Abstract
Deciphering antibody-protein antigen recognition is of fundamental and practical significance. We constructed an antibody structural dataset, partitioned it into human and murine subgroups, and compared it with non-antibody protein-protein complexes. We investigated the physico-chemical properties of regions on and away from the antibody-antigen interfaces, including net-charge, overall antibody charge distributions and their potential role in antigen interaction. We observed that amino acid preference in antibody-protein antigen recognition is entropy driven, with residues having low side-chain entropy appearing to compensate for the high backbone entropy in interaction with protein antigens. Antibodies prefer charged and polar antigen residues, and bridging water molecules. They also prefer positive net-charge, presumably to promote interaction with negatively charged protein antigens, which are common in proteomes. Antibody-antigen interfaces are mostly negatively charged with dominant contribution of Asp and a positively charged region. Here we describe some features of antibody-antigen interfaces and of Fab domains as compared to non-antibody protein-protein interactions. The distributions of interface residues in human and murine antibodies do not differ significantly. Overall, our results provide not only a local but a global anatomy of antibody structures.
Keywords: Protein-Protein interaction, Antibody-Antigen recognition, Allosteric regulation, mAb drugs, Binding epitopes
Introduction
The recognition of foreign antigens by antibodies and T-cell antigen receptors is the hallmark of specific adaptive immune response. The limited repertoire – with six main variable loops in the binding region, and a limited number of antibody classes and isotypes – can be specifically elicited against an unlimited number of possible antigens, including protein antigens1–4. Deciphering antibody-protein antigen recognition is of both fundamental and practical significance. Protein-protein interactions (PPI) constitute important networks that maintain biological functions. Their structural features, concentrations and cellular environments vary5–8. Here we ask if there are features in antibody- protein antigen recognition which differ from other protein-protein interactions. Understanding and exploiting interactions between antibody and protein antigens may help in therapeutic design9. Monoclonal antibodies (mAb) have become promising therapeutics against infections, cancer, and immune disorders10. They can also bind to biologically active targets for clinical applications2. Hundreds of mAb candidates have been clinically tested11, with about fifty mAb drugs approved and listed in drug databases12. While these antibody drugs have shown successes in some diseases, such as certain cancers, it has been less so in neurodegenerative diseases, which are caused by protein aggregates. Among these, therapies aimed at reducing protein processing and clearance for AD have been unsuccessful in clinical trials 13, 14.
Large scale analysis of antibody-antigen structures will help to identify valid antibody-epitope pairs and to predict biologically-relevant epitopes. MacCallum, Martin, and Thornton pioneered the structural analysis of antibody-antigen interactions using 26 complex structures with different antigens, including haptens, carbohydrates, DNA, peptides, and proteins15. In our earlier study of protein-protein interactions, we noticed different amino acids are conserved in antibody-antigen versus other protein-protein interactions16. With more and more antibody-antigen structures becoming available in the last decade, increasing efforts have focused on analysis of antibody-protein interactions to obtain insights into their recognition and applications in design15, 17, 18. Ponomarenko and Bourne collected a dataset of 82 antibody-protein complexes containing different structural epitopes and observed that it is difficult to decipher B-cell epitopes as an intrinsic feature of the protein19. Ramaraj et al. studied antigen-antibody interface composition, residue interactions, and additional features of 53 non-redundant structures20. They confirmed the dominance of Tyr in the Ab paratope-containing surface, with almost two-fold greater abundance than any other residue20. Based on a dataset of 107 dissimilar antibody-protein structures, the B-cell epitope was suggested to be oval-shaped consisting of predominantly hydrophobic amino acids in the center flanked by charged residues21. In a more detailed study of 111 representative antibody–protein complex structures, Peng et al. found that Tyr and other aromatic residues are surrounded by short-chain hydrophilic side chains (Asp, Asn, Ser, Thr, and Gly) in the structural paratopes1. The tyrosyl side chains on the functional paratopes recognize the corresponding epitopes by interacting with backbone atoms and side-chain carbons, which contribute the most to the binding energy as interface hot spots1.
Despite progress, several important questions remain to be answered. First, even though it has been noticed that antibody-protein antigen interactions differ from other protein-protein interactions, there has been no systematic comparison. What is the role of interface water molecules in the recognition? Do human and mouse antibodies differ in their antigen recognition preference? Are allosteric long range interactions important?
Water molecules play an important role in protein folding and binding22–26 as well as in antibody-antigen recognition. A recently solved crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1 revealed almost equal contribution of water bridged interaction and direct polar interactions27. Reichmann et al. analyzed the degree of water conservation between the bound and unbound states in protein-protein complexes, and found that about 20% buried or partially buried waters in the monomer structures were conserved at the same location in the bound structures25. Ahmed et al. found that about 20% of bound water at protein-protein interfaces bridge interactions with both proteins. The buried waters can locate within predominantly hydrophobic environments and form “hydrophobic bubbles” that modulate the interaction22. Hong and Kim studied the interaction between bound water molecules and local protein structures, and suggested that proteins may have evolved to accommodate interacting water molecules that are energetically stable, but result in little entropic cost at room temperature23.
Earlier approaches to therapeutic antibody development used murine antibodies followed by humanization of the lead. More recently, human monoclonal mAb therapeutics has been taken up11; however, few human mAbs entered the clinic. Murine antibodies are easier to produce, but are limited by safety issues and diminished efficacy owing to the immunogenicity of the mouse-derived protein sequences11, which can result in elimination of the therapeutic antibodies from the host and formation of immune complexes that can damage the kidneys. This problem has been reduced to some extent by genetic engineering which develops mAbs that contain a combination of rodent-derived and human-derived sequences, resulting in chimeric and humanized antibodies28. These constitute the majority of candidates in clinical studies during the 1990s 11, and 27 of the 39 mAbs currently approved or in review are either chimeric or humanized products29. Thus, understanding the similarity and differences of the paratopes’ structures and biochemical properties of mouse and human antibodies can be significant for drug/biological design.
While most previous studies focused on the local features of antibody antigen recognition, how the antigen binding sites are coupled with the global antibody structure to differentiate among antigens and communicate with the allosteric receptor binding site was overlooked. All proteins can be allosterically regulated30, and allosteric communication in proteins can define proteins’ functions31. It has been demonstrated that two antibodies with identical variable domains, but with slightly different constant regions, bind antigens differently32–34. A systematic comparison of free and bound structures of all 141 crystal structures of the 49 Abs of these two forms has shown that beyond the binding site, Ag binding may be associated with changes in the relative orientation of the H and L chains in both the variable and constant domains35.
In this work, we thoroughly examined antibody-antigen recognition and compared it with non-antibody protein-protein interactions, including binding site and regions away from the interfaces. We constructed a structural antibody dataset and partitioned it into human and murine subgroups and compared it with 298 non-antibody protein-protein complexes. Starting from the antigen binding interface, we systematically examine amino-acid distributions including away from the interacting antigen, the roles of water molecules, and the similarity/differences between the human and mouse/rat antibodies. We also examine possible antibody net-charge from VDJ recombination aiming to understand the overall antibody charge distributions and their possible role in antigen interaction. We found that antibodies prefer positive net-charge presumably to help antibody proteins approach negatively charged protein antigens, which are common in proteomes36, 37. The antibody binding surfaces are mostly negatively charged with dominant contribution of Asp residue followed by a positively charged region. This residue distribution beyond the antigen binding interfaces may affect allosteric signaling. Our results provide the local and global anatomy of antibody structures in protein antigen recognition.
Materials and Methods
Dataset
The experimental structural datasets of protein-protein complexes were downloaded from the RSCB Protein Data Bank. We merged two previously published datasets to collect non-antibody-protein interaction complexes. The first is the dataset used in a study of water distribution in protein-protein interfaces22, and the second is the Protein-Protein Interaction Affinity Database 38. The antibody complexes in the two datasets were moved to antibody-protein complexes dataset. The antibody-protein complex dataset is mainly from two previous antibody structural studies1, 21. Additional entries were from searching the pdb database for therapeutic antibody, and the antibodies we have previously analyzed. pdb entries with over 95% sequence identities were removed, and we only retained Fab structures. When partitioning the antibody-protein complex dataset into human and mouse subgroups, some chimeric antibodies are repeated in both subgroups. The final curated datasets are listed in supporting material Tables 1–3.
Interface contact and Interface side-chain entropy
We calculated residue-residue contacts across antibody-antigen interfaces with three distance cutoffs. The closest contact for two heavy atoms (within 3 Å), correspond to distances of hydrogen bonds and salt bridges. The next range is 4 Å, which may include hydrophobic interactions. Finally, a 5 Å cutoff, which contains interface residues interacting directly or supporting direct interactions across antibody-antigen interfaces. We examined the water molecules that bridge interactions between two interacting proteins in protein-protein complexes. A water molecule is defined as bridging water if it is within the cutoff distances to the heavy atoms of two interacting proteins. Again, we used 3 Å, 4 Å, and 5 Å cutoffs. We also calculate similar propensities for non-antibody protein-protein interactions, using receptor-ligand complexes.
We count the frequencies (ni) of residues within the cutoff distances or the frequencies (ni) of residues interacting with other residues across interface. The propensity of a residue in interaction is calculated as:
| (1) |
Amino acids are divided into different groups in our analysis: (1) Charged amino acids: Asp, Glu, Lys, Arg; (2) Polar amino acids: Ser, Thr, Gln, Asn, His, and Cys; Hydrophobic group (HP): Ala, Val, Leu, Ile, Phe, Trp, Met, and Pro; Tyr and Gly are not included in any group to consider antibodies’ unique amino acid propensity.
The amino-acid side-chain entropies were taken from Abagyan and Totrov, who have calculated the conformational entropy differences between free and buried states of amino-acid side-chains39. Thus, the surface entropy contributions of antigen binding at 300K were calculated as:
| (2) |
Pi is the propensity of residue i at the interface and the TSi is the conformational entropy differences between free and buried state for residue i.
Statistical significance of amino acid preference and related properties were obtained by bootstrapping using100,000 bootstrapped datasets, which were produced by maintaining the same number of protein structures in each bootstrapped dataset.
Amino acid charge and protein net charge calculation
Protein net-charges are calculated based on Henderson-Hasselbalch formula. The pK values for ionizable groups are taken from reference 40, with following values: Arg 12.5; Lys 10.8; His 6.5; Asp 3.9, and Glu 4.1. The pK value can be perturbed by various factors like protein folding41, dehydration, charge-charge interactions, and charge-dipole interactions40. The pK values used in this study were averaged from fitting the Henderson-Hasselbalch formula42, which only considers pH effects. It is possible to calculated the pK shifts from three dimensional structures to include additional factors. However, such approach is prohibitive for estimation of antibody charges on a large scale when only using sequence information.
Antibody sequences were downloaded from the IMGT/GENE Database (www.imgt.org/vquest/refseqh.html)43. Each sequence for the groups (IGHV, IGHD, IGHJ, IGHC, IGKV, IGKJ, and IGKC) was saved separately. These sequences are for the entire chain, including all fragments. To find the charge variations in the IgG, the sequences of IGHV, IGHD, IGHJ, IGHC, IGKV, IGKJ, and IGKC, were combined to generate possible antibody sequences from VDJ recombination.
Results
Antibodies prefer using low entropy sidechains antibody-antigen interactions
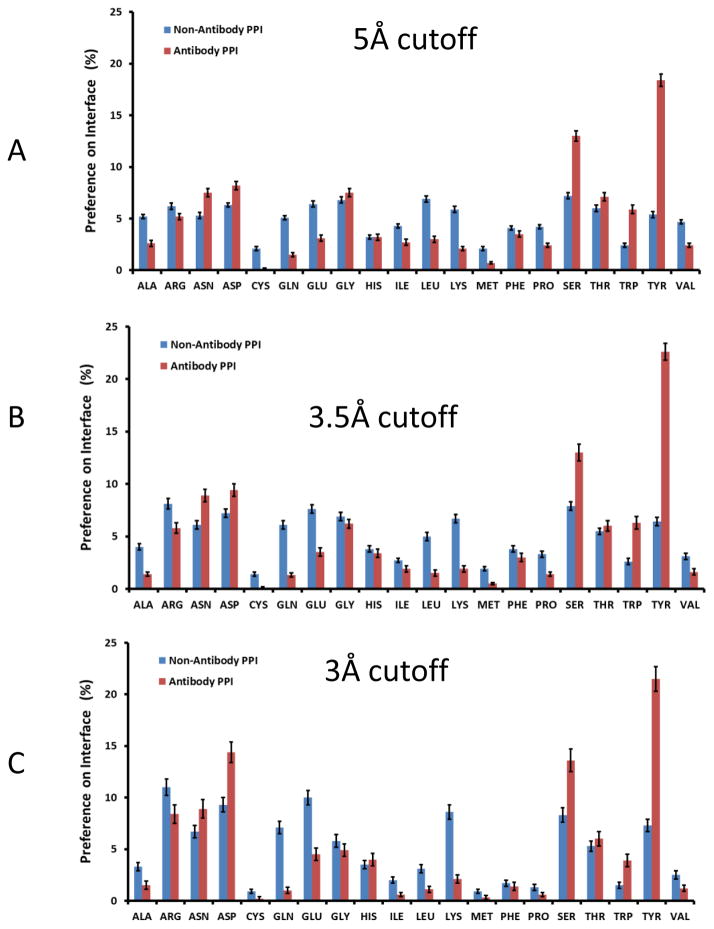
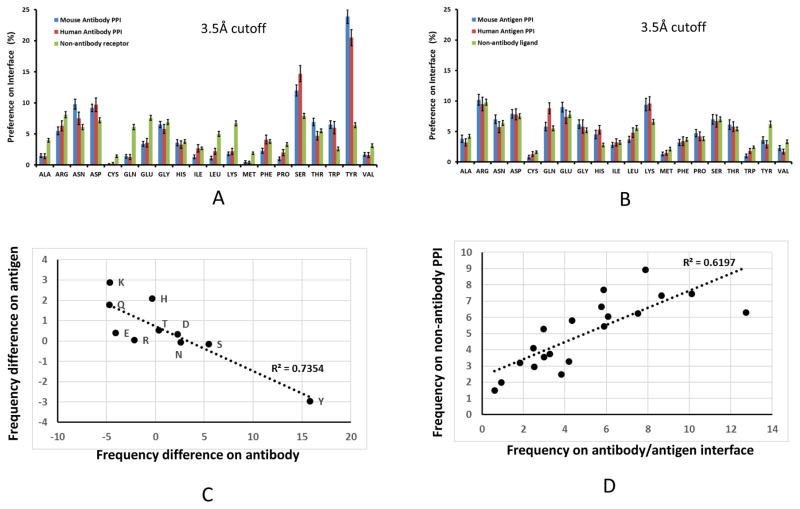
The percentages of amino acids on the antibody-antigen interfaces are reported in Figure 1. The propensities with 5 Å cutoff are close to those obtained with 3.5 Å. The top two enriched amino acids in antibody are Tyr and Ser, as has been observed in previous studies1, 17, 18, 20. The amino acid contact percentage at 3 Å cutoff revealed that while Tyr still has the highest propensity on antibody-antigen interface, Asp and Ser are the second and third enriched amino acid to strongly interact with antigens, respectively (Figure 1).
Figure 1.
Antibodies use less charged residue and less hydrophobic residues in protein-protein interaction. Amino acid interaction propensities at different cutoff distances for protein-protein interactions. A. 5 Å; B. 3.5 Å; and C. 3.0 Å. Blue bar: Non-antibody proteins-protein interactions, red bar: Antibody amino acids in antibody-protein antigen interactions.
Comparing with non-antibody protein-protein interactions, first, while antibodies have slightly lower Phe percentage, their propensities of Trp and Tyr residues are much higher. Thus, antibody recognition surfaces are much more aromatic than non-antibody protein receptors. Secondly, antibodies use less hydrophobic residues to recognize antigens (Table 1). With 5 Å cutoff, the percentage of hydrophobic-hydrophobic interaction in antibody-antigen interaction is 8.4%, and 15.5% for non-antibody PPI. The total interactions by hydrophobic antibody residues are 23.4% and 9.8%, for 5 Å and 3 Å cutoffs respectively. The comparable interactions in non-antibody PPI are 35.0% and 15.3%.
Table 1.
The percentage classification of amino acids on protein-protein recognition interfaces
| Human 5Å | Ag charged | Ag polar | Ag Hydrophobic | Ag Gly | Ag Tyr | Total |
|---|---|---|---|---|---|---|
| Ab Charged | 7.5±0.7 | 5.8±0.6 | 4.3±0.5 | 1.1±0.3 | 0.7±0.1 | 19.4±1.4 |
| Ab polar | 8.3±0.6 | 8.9±0.8 | 7.1±0.6 | 1.6±0.3 | 0.8±0.2 | 26.8±1.1 |
| Ab HP | 6.3±0.5 | 7.5±0.7 | 11.0±0.9 | 1.7±0.4 | 1.1±0.3 | 27.6±1.7 |
| Ab Gly | 1.9±0.3 | 1.7±0.2 | 1.5±0.2 | 0.3±0.1 | 0.3±0.1 | 5.8±0.5 |
| Ab Tyr | 5.8±0.5 | 6.4±0.6 | 6.2±0.5 | 1.2±0.2 | 0.8±0.2 | 20.5±1.2 |
| Total | 29.8±1.6 | 30.3±1.4 | 30.2±1.5 | 5.9±1.0 | 3.8±0.6 | 100 |
| Murine 5Å | Ag charged | Ag polar | Ag Hydrophobic | Ag Gly | Ag Tyr | Total |
| Ab Charged | 7.3±0.5 | 4.8±0.3 | 3.4±0.3 | 1.0±0.1 | 0.9±0.1 | 17.4±0.8 |
| Ab polar | 9.7±0.5 | 9.4±0.6 | 7.6±0.5 | 1.4±0.2 | 1.0±0.2 | 29.1±1.0 |
| Ab HP | 6.1±0.5 | 5.7±0.7 | 6.6.0±0.6 | 1.1±0.2 | 1.0±0.1 | 20.5±0.9 |
| Ab Gly | 2.0±0.2 | 2.2±0.2 | 1.8±0.2 | 0.4±0.1 | 0.3±0.1 | 6.6±0.4 |
| Ab Tyr | 8.7±0.5 | 6.5±0.6 | 8.1±0.6 | 1.8±0.2 | 1.0±0.2 | 26.4±1.3 |
| Total | 33.8±1.3 | 28.6±1.3 | 27.6±1.2 | 5.9±0.6 | 4.1±0.5 | 100 |
| Non-antibody 5Å | Ag charged | Ag polar | Ag Hydrophobic | Ag Gly | Ag Tyr | Total |
| Ab Charged | 9.0±0.3 | 5.9±0.3 | 7.2±0.3 | 1.1±0.1 | 1.4±0.1 | 24.7±0.7 |
| Ab polar | 7.4±0.3 | 7.4±0.3 | 9.5±0.3 | 1.3±0.1 | 1.8±0.2 | 27.4±0.6 |
| Ab HP | 7.0±0.3 | 8.6±0.3 | 15.5±0.5 | 1.6±0.1 | 2.3±0.2 | 35.0±0.7 |
| Ab Gly | 1.5±0.1 | 1.6±0.1 | 2.2±0.2 | 0.4±0.1 | 0.4±0.1 | 6.1±0.3 |
| Ab Tyr | 1.8±0.1 | 1.6±0.1 | 2.8±0.2 | 0.3±0.0 | 0.3±0.1 | 6.8±0.4 |
| Total | 26.6±0.6 | 25.2±0.6 | 37.3±0.7 | 4.7±0.3 | 6.2±0.4 | 100 |
| Human 3Å | Ag charged | Ag polar | Ag Hydrophobic | Ag Gly | Ag Tyr | Total |
| Ab Charged | 20.6±2.3 | 9.3±1.4 | 2.7±0.8 | 1.3±0.6 | 1.1±0.4 | 35.0±2.7 |
| Ab polar | 15.7±1.7 | 11.4±1.6 | 2.9±0.8 | 1.3±0.6 | 0.4±0.3 | 31.6±2.4 |
| Ab HP | 4.2±0.9 | 1.7±0.6 | 1.7±0.6 | 0.2±0.2 | 0.4±0.3 | 9.9±1.4 |
| Ab Gly | 2.5±0.6 | 1.3±0.5 | 0.2±0.2 | 0 | 0 | 4.0±0.9 |
| Ab Tyr | 8.9±1.3 | 8.5±1.5 | 1.1±0.5 | 0.6±0.3 | 0.4±0.3 | 19.5±2.0 |
| Total | 51.1±3.1 | 34.7±2.8 | 8.6±1.5 | 3.4±1.0 | 2.3±0.6 | 100 |
| Murine 3Å | Ag charged | Ag polar | Ag Hydrophobic | Ag Gly | Ag Tyr | Total |
| Ab Charged | 18.4±1.5 | 7.9±1.0 | 1.7±0.6 | 0.9±0.4 | 0.9±0.3 | 29.7±1.8 |
| Ab polar | 15.3±1.4 | 11.1±1.3 | 4.9±0.8 | 0.7±0.3 | 0.5±0.2 | 32.5±1.6 |
| Ab HP | 3.9±0.6 | 3.1±0.7 | 1.7±0.4 | 0.4±0.2 | 0.6±0.3 | 9.8±1.1 |
| Ab Gly | 2.3±0.5 | 1.8±0.4 | 0.4±0.2 | 0.4±0.2 | 0.3±0.2 | 5.0±0.7 |
| Ab Tyr | 12.1±1.3 | 6.6±1.5 | 2.6±0.5 | 1.5±0.4 | 0.1±0.1 | 23.0±1.9 |
| Total | 51.9±2.5 | 30.5±2.4 | 11.4±1.3 | 3.9±1.0 | 2.4±0.6 | 100 |
| Non-antibody 3Å | Ag charged | Ag polar | Ag Hydrophobic | Ag Gly | Ag Tyr | Total |
| Ab Charged | 21.9±1.1 | 11.2±1.0 | 4.7±0.5 | 1.4±0.3 | 2.2±0.4 | 41.4±1.4 |
| Ab polar | 12.0±0.8 | 10.1±1.3 | 5.6±0.6 | 0.9±0.2 | 2.3±0.3 | 30.9±1.1 |
| Ab HP | 4.8±0.5 | 6.0±0.6 | 2.6±0.4 | 0.8±0.2 | 1.1±0.2 | 15.3±0.9 |
| Ab Gly | 1.7±0.3 | 1.6±0.3 | 1.7±0.3 | 0.1±0.1 | 0.2±0.1 | 5.4±0.6 |
| Ab Tyr | 3.1±0.4 | 1.7±0.3 | 1.5±0.3 | 0.4±0.1 | 0.3±0.1 | 7.0±0.6 |
| Total | 43.6±1.3 | 30.7±1.3 | 16.1±0.9 | 3.5±0.4 | 6.1±0.6 | 100 |
The total charge-charge interactions (mostly salt bridges) in antibody-antigen interactions are 7.4% and 19.3% for 5 Å and 3 Å cutoffs respectively. The corresponding percentages for non-antibody PPI are 9.0% and 21.9%. Antibodies recognize higher percentage of charged residues on the antigen interfaces. The ratio of charged residues on the antigen interfaces are 32.2% (5 Å cutoff) and 51.6% (3 Å cutoff), and the corresponding ratios are 26.6% and 43.6% for non-antibody protein-protein interaction. The interaction matrix revealed that a higher percentage of Tyr and Ser residues on the antibody surface interact with mostly negatively charged residues on the antigen surface. As can be seen also in Figure 1, antibodies use very little Lys residues to interact with antigens and Glu is also much lower than in non-antigen PPI.
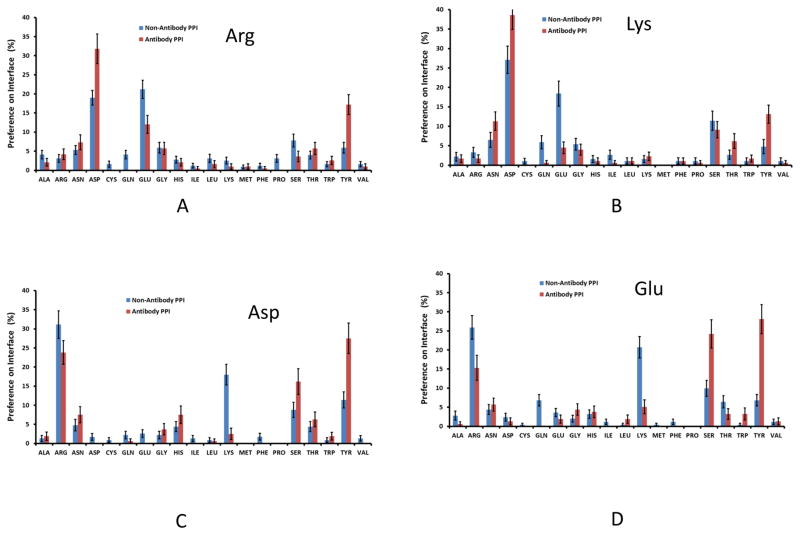
Further details can be seen in Figure 2. For antigen Arg/Lys interactions (Fig 2A and 2B), 35–40% of the antibody residues are Asp, much higher than in non-antibody proteins interacting with Arg residues in ligands. Interestingly, antibodies use less Glu than Asp. Surprisingly, Tyr dominates antibody interactions with negatively charged Asp/Glu residues. Even though a high percentage of Arg residues interact with Asp/Glu, antibody Lys has very little contribution to recognizing Asp/Glu. Possibly antibodies tend to avoid Glu/Lys to minimize the entropy cost, since they have longer sidechains. We used the sidechain entropy values (TS, T=300K) based on the study of Abagyan and Totrov39 to compare residues with similar chemical function and interactions in protein-protein recognition. For example, Asn (TS: 0.81 kcal/mol) has a shorter side chain than Gln (TS: 2.02 kcal/mol), so Asn (7.5%) is preferred over Gln (1.5%) in antibody-antigen interactions. Similarly, Asp (8.2%) is preferred over Glu (3.1%). Tyr (TS: 0.99 kcal/mol) is heavily used to replace Arg (TS: 2.13 kcal/mol) and Lys (TS: 2.21 kcal/mol) to interact with Asp and Glu. Therefore, we propose that low entropy sidechains are preferred. Gly, Ala, and Ser also have overall side-chain entropy. Indeed, Ser is also a preferred residue. Gly has similar distribution in both antibody and non-antibody protein-protein interactions.
Figure 2.
Antibodies use Tyr and Ser, instead of Lys to interact with Asp/Glu on antigen interface. (A)–(D). Amino acid propensities on antibody interface to recognize Arg, Lys, Asp, and Glu residues on antigen interface, respectively.
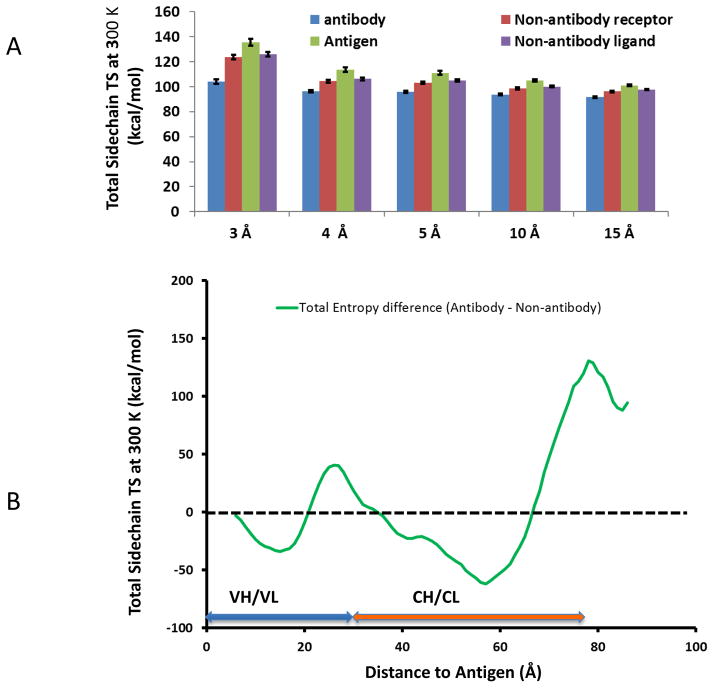
To check if sidechain entropy is a factor affecting amino-acid frequencies in protein-protein interaction, we calculated the interface amino-acid sidechain entropies using equation 3. As can be seen in Figure 3A, the total sidechain entropies for antibody-antigen interaction within 3 Å cutoff is lower than for non-antibody PPI, and the trend appears to continue, albeit diminishing. We further examined the global sidechain entropy distributions based on the residue distances to the heavy atom of the antigens. As we can see in Figure 3B, the antibody residue compositions within a 15 Å shell from the antigen have lower side-chain entropies than non-antibody proteins. Around junctions connecting variable and constant regions (around 30 Å), antibodies have higher side-chain entropies than non-antibody proteins within comparable distances to protein ligands.
Figure 3.
Antibodies minimize side-chain entropy in antigen recognition. (A) Side-chain entropies for the residues within different distances to antigen heavy atoms. (B) Density distributions of sidechain entropy for the amino-acids in antibody proteins mostly lower than non-antibody proteins, except the junction region between variable and constant regions.
Water bridge hot spots in antibody-protein interactions
High resolution crystal structures provide important information about water on protein-protein interaction surfaces. As shown in Figure S1, some antibody-antigen interfaces are extensively coordinated with water molecules. A 1.2-Å structure of the H10L10–HEL (lysozyme) complex has shown about 12 water molecules mediating the antibody and antigen within the 3Å cutoff. While there are 13 directly interacting atomic pairs, as many as 20 interactions are water bridged.
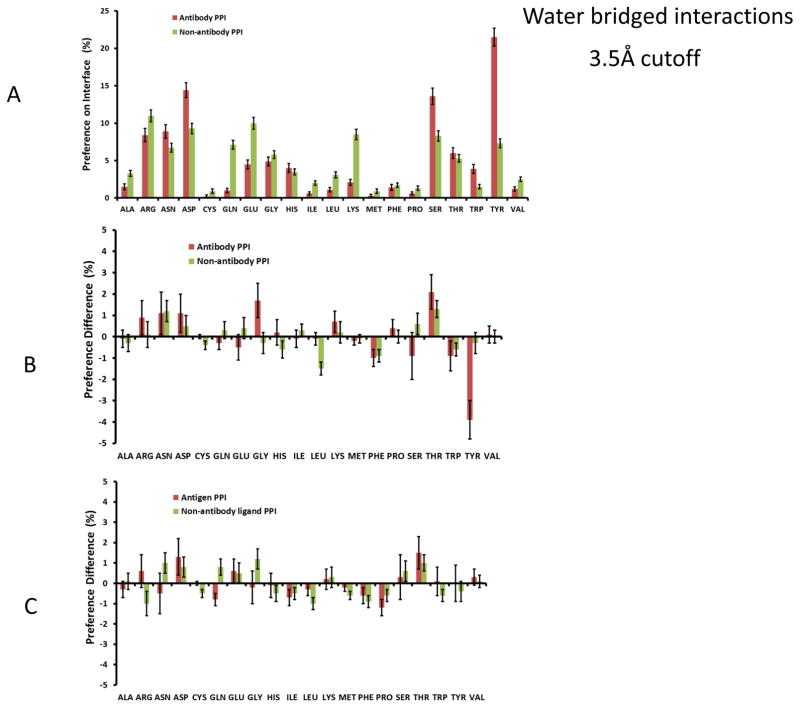
We searched for water-bridged interactions in the interfaces in the antibody and non-antibody datasets. Generally, the propensity of an amino acid to coordinate with bridged water molecules follows the overall propensity of its occurrence on the interfaces. As shown in Figure 4A, the frequencies of coordinated water molecules between antibody-antigen interfaces revealed three peaks at Asp, Ser, and Tyr residues. We compare the change in the frequency of water-bridged amino acids with that of direct interface contacts. As shown in Figure 4B, there is slight reduction around hydrophobic residues and certain increase around charged/polar residues. The frequency of solvated Tyr interaction dropped significantly. We noted that Gly and Thr are two residues with normal frequencies and relatively high preference to have water bridged interactions, with similar trends for both antibody and non-antibody complexes. It is easy to understand that Gly backbone nitrogen and oxygen atoms are preferred binding sites for bridged water molecules since it has no sidechain atoms. The enrichment of Thr water-bridged interaction could reflect the Cγ methyl group.
Figure 4.
Tyr, Ser, and Asp residues are water bridge hot spots in antibody-protein interactions. (A) antibody amino acid propensities to have water bridged interaction with antigen at 3.5Å cutoff distance. (B) and (C) Change of amino acid frequencies between water bridged interaction and direct interaction for antibody (B) and antigen (C).
Human and murine antibody-antigen recognition
Figure 5 and Sup-Fig2, present the analysis of the dominant recognition features in human and murine antibodies. Tyr and Ser frequencies on human antibody interfaces are 16.1% and 14.7%, respectively (Sup-Figure 2A). In comparison, the corresponding propensities are 20.0% and 11.8% for murine antibodies. Within the 3Å cutoff of strong interactions, the Tyr and Ser propensities on human antibody interfaces are 19.8% and 14.1%, respectively, vs 22.5% and 13.2% for murine antibodies. Overall, murine and human antibodies use similar polar residues (Figure 5, Sup Table 4 and Table 1). To compensate for the smaller percentage of Tyr, human antibodies use more charged groups and hydrophobic residues in antigen interactions. The percentage of charged residues are 19.4 % for human antibody vs 17.4% for murine with 5 Å cutoff; with 3 Å cutoff 35.0% vs 29.7% for human and murine antibodies, respectively. Human antibodies have more hydrophobic contacts at 5 Å cutoff, however, both types of antibodies use similar hydrophobic residues in the 3 Å cutoff. The trends are similar when considering the water bridged interactions (Sup Table 4).
Figure 5.
Human antibodies use less tyrosine and more serine than murine antibody in antigen recognition. Green bar: Non-antibody proteins-protein interactions; red bar: Human antibodies; and blue bar: murine antibodies. (A) and (B) Comparison of human and murine amino acid propensities on antibody (A) and antigen (B) interfaces. (C) Antibody and antigen have opposite residue propensity in comparison with non-antibody related protein-protein interaction. X-axis: Pi (Antibody) - Pi (nonantibody receptor), Y-axis: Pi (Antigen) - Pi (nonantibody ligand). (D) Total amino acid frequencies on the antibody-antigen interfaces follow the same trend of general protein-protein interaction. X-axis: (Pi (Antibody) + Pi (antigen))/2, Y-axis: (Pi (protein receptor) + Pi (protein ligand))/2.
We next compared amino-acids on antigen interfaces for human-, murine-, and non-antibodies PPI. Human antibodies recognize more polar and hydrophobic antigen residues than murine (Figure 5, Sup-Figure 2, and Sup Tables 4 and Table 1). However, non-antibody proteins interact with less charged and polar, but more hydrophobic residues. As can also be seen in Figure 5B, Antigen interfaces have more Lys residues than the non-antibody protein ligands, unlike the low Lys content on antibodies interfaces (Figure1). With already high content of antibody Tyr residues on the interface, the antigen epitopes have relatively lower Tyr residues than non-antibody protein-protein interactions. Other polar residues also have similar complementary trend. While antibody binding region is mostly negatively charged, the antigen interface provides positively charged residues. Figure 5C shows the anticorrelation of residues frequencies on the antibody vs antigen interface for polar residues in relative to their frequencies on normal protein-protein interaction.
The anticorrelation of polar residues on antibody-antigen interfaces leads to invariance of total residue frequencies on antibody/antigen interfaces in comparison to non-antibody protein-protein interactions. All these protein-protein interfaces have similar combined percentages of Arg and Lys. As can be seen in Sup-Figure 2, the amino acid propensities on the human- murine- and non-antibody interfaces are very close, except Leu, Ser, and Tyr residues. While the residue frequencies for antibodies differ from non-antibody proteins, the total frequencies on the antibody-antigen interface correlated well with non-antibody protein-protein interaction (Figure 5D). When including water bridged interactions, the correlation of antibody-antigen interface residues with non-antibody proteins is around R2=0.72. Essentially, even though human antibody, mouse antibody, and non-antibody protein receptors have different residue preferences, the overall protein-protein interaction tendencies follow the same trends.
The global anatomy: residue organization and charge distributions in antibodies fab domains
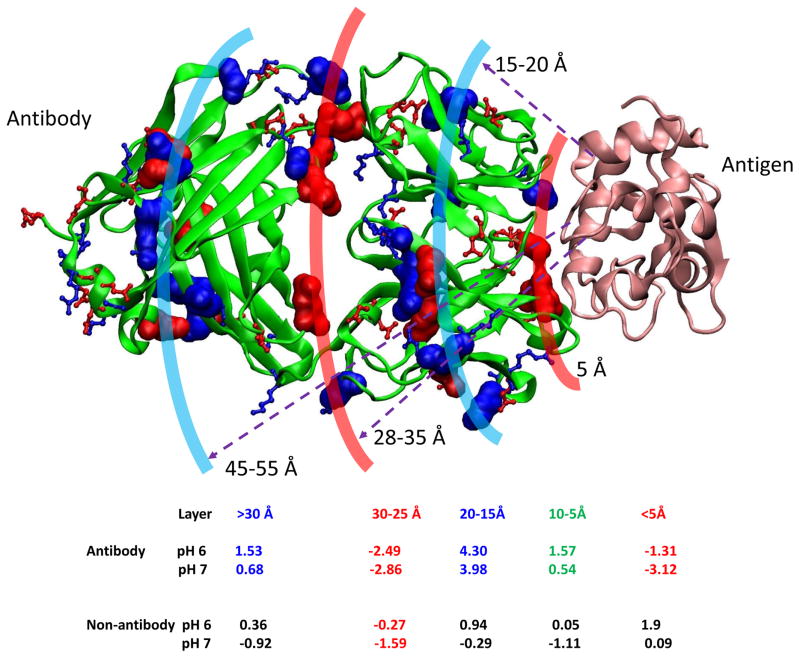
We have shown that antibody-antigen interactions have different charge interaction pattern as compared to non-antibody protein-protein interactions. Therefore, the overall charge distributions in antibodies may affect the electrostatic interactions in antigen recognition. We characterized the net charge distribution based on the radial distance away from antibody-antigen interfaces. In Figure 6, we show the radial distribution of charged residues in Fab domains. The antigen contact layer within 5Å distance to antigen interface is negatively charged at both pH 6 and pH 7. This layer is supported by a positively charged region (5–25 Å), peaking about 20 Å away from the antigen interface. The regions which connect CH1/CL domain become negatively charged again (red color, 30-25 Å to antigen, Figure 6).
Figure 6.
Antibodies have organized charge distributions based on the radial distance away from antigen heavy atoms. Antigen is represented as orange ribbon. The contact layer within 5 Å to antigen is highly negative, followed by positively charged layer. The non-antibody proteins have different distribution pattern.
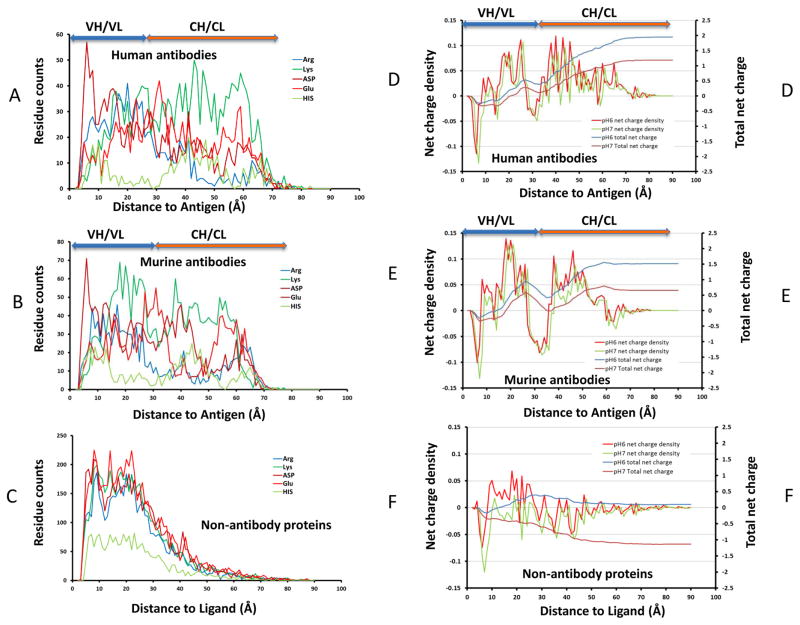
We examine the continuous distributions of the Cα atoms of antibody residues from their shortest distance to antigen Cα atoms. Figure 7 plots these shortest distances and the resulting net-charges distributions. For human and murine antibodies, Asp peaks close to the antigen binding interfaces; Glu around 30 Å, at the J regions connection VH/VL and CH1/CL domains (Figures 7A, 7B). In human and murine antibodies, Arg and His start to populate the structures at around 10 Å away from the antigen. Arg and His are most populated at around 20 Å in the variable and 45 Å in the constant domains, respectively. In human antibodies Lys residues are highly populated at around 45 Å in constant domains, while in murine antibodies it is around 20 Å in the variable domains (Figures 7A and 7B). In non-antibody proteins Asp, Glu, Arg, and Lys all have similar distribution, while His’ population is lower (Figure 7C).
Figure 7.
Global distribution of antibody residues based on their distances from antigen Cα-carbon underlies structural aspect of allosteric communication across Fab domain. (A–C): residue distribution density away from antigen for ionizable residues for human antibody, mouse/rat antibody, and non-antibody proteins, respectively. (D–F) The corresponding charge distributions residues for human antibody, mouse/rat antibody, and non-antibody proteins, respectively.
These distribution patterns of charged residues correspond to the layered net-charge distributions in Figure 6. Figure 7 indicates the net-charge density and total net-charges distributions. The antibody net-charge density measures the net-charge on the semi-spherical surfaces 0–80 Å away from antigen Cα atoms; and the total net-charges distribution is the sum of the total net charge within the volume enclosed by the semi-spherical surfaces. All protein-protein interaction surfaces have a sharp drop of negative charge within 10 Å to antigen/ligand Cα atoms, followed by a positively charged region around 10–15 Å (Figures 7D–7F). However, for non-antibody protein, the positive charge region is pH sensitive and it almost disappears at pH7. Except the brief dip into negative charge around 15 Å, the charges for antibody proteins in the range of 10–25 Å are mostly positive (Figures 7D and 7E), with Lys residues contribute the most for murine antibodies (Figures 7B and 7E). As can be seen in Figures 7D and 7E, after a broad negative charged joint region, the antibody constant regions are dominated by positive charge. Overall, the cumulative charge for antibody structures studied here appears positive at both pH6 and pH7. The total charges for non-antibody proteins appear close to neutral at pH7 and negative at pH6. Consistent with distributions of the charged residues, other non-charged residues also have the radial distribution pattern different from non-antibody protein-protein interactions (Sup-Figure 3).
Germline VDJ recombination suggests that the antibodies may be mostly positively charged
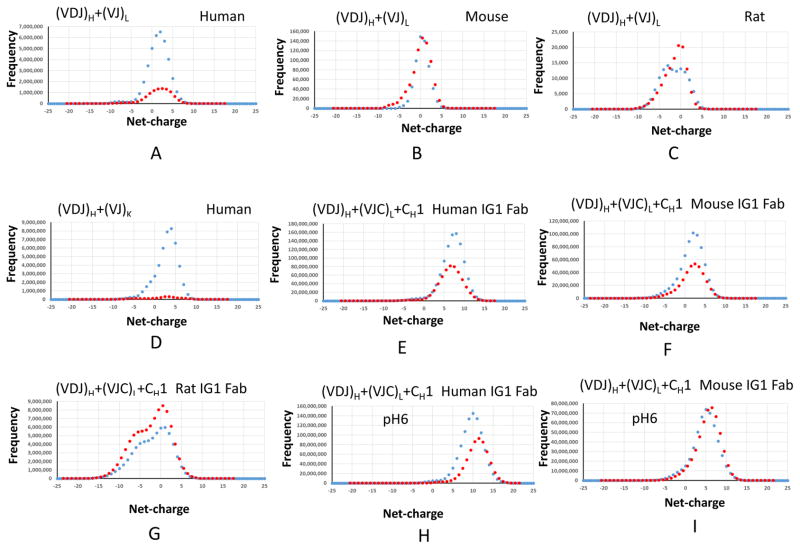
The organized three dimensional charged distributions of antibody Fab domains prompted us to examine possible net-charges carried on the antibody. Using the IMGT IG “V-REGION”, “D-REGION”, “J-REGION”, “C-GENE exon” sets, we calculated all possible antibody net-charges from exhaustive VDJ recombination of both heavy and light chain variable regions for human, mouse, and rat. We initially used 0.1 increments to search the charge distributions. The net-charge distributions are not monotonic. Even with 0.5 increments, we still see two distribution curves for VDJ recombination (Figure 8). Within each curve, there is a regular ±1 space between points. Therefore, the difference curves could reflect different His combinations. The peak for the antibody net-charge distribution of human VDJ combinations is around +2 (Figure 8A), and the distributions span from −20 to +12. Mouse antibodies appear to have narrower charge distribution, ranging from −9.5 to +7.5 and peaking at neutral state (zero charged) (Figure 8B). Rat antibodies appear to have less variability, with peaks at similar points as the mouse antibodies (Figure 8C).
Figure 8.
Germline VDJ recombination of antibody indicates that the antibodies are mostly positively charged.
Compared with the L-chain, K-chain combinations provided more positive charges. The VDJ recombination from H and K chains has the peak at +4 (Figure 8D), more positive than the VDJ recombination from H/L chains. Histidine residues have smaller effect on the charge distributions in the H/K than the H/L combinations, which can be seen from the larger deviation of red and blue lines in Figure 8D. For human antibody genes, since the constant regions of kappa/lambda light chain and CH1 domain are all positively charged, the charge distributions in antibody Fabs shift further to positive charged regions (Figure 8E). The constant regions of kappa/lambda light chain of both mouse and rat are negatively charged. However, their CH1 domains can be either positively or negatively charged. Thus, the constant regions make their charge distributions much broader than in the variable regions (Figures 8F and 8G). Under more acidic conditions (pH=6), the human antibody charge peak shifts to +10 (Figure 8H), and mouse has dominant histidine contributions.
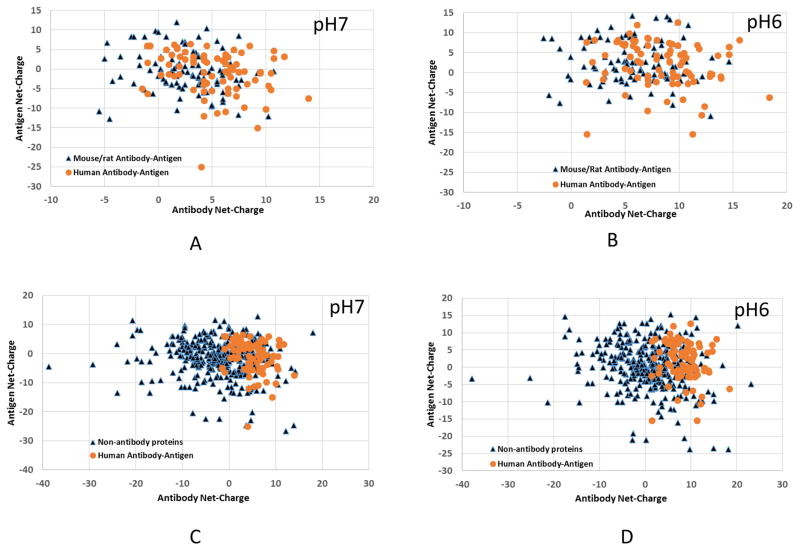
Finally, human antibody structures appear more positively charged than the mouse/rat (Figure 9), in line with the trends from VDJ recombination (Figure 9). Antigens binding to mouse/rat antibodies appear more negatively charged (67 negatively charged vs 44 positively charged), and the human antibodies have about equal number of positively or negatively charged antigens (42 negatively charged vs 40 positively charged). There are only a few negatively charged human antibodies at pH 7, and all human Fabs in our dataset become positively charge at pH 6. The overall trend appears to be that positively charged antibodies recognize negatively charged antigens. Non-antibody protein dataset shows no correlation at both pH 7 and pH 6 (Figure 9C and 9D). Thus, overall, it is still unclear whether the overall net-charge of an antibody is correlated with its antigen binding specificity or affinity.
Figure 9.
Protein net-charge distributions for antibody-antigen complex and non-antibody protein complex indicate that human antibody have more charge interaction correlation.
Discussion and Conclusions
Are there unique sequence and structural characteristics of antibody-protein antigen interactions, as compared with general non-antibody protein-protein interaction that would help in antibody design? We found that the total amino acid percentages are similar for antibody-antigen and non-antibody protein-protein interfaces, indicating all protein-protein interaction may follow similar physico-chemical principles. However, the amino acid preferences on antibody paratopes differ from antigen epitopes and other non-antibody proteins, and the antibody-antigen preference could be entropy driven. Protein conformational dynamic is important in protein-protein interaction, permitting promiscuity and specificity5, 7, 31, 44–47. Protein complex formation leads to a redistribution of dynamics48, 49, but does not restrict the conformational freedom of the partner proteins50. Antibody-antigen recognition, which is associated with structural transitions through inherent conformational flexibility51–53, involves conformational selection54. In terms of side-chain conformational entropy, protein-protein interfacial regions are often less flexible than the rest of the surface55. The antibody appears to further reduce the side-chain conformational entropy by amino acids with shorter side-chains.
We confirmed previous finding that that Tyr and Ser are the two most enriched amino acids. However, human antibodies have less Tyr and more Ser residues than mouse. In addition, Asp is also preferred and Lys is avoided to interact with charged and polar antigens. Enrichment of Tyr as hot spot in antibody-antigen interaction was thought to be evolution driven16. Previous work found that antibody tyrosyl side chains recognize backbone atoms and side-chain carbons1. Our current results reveal that the primary role of Tyr is forming hydrogen bonds with polar and charged residues, with less water bridged interactions. The sidechain entropy cost for Tyr interaction is 0.99 kcal/mol at 300K, much smaller than that for Arg (2.13 kcal/mol) and Lys (2.21 kcal/mol). Therefore, a thermodynamic reason for highly enriched Tyr residues in antibody recognition could be entropy reduction. Recently, it has been shown that there is a strong enrichment of aromatic residues W, Y and F in rcSso7d-based binders, suggesting that the rigidity of this amino acid may mimic the energetic core of antibody paratopes56.
In our study of water-bridged protein-protein interactions, we found that antibody and non-antibody proteins generally follow similar trends, with water-bridged interactions co-existing with direct protein-protein interactions, except that Tyr has less water-bridged interactions in antibody-antigen recognition. Still, both antibody and non-antibody proteins have more water-bridged interactions involving Gly and Thr, comparing with amino acid propensities in direct protein-protein interactions. Water interactions can be considered in antibody design.
To regulate the immune response, antibody-antigen interaction should send a signal to allosteric sites for complement activation and Fc receptors binding. Signal transduction pathways could depend on sequences of the variable regions and allotypes, possibly through hydrogen bonding network, electrostatic interactions, resulting from population shift of dynamic conformations57. The CH1 region affects antigen and other constant regions, most likely through interactions with its neighboring CL domain as well as by transmitting structural and dynamics perturbations to the Ig hinge region and back to the VH domain58. We observed apparent organized residue distributions from the antigen binding site to constant regions, which may provide the structural basis for allosteric signaling. While non-antibody proteins have randomized residue distributions from the binding site, antibodies may have distinct residue distribution patterns of the distances from the antigen binding site. For example, there are three peaks of Tyr residues on the antigen binding surface, about 18 Å away from the binding site on the variable regions, and around 40 A on the constant regions. Gln has the highest percentage around 20 Å away from binding site (VH/VL region) and Val appears mostly at 50 Å away in the CH/CL region (Sup-Figure 3). These patterns reflect conservation of specific residues that are the determinants in the folding and assembly of antibody structure59, as well as other antibody functions.
Electrostatic interactions are important for local interaction and long range signal transduction. Coupled with protein dynamics modulated by antigen binding, organized distributions of charged residues could play a role in communication. This could be the case in HIV, where the net positive charge acts in regulating sensitivity to humoral immunity 60 and broadly neutralizing antibodies of HIV-1 variants61. Several protein-protein complexes are directly controlled by electrostatic attraction. For example, the change in net charge of calmodulin from approximately −5 at pH 4.5 to −15 at pH 7.5 leaves the binding constant with positively charged peptides virtually unchanged62. In our non-antibody dataset, the complex of the gating domain of a Ca2+-activated K+ channel with Ca2+/calmodulin also has strong electrostatic attraction (18 and −23.8 at pH 6, pdb:1G4Y)63. Other examples of protein complexes with strong electrostatic attraction are ribonuclease inhibitor (1a4y and 2q4g). However, the role of electrostatic interaction could be largely to discriminate against unbound proteins rather than to increase the affinity for any target protein62.
Proteins have no net charge at their Isoelectric points (pI). Most of the proteins’ pI values are not near physiological pH (around pH 7). Our previous study suggested that ‘proper’ populations of negatively and positively charged proteins are important for maintaining a protein-protein interaction network in a proteome64. The universal distributions of the proteins’ pI values in the proteomes for all organisms have two peaks, one set for acidic proteins and another for basic proteins, probably due to amino acid pKa values65, 66 or organism function36, 37, 67. The relative abundance of acidic and basic proteins differs among species. While small proteomes tend to have more basic proteins36, 37, most human proteins are negatively charged at pH 7. In this sense, it is not surprising that most antibodies are positively charged, which might enable antibody proteins to approach negative charged antigens through long range electrostatic attraction. In phage display selection of specific antibodies, there is an increased average positive charge among selected proteins in comparison to proteins that are harbored in the library before selection68.
In conclusion, our study revealed features of antibody-antigen recognition in the interface as well as in the whole Fab domain. To recognize an unlimited number of possible antigens, antibody complementarity-determining regions need to be flexible with higher conformational entropy. However, antibodies appear to select amino acids with low side-chain entropy. Therefore, amino acid preference in antibody-protein antigen recognition can be entropy-driven, with large percentages of Tyr, Ser, and Asp, but little Lys. With unique chemistry, Tyr interacts with diverse antigen residues, especially negatively charged Asp and Glu. Electrostatic and hydrophobic interactions in the Ag binding sites might be coupled with Fab domains through organized charge and residue distributions away from the binding interfaces. Our comprehensive global and local analysis provides clues and highlights the challenge in antibody design, such as minimizing Lys in CDR regions. Humanized mouse antibodies appear to have more Lys residues in their constant domain than in their variable domain.
Supplementary Material
Acknowledgments
We thank the financial support from NCI, NIH, under contract number HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Computations were performed at the NIH biowulf supercomputer cluster. We thank Mr. Mingzhen Zhang for helpful discussion.
References
- 1.Peng HP, Lee KH, Jian JW, Yang AS. Origins of specificity and affinity in antibody–protein interactions. Proceedings of the National Academy of Sciences. 2014;111:E2656–E2665. doi: 10.1073/pnas.1401131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimitrov DS. Therapeutic antibodies, vaccines and antibodyomes. MAbs. 2010;2:347–56. doi: 10.4161/mabs.2.3.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O’Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Program NCS, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, Georgiev IS, Chuang GY, Longo NS, Lynch RM, Saunders KO, Soto C, Srivatsan S, Yang Y, Bailer RT, Louder MK, Program NCS, Mullikin JC, Connors M, Kwong PD, Mascola JR, Shapiro L. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–85. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of Protein-Protein Interactions: What are the Preferred Ways For Proteins To Interact? Chem Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 6.Keskin O, Ma B, Rogale K, Gunasekaran K, Nussinov R. Protein-protein interactions: organization, cooperativity and mapping in a bottom-up Systems Biology approach. Phys Biol. 2005;2:S24–35. doi: 10.1088/1478-3975/2/2/S03. [DOI] [PubMed] [Google Scholar]
- 7.Wei G, Xi W, Nussinov R, Ma B. Protein Ensembles: How Does Nature Harness Thermodynamic Fluctuations for Life? The Diverse Functional Roles of Conformational Ensembles in the Cell. Chem Rev. 2016 doi: 10.1021/acs.chemrev.5b00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehermayr C, Mahler HC, Mader K, Fischer S. Assessment of net charge and protein-protein interactions of different monoclonal antibodies. J Pharm Sci. 2011;100:2551–62. doi: 10.1002/jps.22506. [DOI] [PubMed] [Google Scholar]
- 9.Brooks BD. The importance of epitope binning for biological drug discovery. Curr Drug Discov Technol. 2014;11:109–12. doi: 10.2174/1570163810666131124233827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JY, Lee HT, Shin W, Chae J, Choi J, Kim SH, Lim H, Won Heo T, Park KY, Lee YJ, Ryu SE, Son JY, Lee JU, Heo YS. Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy. Nat Commun. 2016;7:13354. doi: 10.1038/ncomms13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9:767–74. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 12.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panza F, Logroscino G, Imbimbo BP, Solfrizzi V. Is there still any hope for amyloid-based immunotherapy for Alzheimer’s disease? Curr Opin Psychiatry. 2014;27:128–37. doi: 10.1097/YCO.0000000000000041. [DOI] [PubMed] [Google Scholar]
- 14.Wood LB, Winslow AR, Strasser SD. Systems biology of neurodegenerative diseases. Integrative Biology. 2015;7:758–775. doi: 10.1039/c5ib00031a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J Mol Biol. 1996;262:732–45. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 16.Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci U S A. 2003;100:5772–7. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams JJ, Sidhu SS. Synthetic antibody technologies. Curr Opin Struct Biol. 2014;24:1–9. doi: 10.1016/j.sbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Hackel BJ, Ackerman ME, Howland SW, Wittrup KD. Stability and CDR composition biases enrich binder functionality landscapes. J Mol Biol. 2010;401:84–96. doi: 10.1016/j.jmb.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponomarenko JV, Bourne PE. Antibody-protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct Biol. 2007;7:64. doi: 10.1186/1472-6807-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramaraj T, Angel T, Dratz EA, Jesaitis AJ, Mumey B. Antigen-antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures. Biochim Biophys Acta. 2012;1824:520–32. doi: 10.1016/j.bbapap.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kringelum JV, Nielsen M, Padkjaer SB, Lund O. Structural analysis of B-cell epitopes in antibody:protein complexes. Mol Immunol. 2013;53:24–34. doi: 10.1016/j.molimm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed MH, Spyrakis F, Cozzini P, Tripathi PK, Mozzarelli A, Scarsdale JN, Safo MA, Kellogg GE. Bound water at protein-protein interfaces: partners, roles and hydrophobic bubbles as a conserved motif. PLoS One. 2011;6:e24712. doi: 10.1371/journal.pone.0024712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong S, Kim D. Interaction between bound water molecules and local protein structures: A statistical analysis of the hydrogen bond structures around bound water molecules. Proteins. 2016;84:43–51. doi: 10.1002/prot.24953. [DOI] [PubMed] [Google Scholar]
- 24.Bellissent-Funel MC, Hassanali A, Havenith M, Henchman R, Pohl P, Sterpone F, van der Spoel D, Xu Y, Garcia AE. Water Determines the Structure and Dynamics of Proteins. Chem Rev. 2016 doi: 10.1021/acs.chemrev.5b00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reichmann D, Phillip Y, Carmi A, Schreiber G. On the contribution of water-mediated interactions to protein-complex stability. Biochemistry. 2008;47:1051–60. doi: 10.1021/bi7019639. [DOI] [PubMed] [Google Scholar]
- 26.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 27.Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016;6:35297. doi: 10.1038/srep35297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apgar JR, Mader M, Agostinelli R, Benard S, Bialek P, Johnson M, Gao Y, Krebs M, Owens J, Parris K, St Andre M, Svenson K, Morris C, Tchistiakova L. Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody. MAbs. 2016;8:1302–1318. doi: 10.1080/19420862.2016.1215786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parren PW, Burton DR, Bradbury A, Huston JS, Carter PJ, Veldman T, Chester KA, Larrick JW, Alfenito MR, Scott JK, Weiner LM, Adams GP, Reichert JM. Antibody Engineering & Therapeutics 2015: The Antibody Society’s annual meeting December 7–10, 2015, San Diego, CA. MAbs. 2015;7:981–8. doi: 10.1080/19420862.2015.1089707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–43. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 31.Ma B, Tsai CJ, Haliloglu T, Nussinov R. Dynamic allostery: linkers are not merely flexible. Structure. 2011;19:907–17. doi: 10.1016/j.str.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pritsch O, Hudry-Clergeon G, Buckle M, Petillot Y, Bouvet JP, Gagnon J, Dighiero G. Can immunoglobulin C(H)1 constant region domain modulate antigen binding affinity of antibodies? J Clin Invest. 1996;98:2235–43. doi: 10.1172/JCI119033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janda A, Casadevall A. Circular Dichroism reveals evidence of coupling between immunoglobulin constant and variable region secondary structure. Mol Immunol. 2010;47:1421–5. doi: 10.1016/j.molimm.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tudor D, Yu H, Maupetit J, Drillet AS, Bouceba T, Schwartz-Cornil I, Lopalco L, Tuffery P, Bomsel M. Isotype modulates epitope specificity, affinity, and antiviral activities of anti-HIV-1 human broadly neutralizing 2F5 antibody. Proc Natl Acad Sci U S A. 2012;109:12680–5. doi: 10.1073/pnas.1200024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sela-Culang I, Alon S, Ofran Y. A systematic comparison of free and bound antibodies reveals binding-related conformational changes. J Immunol. 2012;189:4890–9. doi: 10.4049/jimmunol.1201493. [DOI] [PubMed] [Google Scholar]
- 36.Kiraga J, Mackiewicz P, Mackiewicz D, Kowalczuk M, Biecek P, Polak N, Smolarczyk K, Dudek MR, Cebrat S. The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms. BMC Genomics. 2007;8:163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight CG, Kassen R, Hebestreit H, Rainey PB. Global analysis of predicted proteomes: functional adaptation of physical properties. Proc Natl Acad Sci U S A. 2004;101:8390–5. doi: 10.1073/pnas.0307270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kastritis PL, Moal IH, Hwang H, Weng Z, Bates PA, Bonvin AM, Janin J. A structure-based benchmark for protein-protein binding affinity. Protein Sci. 2011;20:482–91. doi: 10.1002/pro.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 40.Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem. 2009;284:13285–9. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradley J, O’Meara F, Farrell D, Nielsen JE. Highly perturbed pKa values in the unfolded state of hen egg white lysozyme. Biophys J. 2012;102:1636–45. doi: 10.1016/j.bpj.2012.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein Sci. 2006;15:1214–8. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–61. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nussinov R, Ma B, Tsai CJ. Multiple conformational selection and induced fit events take place in allosteric propagation. Biophys Chem. 2014;186:22–30. doi: 10.1016/j.bpc.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai D, Huang Q, Nussinov R, Ma B. Promiscuous and specific recognition among ephrins and Eph receptors. Biochim Biophys Acta. 2014;1844:1729–40. doi: 10.1016/j.bbapap.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai CJ, Ma B, Nussinov R. Protein-protein interaction networks: how can a hub protein bind so many different partners? Trends Biochem Sci. 2009;34:594–600. doi: 10.1016/j.tibs.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci. 2002;11:184–97. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haliloglu T, Keskin O, Ma B, Nussinov R. How similar are protein folding and protein binding nuclei? Examination of vibrational motions of energy hot spots and conserved residues. Biophys J. 2005;88:1552–9. doi: 10.1529/biophysj.104.051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma B, Tsai CJ, Nussinov R. A systematic study of the vibrational free energies of polypeptides in folded and random states. Biophys J. 2000;79:2739–53. doi: 10.1016/S0006-3495(00)76513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grunberg R, Nilges M, Leckner J. Flexibility and conformational entropy in protein-protein binding. Structure. 2006;14:683–93. doi: 10.1016/j.str.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 51.Keskin O. Binding induced conformational changes of proteins correlate with their intrinsic fluctuations: a case study of antibodies. BMC Struct Biol. 2007;7:31. doi: 10.1186/1472-6807-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thielges MC, Zimmermann, Yu W, Oda M, Romesberg FE. Exploring the energy landscape of antibody– antigen complexes: protein dynamics, flexibility, and molecular recognition. Biochemistry. 2008;47:7237–7247. doi: 10.1021/bi800374q. [DOI] [PubMed] [Google Scholar]
- 53.Li T, Tracka MB, Uddin S, Casas-Finet J, Jacobs DJ, Livesay DR. Redistribution of flexibility in stabilizing antibody fragment mutants follows Le Chatelier’s principle. PLoS One. 2014;9:e92870. doi: 10.1371/journal.pone.0092870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma B, Zhao J, Nussinov R. Conformational selection in amyloid-based immunotherapy: Survey of crystal structures of antibody-amyloid complexes. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbagen.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cole C, Warwicker J. Side-chain conformational entropy at protein-protein interfaces. Protein Sci. 2002;11:2860–70. doi: 10.1110/ps.0222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traxlmayr MW, Kiefer JD, Srinivas RR, Lobner E, Tisdale AW, Mehta NK, Yang NJ, Tidor B, Wittrup KD. Strong Enrichment of Aromatic Residues in Binding Sites from a Charge-Neutralized Hyperthermostable Sso7d Scaffold Library. J Biol Chem. 2016 doi: 10.1074/jbc.M116.741314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srivastava A, Tracka MB, Uddin S, Casas-Finet J, Livesay DR, Jacobs DJ. Mutations in Antibody Fragments Modulate Allosteric Response Via Hydrogen-Bond Network Fluctuations. Biophys J. 2016;110:1933–42. doi: 10.1016/j.bpj.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janda A, Bowen A, Greenspan NS, Casadevall A. Ig Constant Region Effects on Variable Region Structure and Function. Front Microbiol. 2016;7:22. doi: 10.3389/fmicb.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campion SR. Conserved aromatic residues as determinants in the folding and assembly of immunoglobulin variable domains. Mol Immunol. 2016;70:63–71. doi: 10.1016/j.molimm.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 60.Naganawa S, Yokoyama M, Shiino T, Suzuki T, Ishigatsubo Y, Ueda A, Shirai A, Takeno M, Hayakawa S, Sato S, Tochikubo O, Kiyoura S, Sawada K, Ikegami T, Kanda T, Kitamura K, Sato H. Net positive charge of HIV-1 CRF01_AE V3 sequence regulates viral sensitivity to humoral immunity. PLoS One. 2008;3:e3206. doi: 10.1371/journal.pone.0003206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borggren M, Repits J, Sterjovski J, Uchtenhagen H, Churchill MJ, Karlsson A, Albert J, Achour A, Gorry PR, Fenyo EM, Jansson M. Increased sensitivity to broadly neutralizing antibodies of end-stage disease R5 HIV-1 correlates with evolution in Env glycosylation and charge. PLoS One. 2011;6:e20135. doi: 10.1371/journal.pone.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andre I, Kesvatera T, Jonsson B, Akerfeldt KS, Linse S. The role of electrostatic interactions in calmodulin-peptide complex formation. Biophys J. 2004;87:1929–38. doi: 10.1529/biophysj.104.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–4. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 64.Xu Y, Wang H, Nussinov R, Ma B. Protein charge and mass contribute to the spatio-temporal dynamics of protein-protein interactions in a minimal proteome. Proteomics. 2013 doi: 10.1002/pmic.201100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiller GF, Caraux G, Sylvester N. The modal distribution of protein isoelectric points reflects amino acid properties rather than sequence evolution. Proteomics. 2004;4:943–9. doi: 10.1002/pmic.200200648. [DOI] [PubMed] [Google Scholar]
- 66.Wu S, Wan P, Li J, Li D, Zhu Y, He F. Multi-modality of pI distribution in whole proteome. Proteomics. 2006;6:449–55. doi: 10.1002/pmic.200500221. [DOI] [PubMed] [Google Scholar]
- 67.Schwartz R, Ting CS, King J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11:703–9. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- 68.Persson H, Persson J, Danielsson L, Ohlin M. Charges drive selection of specific antibodies by phage display. J Immunol Methods. 2010;353:24–30. doi: 10.1016/j.jim.2009.11.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.