and 7.7, respectively (Yoshida et al. 1995). By comparing reflective spectra of the petal with absorption spectra of the homogenates in various pH solutions, the vacuolar pH of I. nil flowers was estimated to be around 8.0 (Yamaguchi et al. 2001).
Fig. 4.
H+ transporters regulate vacuolar pH, affecting the flower color of petunias (A–E) and I. nil (F–H). (A) In wild-type petunias, PH3 activates PH1 and PH5 expression, and the heteromeric complex of two P-ATPases, PH1 and PH5, mediate hyperacidification. (B) In the ph3 mutant, PH1 and PH5 are not expressed, resulting in an increase of vacuolar pH. This mutant line accumulates cyanidin derivatives that exhibit dull gray flowers. (C) PH1 is necessary for the H+ pump activity of PH5, and PH5 alone cannot rescue the ph3 phenotype. (D) The ph3 phenotype is rescued by the co-expression of PH1 and PH5. (E) The rescued phenotype in (D) is canceled by the expression of 35S:NHX1. NHX exchanges cations and H+, resulting in an increase in vacuolar pH. (F) Flower buds of the wild-type I. nil show lower vacuolar pH and red petals. (G) During flower opening, flower color changes from red to blue. In the same stage, PURPLE/InNHX1 (purple circle) is accumulated and mediates vacuolar alkalization. (H) The pr mutant shows partial vacuolar alkalization, and red flower buds change into purple flowers. The pH values of petal homogenates are presented, and those estimated from spectra are shown in parentheses. Scale bars represent 1 cm.
The H+ transporters controlling the vacuolar pH in petunia and I. nil flowers were uncovered by using the flower color mutants. In petunias, seven PH genes that affect vacuolar pH have been reported (de Vlaming et al. 1983, van Houwelingen et al. 1998). Recessive mutants of these genes show bluish flowers, due to increased vacuolar pH. Color shift due to the ph mutations depends on anthocyanin structure. The ph mutants that accumulate malvidin or petunidin derivatives show purple flowers, and those with cyanidin derivatives bloom dull gray flowers (Fig. 4B, Tornielli et al. 2009). Among the seven genes, PH5 and PH1 encode P-ATPases belonging to the 3A and 3B subfamilies, respectively (Fig. 4A, Faraco et al. 2014, Verweij et al. 2008). The P-ATPase superfamily is comprised of five subfamilies of ATP-driven membrane transporters that translocate distinct cations; each subfamily is divided into subgroups (Palmgren and Nissen 2011). The 3A subfamily (P3A-ATPases) is comprised of H+ pumps on the plasma membrane of plants, fungi, and other unicellular eukaryotes. Of the characterized P3A-ATPases, PH5 is the only one residing on the tonoplast (all others were shown to be on the plasma membrane); it transports H+ into the vacuolar lumen (Faraco et al. 2014, Verweij et al. 2008). PH1 is required for strong H+ pumping activity. The 3B subfamily (P3B-ATPase) includes Mg2+ prokaryote pumps. However, PH1 is not considered an Mg2+ pump, since it lacks the conserved aspartate residue that is thought to be essential for cation binding and translocation (Buch-Pedersen et al. 2000). PH1 can make a heteromeric complex with PH5 on the tonoplast and boosts the H+ pumping activity of PH5 (Fig. 4A, 4C, 4D, Faraco et al. 2014). It has been proposed that the interaction of PH1 and PH5 decreases H+/ATP stoichiometry hypothetically from 1.0 to 0.5 H+/ATP, which enables hyperacidification (Eisenach et al. 2014). Recently, Faraco et al. (2017) showed that PH1 has a role in the fusion process of the small vacuoles and the central vacuoles in petunia petal cells. The relationship between the fusion process and vacuolar hyperacidification is still obscure; however, PH5 as well as the vacuolar hyperacidification itself is not necessary for the fusion process. PH3, PH4, and PH6 encode transcriptional regulators that are required for the expression of PH1 and PH5 (Quattrocchio et al. 2006b, Spelt et al. 2002, Verweij et al. 2016, see below).
In I. nil, the Purple gene encoding a cation/H+ exchanger, InNHX1 (Fig. 4G, Fukada-Tanaka et al. 2000), is the only known H+ transporter gene controlling vacuolar pH and flower color in this species. Plant NHX proteins passively exchange H+ with K+ and Na+, consuming the H+ gradient (Bassil and Blumwald 2014). They are classified into three types: vacuolar, vesicular, and plasma membrane NHXs. InNHX1 belongs to vacuolar NHXs that mediate H+ efflux from vacuolar lumen. The InNHX1-deficient purple (pr) mutant shows purple flowers due to the decreased vacuolar pH (Yamaguchi et al. 2001). The pr mutant can increase vacuolar pH partially during flower opening, and reddish-purple flower buds change to purple open flowers (Fig. 4H). Therefore, InNHX1 is responsible for part of the vacuolar alkalization that results in flower bluing. It was suggested that other proteins, including InNHX2, take part in vacuolar alkalization and flower bluing (Ohnishi et al. 2005).
In I. tricolor, an InNHX1 homologue, ItNHX1, is expressed in petal cells, and its temporal expression pattern is similar to that of InNHX1 (Yoshida et al. 2005). Protoplast cells prepared from bud petal cells 7.5 hours before flower opening are able to change their color from red to blue when the cells are treated with 50 mM KCl or NaCl. Petal cells accumulate K+ rather than Na+ along with ItNHX1 mRNA during flower opening (Yoshida et al. 2009a). From these observations, it has been thought that ItNHX1 acts as a K+/H+ exchanger in the flower petals of I. tricolor. The vacuolar pH in opened flower is 7.7, which seems to be higher than the general pH (around 7.2) of plant cytosol. In other words, the concentration of cytosolic H+ is higher than that of vacuolar H+. Therefore, we can speculate that ItNHX1, and probably InNHX1, uses K+ gradient rather than H+ gradient to exchange K+ and H+ mediating vacuolar alkalization. In fact, tomato LeNHX2 recombinant protein reconstituted in liposomes could show H+ efflux activity in vitro using a cation gradient in the absence of a preimposed pH gradient (Venema et al. 2003). The further characterization of Ipomoea NHX activity is essential for better understanding vacuolar alkalization mechanisms.
Vacuolar pH control–regulatory systems of the proton transporters
In the petunia, the regulatory system’s activating transcription of PH1 and PH5 has been well characterized. Transcriptional regulatory complexes composed of proteins that contain MYB, bHLH (basic helix-loop-helix), and WD40 (or WDR) domains (so-called MBW complexes) control epidermal cell diversity, including anthocyanin biosynthesis, trichome and root hair formation, and seed coat pigmentation in angiosperms (Koes et al. 2005, Lloyd et al. 2017, Ramsay and Glover 2005). The transcriptional regulators of the MBW complexes that involve anthocyanin biosynthesis have been isolated in both petunias and Ipomoea (deVetten et al. 1997, Morita et al. 2006, Park et al. 2004, 2007, Quattrocchio et al. 1999, Spelt et al. 2000). Mutants of the genes for transcriptional regulators show white or pale-colored flowers. Of these, AN1 and AN11 are petunia bHLH and WD40 proteins, respectively. These proteins induce vacuolar acidification through the activation of PH1 and PH5 transcription. The MYB protein AN2 is also involved in vacuolar acidification, and an2 mutants show only a partial hyperacidification defect (Tornielli et al. 2009). AN1 and PH6 are two names for the same gene, and the ph6 allele expresses truncated AN1 proteins that can activate anthocyanin pigmentation but not vacuolar acidification (Spelt et al. 2002). AN1 and AN11 make an MBW complex with an MYB protein of PH4 (Quattrocchio et al. 2006b, Spelt et al. 2002). The MBW complex activates the expression of the gene for PH3 of a WRKY protein (Verweij et al. 2016). PH3 can bind to AN11 and is necessary for the transcription of PH5. It is still unclear whether PH3 is a component of the MBW complex or a component of another complex that includes AN11 but not AN1 and PH4 (Lloyd et al. 2017).
To date, neither transcriptional regulators for Ipomoea NHXs nor transcriptional regulators for anthocyanin biosynthesis involving vacuolar pH alkalization have been reported. The transcriptional activation of Ipomoea NHXs seems to be connected with flower opening when a large influx of water and ion into petal cells and cell expansion are caused. A regulatory system for such an influx and cell expansion process may be involved in the activation of Ipomoea NHX. Yoshida et al. (2009a) suggested that Ipomoea NHXs contribute to the increase in vacuolar osmotic pressure through K+ accumulation, inducing water influx for cell expansion growth and flower opening (Fig. 4F, 4G). However, since the pr mutant lacking InNHX1 still shows normal cell expansion and flower opening (Fig. 4G), InNHX1 is not essential for such processes (Fukada-Tanaka et al. 2000, Pittman 2012, Yamaguchi et al. 2001).
Diversity and universality of the vacuolar pH regulatory systems in flower color
Vacuolar pH regulation by P3A-ATPase/P3B-ATPase and NHX proteins is thought to be not specific to the flowers of petunias and Ipomoea, respectively. Functional PH5 and PH1 homologues are widely spread in angiosperms; however, independent losses of these homologues have occurred in many angiosperms (Li et al. 2016). We found that these homologues are absent in the I. nil genome (data not shown, Hoshino et al. 2016). PH5 homologues were present not only in angiosperms but also in gymnosperms, and they were shown to have evolved from plasma membrane P3A-ATPases. The constitutive expression of Arabidopsis, carnation, grape, and rose PH5 homologues could complement the ph5 mutation of the petunia (Appelhagen et al. 2015, Li et al. 2016). However, PH5 homologues are absent in a number of angiosperms, even in Solanaceae plants (e.g., tomato, potato, pepper). PH1 homologues are found in some groups of bacteria and fungi. Since PH1 homologues are absent in most algae, it was suggested that several cases of gene loss and horizontal transfer events have been involved in their evolution (Li et al. 2016). Grape and rose PH1 homologues could complement the petunia ph1 mutant. Takahashi et al. (2013) showed that soybean mutants that exhibit purple-blue flowers have a nonsense mutation in the apparent PH4 homologues. These observations suggest that the vacuolar hyperacidification systems, including P-ATPases as well as MBW complexes, are partially conserved among angiosperms and that some plants use the systems to create red flowers.
In contrast to the distribution of PH5 and PH1 homologues, vacuolar NHXs are ubiquitous in plants (Bassil and Blumwald 2014). Arabidopsis has two vacuolar NHX genes, and double mutants of the genes exhibit the acidification of vacuoles in the root cells (Bassil et al. 2011). PhNHX1 of the petunia is expressed in flower petals (Yamaguchi et al. 2001), and PhNHX1 can reduce the pH gradient across the tonoplast (Faraco et al. 2014). The dull gray flower phenotype of the ph3 mutant (Fig. 4B) was rescued by the co-expression of PH1 and PH5 (Fig. 4D). The rescue was canceled by the overexpression of PhNHX1 in the rescued mutant, and the ph3 mutant co-expressing PH1, PH5, and PhNHX1 showed dull gray flowers like those of the ph3 mutant (Fig. 4E). Interestingly, the pH value (around 6.2) of the petal homogenate of the PH1-, PH5-, and PhNHX1-expressed ph3 mutant is higher than that of the ph3 mutant (around 5.8).
Future perspectives
In I. nil, flower color modification using the genome editing tool, CRISPR/Cas9, has recently been reported (Watanabe et al. 2017). In one of the three DFR genes, DFR-B, in the I. nil cv. Violet CRISPR/Cas9 induced mutations, and the flower color of the cultivar changed from white from red. The genome editing technology will be widely applied to ornamental flower breeding in the near future. Disruption of EFP or 3GT genes in ornamental plants seems to be an ideal method to create pale flower varieties.
As in the case of I. nil dk-2 mutants (Fig. 3A), transposons can epigenetically control gene expression in land plants. Transposons are generally distributed among angiosperms, and sometimes act as endogenous mutagens and generate floral color variations in ornamental plants. As the dk-2 mutant, transposons integrated within promoter sequences of flower pigmentation genes might be involved in particular flower variegation in ornamental plants other than I. nil.
Bicolor floral traits of P. hybrida Picotee and Star are regulated by spatiotemporal production of siRNA molecules from tandemly orientated flower pigmentation genes. It is considered that trans-acting regulatory factors control the spatiotemporal production of siRNA molecules and the regulatory locus is introduced by interspecific hybridization breeding. Characterization of the trans-acting regulatory factors is important for understanding of the RNA-silencing mechanism in flower pigmentation patterns and utilization in breeding of ornamental plants.
The creation of blue flowers in a species with no blue variation is one of the biggest goals of plant breeders. The overexpression of vacuolar NHXs and the suppression of PH5 homologues could be a useful strategy for creating blue flowers through molecular breeding. The first trial has been reported by Kasajima and Sasaki (2016). They introduced expression cassettes of InNHX1, InNHX2, and a chimera repressor of petunia PH4 into torenia. However, neither blue flowers nor the pH increase of petal exudates was observed in transgenic plants. It is possible that the species specificity of the proteins resulted in inhibited H+ transporter activities. The modification of endogenous vacuolar NHX genes and/or endogenous homologues of petunia PH genes are possible ways to overcome such species specificity problems.
Acknowledgments
We thank Francesca Quattrocchio for providing petunia photographs and valuable comments about the manuscript. We also thank Toshio Yamaguchi for his valuable participation in discussions.
Literature Cited
- Albert, N.W., Davies, K.M., Lewis, D.H., Zhang, H., Montefiori, M., Brendolise, C., Boase, M.R., Ngo, H., Jameson, P.E. and Schwinn, K.E. (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhagen, I., Nordholt, N., Seidel, T., Spelt, K., Koes, R., Quattrochio, F., Sagasser, M. and Weisshaar, B. (2015) TRANSPARENT TESTA 13 is a tonoplast P3A-ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 82: 840–849. [DOI] [PubMed] [Google Scholar]
- Bassil, E., Tajima, H., Liang, Y.C., Ohto, M.A., Ushijima, K., Nakano, R., Esumi, T., Coku, A., Belmonte, M. and Blumwald, E. (2011) The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23: 3482–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil, E. and Blumwald, E. (2014) The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 22: 1–6. [DOI] [PubMed] [Google Scholar]
- Bombarely, A., Moser, M., Amrad, A., Bao, M., Bapaume, L., Barry, C.S., Bliek, M., Boersma, M.R., Borghi, L., Bruggmann, R. et al. (2016) Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2: 16074. [DOI] [PubMed] [Google Scholar]
- Buch-Pedersen, M.J., Venema, K., Serrano, R. and Palmgren, M.G. (2000) Abolishment of proton pumping and accumulation in the E1P conformational state of a plant plasma membrane H+-ATPase by substitution of a conserved aspartyl residue in transmembrane segment 6. J. Biol. Chem. 275: 39167–39173. [DOI] [PubMed] [Google Scholar]
- Burbulis, I.E. and Winkel-Shirley, B. (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA 96: 12929–12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y.B., Jones, S.I. and Vodkin, L.O. (2017) Mutations in Argonaute5 illuminate epistatic interactions of the K1 and I loci leading to saddle seed color patterns in Glycine max. Plant Cell 29: 708–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, S., Hoshino, A., Boddu, J. and Iida, S. (2006) Flavonoid pigments as tools in molecular genetics. In: Grotewold, E. (ed.) The Science of Flavonoids, Springer, New York, pp. 147–173. [Google Scholar]
- de Vetten, N., Quattrocchio, F., Mol, J. and Koes, R. (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11: 1422–1434. [DOI] [PubMed] [Google Scholar]
- de Vlaming, P., Schram, A.W. and Wiering, H. (1983) Genes affecting flower colour and pH of flower limb homogenates in Petunia hybrida. Theor. Appl. Genet. 66: 271–278. [DOI] [PubMed] [Google Scholar]
- Eisenach, C., Baetz, U. and Martinoia, E. (2014) Vacuolar proton pumping: more than the sum of its parts? Trends Plant Sci. 19: 344–346. [DOI] [PubMed] [Google Scholar]
- Ewart, L. (1984) Plant breeding. In: Sink, K.C. (ed.) Petunia, Springer, Berlin, pp. 180–202. [Google Scholar]
- Faraco, M., Spelt, C., Bliek, M., Verweij, W., Hoshino, A., Espen, L., Prinsi, B., Jaarsma, R., Tarhan, E., de Boer, A.H. et al. (2014) Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep. 6: 32–43. [DOI] [PubMed] [Google Scholar]
- Faraco, M., Li, Y.B., Li, S.J., Spelt, C., Di Sansebastiano, G.P., Reale, L., Ferranti, F., Verweij, W., Koes, R. and Quattrocchio, F.M. (2017) A tonoplast P3B-ATPase mediates fusion of two types of vacuoles in petal cells. Cell Rep. 19: 2413–2422. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V., Furtek, D.B. and Nelson, O.E. (1984) Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc. Natl. Acad. Sci. USA 81: 3825–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada-Tanaka, S., Inagaki, Y., Yamaguchi, T., Saito, N. and Iida, S. (2000) Colour-enhancing protein in blue petals. Nature 407: 581. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A., Palmgren, M.G. and Schumacher, K. (2007) Plant proton pumps. FEBS Lett. 581: 2204–2214. [DOI] [PubMed] [Google Scholar]
- Holton, T.A., Brugliera, F., Lester, D.R., Tanaka, Y., Hyland, C.D., Menting, J.G., Lu, C.Y., Farcy, E., Stevenson, T.W. and Cornish, E.C. (1993) Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366: 276–279. [DOI] [PubMed] [Google Scholar]
- Hoshino, A., Johzuka-Hisatomi, Y. and Iida, S. (2001) Gene duplication and mobile genetic elements in the morning glories. Gene 265: 1–10. [DOI] [PubMed] [Google Scholar]
- Hoshino, A., Park, K.I. and Iida, S. (2009) Identification of r mutations conferring white flowers in the Japanese morning glory (Ipomoea nil). J. Plant Res. 122: 215–222. [DOI] [PubMed] [Google Scholar]
- Hoshino, A., Jayakumar, V., Nitasaka, E., Toyoda, A., Noguchi, H., Itoh, T., Shin-I, T., Minakuchi, Y., Koda, Y., Nagano, A.J. et al. (2016) Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 7: 13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, S., Morita, Y., Choi, J.D., Park, K.I. and Hoshino, A. (2004) Genetics and epigenetics in flower pigmentation associated with transposable element in morning glories. Adv. Biophys. 38: 141–159. [PubMed] [Google Scholar]
- Imai, Y. (1931) Analysis of flower colour in Pharbitis nil. J. Genet. 24: 203–224. [Google Scholar]
- Imai, Y. (1935) Recurrent reversible mutations in the duskish allelomorphs of Pharbitis Nil. Z. Indukt. Abstamm. Vererbungsl. 68: 242–264. [Google Scholar]
- Inagaki, Y., Hisatomi, Y., Suzuki, T., Kasahara, K. and Iida, S. (1994) Isolation of a Suppressor-mutator/Enhancer-like transposable element, Tpn1, from Japanese morning glory bearing variegated flowers. Plant Cell 6: 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka-Hisatomi, Y., Noguchi, H. and Iida, S. (2011) The molecular basis of incomplete dominance at the A locus of CHS-D in the common morning glory, Ipomoea purpurea. J. Plant Res. 124: 299–304. [DOI] [PubMed] [Google Scholar]
- Jorgensen, K., Rasmussen, A.V., Morant, M., Nielsen, A.H., Bjarnholt, N., Zagrobelny, M., Bak, S. and Moller, B.L. (2005) Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8: 280–291. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A. (1995) Cosuppression, flower color patterns, and metastable gene-expression states. Science 268: 686–691. [DOI] [PubMed] [Google Scholar]
- Kasajima, I. and Sasaki, K. (2016) A chimeric repressor of petunia PH4 R2R3-MYB family transcription factor generates margined flowers in torenia. Plant Signal. Behav. 11: e1177693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes, R.E., Spelt, C.E., Mol, J.N.M. and Gerats, A.G.M. (1987) The chalcone synthase multigene family of Petunia hybrida (V30): sequence homology, chromosomal localization and evolutionary aspects. Plant Mol. Biol. 10: 375–389. [DOI] [PubMed] [Google Scholar]
- Koes, R., Verweij, W. and Quattrocchio, F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10: 236–242. [DOI] [PubMed] [Google Scholar]
- Koseki, M., Goto, K., Masuta, C. and Kanazawa, A. (2005) The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol. 46: 1879–1883. [DOI] [PubMed] [Google Scholar]
- Kroon, J., Souer, E., de Graaff, A., Xue, Y.B., Mol, J. and Koes, R. (1994) Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J. 5: 69–80. [DOI] [PubMed] [Google Scholar]
- Li, Y.B., Provenzano, S., Bliek, M., Spelt, C., Appelhagen, I., de Faria, L.M., Verweij, W., Schubert, A., Sagasser, M., Seidel, T. et al. (2016) Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification. New Phytol. 211: 1092–1107. [DOI] [PubMed] [Google Scholar]
- Lloyd, A., Brockman, A., Aquirre, L., Campbell, A., Bean, A., Cantero, A. and Gonzalez, A. (2017) Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58: 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, Y., Hoshino, A., Kikuchi, Y., Okuhara, H., Ono, E., Tanaka, Y., Fukui, Y., Saito, N., Nitasaka, E., Noguchi, H. et al. (2005) Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O- glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J. 42: 353–363. [DOI] [PubMed] [Google Scholar]
- Morita, Y., Saitoh, M., Hoshino, A., Nitasaka, E. and Iida, S. (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 47: 457–470. [DOI] [PubMed] [Google Scholar]
- Morita, Y., Saito, R., Ban, Y., Tanikawa, N., Kuchitsu, K., Ando, T., Yoshikawa, M., Habu, Y., Ozeki, Y. and Nakayama, M. (2012) Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J. 70: 739–749. [DOI] [PubMed] [Google Scholar]
- Morita, Y., Takagi, K., Fukuchi-Mizutani, M., Ishiguro, K., Tanaka, Y., Nitasaka, E., Nakayama, M., Saito, N., Kagami, T., Hoshino, A. et al. (2014) A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 78: 294–304. [DOI] [PubMed] [Google Scholar]
- Morita, Y., Ishiguro, K., Tanaka, Y., Iida, S. and Hoshino, A. (2015a) Spontaneous mutations of the UDP-glucose:flavonoid 3-O-glucosyltransferase gene confers pale- and dull-colored flowers in the Japanese and common morning glories. Planta 242: 575–587. [DOI] [PubMed] [Google Scholar]
- Morita, Y., Tanase, K., Ohmiya, A., Hisamatsu, T. and Nakayama, M. (2015b) Isolation and characterization of enhancer of flavonoid production genes from floricultural crops. Sci. Rep. Fac. Agric. Meijo Univ. 51: 35–41. [Google Scholar]
- Napoli, C., Lemieux, C. and Jorgensen, R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngaki, M.N., Louie, G.V., Philippe, R.N., Manning, G., Pojer, F., Bowman, M.E., Li, L., Larsen, E., Wurtele, E.S. and Noel, J.P. (2012) Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 485: 530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi, M., Fukada-Tanaka, S., Hoshino, A., Takada, J., Inagaki, Y. and Iida, S. (2005) Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory. Plant Cell Physiol. 46: 259–267. [DOI] [PubMed] [Google Scholar]
- Palmgren, M.G. and Nissen, P. (2011) P-TypeATPases. Annu. Rev. Biophys. 40: 243–266. [DOI] [PubMed] [Google Scholar]
- Park, K.I., Choi, J.D., Hoshino, A., Morita, Y. and Iida, S. (2004) An intragenic tandem duplication in a transcriptional regulatory gene for anthocyanin biosynthesis confers pale-colored flowers and seeds with fine spots in Ipomoea tricolor. Plant J. 38: 840–849. [DOI] [PubMed] [Google Scholar]
- Park, K.I., N. Ishikawa, Y. Morita, J.D. Choi, A. Hoshino and S. Iida (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J. 49: 641–654. [DOI] [PubMed] [Google Scholar]
- Pittman, J. (2012) Multiple transport pathways for mediating intracellular pH homeostasis: the contribution of H+/ion exchangers. Front. Plant Sci. 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J. and Koes, R. (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., Baudry, A., Lepiniec, L. and Grotewold, E. (2006a) The regulation of flavonoid biosynthesis. In: Grotewold, E. (ed.) The Science of Flavonoids, Springer, New York, pp. 97–122. [Google Scholar]
- Quattrocchio, F., Verweij, W., Kroon, A., Spelt, C., Mol, J. and Koes, R. (2006b) PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston, L., Subramanian, S., Matsuno, M. and Yu, O. (2005) Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol. 137: 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, N.A. and Glover, B.J. (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70. [DOI] [PubMed] [Google Scholar]
- Saito, N., Cheng, J., Ichimura, M., Yokoi, M., Abe, Y. and Honda, T. (1994) Flavonoids in the acyanic flowers of Pharbitis nil. Phytochemistry 35: 687–691. [Google Scholar]
- Saito, N., Tatsuzawa, F., Kasahara, K., Iida, S. and Honda, T. (1998) Acylated cyanidin 3-sophorosides in the brownish-red flowers of Ipomoea purpurea. Phytochemistry 49: 875–880. [Google Scholar]
- Saito, N., Tatsuzawa, F., Hoshino, A., Abe, Y., Ichimura, M., Yokoi, M., Toki, K., Morita, Y., Iida, S. and Honda, T. (2011) Anthocyanin pigmentation controlled by speckled and c-1 mutations of Japanese morning glory. J. Japan. Soc. Hort. Sci. 80: 452–460. [Google Scholar]
- Saito, R., Fukuta, N., Ohmiya, A., Itoh, Y., Ozeki, Y., Kuchitsu, K. and Nakayama, M. (2006) Regulation of anthocyanin biosynthesis involved in the formation of marginal picotee petals in Petunia. Plant Sci. 170: 828–834. [Google Scholar]
- Saito, R., Kuchitsu, K., Ozeki, Y. and Nakayama, M. (2007) Spatiotemporal metabolic regulation of anthocyanin and related compounds during the development of marginal picotee petals in Petunia hybrida (Solanaceae). J. Plant Res. 120: 563–568. [DOI] [PubMed] [Google Scholar]
- Sink, K.C. (1984) Taxonomy. In: Sink, K.C. (ed.) Petunia, Springer, Berlin, pp. 3–9. [Google Scholar]
- Spelt, C., Quattrocchio, F., Mol, J.N. and Koes, R. (2000) anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt, C., Quattrocchio, F., Mol, J. and Koes, R. (2002) ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 14: 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmann, J.R., Lorenz-Lemke, A.P., Freitas, L.B. and Semir, J. (2009) The genus petunia. In: Gerats, T. and Stommer J. (eds.) Petunia. Evolutionaly, Developmental and Physiological Genetics, Springer, New York, pp. 1–28. [Google Scholar]
- Takahashi, R., Yamagishi, N. and Yoshikawa, N. (2013) A MYB transcription factor controls flower color in soybean. J. Hered. 104: 149–153. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Sasaki, N. and Ohmiya, A. (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54: 733–749. [DOI] [PubMed] [Google Scholar]
- Tohge, T., Nishiyama, Y., Hirai, M.Y., Yano, M., Nakajima, J., Awazuhara, M., Inoue, E., Takahashi, H., Goodenowe, D.B., Kitayama, M. et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235. [DOI] [PubMed] [Google Scholar]
- Toki, K., Saito, N., Iida, S., Hoshino, A., Shigihara, A. and Honda, T. (2001) A novel acylated pelargonidin 3-sophoroside-5-glucoside from greyish-purple flowers of the Japanese morning glory. Heterocycles 55: 2261–2267. [Google Scholar]
- Tornielli, G., Koes, R. and Quattrocchio, F. (2009) The genetics of flower color. In: Gerats, T. and Strommer J. (eds.) Petunia, Springer, New York, pp. 269–299. [Google Scholar]
- van der Krol, A.R., Mur, L.A., Beld, M., Mol, J.N. and Stuitje, A.R. (1990) Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houwelingen, A., Souer, E., Spelt, K., Kloos, D., Mol, J. and Koes, R. (1998) Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J. 13: 39–50. [DOI] [PubMed] [Google Scholar]
- Vandenbussche, M., Chambrier, P., Bento, S.R. and Morel, P. (2016) Petunia, your next supermodel? Front. Plant Sci. 7: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, K., Belver, A., Marin-Manzano, M.C., Rodriguez-Rosales, M.P. and Donaire, J.P. (2003) A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J. Biol. Chem. 278: 22453–22459. [DOI] [PubMed] [Google Scholar]
- Verweij, W., Spelt, C., Di Sansebastiano, G.P., Vermeer, J., Reale, L., Ferranti, F., Koes, R. and Quattrocchio, F. (2008) An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 10: 1456–1462. [DOI] [PubMed] [Google Scholar]
- Verweij, W., Spelt, C.E., Bliek, M., de Vries, M., Wit, N., Faraco, M., Koes, R. and Quattrocchio, F.M. (2016) Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in arabidopsis. Plant Cell 28: 786–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., Kobayashi, A., Endo, M., Sage-Ono, K., Toki, S. and Ono, M. (2017) CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 7: 10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107: 142–149. [Google Scholar]
- Winkel-Shirley, B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, L., Rizzini, L., Stracke, R., Ulm, R. and Rensing, S.A. (2010) The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiol. 153: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., Fukada-Tanaka, S., Inagaki, Y., Saito, N., Yonekura-Sakakibara, K., Tanaka, Y., Kusumi, T. and Iida, S. (2001) Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol. 42: 451–461. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Tohge, T., Matsuda, F., Nakabayashi, R., Takayama, H., Niida, R., Watanabe-Takahashi, A., Inoue, E. and Saito, K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Fukushima, A., Nakabayashi, R., Hanada, K., Matsuda, F., Sugawara, S., Inoue, E., Kuromori, T., Ito, T., Shinozaki, K. et al. (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 69: 154–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., Kondo, T., Okazaki, Y. and Katou, K. (1995) Cause of blue petal color. Nature 373: 291–291. [Google Scholar]
- Yoshida, K., Kawachi, M., Mori, M., Maeshima, M., Kondo, M., Nishimura, M. and Kondo, T. (2005) The involvement of tonoplast proton pumps and Na+(K+)/H+ exchangers in the change of petal color during flower opening of morning glory, Ipomoea tricolor cv. Heavenly Blue. Plant Cell Physiol. 46: 407–415. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., Miki, N., Momonoi, K., Kawachi, M., Katou, K., Okazaki, Y., Uozumi, N., Maeshima, M. and Kondo, T. (2009a) Synchrony between flower opening and petal-color change from red to blue in morning glory, Ipomoea tricolor cv. Heavenly Blue. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 85: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., Mori, M. and Kondo, T. (2009b) Blue flower color development by anthocyanins: from chemical structure to cell physiology. Nat. Prod. Rep. 26: 884–915. [DOI] [PubMed] [Google Scholar]