Abstract
Purpose
Advanced anaplastic lymphoma kinase (ALK) fusion-positive non–small-cell lung cancers (NSCLCs) are effectively treated with ALK tyrosine kinase inhibitors (TKIs). However, clinical outcomes in these patients vary, and the benefit of TKIs is limited as a result of acquired resistance. Emerging data suggest that the ALK fusion variant may affect clinical outcome, but the molecular basis for this association is unknown.
Patients and Methods
We identified 129 patients with ALK-positive NSCLC with known ALK variants. ALK resistance mutations and clinical outcomes on ALK TKIs were retrospectively evaluated according to ALK variant. A Foundation Medicine data set of 577 patients with ALK-positive NSCLC was also examined.
Results
The most frequent ALK variants were EML4-ALK variant 1 in 55 patients (43%) and variant 3 in 51 patients (40%). We analyzed 77 tumor biopsy specimens from patients with variants 1 and 3 who had progressed on an ALK TKI. ALK resistance mutations were significantly more common in variant 3 than in variant 1 (57% v 30%; P = .023). In particular, ALK G1202R was more common in variant 3 than in variant 1 (32% v 0%; P < .001). Analysis of the Foundation Medicine database revealed similar associations of variant 3 with ALK resistance mutation and with G1202R (P = .010 and .015, respectively). Among patients treated with the third-generation ALK TKI lorlatinib, variant 3 was associated with a significantly longer progression-free survival than variant 1 (hazard ratio, 0.31; 95% CI, 0.12 to 0.79; P = .011).
Conclusion
Specific ALK variants may be associated with the development of ALK resistance mutations, particularly G1202R, and provide a molecular link between variant and clinical outcome. ALK variant thus represents a potentially important factor in the selection of next-generation ALK inhibitors.
INTRODUCTION
Anaplastic lymphoma kinase (ALK) gene rearrangements encode driver fusion oncoproteins and are found in approximately 5% of non–small-cell lung cancers (NSCLCs).1 Since crizotinib,2-5 multiple second-generation (eg, ceritinib, alectinib, brigatinib)6-14 and third-generation (eg, lorlatinib)15 ALK tyrosine kinase inhibitors (TKIs) have been developed for patients with ALK-positive NSCLC, all with higher potency and greater CNS penetration than crizotinib. Although these ALK inhibitors have dramatically expanded the therapeutic landscape of ALK-positive NSCLC, clinical outcomes in patients can vary widely, and the biologic mechanisms that underpin such heterogeneous outcomes are unknown. Moreover, at some point, essentially all patients experience a relapse while receiving TKI therapy as a result of acquired drug resistance.1,4,5 The elucidation of resistance mechanisms in patients has proven critical in efforts to rationally select subsequent therapies.1,16
Emerging data indicate that ALK fusion variants may have biologic and clinical implications in ALK-positive lung cancer. The predominant ALK fusion partner in NSCLC is the echinoderm microtubule–associated protein-like 4 (EML4) gene.17,18 Among > 15 EML4-ALK variants identified to date, the most common are variant 1 (v1; exon 13 of EML4 fused to exon 20 of ALK [E13;A20]) and v3a/b (exon 6a/b of EML4 fused to exon 20 of ALK [E6a/b;A20]).17,19-22 All variants retain the entire tyrosine kinase domain of ALK and the N-terminal coiled-coil region of EML4, which is necessary and sufficient for the dimerization and constitutive activation of ALK.17 Different variants may have different protein stabilities, which affects sensitivity to crizotinib in vitro.23-26
Recent studies have suggested differential responses to crizotinib according to ALK variant in patients.27,28 For example, longer responses to crizotinib were reported with v1 compared with non-v127 or with non-v3 compared with v3,28 yet two other studies found no difference in clinical response to crizotinib on the basis of ALK variant, which highlights the need for additional investigation.29,30 Moreover, the potential effect of ALK variants on the efficacy of next-generation ALK TKIs or the development of resistance mechanisms, which can influence responses to subsequent therapies, has not been examined. We evaluated the frequency and spectrum of ALK resistance mutations according to fusion variant in patients with ALK-positive NSCLC with acquired TKI resistance and clinical outcomes of these patients who received various generations of ALK inhibitors.
PATIENTS AND METHODS
Study Population
Between January 2008 and January 2017, 129 patients with ALK-positive NSCLC and known ALK variant were identified at the Massachusetts General Hospital (MGH; n = 113) and University of California, Irvine (n = 16; Data Supplement). The study was approved by the institutional review boards (IRBs) at each site.
In addition, a separate group of 577 patients with ALK-positive NSCLC and known ALK variant identified during routine clinical care from August 2012 to December 2016 with FoundationOne next-generation sequencing (NGS) assays at Foundation Medicine were analyzed for the frequency and distribution of ALK resistance mutations. Approval for the study of this cohort was obtained from the Western IRB (protocol no. 20152817).
Data Collection
For the 129 patients included in the main study cohort, data on clinicopathologic features and treatment histories were extracted from medical records. Progression-free survival (PFS) and overall survival (OS) outcomes were measured as detailed in the Data Supplement. Data were updated as of November 15, 2017.
Identification of ALK Variant
ALK fusion variants were detected by using the MGH fusion panel, which uses targeted RNA sequencing with anchored multiplex polymerase chain reaction (PCR) to detect fusion transcripts that involve known oncogenes, including ALK31; the FoundationOne platform32; targeted NGS platforms at outside institutions as previously described33,34; or reverse transcription PCR (commercial or as previously described35) and Sanger sequencing of cDNA (Data Supplement).
Genotyping for ALK Resistance Mutation
Postprogression tumor biopsy specimens were analyzed for the presence of ALK resistance mutations under an IRB-approved tissue collection protocol. Methodologies to detect ALK resistance mutations included the MGH SNaPshot NGS platform (which uses anchored PCR to detect single-nucleotide variants and insertions/deletions in cancer-related genes, including ALK)31, the FoundationOne NGS32, and the OncoPanel NGS.33 A subset of specimens underwent Sanger sequencing of cDNA for the entire ALK domain,36 and one specimen underwent whole-exome sequencing as previously reported.16
Statistical Analysis
Detailed statistical methods are provided in the Data Supplement. PFS and OS curves were estimated using the Kaplan-Meier method, and the Cox proportional hazards regression model was used to estimate the hazard ratio (HR) to express PFS and OS differences between variant groups. All statistical analyses were performed with SAS 9.4 software (SAS Institute, Cary, NC).
RESULTS
Patient Characteristics
Baseline patient characteristics are listed in Table 1. The majority of patients received crizotinib (121 [94%] of 129)—67 (52%) as first-line, 31 (24%) as second-line, and 23 (18%) as third-line or higher-line treatment. Over the course of their disease, 45 patients (35%) received two ALK inhibitors, and 56 (43%) received three or more ALK inhibitors.
Table 1.
Clinical Characteristics According to ALK Variant
Among the 129 patients, 123 (95%) had an EML4-ALK fusion. The most frequent EML4-ALK variants were v1 in 55 patients (43%), and v3 in 51 patients (40%; Fig 1). No differences were found in clinicopathologic features between patients with v1 and v3 (Table 1). The remaining EML4-ALK fusions consisted of v2 (6% [E20;A20]), v5′ (4% [E18;A20]), v5 (2% [E2;A20]), and v7 (1% [E14;A20]). Among non–EML4-ALK fusions detected in six patients (5%), the fusion partner genes included HIP1 (n = 3),37-39 KIF5B (n = 1),40 PRKAR1A (n = 1),41 and MTA3 (n = 1; not previously reported). Comparable baseline characteristics also were observed across these variant groups (Data Supplement).
Fig 1.
Frequency of ALK variants in the study cohort (N = 129). Schematic key: blue, coiled-coil region of EML4; gold, tandem atypical propeller EML domain of EML4; gray, tyrosine kinase domain of ALK. Note that v3 and v5 exist as isoforms (v3a and v3b and v5a and v5b, respectively) generated by alternative splicing.24 A, ALK exon; E, EML4 exon; v, variant.
ALK Resistance Mutations by Variant
To determine whether the ALK variant affects the development of molecular mechanisms of resistance, we identified patients in the study cohort who underwent a repeat biopsy after progression on a first- or second-generation ALK inhibitor. Seventy-seven patients (60%) had a postprogression tumor biopsy. Of these, 12 had two repeat biopsies, and two underwent three serial biopsies (Data Supplement). A total of 93 tumor biopsies were performed. The Data Supplement shows the distribution of post-TKI biopsies according to ALK fusion variant and timing of biopsy.
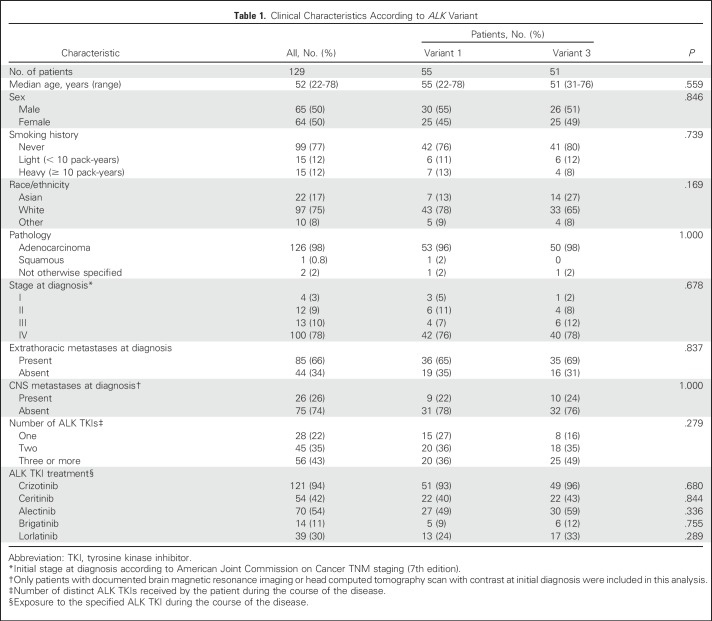
Because the number of patients with non-v1 and/or -v3 was small (Data Supplement; Fig 1), the most common variant groups, v1 and v3, were selected for additional analysis. Overall, 33 v1 and 44 v3 repeat biopsies were performed after progression on an ALK inhibitor. ALK resistance mutations were identified in 10 patients with v1 (30%) compared with 25 with v3 (57%; P = .023; Fig 2). Of note, the ALK G1202R solvent-front mutation, which causes steric interference with drug binding16,36,42 and confers high-level resistance to first- and second-generation ALK inhibitors,16 was detected in zero (0%) of 33 patients with v1 versus 14 (32%) of 44 with v3 (P < .001).
Fig 2.
ALK resistance mutations in tumor biopsy specimens obtained after progression on an ALK inhibitor according to EML4-ALK variant.
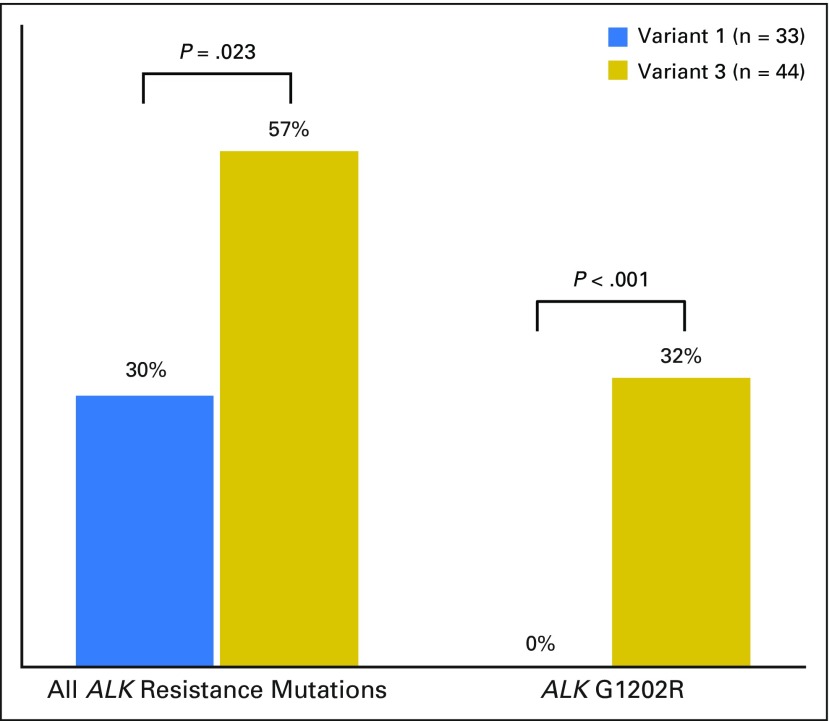
Our group has previously shown that ALK resistance mutations as a whole and ALK G1202R specifically occur more frequently after second-generation ALK TKIs (50% to 60% and 20% to 40%, respectively) compared with crizotinib (20% to 30% and 2%, respectively).16 To address imbalances in postcrizotinib versus post–second-generation ALK TKI samples that could potentially confound the comparison of resistance mutations in v1 versus v3, we focused on the distribution of ALK mutations in biopsy specimens obtained after progression on a second-generation ALK inhibitor. ALK resistance mutations were more common in v3 (21 [66%] of 32) than v1 (eight [42%] of 19), although this difference was not statistically significant (P = .145). ALK G1202R was significantly more common in v3 (14 [44%] of 32) than in v1 (zero [0%] of 19; P = .001; Fig 3). Of note, no statistically significant difference was found between v1 and v3 in the prebiopsy second-generation ALK inhibitor administered, although more v3 than v1 specimens were postalectinib biopsies. Similarly, the cumulative number of prior TKIs was not significantly different (Data Supplement). The sequencing methods used to detect an ALK resistance mutation for v1 and v3 are shown in the Data Supplement.
Fig 3.
Distribution of ALK resistance mutations in tumor biopsy specimens obtained after disease progression on a second-generation ALK inhibitor by EML4-ALK variant. WT, wild-type (nonmutated) ALK.
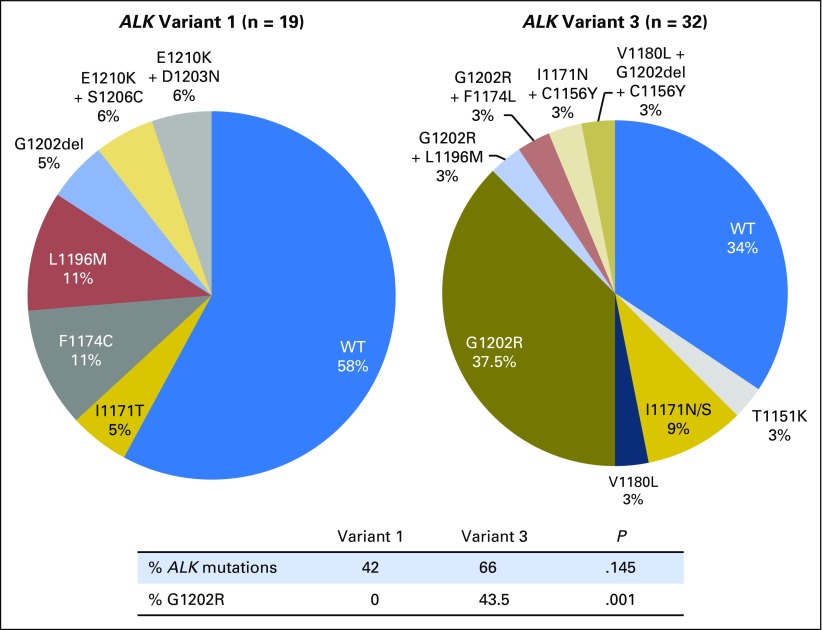
Among postcrizotinib tumor biopsy specimens, ALK resistance mutations were detected in two (14%) of 14 samples of v1 and four (33%) of 12 samples of v3 (P = .365; Fig 4). None of the samples harbored an ALK G1202R mutation. The overall lower frequency of ALK resistance mutations (23%; P = .007) and specifically of ALK G1202R (0%; P = .002) after crizotinib versus a second-generation ALK inhibitor (57% and 27%, respectively) is consistent with our previous report.16
Fig 4.
Distribution of ALK resistance mutations in tumor biopsy specimens obtained after disease progression on crizotinib by EML4-ALK variant. WT, wild-type ALK.
Analysis of 577 ALK-Positive NSCLCs
To validate the associations between variant and ALK resistance mutations, we subsequently examined a database of ALK-positive NSCLCs with known ALK variants sequenced at Foundation Medicine. Among 577 patients with ALK-positive NSCLC, the most common variants were EML4-ALK v3 (n = 186 [32%]) and v1 (n = 182 [32%]) followed by v2 (n = 47 [8%]), other EML4-ALK variants (n = 69 [12%]), and non–EML4-ALK variants (n = 93 [16%]; Data Supplement). A total of 624 tumor tissue biopsy specimens were taken from these patients (v3, n = 201; v1, n = 199; v2, n = 49; other EML4-ALK, n = 74; non–EML4-ALK, n = 101). Although the frequencies of EML4-ALK v1 and v3 were almost identical, ALK resistance mutations (n = 30) and specifically ALK G1202R (n = 9) were significantly more common for v3 than for v1 (ALK mutation, 8% v 2% [P = .010]; G1202R, 3.5% v 0% [P = .015]; Data Supplement). Of note, clinical information, including prebiopsy treatment history, was not available for the patients included in the Foundation Medicine data set, and therefore, the distribution of patients who were treatment naive, or treated with crizotinib or next-generation ALK inhibitors, is not known. Nevertheless, these findings collectively support the notion that EML4-ALK v3 is associated with the development of ALK resistance mutations and, in particular, the highly refractory G1202R mutation.
Clinical Outcomes on ALK TKIs With v1 and v3
We next evaluated the effect of ALK variants on clinical responses to various ALK inhibitors. Again, we focused this analysis on the most common variants v1 and v3. Treatment histories of patients in the v1 and v3 cohorts were overall well balanced (Table 1). The median OS from the time of diagnosis of advanced disease was 5.0 years and 3.6 years for v1 and v3 cohorts, respectively (HR, 1.16; 95% CI, 0.67 to 2.01; P = .584; Data Supplement), similar to the previously reported OS for patients with ALK-positive NSCLC who received sequential ALK TKIs.43 Of note, this OS analysis is relatively immature, with only 52 deaths (49%) at the time of data cutoff.
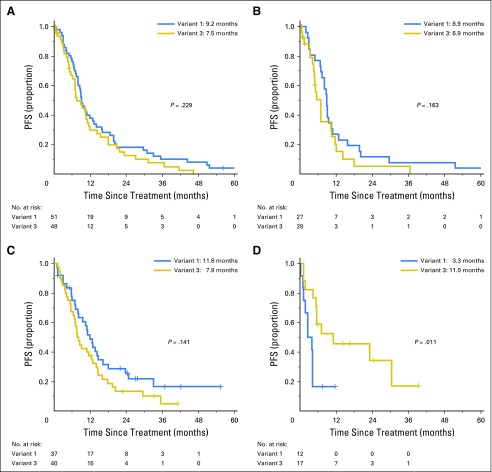
Among 99 patients treated with crizotinib as the first ALK inhibitor, no significant difference was found in PFS in patients with EML4-ALK v1 (n = 51) versus v3 (n = 48; HR, 1.30; 95% CI, 0.85 to 1.98; P = .229; Fig 5A). To address whether these results were confounded by prior lines of chemotherapy, we also examined PFS among 55 patients who received crizotinib as first-line therapy. Again, no difference in PFS between v1 (n = 27) and v3 (n = 28) were found (HR, 1.61; 95% CI, 0.84 to 2.75; P = .163; Fig 5B). Similarly, no significant difference was found in PFS after second-generation ALK TKIs (ie, ceritinib, alectinib, brigatinib) given as the second ALK inhibitor after crizotinib among 77 patients with v1 (n = 37) versus v3 (n = 40; HR, 1.45; 95% CI, 0.88 to 2.38; P = .141; Fig 5C). For patients in each cohort who experienced disease progression, the pattern of progression (ie, CNS only, both intra- and extracranially, or extracranially only) was not different between v1 and v3 (Data Supplement).
Fig 5.
Kaplan-Meier curves for progression-free survival (PFS) for (A) crizotinib as first ALK tyrosine kinase inhibitor (TKI) in patients with EML4-ALK variant 1 (v1; n = 51) versus v3 (n = 48), (B) crizotinib as first-line therapy in v1 (n = 27) versus v3 (n = 28), (C) second-generation ALK TKI administered as the second ALK inhibitor after crizotinib in v1 (n = 37) versus v3 (n = 40), and (D) lorlatinib administered after crizotinib and at least one second-generation ALK TKI in v1 (n = 12) and v3 (n = 17).
In an exploratory analysis of 12 patients with v1 and 17 with v3 who received the third-generation ALK TKI lorlatinib after experiencing treatment failure with both crizotinib and at least one second-generation ALK inhibitor, v3 was associated with significantly longer PFS than v1 (median, 11.0 v 3.3 months; HR, 0.31; 95% CI, 0.12 to 0.79; P = .011; Fig 5D). By univariable analysis, no other baseline features, including sex, age, race/ethnicity, smoking history, initial stage at diagnosis, presence of extrathoracic disease, and presence of CNS metastasis at the diagnosis of advanced disease, was significantly associated with PFS on lorlatinib. Lorlatinib has previously been shown to retain potent activity against all known crizotinib-resistant ALK mutations, including G1202R, in vitro16,44 and has demonstrated activity in patients with ALK-positive NSCLC previously treated with two or more ALK inhibitors, including those with ALK G1202R.15 In the current cohort, six patients with v1 and 15 with v3 who received lorlatinib underwent a prelorlatinib tumor biopsy, and an ALK resistance mutation was present in one v1 (17%; G1269A) and 13 v3 (87%) samples, respectively (Data Supplement). Of the 13 v3 samples with a prelorlatinib ALK mutation, 12 harbored ALK G1202R. Finally, no difference in PFS was noted in patients with v1 versus v3 who received pemetrexed (HR, 1.11; 95% CI, 0.60 to 2.07; P = .742) or platinum and pemetrexed combination (HR, 0.84; 95% CI, 0.40 to 1.78; P = .649; Data Supplement).
DISCUSSION
To our knowledge, we present the largest analysis to date to examine the clinical effect of ALK variants in ALK-positive NSCLC and the first study to evaluate ALK resistance mutations according to EML4-ALK variant. We found that EML4-ALK v3 was significantly associated with the development of ALK resistance mutations, particularly ALK G1202R, which suggests that patients with v3 may be more likely to acquire distinct secondary ALK resistance mutations as they undergo treatment with ALK TKIs.
Previously, we demonstrated the unique spectrum of activity of each currently available ALK inhibitor against different ALK resistance mutations.16 Of note, the solvent-front ALK G1202R mutation conferred high-level resistance to all first- and second-generation ALK inhibitors but retained sensitivity to lorlatinib.16 The strong association between v3 and ALK G1202R suggests that the establishment of ALK variant status may have important therapeutic implications. Indeed, we observed a significantly longer PFS among patients with EML4-ALK v3 versus v1 who received lorlatinib. A priori knowledge of the ALK variant status thus could help to select patients more likely to achieve a durable response to lorlatinib. We speculate that the greater propensity of v3 to develop ALK mutations, particularly G1202R, likely underlies these different outcomes. Supportive of this notion, the activity of lorlatinib was found to correlate tightly with the presence of an ALK resistance mutation (which suggests continued ALK dependency) in a small series of ceritinib-resistant patient-derived cell lines.16 Several ongoing and completed studies of second- and third-generation ALK TKIs, including the phase III study of first-line lorlatinib versus crizotinib (ClinicalTrials.gov identifier: NCT03052608), are retrospectively evaluating the association between efficacy and ALK variant and/or preexisting ALK resistance mutations and may further enhance our understanding of the molecular variables that contribute to heterogeneous clinical outcomes.
We also examined the effect of ALK variants on clinical responses to first- and second-generation ALK inhibitors. In this study, no statistically significant difference in PFS was observed among patients with v1 versus v3 treated with crizotinib or a second-generation ALK TKI. However, the median PFS in all contexts was numerically shorter for v3 than for v1, which suggests that larger patient cohorts may be needed to achieve sufficient power to detect a statistically significant difference. In light of the Alectinib Versus Crizotinib in Untreated ALK-Positive NSCLC study that demonstrated superiority of front-line alectinib over crizotinib,12 continuing the investigation of the effect of specific ALK variants on resistance patterns and clinical outcomes in patients treated with first-line alectinib will be of particular interest.
Four previously published studies evaluated clinical responses to ALK TKIs according to ALK variant. All were centered on crizotinib, and conflicting findings have been reported (eg, PFS longer for patients with v1 v non-v1,27 longer for those with non-v3 v v3,28 not different on the basis of variant29,30). Several limitations may account for the discordant findings, including small sample size, lack of distinction in crizotinib line of therapy, and clustering of variants into groups that may not be biologically or clinically relevant. To address these limitations, we analyzed a larger cohort of patients with a known ALK variant, and focused on the most common variants v1 and v3 (to avoid the confounding effect of arbitrary variant clustering) and on crizotinib administered as the first ALK TKI or as first-line therapy (to minimize the effect of treatment lines).
Nonetheless, the current study had limitations. First, it was a retrospective study and still limited in the number of patients. Variant analysis in larger, prospective studies will be needed to validate and expand on our findings. Second, this study focused on the most common EML4-ALK variants v1 and v3, and therefore, the potential effect of non-v1/v3 variants on ALK TKI resistance mechanisms and clinical outcomes remains unknown. Moreover, the effect of ALK variants on OS of patients with ALK-positive NSCLC requires more investigation because the OS data in this study were not mature. Of note, as shown in the Data Supplement, some differences were found in the sequencing methods used to detect the ALK resistance mutations in v1 and v3 post-TKI biopsies. To our knowledge, the particular methods used in this study are not limited in the ability to detect specific ALK fusion variants or resistance mutations such as G1202R.
Future studies will need to explore the mechanisms that underlie the differential association between ALK variant and resistance mutations. Prior studies have shown that ALK variants retain varying portions of the EML4 tandem atypical propeller EML (TAPE) domain, which results in differential fusion protein stability in vitro.25,26 In particular, shorter EML4-ALK variants (eg, v3) that lack the entire TAPE domain have been found to be more stable than longer variants (eg, v1) that retain a partial TAPE domain.23-26 In theory, ALK-rearranged tumor cells with the more stable v3 could be more ALK addicted, which necessitates the development of potent on-target ALK mutations (eg, G1202R) to mediate acquired resistance. Alternatively, an assessment of whether certain variants such as v3 are more structurally vulnerable to developing particular ALK resistance mutations and whether mutations confer differential levels of resistance depending on the variant would be interesting.
In summary, the findings suggest that EML4-ALK v3 is associated with a significantly higher incidence of ALK resistance mutations, particularly G1202R, and provide a potential molecular link between variant and clinical outcome. Thus, ALK variant status may represent an important emerging factor in guiding the treatment strategy for ALK-positive NSCLC.
Footnotes
Supported by grants from the National Cancer Institute (R01CA164273 to A.T.S.) and the National Foundation for Cancer Research (to A.T.S.), by Be a Piece of the Solution, and by the Targeting a Cure for Lung Cancer Research Fund at Massachusetts General Hospital.
Presented at the 53rd Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, June 2-6, 2017.
See accompanying article on page 1257
AUTHOR CONTRIBUTIONS
Conception and design: Jessica J. Lin, Jeffrey S. Ross, Alice T. Shaw, Sai-Hong Ignatius Ou
Financial support: Alice T. Shaw
Provision of study materials or patients: Jessica J. Lin, Alexa B. Schrock, Alice T. Shaw, Sai-Hong Ignatius Ou
Collection and assembly of data: Jessica J. Lin, Viola W. Zhu, Satoshi Yoda, Alexa B. Schrock, Nicholas A. Jessop, Ginger Y. Jiang, Long P. Le, Jeffrey S. Ross, Alice T. Shaw, Sai-Hong Ignatius Ou
Data analysis and interpretation: Jessica J. Lin, Satoshi Yoda, Beow Y. Yeap, Alexa B. Schrock, Ibiayi Dagogo-Jack, Long P. Le, Kyle Gowen, Philip J. Stephens, Jeffrey S. Ross, Siraj M. Ali, Vincent A. Miller, Melissa L. Johnson, Christine M. Lovly, Aaron N. Hata, Justin F. Gainor, Anthony J. Iafrate, Alice T. Shaw, Sai-Hong Ignatius Ou
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Jessica J. Lin
Honoraria: Chugai Pharmaceutical
Consulting or Advisory Role: Boehringer Ingelheim
Viola W. Zhu
Stock or Other Ownership: TP Therapeutics (I)
Honoraria: AstraZeneca
Consulting or Advisory Role: AstraZeneca, Bayer AG, TP Therapeutics, Biocept
Speakers’ Bureau: AstraZeneca, Roche, Genentech, Takeda Pharmaceuticals
Satoshi Yoda
No relationship to disclose
Beow Y. Yeap
Stock or Other Ownership: SISCAPA Assay Technologies (I)
Consulting or Advisory Role: Abcodia (I)
Alexa B. Schrock
Employment: Foundation Medicine
Stock or Other Ownership: Foundation Medicine
Ibiayi Dagogo-Jack
Honoraria: Foundation Medicine
Consulting or Advisory Role: Boehringer Ingelheim
Nicholas A. Jessop
No relationship to disclose
Ginger Y. Jiang
Employment: Novartis (I)
Long P. Le
Stock or Other Ownership: ArcherDx
Consulting or Advisory Role: ArcherDx
Patents, Royalties, Other Intellectual Property: Anchored multiplex polymerase chain reaction patent licensed to ArcherDx
Kyle Gowen
Employment: Foundation Medicine
Stock or Other Ownership: Foundation Medicine
Travel, Accommodations, Expenses: Foundation Medicine
Philip J. Stephens
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock or Other Ownership: Foundation Medicine
Jeffrey S. Ross
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock or Other Ownership: Foundation Medicine
Siraj M. Ali
Employment: Foundation Medicine
Stock or Other Ownership: Exelixis, Otonomy, OncoSec Medical, Genocea Biosciences
Consulting or Advisory Role: Incysus
Patents, Royalties, Other Intellectual Property: Patents through Foundation Medicine, and patents through Seres Health on microbiomes in non-neoplastic disease (I)
Vincent A. Miller
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock or Other Ownership: Foundation Medicine
Consulting or Advisory Role: Revolution Medicine
Patents, Royalties, Other Intellectual Property: Receives periodic royalties related to T790M patent awarded to Memorial Sloan Kettering Cancer Center
Melissa L. Johnson
Consulting or Advisory Role: Astellas Pharma (I), Otsuka (I), Genentech (Inst), Roche (Inst), Celgene (Inst), Boehringer Ingelheim (Inst), AbbVie (Inst)
Research Funding: OncoMed Pharmaceuticals (Inst), BerGenBio (Inst), Eli Lilly (Inst), EMD Serono (Inst), Kadmon Holdings (Inst), Janssen Pharmaceuticals (Inst), Mirati Therapeutics (Inst), Genmab (Inst), Pfizer (Inst), AstraZeneca (Inst), Genentech (Inst), Roche (Inst), Stemcentrx (Inst), Novartis (Inst), Checkpoint Therapeutics (Inst), Array BioPharma (Inst), Regeneron Pharmaceuticals (Inst)
Christine M. Lovly
Honoraria: Novartis, Sequenom, QIAGEN, Pfizer, National Comprehensive Cancer Network, Takeda Pharmaceuticals
Consulting or Advisory Role: ARIAD Pharmaceuticals, Clovis Oncology, Genoptix, Novartis, ARIAD Pharmaceuticals, Foundation Medicine, Cepheid
Research Funding: AstraZeneca, Novartis
Travel, Accommodations, Expenses: Pfizer, Takeda Pharmaceuticals, Roche, Cepheid
Aaron N. Hata
Research Funding: Amgen, Novartis (Inst), Relay Therapeutics
Justin F. Gainor
Honoraria: Merck, Incyte, ARIAD Pharmaceuticals, Pfizer, Novartis
Consulting or Advisory Role: Genentech, Bristol-Myers Squibb, Theravance Biopharma, Loxo Oncology, Takeda Pharmaceuticals
Research Funding: Merck (Inst), Novartis (Inst), Genetech (Inst), Bristol-Myers Squibb (Inst), Adaptimmune (Inst), AstraZeneca (Inst), ARIAD Pharmaceuticals (Inst), Jounce Therapeutics (Inst), Moderna Therapeutics (Inst)
Travel, Accommodations, Expenses: Affymetrix
Anthony J. Iafrate
Stock or Other Ownership: ArcherDx
Consulting or Advisory Role: Debiopharm Group, Constellation Pharmaceuticals, Chugai Pharmaceutical, Roche
Research Funding: Blueprint Medicines
Patents, Royalties, Other Intellectual Property: ArcherDx exclusive license to AMP technology
Alice T. Shaw
Honoraria: Pfizer, Novartis, Roche, Genentech, Foundation Medicine
Consulting or Advisory Role: Pfizer, Novartis, Genentech, Roche, ARIAD Pharmaceuticals, Ignyta, Blueprint Medicines, Daiichi Sankyo, EMD Serono, Taiho Pharmaceutical, KSQ Therapeutics, Natera
Research Funding: Pfizer, Novartis, Roche, Genentech
Sai-Hong Ignatius Ou
Honoraria: Novartis, Pfizer, Roche, Genentech, ARIAD Pharmaceuticals, Takeda Pharmaceuticals, AstraZeneca
Consulting or Advisory Role: ARIAD Pharmaceuticals, Takeda Pharmaceuticals, Pfizer, Novartis, AstraZeneca, Roche, Genentech
Speakers’ Bureau: AstraZeneca, Genentech, Takeda Pharmaceuticals, ARIAD Pharmaceuticals
Research Funding: AstraZeneca (Inst), Pfizer (Inst), Roche (Inst), AstraZeneca (Inst), MedImmune (Inst), Clovis Oncology (Inst), ARIAD Pharmaceuticals (Inst), Ignyta (Inst), Peregrine Pharmaceuticals (Inst), GlaxoSmithKline (Inst), Astellas Pharma (Inst), Chugai Pharmaceutical (Inst), TP Therapeutics (Inst), Novartis (Inst), Blueprint Medicines (Inst)
REFERENCES
- 1.Lin JJ, Riely GJ, Shaw AT: Targeting ALK: Precision medicine takes on drug resistance. Cancer Discov 7:137-155, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang YJ, Camidge DR, et al. : Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363:1693-1703, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camidge DR, Bang YJ, Kwak EL, et al. : Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: Updated results from a phase 1 study. Lancet Oncol 13:1011-1019, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AT, Kim DW, Nakagawa K, et al. : Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 368:2385-2394, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Solomon BJ, Mok T, Kim DW, et al. : First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 371:2167-2177, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Shaw AT, Kim DW, Mehra R, et al. : Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 370:1189-1197, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DW, Mehra R, Tan DS, et al. : Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): Updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol 17:452-463, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AT, Kim TM, Crinò L, et al. : Ceritinib versus chemotherapy in patients with ALK-rearranged non-small-cell lung cancer previously given chemotherapy and crizotinib (ASCEND-5): A randomised, controlled, open-label, phase 3 trial. Lancet Oncol 18:874-886, 2017 [DOI] [PubMed] [Google Scholar]
- 9.Soria JC, Tan DSW, Chiari R, et al. : First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): A randomised, open-label, phase 3 study. Lancet 389:917-929, 2017 [DOI] [PubMed] [Google Scholar]
- 10.Ou SH, Ahn JS, De Petris L, et al. : Alectinib in crizotinib-refractory ALK-rearranged non–small-cell lung cancer: A phase II global study. J Clin Oncol 34:661-668, 2016 [DOI] [PubMed] [Google Scholar]
- 11.Shaw AT, Gandhi L, Gadgeel S, et al. : Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: A single-group, multicentre, phase 2 trial. Lancet Oncol 17:234-242, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters S, Camidge DR, Shaw AT, et al. : Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med 377:829-838, 2017 [DOI] [PubMed] [Google Scholar]
- 13.Gettinger SN, Bazhenova LA, Langer CJ, et al. : Activity and safety of brigatinib in ALK-rearranged non-small-cell lung cancer and other malignancies: A single-arm, open-label, phase 1/2 trial. Lancet Oncol 17:1683-1696, 2016 [DOI] [PubMed] [Google Scholar]
- 14.Kim DW, Tiseo M, Ahn MJ, et al. : Brigatinib in patients with crizotinib-refractory anaplastic lymphoma kinase-positive non–small-cell lung cancer: A randomized, multicenter phase II trial. J Clin Oncol 35:2490-2498, 2017 [DOI] [PubMed] [Google Scholar]
- 15.Shaw AT, Felip E, Bauer TM, et al. : Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: An international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 18:1590-1599, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gainor JF, Dardaei L, Yoda S, et al. : Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 6:1118-1133, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soda M, Choi YL, Enomoto M, et al. : Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448:561-566, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Shaw AT, Engelman JA: ALK in lung cancer: Past, present, and future. J Clin Oncol 31:1105-1111, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi K, Choi YL, Soda M, et al. : Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 14:6618-6624, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Choi YL, Takeuchi K, Soda M, et al. : Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res 68:4971-4976, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Koivunen JP, Mermel C, Zejnullahu K, et al. : EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 14:4275-4283, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki T, Rodig SJ, Chirieac LR, et al. : The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer 46:1773-1780, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards MW, Law EW, Rennalls LP, et al. : Crystal structure of EML1 reveals the basis for Hsp90 dependence of oncogenic EML4-ALK by disruption of an atypical β-propeller domain. Proc Natl Acad Sci U S A 111:5195-5200, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayliss R, Choi J, Fennell DA, et al. : Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell Mol Life Sci 73:1209-1224, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabir SR, Yeoh S, Jackson G, et al. : EML4-ALK variants: Biological and molecular properties, and the implications for patients. Cancers (Basel) 9:9, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heuckmann JM, Balke-Want H, Malchers F, et al. : Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res 18:4682-4690, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Yoshida T, Oya Y, Tanaka K, et al. : Differential crizotinib response duration among ALK fusion variants in ALK-positive non–small-cell lung cancer. J Clin Oncol 34:3383-3389, 2016 [DOI] [PubMed] [Google Scholar]
- 28.Woo CG, Seo S, Kim SW, et al. : Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 28:791-797, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Cha YJ, Kim HR, Shim HS: Clinical outcomes in ALK-rearranged lung adenocarcinomas according to ALK fusion variants. J Transl Med 14:296, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei YY, Yang JJ, Zhang XC, et al. : Anaplastic lymphoma kinase variants and the percentage of ALK-positive tumor cells and the efficacy of crizotinib in advanced NSCLC. Clin Lung Cancer 17:223-231, 2016 [DOI] [PubMed] [Google Scholar]
- 31.Zheng Z, Liebers M, Zhelyazkova B, et al. : Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 20:1479-1484, 2014 [DOI] [PubMed] [Google Scholar]
- 32.Frampton GM, Fichtenholtz A, Otto GA, et al. : Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023-1031, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagle N, Berger MF, Davis MJ, et al. : High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2:82-93, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard CC, Salipante SJ, Koehler K, et al. : Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 16:56-67, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw AT, Yeap BY, Mino-Kenudson M, et al. : Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27:4247-4253, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayama R, Shaw AT, Khan TM, et al. : Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 4:120ra17, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang DD, Zhang B, Gu Q, et al. : HIP1-ALK, a novel ALK fusion variant that responds to crizotinib. J Thorac Oncol 9:285-294, 2014 [DOI] [PubMed] [Google Scholar]
- 38.Hong M, Kim RN, Song JY, et al. : HIP1-ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol 9:419-422, 2014 [DOI] [PubMed] [Google Scholar]
- 39.Ou SH, Klempner SJ, Greenbowe JR, et al. : Identification of a novel HIP1-ALK fusion variant in non-small-cell lung cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to alectinib. J Thorac Oncol 9:1821-1825, 2014 [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi K, Choi YL, Togashi Y, et al. : KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 15:3143-3149, 2009 [DOI] [PubMed] [Google Scholar]
- 41.Ali SM, Hensing T, Schrock AB, et al. : Comprehensive genomic profiling identifies a subset of crizotinib-responsive ALK-rearranged non-small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist 21:762-770, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friboulet L, Li N, Katayama R, et al. : The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 4:662-673, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gainor JF, Tan DS, De Pas T, et al. : Progression-free and overall survival in ALK-positive NSCLC patients treated with sequential crizotinib and ceritinib. Clin Cancer Res 21:2745-2752, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou HY, Friboulet L, Kodack DP, et al. : PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell 28:70-81, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]