Abstract
In vitro models of the human liver are important for the following: (1) mitigating the risk of drug-induced liver injury to human beings, (2) modeling human liver diseases, (3) elucidating the role of single and combinatorial microenvironmental cues on liver cell function, and (4) enabling cell-based therapies in the clinic. Methods to isolate and culture primary human hepatocytes (PHHs), the gold standard for building human liver models, were developed several decades ago; however, PHHs show a precipitous decline in phenotypic functions in 2-dimensional extracellular matrix–coated conventional culture formats, which does not allow chronic treatment with drugs and other stimuli. The development of several engineering tools, such as cellular microarrays, protein micropatterning, microfluidics, biomaterial scaffolds, and bioprinting, now allow precise control over the cellular microenvironment for enhancing the function of both PHHs and induced pluripotent stem cell–derived human hepatocyte-like cells; long-term (4+ weeks) stabilization of hepatocellular function typically requires co-cultivation with liver-derived or non–liver-derived nonparenchymal cell types. In addition, the recent development of liver organoid culture systems can provide a strategy for the enhanced expansion of therapeutically relevant cell types. Here, we discuss advances in engineering approaches for constructing in vitro human liver models that have utility in drug screening and for determining microenvironmental determinants of liver cell differentiation/function. Design features and validation data of representative models are presented to highlight major trends followed by the discussion of pending issues that need to be addressed. Overall, bioengineered liver models have significantly advanced our understanding of liver function and injury, which will prove useful for drug development and ultimately cell-based therapies.
Keywords: Microfluidics, Hepatocytes, Micropatterned Co-Cultures, Spheroids, Bioprinting, Cellular Microarrays
Abbreviations used in this paper: BAL, bioartificial liver; CRP, C-reactive protein; CYP450, cytochrome P450; DILI, drug-induced liver injury; ECM, extracellular matrix; HSC, hepatic stellate cell; iHep, induced pluripotent stem cell-derived human hepatocyte-like cell; IL, interleukin; iPS, induced pluripotent stem; KC, Kupffer cell; LSEC, liver sinusoidal endothelial cell; MPCC, micropatterned co-culture; NPC, nonparenchymal cell; PEG, polyethylene glycol; PHH, primary human hepatocyte; 3D, 3-dimensional
Summary.
This review discusses advances in engineering approaches for constructing liver models with utility in drug screening and for determining microenvironmental determinants of liver cell differentiation/function. Design features and validation of representative models are discussed as well as anticipated future trends.
Drug-induced liver injury (DILI) is a leading cause of preclinical and clinical drug attrition, black-box warnings on marketed drugs, and acute liver failures in the United States alone.1 Almost 1000 marketed drugs can cause either cell necrosis, hepatitis, cholestasis, fibrosis, or a mixture of injury types.2 DILI severity can be exacerbated by states of stress (ie, inflammation), patient-specific risk factors (ie, genetics, age, sex, and diet), and underlying disease states (ie, hepatitis, cholestasis, and fibrosis). Unfortunately, the live animal testing required by the Food and Drug Administration during preclinical drug development is only capable of identifying <50% of human DILI, largely owing to differences in species-specific drug metabolism pathways and the inability to accurately capture human genetics and disease backgrounds.3 Given such challenges with screening drugs in animals, the field of in vitro human liver cultures has gained ever-increasing importance in the past 10–15 years.4 The utilization of engineering tools such as high-throughput microarrays, protein micropatterning, microfluidics, specialized plates, biomaterial scaffolds, and bioprinting has enabled greater control over the cellular microenvironment, which has increased the longevity and reproducibility of cell functions in vitro as well as enabled de-coupling of cues that modulate cellular responses. In this review, we discuss the design features and utility of bioengineered liver models for drug testing and cell differentiation studies because these applications are intricately tied toward enabling accurate prediction of clinically relevant outcomes. We highlight representative platforms, some in commercial practice, to demonstrate key points of emphasis. Finally, we discuss pending issues that will need to be addressed moving forward in the field of liver tissue engineering for in vitro applications.
High-Throughput Cell Microarrays
In early drug development, when many compounds need to be tested and the amount of compounds is limiting, culture platforms need to be high throughput, relatively low cost, and provide actionable data quickly (within 24–48 h). Similarly, high-throughput platforms can enable the systematic analysis of hundreds of microenvironmental signals (using a high number of replicates), and, thereby, the identification and optimization of culture conditions for hepatocellular differentiation/function. Several high-throughput culture platforms have been developed for the culture of multiple sources of liver cells. These cells include cancerous and immortalized cell lines that can serve as cheaper and more sustainable sources of liver cells compared with primary human hepatocytes (PHHs), but can suffer from abnormal functions.5 For instance, Kwon et al6 designed a microchip platform for transducing 3-dimensional (3D) liver cell cultures with genes for drug metabolism enzymes. The platform features 532 reaction vessels (micropillars and corresponding microwells) on a 75 × 25 mm slide outline. Cells are suspended in a Matrigel (Corning Inc, Corning, NY) droplet (∼60 nL), which is spotted on a micropillar. The micropillar then is placed into a corresponding microwell containing recombinant adenoviruses. Transformed human liver epithelial cells were transduced with adenoviruses to manipulate the expression of human drug metabolism enzyme genes. A single microarray was used to create 84 combinations of metabolic gene perturbations, which provided information on which enzyme combinations led to drug toxicity in cells. In another example, a 3D Hep3B microarray was coupled with a microarray containing various combinations of recombinant drug metabolism enzymes to evaluate the metabolism-mediated toxicity of drugs.7 In contrast to using cell lines, Fukuda and Nakazawa8 designed a microarray that allowed stable immobilization of 100 μm rat hepatocyte spheroids in microwells for probing cytochrome P450 (CYP450) activities after drug treatment. A row of microwells also could be connected to a microchannel for simultaneous detection of different CYP450 enzyme activities on a single chip.
To optimize hepatocellular culture conditions, including defined conditions for facilitating hepatocyte differentiation of stem cells, investigators have sought to systematically evaluate the effects of a range of microenvironmental signals. Specifically, by incorporating advances in biomaterial chemistries and microfabrication techniques, these methods have emphasized the reduction of multicomponent cellular microenvironments into distinct individual signals that can be tightly controlled within engineered systems. Moving forward, to more effectively study and manipulate stem cell microenvironments, it has been increasingly recognized that larger-scale analyses are required. As an analogy, recent improvements in high-throughput sequencing and expression profiling methods have revolutionized genomics. Robust high-throughput strategies for probing cell microenvironments could similarly advance the understanding of cellular processes by enabling unprecedented systematic analysis of combinations of signals and could form the basis for necessary but challenging studies into the details of bidirectional cell–microenvironment interactions.
The fabrication of microwell platforms represents an example of one of the approaches aimed at exploring, in parallel, a range of combinations of signals that cannot be practically evaluated with standard techniques. Microwell arrays typically are fabricated through direct etching of hard materials (eg, glass, silicon) or through a combination of photopolymerization and soft-lithography–based molding of hydrogels. In particular, this approach has been applied to the analysis of individual stem cells to evaluate clonal heterogeneity.9, 10 Hydrogel microwells can be functionalized with biomolecules, as highlighted by a recent strategy that paired microwell molding with protein microarraying to analyze neural stem cells and mesenchymal stem cells within microwell arrays presenting a range of combinatorial stimuli.11 Microfluidic-based approaches, integrating microwells or hydrodynamic traps, also have been used to generate cellular arrays.12, 13, 14, 15 In addition, the emerging field of droplet microfluidics16 represents an attractive approach to increase the throughput of 3D fabrication while maintaining precise control of the environmental components. For example, miniaturized cell-encapsulated hydrogels were fabricated with microfluidic methods to examine co-cultures,17 incrementally modulate material stiffness,18 and generate microscale constructs for the assembly of larger patterned structures.19
Cellular microarrays, in which viable cells are seeded onto printed spots of materials/biomolecules, represent another approach for defining the microenvironment of cells (Figure 1A). In these platforms, the spots typically include adhesive components to retain cells, in addition to combinations of other elements to either deliver factors to cells or to stimulate or measure cellular processes.20, 21, 22, 23, 24 Cellular microarrays based on spotted biomaterial libraries have been applied to several investigations aimed at exploring stem cell functions. Specifically, these studies have examined the effect of polymer backbone chemistries and end group functionalization on pluripotent and multipotent stem cell proliferation and differentiation.25, 26, 27, 28, 29 In particular, microarrays of extracellular matrix (ECM) molecules have shown substantial effects of combinatorial ECM presentation on cellular functions. Initial experiments in this area have shown the capabilities of an ECM microarray approach, focusing on the influence of ECM combinations on hepatocyte adhesion and survival, the early differentiation of embryonic stem cells, and notable synergistic or antagonistic effects of ECM components.20, 30 These experiments, as well as numerous others in both 2-dimensional and 3D contexts,31 suggest that ECM can influence hepatocyte functions such as albumin expression. Subsequent studies have used arrayed ECM proteins for investigating a range of cell types.21, 22, 32, 33, 34 For example, Kaylan et al35 recently used an ECM microarray approach to show that ECM composition has a significant influence on the adhesion and degree of differentiation of liver progenitor cells when they are induced to differentiate. As part of this work, an automated fluorescence imaging and custom analysis pipeline that enables single-cell measurements within the array was established. In addition to ECM molecules, printed arrays containing combinations of growth factors or cell surface ligands have provided clues into how cells respond to complex extracellular signals. For example, the effect of the Notch ligand, Jagged-1, on the differentiation of neural23 or mammary progenitor cells24 was shown to be dependent on the context of the combinatorial stimuli, specifically the presence or absence of Wnt or ECM proteins, respectively. In the analysis of liver progenitor differentiation, a cell microarray-based approach showed that the Notch ligands Jagged-1, Delta-like 1, and Delta-like 4 each can induce an increase in biliary differentiation, and this occurs in the absence of any additional exogenous differentiation-inducing factors.35 Furthermore, cell microarrays can be fabricated on substrates of modular stiffness to explore combinatorial effects of mechanical signals, and recently have been integrated with traction force microscopy for the direct analysis of cell traction stresses within distinct microenvironmental contexts.36 Collectively, these high-throughput approaches can provide a wealth of empiric data toward the evaluation of combinatorial effects that are difficult to predict a priori, and are useful for developing highly functional and long-lasting liver models for drug development and other applications (ie, cell-based therapies).
Figure 1.
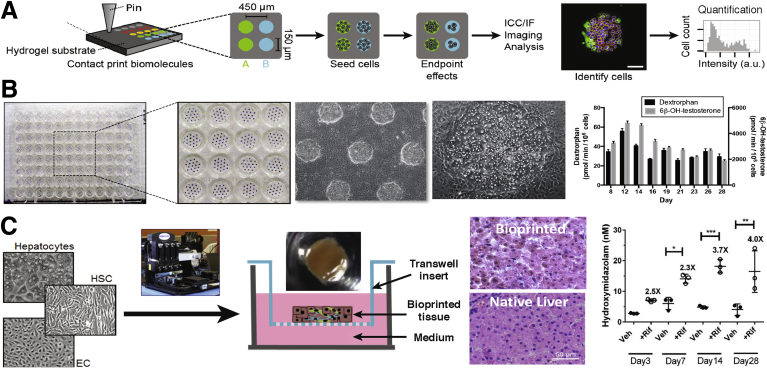
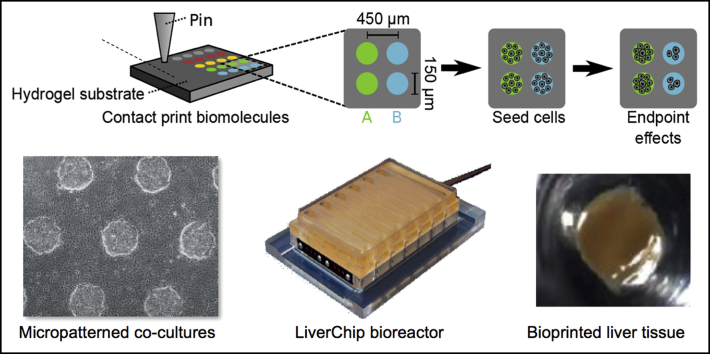
Micropatterned/printed liver culture platforms. (A) High-throughput cellular microarrays for investigating hepatocellular differentiation.35Left to right: Schematic of a microarray experiment for investigating hepatocellular differentiation. Biomolecules and ECM proteins are patterned on a polyacrylamide hydrogel substrate using contact printing. Cells seeded on arrays adhere only to the patterned regions and are exposed to the deposited biomolecules and any experiment-specific soluble factors, fixed at end point, immunolabeled, imaged, and analyzed. Individual cells on islands are automatically identified by nuclear stain (4′,6-diamidino-2-phenylindole) and associated with intensities in other channels, resulting in both single-cell and summary quantifications (eg, percentage of cells positive for a marker) of results by deposited biomolecule and soluble factor treatment. (B) Micropatterned co-cultures (MPCCs).49, 51Left to right: A 96-well plate showing uniform PHH islands micropatterned using semiconductor-driven soft lithography and subsequently surrounded by NPC types (3T3-J2 murine embryonic fibroblasts shown in this example). Phase-contrast images of MPCCs at different magnifications are shown. MPCCs maintain high levels of CYP450 enzyme activities for several weeks (CYP3A4 activity was assessed via metabolism of testosterone into 6β-OH-testosterone, whereas CYP2D6 activity was assessed via metabolism of dextromethorphan into dextrorphan). (C) Bioprinted liver organoids.92, 129Left to right: schematic of transverse cross-section of bioprinted liver organoids containing hepatocytes, endothelial cells (ECs) and HSCs. Gross image of bioprinted human liver organoid with 2.5-mm diameter and 0.5-mm thickness is shown above the schematic. Comparison of H&E-stained bioprinted liver organoid and native human liver. Basal and rifampicin-induced CYP3A4 activity in bioprinted human liver organoids measured by the formation of 4-hydroxymidazolam from midazolam. *P < .05, **P < .01, ***P < .001. ICC/IF, immunocytochemistry/immunofluorescence; +Rif, rifampin; Veh, vehicle.
Micropatterned Co-Cultures
Heterotypic interactions between parenchymal and nonparenchymal cells (NPCs) are important in liver development, physiology, and pathophysiology. In vitro, co-culture with both liver- and non–liver-derived NPC types can transiently induce functions in primary hepatocytes from multiple species, including human beings.37 Although the complete mechanism underlying this so-called co-culture effect remains undefined, liver co-cultures have proven useful for investigating host response to sepsis,38 mutagenesis,39 xenobiotic metabolism and toxicity,40 response to oxidative stress,41 lipid metabolism,42 and induction of the acute phase response43; such co-cultures also have been explored for clinical bioartificial liver devices.44 However, randomly distributed co-cultures do not allow precise modulation of homotypic and heterotypic cell–cell interactions that play critical roles in liver functions. In contrast, Bhatia et al45, 46 used a technique adapted from the semiconductor industry to first micropattern rat hepatocytes on collagen-coated circular domains and then surround the hepatocyte domains with 3T3-J2 murine embryonic fibroblasts, which can secrete molecules present in the liver.47, 48 These so-called micropatterned co-cultures (MPCCs) allowed tuning of homotypic interactions between hepatocytes and the heterotypic interface between hepatocytes and the fibroblasts while keeping cell numbers/ratios constant across the various patterned configurations. Overall, several key findings emerged from these pioneering studies, as follows: (1) circular domains, as opposed to patterns with sharp corners (ie, rectangles), led to better retention of patterning fidelity over several weeks in culture; (2) controlling homotypic interactions between hepatocytes alone was not sufficient to rescue liver-specific functions in the absence of fibroblasts; (3) increasing the heterotypic interface between fibroblasts and hepatocytes via a reduction in the diameter of the collagen-coated domains led to higher hepatocellular functions than when the domain diameter was larger; and (4) contact with fibroblasts was necessary because both fibroblast- and co-culture–conditioned media were not able to rescue the phenotype of hepatocyte-only cultures. However, in contrast to rat hepatocytes, Khetani and Bhatia49 showed that PHHs displayed highest functions on collagen-coated domains of intermediate diameters (∼500 μm domain diameter with 1200 μm center-to-center spacing between domains), suggesting a species-specific balance in homotypic interactions between hepatocytes and their heterotypic interactions with the fibroblasts. Most importantly, PHHs showed high and stable functions in MPCCs for 4–6 weeks as compared with an unstable phenotype observed in randomly distributed co-cultures of the same 2 cell types (Figure 1B).
Human MPCCs miniaturized into a multiwell format have shown utility for several applications in drug development, such as drug clearance predictions,50, 51, 52 drug–drug interactions,49, 51, 53, 54 drug metabolite profiling,55, 56, 57 drug-transporter interactions,54, 58 DILI prediction,59, 60 gluconeogenesis inhibition (for type 2 diabetes mellitus therapies),61 and infection with hepatitis B/C viruses62, 63 and malaria.64, 65 For instance, MPCCs were treated for up to 9 days with 45 drugs, of which 35 have known DILI liabilities in the clinic while 10 generally are considered not toxic to the liver.60 Given interindividual differences in drug concentrations in plasma and within the liver, MPCCs were treated with drug concentrations up to 100-fold of the reported maximum drug concentration in human plasma for each drug, which is also common with other platforms and does not increase the false-positive rate for DILI detection.66 Overall, repeat drug treatment for at least 9 days improved the sensitivity for DILI detection without a reduction in specificity (as assessed via adenosine triphosphate, glutathione, albumin, and urea); MPCCs showed a significantly higher sensitivity than 24-hour treatment of conventional PHH monolayers with the same drugs; and human MPCCs were more sensitive than rat MPCCs for human DILI detection. In another study, human MPCCs, but not conventional PHH monolayers, picked up the toxicity of fialuridine, a nucleoside analog drug for hepatitis B viral infection that caused liver failure and the deaths of 5 patients in clinical trials as a result of lactic acidosis.67
More recently, the MPCC technology was adapted to induced pluripotent stem cell–derived human hepatocyte-like cells (iHeps),68 which afford the opportunity to sustainably evaluate cell responses to drugs and other stimuli across diverse genetic backgrounds.69 In contrast to a severely immature and declining iHep phenotype in conventional monolayers, iHeps in MPCCs showed higher levels of adult-like functions and a reduction in fetal markers (ie, α-fetoprotein) over 4 weeks.68 Furthermore, when iHeps in MPCCs were treated with 47 drugs for 6 days and assessed for hepatotoxicity (adenosine triphosphate) and liver functions (albumin and urea), the sensitivity (65%) and specificity (100%) for DILI detection relative to known clinical outcomes were remarkably similar to the values obtained with MPCCs containing PHHs treated with the same drugs (70% sensitivity and 100% specificity)70; these results suggest that MPCCs containing iHeps may be useful for an initial drug toxicity screen during drug development using a nearly infinite source of liver-like cells.
The MPCC platform was designed to be modular in that the NPC type/population can be modified without significantly affecting the hepatocyte homotypic interactions on the micropatterned domains, which are important for maintaining cell polarity. Nguyen et al71 augmented pre-established MPCCs with primary human Kupffer cells (KCs)/macrophages once the hepatic phenotype was stable after 5–7 days. Stimulating the KCs in MPCCs with bacterial-derived endotoxin, lipopolysaccharide, led to cytokine-mediated down-regulation of CYP450s in PHHs, which can affect DILI outcomes. Davidson et al72 recently augmented MPCCs with activated (fibrogenic) primary human hepatic stellate cells (HSCs) at physiological ratios with PHHs and showed effects on hepatic functions that are reminiscent of a nonalcoholic steatohepatitis/early fibrosis phenotype. Although albumin and urea secretions were relatively similar in HSC-augmented MPCCs and HSC-free MPCCs (suggesting well-differentiated PHHs), over the course of 2 weeks, increasing fibrogenic HSC numbers (1) down-regulated hepatic CYP450 (2A6, 3A4) and transporter activities, (2) caused hepatic steatosis, and (3) enhanced the secretion of proinflammatory interleukin 6 (IL6) and C-reactive protein (CRP); effects that are consistent with clinical findings in patients with early stages of nonalcoholic steatohepatitis/fibrosis.73, 74 Importantly, inhibition of reduced nicotinamide adenine dinucleotide phosphate oxidase (NOX) and/or activation of farnesoid X receptor (FXR) using clinically relevant drugs, GKT137831 and obeticholic acid, respectively, alleviated hepatic dysfunctions owing to fibrogenic HSCs, thereby suggesting platform utility for drug screening. Finally, Ware et al75 co-cultured micropatterned PHH colonies with either liver sinusoidal endothelial cells (LSECs) or human umbilical vein endothelial cells (HUVECs) and found that neither endothelial cell type could maintain PHH morphology and functions to the same magnitude/longevity as the 3T3-J2 fibroblasts. In contrast, both PHHs and endothelial cells (LSECs or HUVECs) showed stable phenotype, including the appearance of fenestrations in LSECs, for at least 3 weeks in PHH/fibroblast/endothelial cell tricultures. Tricultures created with cells in the same plane or the PHH/fibroblast co-cultures separated by the endothelial cells via a protein gel (to mimic the space of Disse) showed similar functions over time.
Liver Spheroids/Organoids
Hepatocytes can be stabilized in 3D spheroids/organoids, which leads to the establishment of homotypic cell–cell interactions and the presence of ECM proteins within and around the cells.76 Hepatocellular functions can be further enhanced via co-cultivation with NPCs and the role of heterotypic cell–cell interactions on modulating outcomes resulting from drugs and other stimuli can be evaluated. Hepatic spheroids can spontaneously form on nontreated culture plates or those coated with various polymers.77, 78 Such spheroids have been shown to display high viability and some functions78; however, it is difficult to control the spheroid size and smaller spheroids can merge to form larger spheroids with necrotic cores owing to poor diffusion of oxygen/nutrients. To mitigate such a challenge, specialized plates and scaffolds have been developed to direct the assembly of uniformly sized spheroids that remain separated for interrogation after drug/stimuli treatment. For instance, Messner et al79 developed a specialized plate for creating hanging liquid drops that allow the formation of hepatocyte–endothelial–KC spheroids (1 per well) of controlled diameters, which remain viable and secrete albumin for approximately 1 month. These spheroids showed increased sensitivity in identifying known hepatotoxic drugs than short-term PHH monolayers (∼60% vs ∼40%), while specificity was similar across both assays (∼80%–85%).80 In another platform, Kostadinova et al81 seeded a mixture of liver NPCs onto a porous nylon scaffold followed by seeding of PHHs onto the pre-established liver NPC/nylon culture. PHHs in this platform secreted liver proteins (albumin, transferrin, and fibrinogen) and showed CYP450 activities for 77–90 days, and were more sensitive to hepatotoxic drugs than monolayers. On the other hand, Takayama et al82 used a nanopillar plate to create iHep spheroids, which were more sensitive to drug toxicity than HepG2 spheroids; however, iHep spheroids showed lower sensitivity than conventional PHH monolayers, suggesting that further maturation of the iHeps is likely required.
The earlier-mentioned spheroids rely on cell-secreted ECM; however, such an approach does not allow precise and reproducible tuning of the biochemical and biomechanical microenvironment around cells. In contrast, naturally derived (ie, alginate, chitosan, and cellulose) and synthetic biomaterials (ie, polyethylene glycol [PEG]) can be used to mitigate such a limitation by presenting an engineered polymer matrix to cells.76 For instance, biocompatible PEG hydrogels provide control over mechanical properties via customization of chain length and control over biochemical properties by the tethering of ligands such as cell adhesion peptides and growth factors.83 Chen et al84 co-cultivated PHHs, 3T3-J2 fibroblasts, and immortalized LSECs in PEG hydrogels modified with cell adhesion ligands and observed relatively stable albumin and urea secretion for at least 8 days in vitro. A microfluidic droplet generator was subsequently used to generate PEG-based hepatic microtissues, which are more amenable to high-throughput drug studies than bulk gels.85 In a study using a naturally derived biomaterial, Tasnim et al86 encapsulated human pluripotent stem cell–derived hepatocyte-like cells in galactosylated cellulosic sponges, which promoted the formation and retention of spheroids; such spheroids were more sensitive to the toxicity of hepatotoxic drugs as compared with conventional monolayers, and responses in stem cell spheroids were similar to those observed in PHHs. Larkin et al87 designed a detachable, nanoscale, and mechanically tunable space of Disse (ie, overlay) to separate rat hepatocyte cultures from a mixture of LSECs and KCs using self-assembled polyelectrolyte multilayers of chitosan and hyaluronic acid. When tuned to show liver-like stiffness, the polymeric space of Disse enabled higher albumin secretion and CYP1A activity in the hepatocytes, while hepatocytes and KCs showed some proliferation as compared with the nonpolymeric controls.
Building on the insights gained from miniaturized 3D liver spheroids, a broad range of recent efforts have been aimed at the development of larger (millimeter to centimeter scale) liver organoids that can further promote cell proliferation and the recapitulation of characteristic cell functions and spatial organization. Liver organoids have been generated from both human pluripotent stem cells88 and adult human liver bipotential cells.89 Notably, induced pluripotent stem (iPS) cell–derived liver bud organoids share numerous features with fetal liver cells, based on single-cell RNA sequencing analysis, and these organoid cultures have highlighted the important role of bidirectional signaling between differentiating hepatoblasts and vascular endothelial cells.90 In the adult bipotential cell system, organoid cultures have been shown to support long-term cell expansion (at least 3 mo) without chromosome abnormalities, and these expanded cells could be successively differentiated into functional hepatocytes in vitro and in vivo.89 Furthermore, toward the development of disease models, organoid cultures were generated from Alagille syndrome and α1-antitrypsin–deficiency patients, and showed abnormal phenotypic markers consistent with the in vivo disease features.89 In addition, recent studies have shown an approach for the generation of primary extrahepatic cholangiocyte organoids, which facilitate substantial in vitro cell expansion as well as subsequent in vivo transplantation, either as cell clusters or postseeding of a biodegradable polyglycolic acid scaffold.91
It is difficult to precisely control the spatial arrangement of different cell types (as in vivo) in randomly distributed spheroids/organoids except for what is induced by the spontaneous sorting of specific cell types. On the other hand, bioprinting methods can position different cell populations in organoids to mimic liver lobule architecture. Norona et al92 developed a bioprinted human liver organoid (centimeter scale) containing a compartment of PHHs next to an NPC compartment containing HSCs and endothelial cells housed in a 24-well Transwell (Corning Inc, Corning, NY) format (Figure 1C). These organoids showed high viability, albumin secretion, and CYP3A4 activity for 28 days, and were more sensitive to the toxicity of trovafloxacin after 7 days of treatment than conventional monolayers. Similarly, Jeon et al93 used a 3D bioprinting system to create organoids containing HepG2 cells in alginate and found better growth and expression of liver-specific genes in the organoids relative to monolayers. Ma et al94 used 3D bioprinting to create liver lobule-like hexagonal organoids containing iHeps, endothelial cells, and adipose-derived stem cells embedded in a hydrogel. Liver gene expression and functions in co-cultured organoids were detected for up to 32 days at higher levels than in iHep-only organoids or monolayers.
Liver-on-a-Chip Devices
In contrast to static platforms, perfusion systems or bioreactors can allow automated control over culture medium pH, temperature, fluid pressures, cell shear stress, nutrient supply, and waste removal. The Griffith group at Massachusetts Institute of Technology has pioneered a perfused liver platform for drug screening, called the LiverChip, in which hepatocyte aggregates adhere to the collagen-coated walls of microchannels (created using silicon or polycarbonate in a multiwell footprint) and are perfused at flow rates that meet both the oxygen demands of the hepatocytes and subject the cells to low shear stress as in vivo (Figure 2A).95, 96 The hepatocyte aggregates maintain functions under perfusion that are an order of magnitude higher than in static controls. Co-culture of rat LSECs with rat hepatocytes in the LiverChip showed that the LSECs demonstrated moderate proliferation and were positive for the prototypical marker, SE-1 (hepatic sinusoidal endothelial cells), whereas LSECs entirely disappeared from conventional monolayers after 13 days in culture.97 Human hepatocyte–KC aggregates in the LiverChip respond appropriately to lipopolysaccharide stimulation by increasing the secretion of 11 different proinflammatory cytokines (ie, IL6, tumor necrosis factor α, hemokine C-C motif ligand 5).98, 99 Stimulating these hepatocyte–KC aggregates with IL6 caused a dose-dependent decrease in CYP3A4 activity, an increase in CRP secretion, and a decrease in shed soluble IL6-receptor (IL6R) levels, which shows an in vivo–like response of PHHs to IL6.100 Further treating the IL6-stimulated co-cultures with tocilizumab, an anti-IL6R monoclonal antibody, led to the recovery of CYP3A4 activity and a reduction in CRP levels after 72 hours of treatment. More recently, major phase I and II metabolites of diclofenac produced from the hepatocyte–KC aggregates were similar to those observed in human beings.101
Figure 2.
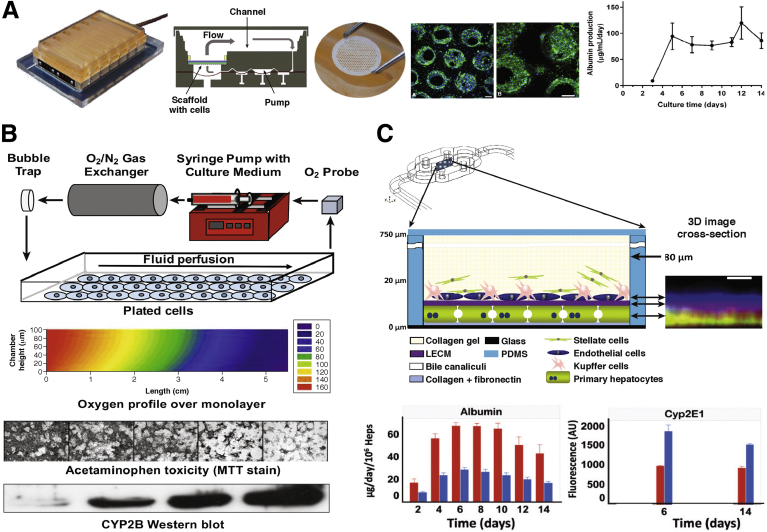
Engineered liver platforms incorporating fluid flow and zonated functions. (A) The LiverChip platform.100Left to right: A cell culture plate is attached to a pneumatic plate forming 12 fluidically isolated bioreactors per plate footprint. Bioreactor cross-section schematic is shown. A collagen-coated polystyrene scaffold (1-cm diameter) containing microchannels is placed into each bioreactor for cell culture. Low-magnification and high-magnification immunofluorescent images showing PHH morphology after 7 days (green, f-actin; blue, Hoescht). Scale bar: 100 μm. Albumin secretion from the LiverChip over time. (B) Zonated hepatocyte cultures in a parallel-plate bioreactor.113, 114Top to bottom: Parallel-plate bioreactor schematic to expose cells to an oxygen gradient. Two-dimensional contour plot of predicted oxygen concentration profile in cross-section of bioreactor. Cells at the bioreactor outlet are exposed to a lower oxygen tension than cells at the bioreactor inlet. Rat hepatocyte bioreactor treated with acetaminophen showed greater (zonal) toxicity near the bioreactor outlet relative to the inlet as assessed by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) stain. Higher (zonal) amount of CYP2B enzyme protein at the outlet of the bioreactor as compared with the inlet, suggesting that acetaminophen was bioactivated by CYP2B into a greater amount of toxic metabolite at the outlet. (C) Microfluidic hepatocyte/nonparenchymal cell co-cultures.107, 108Top to bottom: Structure of a multilayered liver co-culture housed in a commercial microfluidic device. An X–Z projection shows cell layering from confocal images of labeled hepatocytes, the porcine-derived whole-liver extracellular matrix (LECM), and endothelial cells. Scale bar: 10 μm. The device was operated with different perfusion rates (5 μL/h for zone 1, periportal - red bars, and 15 μL/h for zone 3, perivenous - blue bars) to subject the co-cultures to different oxygen tensions as in liver zonation. Albumin level was measured in the efflux at the device outlet, whereas CYP2E1 protein expression level was measured via imaging of a fluorescently labeled antibody. Heps, hepatocytes; PDMS, polydimethylsiloxane.
Other groups have used polydimethylsiloxane (PDMS)–based microfluidic devices to perfuse liver co-cultures for drug screening. PDMS offers the advantages of rapid prototyping of different device designs and is a biocompatible and transparent (for microscopy) material. For instance, Kane et al102 developed an 8 × 8 element nonaddressable array of microfluidic wells containing MPCCs of rat hepatocytes and 3T3-J2 fibroblasts that were independently perfused with culture medium and oxygen. In another study, Novik et al103 showed that the production of drug metabolites was observed at a greater rate in perfused hepatocyte-endothelial co-cultures relative to static controls. Similarly, Esch et al104 found higher albumin and urea secretions in perfused co-cultures of PHHs and a liver NPC mixture (fibroblasts, HSCs, and KCs) compared with static controls.
More recent PDMS-based microfluidic devices use multiple chambers to mimic the sinusoidal architecture of the liver. For instance, Kang et al105 found that primary rat hepatocytes maintained normal morphology and produced urea for 30 days when they were cultured on one side of a Transwell membrane while immortalized bovine aortic endothelial cells were cultured on the other side of the membrane that was subjected to dual-channel microfluidic perfusion. Prodanov et al106 used a polyethylene terephthalate membrane to separate 2 cell culture chambers in a microfluidic device. PHHs were seeded in the bottom chamber and overlaid with a collagen gel containing immortalized HSCs (LX-2 line), while a mixture of EA.hy926 endothelial cell line and U937 monocyte cell line was seeded in the top chamber. The perfused co-cultures showed higher albumin and urea secretions than static co-cultures for approximately 4 weeks. In another device, Vernetti et al107 created layered liver co-cultures in a single-chamber, commercially available microfluidic device (Nortis, Inc, Seattle, WA). PHHs were allowed to first attach overnight, followed by seeding of a mixture of EA.hy926 cells and U937 cells on top of the attached PHHs, and then the co-culture was covered with LX-2 cells embedded in a collagen gel. In this device, approximately 20% of the PHHs were transduced with lentivirus carrying biosensors for apoptosis (cytochrome C) and reactive oxygen species (hydrogen peroxide) toward detecting cell responses to drugs and other stimuli using high-content imaging. A more recent iteration of this device uses a gel composed of porcine-derived whole-liver extracellular matrix instead of rat tail collagen, primary human microvascular endothelial cells instead of the EA.hy926 cell line, THP-1 monocyte cell line instead of the U-937 cell line, and culture medium with reduced serum as well as soluble porcine-derived whole-liver extracellular matrix, which better supports NPC functions.108
In addition to the benefits of perfusion on the functions of liver co-cultures, perfusion also can subject the cells to gradients of oxygen, nutrients, and hormones, which have been shown to lead to zonation or differential functions in hepatocytes across the length of the sinusoid.109, 110 DILI also can manifest itself with a zonal pattern dependent on the mechanism of action of the drug and its metabolism by specific isoenzymes in the hepatocytes.111, 112 Allen and Bhatia113 described a parallel-plate bioreactor with oxygen gradients that was used to induce an in vivo–like zonal pattern of CYP450s and acetaminophen toxicity in rat hepatocyte cultures, and Allen et al114 described hepatocyte-fibroblast co-cultures (Figure 2B). More recently, layered human hepatocyte–NPC co-cultures were subjected to zone 1 oxygen (10%–12%) or zone 3 oxygen (3%–5%) levels via variable culture medium perfusion rates in separate devices (Figure 2C).108 Zone 1 co-cultures showed greater levels of oxidative phosphorylation, albumin secretion, and urea synthesis compared with zone 3 co-cultures, while zone 3 co-cultures showed greater levels of α1-antitrypsin activity, glycolysis, steatosis, CYP2E1 activity, and acetaminophen toxicity compared with zone 1 co-cultures; these findings are consistent with known zonation outcomes in vivo. In contrast to an oxygen gradient, McCarty et al115 generated a gradient of soluble factors (ie, hormones and drugs) onto a rat hepatocyte monolayer using a microfluidic device. Subjecting the cells to glucagon and insulin gradients led to an expected staining pattern for glycogen, such that cells contained less cytoplasmic glycogen in the presence of high levels of glucagon and more glycogen in the presence of high levels of insulin. Furthermore, cultures that were subjected to a gradient of 3-methylcholanthrene, an inducer of glutathione S-transferase and CYP450 enzymes, showed greater hepatotoxicity of allyl alcohol in the low 3-methylcholanthrene region and greater hepatotoxicity of acetaminophen in the high 3-methylcholanthrene region.
Implications For Clinical Liver-Assist Devices
It is expected that progress toward the development of highly functional in vitro culture models will provide a reciprocal benefit for the advancement of bioartificial liver (BAL) devices that can be used as clinical treatments. Similar to the drug screening platforms, the design of an effective BAL device is dependent on the incorporation of the appropriate environmental and organizational cues that enable maximal survival and function of the hepatocellular component. A range of modifications aimed at optimizing cellular performance within BAL devices have been explored. In particular, because of the enhanced function of hepatocyte aggregates relative to single-cell suspensions, many device configurations contain either attached or encapsulated hepatocellular spheroids.116, 117, 118, 119, 120 In the modular extracorporeal liver support system (Charite, Berlin, Germany), hepatocytes are aggregated in co-culture with liver NPCs, resulting in the formation of tissue-like organoid structures.121 Overall, environmental conditions within a BAL device, such as oxygen tension and fluid shear forces, can significantly affect hepatocyte functions.122 In addition, both the convective and diffusive properties of the systems must be optimized to provide vital nutrients to the cells while simultaneously allowing export of therapeutic cellular products. Currently, although clinical efficacy of BAL devices remains limited, improvements in device and trial design continue to be implemented. Many of these efforts leverage insights gained from small-scale in vitro bioreactor systems, which have been used to systematically examine the effects of shear stress and oxygen tension on hepatocyte function.123, 124 Furthermore, a number of ongoing studies are aimed at identifying protein or metabolite biomarkers that can serve as quantitative indicators of BAL device performance.
Conclusions and Future Outlook
In contrast to conventional monolayers, sophisticated engineering tools, such as micropatterning,49 microfluidics,107, 125 specialized plates,79, 126 biomaterial scaffolds,127 and 3D bioprinting,92 allow more precise control over the liver cell microenvironment, which has led to stabilized liver functions for several weeks (Table 1). Such longevity of functions has proven highly useful for chronic treatment with drugs and other stimuli (eg, viruses and cell differentiation cues) to significantly enhance the sensitivity for the prediction of clinical outcomes as compared with short-term (<24 hours) treatment of monolayers.4 Most of the liver models that show high levels of function over several weeks co-cultivate hepatocytes with NPCs; even non–liver-derived NPC types (ie, 3T3-J2 murine embryonic fibroblasts) can induce high levels of function in hepatocytes from multiple species, including human beings, which suggests that the molecular mediators underlying the co-culture effect are relatively well conserved across species.37 Often, the exact liver architecture is not fully recapitulated in engineered liver co-cultures (eg, disorganized spheroids/organoids and circular islands in MPCCs), but still leads to healthy and functioning liver cells, which suggests that the biochemical and biophysical microenvironment around the cells is ultimately more important for generating high-fidelity human liver models than mimicking the macro-architecture of the native liver. Furthermore, optimizing the homotypic and heterotypic cell–cell interactions using technology (eg, micropatterning, specialized plates to create controlled-sized spheroids, and bioprinting) is important to enhance liver functions and enable reproducible data sets across many experiments.
Table 1.
Benefits and Potential Limitations of Different Bioengineered Liver Models for In Vitro Studies
| Model | Benefits | Potential limitations |
|---|---|---|
| High-throughput cell microarrays | Enhanced capabilities to evaluate combinatorial effects of multiple signals Independent control of biochemical and biomechanicalcues Well-defined material properties of substrate and arrayed molecules Low material usage |
Primarily dependent on imaging-based read-outs Limited ability to investigate cell responses to gradients of microenvironmental signals |
| Randomly distributed (conventional) co-cultures |
Can be cultured in high-throughput plate formats No specialized system needed to establish co-cultures Different NPC types can be used to support hepatocytes Easily compatible with high-content imaging read-outs |
Can display variability in induction of hepatocyte functions with the choice of specific NPC type Can display morphologic and functional instability owing to regions of suboptimal cell–cell interactions within the monolayer Are not able to sustain infection with HBV/HCV and malaria owing to potential lack of complete hepatocyte polarity |
| Micropatterned co-cultures | Controlled cell–cell interactions allow for higher and stable functions for 4–6 weeks than randomly distributed co-cultures Modular design allows for the use of different NPC types without significantly altering hepatocyte homotypic interactions Can be infected with HBV, HCV, and malaria Display fatty liver phenotype when treated with hyperglycemic and/or high-fatty-acid–containing culture medium Compatible with high-content imaging read-outs |
Currently rely on collagen alone for hepatocyte attachment as opposed to more complex liver-inspired ECM Currently lack all liver stromal cells Use non–human-supporting fibroblasts Require specialized equipment and devices for patterning collagen |
| Randomly distributed spheroids/organoids | Can be created using a variety of different methods/plates Cell-secreted ECM protein matrix forms around the spheroids Multicellular interactions can be studied Maintenance of major liver functions for several weeks Have been shown to be compatible with multiple applications within the drug development pipeline |
Can be difficult to control disorganized cell type interactions over time Necrosis can occur in the center of larger spheroids Size variability can occur with some methods High-content imaging for entire spheroid may require expensive confocal microscopy depending on the spheroid size |
| Bioprinted organoids |
Precise control of cell placement allows formation of separate hepatocyte and NPC compartments Versatile method to create diverse architectures as desired Multicellular interactions can be studied Maintenance of major liver functions for 1 month Compatible with DILI screening and to model drug-induced fibrosis |
Printing resolution does not always allow placement of individual cells Low-throughput Requires complex and expensive equipment Requires significantly more cells than other higher-throughput/miniaturized methods Potential heterogeneous drug distribution across large printed tissues |
| Liver-on-a-chip (perfusion) devices | Dynamic fluid flow for nutrient and waste exchange Several commercial configurable devices available for cell culture and perfusion Layered architectures can be created with single-chamber or multichamber microfluidic device designs Sustained functionality for 2–4 wk Gradients of oxygen/hormones can be created to model zonal liver phenotypes |
Potential binding of drugs to tubing and materials used Large dead volume requiring higher quantities of novel compounds for the treatment of cell cultures Low-throughput Shear stress may cause lower hepatic functions May wash away built-up beneficial molecules with perfusion |
HBV, hepatitis B virus; HCV, hepatitis C virus.
Some key issues that pertain to bioengineered livers will need to be addressed moving forward. First, it will be useful to rely on similar endpoints and data normalization schemes (eg, based on cell number, protein, and/or RNA levels) when showing functionality and stability of a bioengineered liver so that the data can be compared across different laboratories. Second, consortia led by regulatory agencies will be important to evaluate multiple bioengineered livers using a consistent set of drugs and endpoints. Currently, it remains unclear how to directly compare the performance of bioengineered liver systems owing to a lack of standardized measurements by the same personnel in the same laboratories using the same cell donors. Third, bioengineered livers will need to mimic aspects of innate and adaptive immunity as well as different liver diseases to better predict idiosyncratic (unpredictable) DILI in the clinic. Nonetheless, bioengineered livers coupled with cellular stress markers have been shown to accurately detect the DILI potential of hepatotoxins that were previously thought to be idiosyncratic (eg, troglitazone, diclofenac, clozapine).60, 70, 92, 107 However, it is not currently possible to predict with in vitro approaches which specific individuals will adapt to cell stress and which individual will experience progressive and severe DILI. The differentiation of induced pluripotent stem cells (iPSCs) from thousands of human patients with different genetic backgrounds into multiple types of liver cells may potentially be useful to elucidate interindividual variations in DILI outcomes.69, 128 However, further improvements in the functional maturity of iHeps and other iPSC-derived liver NPC types will be needed to enable the routine use of these cells for drug screening; high-throughput cell microarrays are ideally suited to enable further progress in these goals given the large space of combinatorial microenvironmental cues that will need to be explored. Finally, although liver NPCs such as KCs, HSCs, and LSECs have all been incorporated into engineered liver co-cultures, it remains unclear how to incorporate biliary epithelial cells (ie, cholangiocytes) in models in such a way that they can form bile ducts that drain the contents of the hepatic bile canaliculi into a separate flow compartment than that used to mimic blood flow.
In conclusion, bioengineered liver models of increasing cellular and technologic complexities are available for investigating cell responses to drugs and other stimuli based on the posed hypotheses and throughput requirements. We anticipate that the ongoing development of more sophisticated engineering tools for manipulating cells in culture will lead to continual advances in bioengineered livers that show improving sensitivity for the prediction of clinically relevant drug and disease outcomes.
Acknowledgments
The authors would like to thank Chase Monckton, Christine Lin, and Brenton Ware for their helpful discussions.
Both authors contributed to the drafting and editing of the manuscript.
Footnotes
Conflicts of interest This author discloses the following: Salman R. Khetani is an equity holder in Ascendance Biotechnology, which has licensed the micropatterned co-culture and related systems from Massachusetts Institute of Technology and Colorado State University for commercial distribution. The remaining author discloses no conflicts.
Funding This work was funded by National Institutes of Health grants 1R21ES027622-01 (S.R.K.) and 1R03EB022254-01A1 (G.H.U.).
Contributor Information
Gregory H. Underhill, Email: gunderhi@illinois.edu.
Salman R. Khetani, Email: skhetani@uic.edu.
Supplementary Material
Graphical Summary.
References
- 1.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 2.Abboud G., Kaplowitz N. Drug-induced liver injury. Drug Saf. 2007;30:277–294. doi: 10.2165/00002018-200730040-00001. [DOI] [PubMed] [Google Scholar]
- 3.Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W., Dorato M., Van Deun K., Smith P., Berger B., Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 4.Lin C., Khetani S.R. Advances in engineered liver models for investigating drug-induced liver injury. Biomed Res Int. 2016;2016:1829148. doi: 10.1155/2016/1829148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkening S., Stahl F., Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- 6.Kwon S.J., Lee D.W., Shah D.A., Ku B., Jeon S.Y., Solanki K., Ryan J.D., Clark D.S., Dordick J.S., Lee M.-Y. High-throughput and combinatorial gene expression on a chip for metabolism-induced toxicology screening. Nat Commun. 2014;5:3739. doi: 10.1038/ncomms4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee D.W., Lee M.-Y., Ku B., Yi S.H., Ryu J.-H., Jeon R., Yang M. Application of the DataChip/MetaChip technology for the evaluation of ajoene toxicity in vitro. Arch Toxicol. 2014;88:283–290. doi: 10.1007/s00204-013-1102-9. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda J., Nakazawa K. Hepatocyte spheroid arrays inside microwells connected with microchannels. Biomicrofluidics. 2011;5:22205. doi: 10.1063/1.3576905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutolf M.P., Doyonnas R., Havenstrite K., Koleckar K., Blau H.M. Perturbation of single hematopoietic stem cell fates in artificial niches. Integr Biol (Camb) 2009;1:59–69. doi: 10.1039/b815718a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert P.M., Havenstrite K.L., Magnusson K.E., Sacco A., Leonardi N.A., Kraft P., Nguyen N.K., Thrun S., Lutolf M.P., Blau H.M. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gobaa S., Hoehnel S., Roccio M., Negro A., Kobel S., Lutolf M.P. Artificial niche microarrays for probing single stem cell fate in high throughput. Nat Methods. 2011;8:949–955. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- 12.Di Carlo D., Wu L.Y., Lee L.P. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 13.McKenna B.K., Evans J.G., Cheung M.C., Ehrlich D.J. A parallel microfluidic flow cytometer for high-content screening. Nat Methods. 2011;8:401–403. doi: 10.1038/nmeth.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecault V., Vaninsberghe M., Sekulovic S., Knapp D.J., Wohrer S., Bowden W., Viel F., McLaughlin T., Jarandehei A., Miller M., Falconnet D., White A.K., Kent D.G., Copley M.R., Taghipour F., Eaves C.J., Humphries R.K., Piret J.M., Hansen C.L. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat Methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- 15.King K.R., Wang S., Jayaraman A., Yarmush M.L., Toner M. Microfluidic flow-encoded switching for parallel control of dynamic cellular microenvironments. Lab Chip. 2008;8:107–116. doi: 10.1039/b716962k. [DOI] [PubMed] [Google Scholar]
- 16.Duncanson W.J., Lin T., Abate A.R., Seiffert S., Shah R.K., Weitz D.A. Microfluidic synthesis of advanced microparticles for encapsulation and controlled release. Lab Chip. 2012;12:2135–2145. doi: 10.1039/c2lc21164e. [DOI] [PubMed] [Google Scholar]
- 17.Tumarkin E., Tzadu L., Csaszar E., Seo M., Zhang H., Lee A., Peerani R., Purpura K., Zandstra P.W., Kumacheva E. High-throughput combinatorial cell co-culture using microfluidics. Integr Biol (Camb) 2011;3:653–662. doi: 10.1039/c1ib00002k. [DOI] [PubMed] [Google Scholar]
- 18.Kumachev A., Greener J., Tumarkin E., Eiser E., Zandstra P.W., Kumacheva E. High-throughput generation of hydrogel microbeads with varying elasticity for cell encapsulation. Biomaterials. 2011;32:1477–1483. doi: 10.1016/j.biomaterials.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Li C.Y., Wood D.K., Hsu C.M., Bhatia S.N. DNA-templated assembly of droplet-derived PEG microtissues. Lab Chip. 2011;11:2967–2975. doi: 10.1039/c1lc20318e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaim C.J., Chien S., Bhatia S.N. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 21.Brafman D.A., de Minicis S., Seki E., Shah K.D., Teng D., Brenner D., Willert K., Chien S. Investigating the role of the extracellular environment in modulating hepatic stellate cell biology with arrayed combinatorial microenvironments. Integr Biol (Camb) 2009;1:513–524. doi: 10.1039/b912926j. [DOI] [PubMed] [Google Scholar]
- 22.Brafman D.A., Shah K.D., Fellner T., Chien S., Willert K. Defining long-term maintenance conditions of human embryonic stem cells with arrayed cellular microenvironment technology. Stem Cells Dev. 2009;18:1141–1154. doi: 10.1089/scd.2008.0410. [DOI] [PubMed] [Google Scholar]
- 23.Soen Y., Mori A., Palmer T.D., Brown P.O. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaBarge M.A., Nelson C.M., Villadsen R., Fridriksdottir A., Ruth J.R., Stampfer M.R., Petersen O.W., Bissell M.J. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integr Biol (Camb) 2009;1:70–79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson D.G., Levenberg S., Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 26.Unadkat H.V., Hulsman M., Cornelissen K., Papenburg B.J., Truckenmuller R.K., Post G.F., Uetz M., Reinders M.J., Stamatialis D., van Blitterswijk C.A., de Boer J. An algorithm-based topographical biomaterials library to instruct cell fate. Proc Natl Acad Sci U S A. 2011;108:16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei Y., Saha K., Bogatyrev S.R., Yang J., Hook A.L., Kalcioglu Z.I., Cho S.W., Mitalipova M., Pyzocha N., Rojas F., Van Vliet K.J., Davies M.C., Alexander M.R., Langer R., Jaenisch R., Anderson D.G. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha K., Mei Y., Reisterer C.M., Pyzocha N.K., Yang J., Muffat J., Davies M.C., Alexander M.R., Langer R., Anderson D.G., Jaenisch R. Surface-engineered substrates for improved human pluripotent stem cell culture under fully defined conditions. Proc Natl Acad Sci U S A. 2011;108:18714–18719. doi: 10.1073/pnas.1114854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R., Liberski A., Sanchez-Martin R., Bradley M. Microarrays of over 2000 hydrogels–identification of substrates for cellular trapping and thermally triggered release. Biomaterials. 2009;30:6193–6201. doi: 10.1016/j.biomaterials.2009.07.055. [DOI] [PubMed] [Google Scholar]
- 30.Flaim C.J., Teng D., Chien S., Bhatia S.N. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cell Dev. 2008;17:29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 31.Bhatia S.N., Underhill G.H., Zaret K.S., Fox I.J. Cell and tissue engineering for liver disease. Sci Transl Med. 2014;6:245sr2. doi: 10.1126/scitranslmed.3005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang N.F., Patlolla B., Abilez O., Sharma H., Rajadas J., Beygui R.E., Zarins C.K., Cooke J.P. A matrix micropatterning platform for cell localization and stem cell fate determination. Acta Biomater. 2010;6:4614–4621. doi: 10.1016/j.actbio.2010.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodrow K.A., Wood M.J., Saucier-Sawyer J.K., Solbrig C., Saltzman W.M. Biodegradable meshes printed with extracellular matrix proteins support micropatterned hepatocyte cultures. Tissue Eng Part A. 2009;15:1169–1179. doi: 10.1089/ten.tea.2008.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mei Y., Cannizzaro C., Park H., Xu Q., Bogatyrev S.R., Yi K., Goldman N., Langer R., Anderson D.G. Cell-compatible, multicomponent protein arrays with subcellular feature resolution. Small. 2008;4:1600–1604. doi: 10.1002/smll. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaylan K.B., Ermilova V., Yada R.C., Underhill G.H. Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFbeta, and extracellular matrix. Sci Rep. 2016;6:23490. doi: 10.1038/srep23490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kourouklis A.P., Kaylan K.B., Underhill G.H. Substrate stiffness and matrix composition coordinately control the differentiation of liver progenitor cells. Biomaterials. 2016;99:82–94. doi: 10.1016/j.biomaterials.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Bhatia S.N., Balis U.J., Yarmush M.L., Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 38.West M.A., Manthei R., Bubrick M.P. Autoregulation of hepatic macrophage activation in sepsis. J Trauma. 1993;34:473–480. doi: 10.1097/00005373-199304000-00001. [DOI] [PubMed] [Google Scholar]
- 39.Michalopoulos G., Strom S.C., Kligerman A.D., Irons G.P., Novicki D.L. Mutagenesis induced by procarcinogens at the hypoxanthine-guanine phosphoribosyl transferase locus of human fibroblasts cocultured with rat hepatocytes. Cancer Res. 1981;41:1873–1878. [PubMed] [Google Scholar]
- 40.Guillouzo A., Morel F., Fardel O., Meunier B. Use of human hepatocyte cultures for drug metabolism studies. Toxicology. 1993;82:209–219. doi: 10.1016/0300-483x(93)90065-z. [DOI] [PubMed] [Google Scholar]
- 41.Mertens K., Rogiers V., Vercruysse A. Glutathione dependent detoxication in adult rat hepatocytes under various culture conditions. Arch Toxicol. 1993;67:680–685. doi: 10.1007/BF01973691. [DOI] [PubMed] [Google Scholar]
- 42.De La Vega F.M., Mendoza-Figueroa T. Dimethyl sulfoxide enhances lipid synthesis and secretion by long-term cultures of adult rat hepatocytes. Biochimie. 1991;73:621–624. doi: 10.1016/0300-9084(91)90033-w. [DOI] [PubMed] [Google Scholar]
- 43.Lebreton J.P., Daveau M., Hiron M., Fontaine M., Biou D., Gilbert D., Guguen-Guillouzo C. Long-term biosynthesis of complement component C3 and alpha-1 acid glycoprotein by adult rat hepatocytes in a co-culture system with an epithelial liver cell-type. Biochem J. 1986;235:421–427. doi: 10.1042/bj2350421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen J.W., Hassanein T., Bhatia S.N. Advances in bioartificial liver devices. Hepatology. 2001;34:447–455. doi: 10.1053/jhep.2001.26753. [DOI] [PubMed] [Google Scholar]
- 45.Bhatia S.N., Balis U.J., Yarmush M.L., Toner M. Microfabrication of hepatocyte/fibroblast co-cultures: role of homotypic cell interactions. Biotechnol Prog. 1998;14:378–387. doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- 46.Bhatia S.N., Balis U.J., Yarmush M.L., Toner M. Probing heterotypic cell interactions: hepatocyte function in microfabricated co-cultures. J Biomater Sci Polym Ed. 1998;9:1137–1160. doi: 10.1163/156856298x00695. [DOI] [PubMed] [Google Scholar]
- 47.Khetani S.R., Szulgit G., Del Rio J.A., Barlow C., Bhatia S.N. Exploring interactions between rat hepatocytes and nonparenchymal cells using gene expression profiling. Hepatology. 2004;40:545–554. doi: 10.1002/hep.20351. [DOI] [PubMed] [Google Scholar]
- 48.Khetani S.R., Chen A.A., Ranscht B., Bhatia S.N. T-cadherin modulates hepatocyte functions in vitro. FASEB J. 2008;22:3768–3775. doi: 10.1096/fj.07-105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khetani S.R., Bhatia S.N. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 50.Chan T.S., Yu H., Moore A., Khetani S.R., Tweedie D. Meeting the challenge of predicting hepatic clearance of compounds slowly metabolized by cytochrome P450 using a novel hepatocyte model, HepatoPac. Drug Metab Dispos. 2013;41:2024–2032. doi: 10.1124/dmd.113.053397. [DOI] [PubMed] [Google Scholar]
- 51.Lin C., Shi J., Moore A., Khetani S.R. Prediction of drug clearance and drug-drug interactions in microscale cultures of human hepatocytes. Drug Metab Dispos. 2016;44:127–136. doi: 10.1124/dmd.115.066027. [DOI] [PubMed] [Google Scholar]
- 52.Kratochwil N.A., Meille C., Fowler S., Klammers F., Ekiciler A., Molitor B., Simon S., Walter I., McGinnis C., Walther J., Leonard B., Triyatni M., Javanbakht H., Funk C., Schuler F., Lave T., Parrott N.J. Metabolic profiling of human long-term liver models and hepatic clearance predictions from in vitro data using nonlinear mixed-effects modeling. AAPS J. 2017;19:534–550. doi: 10.1208/s12248-016-0019-7. [DOI] [PubMed] [Google Scholar]
- 53.Dixit V., Moore A., Tsao H., Hariparsad N. Application of micropatterned cocultured hepatocytes to evaluate the inductive potential and degradation rate of major xenobiotic metabolizing enzymes. Drug Metab Dispos. 2016;44:250–261. doi: 10.1124/dmd.115.067173. [DOI] [PubMed] [Google Scholar]
- 54.Moore A., Chothe P.P., Tsao H., Hariparsad N. Evaluation of the interplay between uptake transport and CYP3A4 induction in micropatterned cocultured hepatocytes. Drug Metab Dispos. 2016;44:1910–1919. doi: 10.1124/dmd.116.072660. [DOI] [PubMed] [Google Scholar]
- 55.Ballard T.E., Wang S., Cox L.M., Moen M.A., Krzyzewski S., Ukairo O., Obach R.S. Application of a micropatterned cocultured hepatocyte system to predict preclinical and human-specific drug metabolism. Drug Metab Dispos. 2016;44:172–179. doi: 10.1124/dmd.115.066688. [DOI] [PubMed] [Google Scholar]
- 56.Wang W.W., Khetani S.R., Krzyzewski S., Duignan D.B., Obach R.S. Assessment of a micropatterned hepatocyte coculture system to generate major human excretory and circulating drug metabolites. Drug Metab Dispos. 2010;38:1900–1905. doi: 10.1124/dmd.110.034876. [DOI] [PubMed] [Google Scholar]
- 57.Ramsden D., Tweedie D.J., St George R., Chen L.Z., Li Y. Generating an in vitro-in vivo correlation for metabolism and liver enrichment of a hepatitis C virus drug, faldaprevir, using a rat hepatocyte model (HepatoPac) Drug Metab Dispos. 2014;42:407–414. doi: 10.1124/dmd.113.055947. [DOI] [PubMed] [Google Scholar]
- 58.Ramsden D., Tweedie D.J., Chan T.S., Taub M.E., Li Y. Bridging in vitro and in vivo metabolism and transport of faldaprevir in human using a novel cocultured human hepatocyte system, HepatoPac. Drug Metab Dispos. 2014;42:394–406. doi: 10.1124/dmd.113.055897. [DOI] [PubMed] [Google Scholar]
- 59.Trask O.J., Moore A., LeCluyse E.L. A micropatterned hepatocyte coculture model for assessment of liver toxicity using high-content imaging analysis. Assay Drug Dev Technol. 2014;12:16–27. doi: 10.1089/adt.2013.525. [DOI] [PubMed] [Google Scholar]
- 60.Khetani S.R., Kanchagar C., Ukairo O., Krzyzewski S., Moore A., Shi J., Aoyama S., Aleo M., Will Y. Use of micropatterned cocultures to detect compounds that cause drug-induced liver injury in humans. Toxicol Sci. 2013;132:107–117. doi: 10.1093/toxsci/kfs326. [DOI] [PubMed] [Google Scholar]
- 61.Davidson M.D., Lehrer M., Khetani S.R. Hormone and drug-mediated modulation of glucose metabolism in a microscale model of the human liver. Tissue Eng Part C Methods. 2015;21:716–725. doi: 10.1089/ten.tec.2014.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ploss A., Khetani S.R., Jones C.T., Syder A.J., Trehan K., Gaysinskaya V.A., Mu K., Ritola K., Rice C.M., Bhatia S.N. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shlomai A., Schwartz R.E., Ramanan V., Bhatta A., de Jong Y.P., Bhatia S.N., Rice C.M. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A. 2014;111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.March S., Ng S., Velmurugan S., Galstian A., Shan J., Logan D.J., Carpenter A.E., Thomas D., Sim B.K.L., Mota M.M., Hoffman S.L., Bhatia S.N. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microbe. 2013;14:104–115. doi: 10.1016/j.chom.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng S., March S., Galstian A., Hanson K., Carvalho T., Mota M.M., Bhatia S.N. Hypoxia promotes liver stage malaria infection in primary human hepatocytes in vitro. Dis Model Mech. 2014;7:215–224. doi: 10.1242/dmm.013490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu J.J., Henstock P.V., Dunn M.C., Smith A.R., Chabot J.R., de Graaf D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol Sci. 2008;105:97–105. doi: 10.1093/toxsci/kfn109. [DOI] [PubMed] [Google Scholar]
- 67.Manning F.J., Swartz M., editors. Review of the fialuridine (FIAU) clinical trials. National Academies Press; Washington, DC: 1995. Institute of Medicine (US) Committee to Review the Fialuridine (FIAU/FIAC) Clinical Trials. [PubMed] [Google Scholar]
- 68.Berger D.R., Ware B.R., Davidson M.D., Allsup S.R., Khetani S.R. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- 69.Davidson M.D., Ware B.R., Khetani S.R. Stem cell-derived liver cells for drug testing and disease modeling. Discov Med. 2015;19:349–358. [PMC free article] [PubMed] [Google Scholar]
- 70.Ware B.R., Berger D.R., Khetani S.R. Prediction of drug-induced liver injury in micropatterned co-cultures containing iPSC-derived human hepatocytes. Toxicol Sci. 2015;145:252–262. doi: 10.1093/toxsci/kfv048. [DOI] [PubMed] [Google Scholar]
- 71.Nguyen T.V., Ukairo O., Khetani S.R., McVay M., Kanchagar C., Seghezzi W., Ayanoglu G., Irrechukwu O., Evers R. Establishment of a hepatocyte-Kupffer cell coculture model for assessment of proinflammatory cytokine effects on metabolizing enzymes and drug transporters. Drug Metab Dispos. 2015;43:774–785. doi: 10.1124/dmd.114.061317. [DOI] [PubMed] [Google Scholar]
- 72.Davidson M.D., Kukla D., Khetani S.R. Microengineered cultures containing human hepatic stellate cells and hepatocytes for drug development. Integr Biol (Camb) 2017;9:662–677. doi: 10.1039/c7ib00027h. [DOI] [PubMed] [Google Scholar]
- 73.Hardwick R.N., Fisher C.D., Canet M.J., Scheffer G.L., Cherrington N.J. Variations in ATP-binding cassette transporter regulation during the progression of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2011;39:2395–2402. doi: 10.1124/dmd.111.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merrell M.D., Cherrington N.J. Drug metabolism alterations in nonalcoholic fatty liver disease. Drug Metab Rev. 2011;43:317–334. doi: 10.3109/03602532.2011.577781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ware B.R., Durham M.J., Monckton C.P., Khetani S.R. A cell culture platform to maintain long-term phenotype of primary human hepatocytes and endothelial cells. Cell Molec Gastroenterol Hepatol. 2017 doi: 10.1016/j.jcmgh.2017.11.007. in press. http://doi.org/10.1016/j.jcmgh.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Godoy P., Hewitt N.J., Albrecht U., Andersen M.E., Ansari N., Bhattacharya S., Bode J.G., Bolleyn J., Borner C., Böttger J., Braeuning A., Budinsky R.A., Burkhardt B., Cameron N.R., Camussi G., Cho C.-S., Choi Y.-J., Craig Rowlands J., Dahmen U., Damm G., Dirsch O., Donato M.T., Dong J., Dooley S., Drasdo D., Eakins R., Ferreira K.S., Fonsato V., Fraczek J., Gebhardt R., Gibson A., Glanemann M., Goldring C.E.P., Gómez-Lechón M.J., Groothuis G.M.M., Gustavsson L., Guyot C., Hallifax D., Hammad S., Hayward A., Häussinger D., Hellerbrand C., Hewitt P., Hoehme S., Holzhütter H.-G., Houston J.B., Hrach J., Ito K., Jaeschke H., Keitel V., Kelm J.M., Kevin Park B., Kordes C., Kullak-Ublick G.A., LeCluyse E.L., Lu P., Luebke-Wheeler J., Lutz A., Maltman D.J., Matz-Soja M., McMullen P., Merfort I., Messner S., Meyer C., Mwinyi J., Naisbitt D.J., Nussler A.K., Olinga P., Pampaloni F., Pi J., Pluta L., Przyborski S.A., Ramachandran A., Rogiers V., Rowe C., Schelcher C., Schmich K., Schwarz M., Singh B., Stelzer E.H.K., Stieger B., Stöber R., Sugiyama Y., Tetta C., Thasler W.E., Vanhaecke T., Vinken M., Weiss T.S., Widera A., Woods C.G., Xu J.J., Yarborough K.M., Hengstler J.G. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling, and ADME. Arch Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Acikgoz A., Giri S., Cho M.G., Bader A. Morphological and functional analysis of hepatocyte spheroids generated on poly-HEMA-treated surfaces under the influence of fetal calf serum and nonparenchymal cells. Biomolecules. 2013;3:242–269. doi: 10.3390/biom3010242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bell C.C., Hendriks D.F.G., Moro S.M.L., Ellis E., Walsh J., Renblom A., Fredriksson Puigvert L., Dankers A.C.A., Jacobs F., Snoeys J., Sison-Young R.L., Jenkins R.E., Nordling Å., Mkrtchian S., Park B.K., Kitteringham N.R., Goldring C.E.P., Lauschke V.M., Ingelman-Sundberg M. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep. 2016;6:25187. doi: 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Messner S., Agarkova I., Moritz W., Kelm J.M. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch Toxicol. 2013;87:209–213. doi: 10.1007/s00204-012-0968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Proctor W.R., Foster A.J., Vogt J., Summers C., Middleton B., Pilling M.A., Shienson D., Kijanska M., Strobel S., Kelm J.M., Morgan P., Messner S., Williams D. Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch Toxicol. 2017;91:2849–2863. doi: 10.1007/s00204-017-2002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kostadinova R., Boess F., Applegate D., Suter L., Weiser T., Singer T., Naughton B., Roth A. A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol Appl Pharmacol. 2013;268:1–16. doi: 10.1016/j.taap.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 82.Takayama K., Kawabata K., Nagamoto Y., Kishimoto K., Tashiro K., Sakurai F., Tachibana M., Kanda K., Hayakawa T., Furue M.K., Mizuguchi H. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 83.Lutolf M.P., Hubbell J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 84.Chen A.A., Thomas D.K., Ong L.L., Schwartz R.E., Golub T.R., Bhatia S.N. Humanized mice with ectopic artificial liver tissues. Proc Natl Acad Sci U S A. 2011;108:11842–11847. doi: 10.1073/pnas.1101791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li C.Y., Stevens K.R., Schwartz R.E., Alejandro B.S., Huang J.H., Bhatia S.N. Micropatterned cell-cell interactions enable functional encapsulation of primary hepatocytes in hydrogel microtissues. Tissue Eng Part A. 2014;20:2200–2212. doi: 10.1089/ten.tea.2013.0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tasnim F., Toh Y.-C., Qu Y., Li H., Phan D., Narmada B.C., Ananthanarayanan A., Mittal N., Meng R.Q., Yu H. Functionally enhanced human stem cell derived hepatocytes in galactosylated cellulosic sponges for hepatotoxicity testing. Mol Pharm. 2016;13:1947–1957. doi: 10.1021/acs.molpharmaceut.6b00119. [DOI] [PubMed] [Google Scholar]
- 87.Larkin A.L., Rodrigues R.R., Murali T.M., Rajagopalan P. Designing a multicellular organotypic 3D liver model with a detachable, nanoscale polymeric Space of Disse. Tissue Eng Part C Methods. 2013;19:875–884. doi: 10.1089/ten.tec.2012.0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.R., Ueno Y., Zheng Y.W., Koike N., Aoyama S., Adachi Y., Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 89.Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M., Ellis E., van Wenum M., Fuchs S.A., de Ligt J., van de Wetering M., Sasaki N., Boers S.J., Kemperman H., de Jonge J., Ijzermans J.N., Nieuwenhuis E.E., Hoekstra R., Strom S., Vries R.R., van der Laan L.J., Cuppen E., Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Camp J.G., Sekine K., Gerber T., Loeffler-Wirth H., Binder H., Gac M., Kanton S., Kageyama J., Damm G., Seehofer D., Belicova L., Bickle M., Barsacchi R., Okuda R., Yoshizawa E., Kimura M., Ayabe H., Taniguchi H., Takebe T., Treutlein B. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 91.Sampaziotis F., Justin A.W., Tysoe O.C., Sawiak S., Godfrey E.M., Upponi S.S., Gieseck R.L., 3rd, de Brito M.C., Berntsen N.L., Gomez-Vazquez M.J., Ortmann D., Yiangou L., Ross A., Bargehr J., Bertero A., Zonneveld M.C.F., Pedersen M.T., Pawlowski M., Valestrand L., Madrigal P., Georgakopoulos N., Pirmadjid N., Skeldon G.M., Casey J., Shu W., Materek P.M., Snijders K.E., Brown S.E., Rimland C.A., Simonic I., Davies S.E., Jensen K.B., Zilbauer M., Gelson W.T.H., Alexander G.J., Sinha S., Hannan N.R.F., Wynn T.A., Karlsen T.H., Melum E., Markaki A.E., Saeb-Parsy K., Vallier L. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat Med. 2017;23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
- 92.Norona L.M., Nguyen D.G., Gerber D.A., Presnell S.C., LeCluyse E.L. Editor's highlight: modeling compound-induced fibrogenesis in vitro using three-dimensional bioprinted human liver tissues. Toxicol Sci. 2016;154:354–367. doi: 10.1093/toxsci/kfw169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jeon H., Kang K., Park S.A., Kim W.D., Paik S.S., Lee S.H., Jeong J., Choi D. Generation of multilayered 3D structures of HepG2 cells using a bio-printing technique. Gut Liver. 2017;11:121–128. doi: 10.5009/gnl16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma X., Qu X., Zhu W., Li Y.-S., Yuan S., Zhang H., Liu J., Wang P., Lai C.S.E., Zanella F., Feng G.-S., Sheikh F., Chien S., Chen S. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A. 2016;113:2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Powers M.J., Domansky K., Kaazempur-Mofrad M.R., Kalezi A., Capitano A., Upadhyaya A., Kurzawski P., Wack K.E., Stolz D.B., Kamm R., Griffith L.G. A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol Bioeng. 2002;78:257–269. doi: 10.1002/bit.10143. [DOI] [PubMed] [Google Scholar]
- 96.Domansky K., Inman W., Serdy J., Dash A., Lim M.H., Griffith L.G. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10:51–58. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hwa A.J., Fry R.C., Sivaraman A., So P.T., Samson L.D., Stolz D.B., Griffith L.G. Rat liver sinusoidal endothelial cells survive without exogenous VEGF in 3D perfused co-cultures with hepatocytes. FASEB J. 2007;21:2564–2579. doi: 10.1096/fj.06-7473com. [DOI] [PubMed] [Google Scholar]
- 98.Tsamandouras N., Kostrzewski T., Stokes C.L., Griffith L.G., Hughes D.J., Cirit M. Quantitative assessment of population variability in hepatic drug metabolism using a perfused three-dimensional human liver microphysiological system. J Pharmacol Exp Ther. 2017;360:95–105. doi: 10.1124/jpet.116.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sarkar U., Rivera-Burgos D., Large E.M., Hughes D.J., Ravindra K.C., Dyer R.L., Ebrahimkhani M.R., Wishnok J.S., Griffith L.G., Tannenbaum S.R. Metabolite profiling and pharmacokinetic evaluation of hydrocortisone in a perfused three-dimensional human liver bioreactor. Drug Metab Dispos. 2015;43:1091–1099. doi: 10.1124/dmd.115.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Long T.J., Cosgrove P.A., Dunn R.T., 2nd, Stolz D.B., Hamadeh H., Afshari C., McBride H., Griffith L.G. Modeling therapeutic antibody-small molecule drug-drug interactions using a three-dimensional perfusable human liver coculture platform. Drug Metab Dispos. 2016;44:1940–1948. doi: 10.1124/dmd.116.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarkar U., Ravindra K.C., Large E., Young C.L., Rivera-Burgos D., Yu J., Cirit M., Hughes D.J., Wishnok J.S., Lauffenburger D.A., Griffith L.G., Tannenbaum S.R. Integrated assessment of diclofenac biotransformation, pharmacokinetics, and omics-based toxicity in a 3D human liver-immunocompetent co-culture system. Drug Metab Dispos. 2017;45:855–866. doi: 10.1124/dmd.116.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kane B.J., Zinner M.J., Yarmush M.L., Toner M. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem. 2006;78:4291–4298. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- 103.Novik E., Maguire T.J., Chao P., Cheng K.C., Yarmush M.L. A microfluidic hepatic coculture platform for cell-based drug metabolism studies. Biochem Pharmacol. 2010;79:1036–1044. doi: 10.1016/j.bcp.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Esch M.B., Prot J.M., Wang Y.I., Miller P., Llamas-Vidales J.R., Naughton B.A., Applegate D.R., Shuler M.L. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab Chip. 2015;15:2269–2277. doi: 10.1039/c5lc00237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang Y.B.A., Sodunke T.R., Lamontagne J., Cirillo J., Rajiv C., Bouchard M.J., Noh M. Liver sinusoid on a chip: long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol Bioeng. 2015;112:2571–2582. doi: 10.1002/bit.25659. [DOI] [PubMed] [Google Scholar]
- 106.Prodanov L., Jindal R., Bale S.S., Hegde M., McCarty W.J., Golberg I., Bhushan A., Yarmush M.L., Usta O.B. Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol Bioeng. 2016;113:241–246. doi: 10.1002/bit.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vernetti L.A., Senutovitch N., Boltz R., Debiasio R., Shun T.Y., Gough A., Taylor D.L. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med. 2016;241:101–114. doi: 10.1177/1535370215592121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee-Montiel F.T., George S.M., Gough A.H., Sharma A.D., Wu J., DeBiasio R., Vernetti L.A., Taylor D.L. Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Exp Biol Med (Maywood) 2017;242:1617–1632. doi: 10.1177/1535370217703978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kietzmann T. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol. 2017;11:622–630. doi: 10.1016/j.redox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jungermann K., Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Ann Rev Nutr. 1996;16:179–203. doi: 10.1146/annurev.nu.16.070196.001143. [DOI] [PubMed] [Google Scholar]
- 111.Anundi I., Lähteenmäki T., Rundgren M., Moldeus P., Lindros K.O. Zonation of acetaminophen metabolism and cytochrome P450 2E1-mediated toxicity studied in isolated periportal and perivenous hepatocytes. Biochem Pharmacol. 1993;45:1251–1259. doi: 10.1016/0006-2952(93)90277-4. [DOI] [PubMed] [Google Scholar]
- 112.Soto-Gutierrez A., Gough A., Vernetti L.A., Taylor D.L., Monga S.P. Pre-clinical and clinical investigations of metabolic zonation in liver diseases: the potential of microphysiology systems. Exp Biol Med (Maywood) 2017;242:1605–1616. doi: 10.1177/1535370217707731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Allen J.W., Bhatia S.N. Formation of steady-state oxygen gradients in vitro: application to liver zonation. Biotechnol Bioeng. 2003;82:253–262. doi: 10.1002/bit.10569. [DOI] [PubMed] [Google Scholar]
- 114.Allen J.W., Khetani S.R., Bhatia S.N. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol Sci. 2005;84:110–119. doi: 10.1093/toxsci/kfi052. [DOI] [PubMed] [Google Scholar]
- 115.McCarty W.J., Usta O.B., Yarmush M.L. A microfabricated platform for generating physiologically-relevant hepatocyte zonation. Sci Rep. 2016;6:26868. doi: 10.1038/srep26868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rozga J., Holzman M.D., Ro M.S., Griffin D.W., Neuzil D.F., Giorgio T., Moscioni A.D., Demetriou A.A. Development of a hybrid bioartificial liver. Ann Surg. 1993;217:502–511. doi: 10.1097/00000658-199305010-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van de Kerkhove M.P., Hoekstra R., Chamuleau R.A., van Gulik T.M. Clinical application of bioartificial liver support systems. Ann Surg. 2004;240:216–230. doi: 10.1097/01.sla.0000132986.75257.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Park J.K., Lee D.H. Bioartificial liver systems: current status and future perspective. J Biosci Bioeng. 2005;99:311–319. doi: 10.1263/jbb.99.311. [DOI] [PubMed] [Google Scholar]
- 119.Coward S.M., Legallais C., David B., Thomas M., Foo Y., Mavri-Damelin D., Hodgson H.J., Selden C. Alginate-encapsulated HepG2 cells in a fluidized bed bioreactor maintain function in human liver failure plasma. Artif Organs. 2009;33:1117–1126. doi: 10.1111/j.1525-1594.2009.00821.x. [DOI] [PubMed] [Google Scholar]
- 120.Rahman T.M., Selden C., Khalil M., Diakanov I., Hodgson H.J. Alginate-encapsulated human hepatoblastoma cells in an extracorporeal perfusion system improve some systemic parameters of liver failure in a xenogeneic model. Artif Organs. 2004;28:476–482. doi: 10.1111/j.1525-1594.2004.07259.x. [DOI] [PubMed] [Google Scholar]
- 121.Sauer I.M., Neuhaus P., Gerlach J.C. Concept for modular extracorporeal liver support for the treatment of acute hepatic failure. Metab Brain Dis. 2002;17:477–484. doi: 10.1023/a:1021938708670. [DOI] [PubMed] [Google Scholar]
- 122.Wang Y., Susando T., Lei X., Anene-Nzelu C., Zhou H., Liang L.H., Yu H. Current development of bioreactors for extracorporeal bioartificial liver (review) Biointerphases. 2010;5:FA116–FA131. doi: 10.1116/1.3521520. [DOI] [PubMed] [Google Scholar]
- 123.Park J., Berthiaume F., Toner M., Yarmush M.L., Tilles A.W. Microfabricated grooved substrates as platforms for bioartificial liver reactors. Biotechnol Bioeng. 2005;90:632–644. doi: 10.1002/bit.20463. [DOI] [PubMed] [Google Scholar]
- 124.Tilles A.W., Baskaran H., Roy P., Yarmush M.L., Toner M. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a microchannel flat-plate bioreactor. Biotechnol Bioeng. 2001;73:379–389. doi: 10.1002/bit.1071. [DOI] [PubMed] [Google Scholar]
- 125.Sivaraman A., Leach J.K., Townsend S., Iida T., Hogan B.J., Stolz D.B., Fry R., Samson L.D., Tannenbaum S.R., Griffith L.G. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr Drug Metab. 2005;6:569–591. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- 126.Miyamoto Y., Ikeuchi M., Noguchi H., Yagi T., Hayashi S. Spheroid formation and evaluation of hepatic cells in a three-dimensional culture device. Cell Med. 2015;8:47–56. doi: 10.3727/215517915X689056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Tsang V., Chen A.A., Cho L.M., Jadin K.D., Sah R.L., DeLong S., West J.L., Bhatia S.N. Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels. FASEB J. 2007;21:790–801. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 128.Scott C.W., Peters M.F., Dragan Y.P. Human induced pluripotent stem cells and their use in drug discovery for toxicity testing. Toxicol Lett. 2013;219:49–58. doi: 10.1016/j.toxlet.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 129.Nguyen D.G., Funk J., Robbins J.B., Crogan-Grundy C., Presnell S.C., Singer T., Roth A.B. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS One. 2016;11:e0158674. doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]