Abstract
Heat shock proteins (HSPs), also known as molecular chaperones, participate in important cellular processes, such as protein aggregation, disaggregation, folding, and unfolding. HSPs have cytoprotective functions that are commonly explained by their antiapoptotic role. Their involvement in anticancer drug resistance has been the focus of intense research efforts, and the relationship between HSP induction and DNA repair mechanisms has been in the spotlight during the past decades. Because DNA is permanently subject to damage, many DNA repair pathways are involved in the recognition and removal of a diverse array of DNA lesions. Hence, DNA repair mechanisms are key to maintain genome stability. In addition, the interactome network of HSPs with DNA repair proteins has become an exciting research field and so their use as emerging targets for cancer therapy. This article provides a historical overview of the participation of HSPs in DNA repair mechanisms as part of their molecular chaperone capabilities.
Keywords: Heat shock proteins, DNA repair, DNA damage response, Molecular chaperones
Introduction
Heat shock proteins (HSPs) have been extensively studied since their discovery in the early 1960s by Ferruccio Ritossa (Ritossa and Vonborstel 1964). These highly conserved molecular chaperones are present in all living cells and have been implicated in multiple physiological processes. They are known as key players in processes associated with development, differentiation, survival, aging, and death. At the cellular level, they participate in protein homeostasis, preventing protein denaturation and unfolding under normal and stressful conditions (e.g., high temperature, hypoxia, heavy metals, chemical agents, and antineoplastic drugs) (Macario and Conway de Macario 2007). On the other hand, HSPs are commonly overexpressed in tumor cells, and they are known as “chaperones of tumorigenesis” because of their functions in promoting cancer cell proliferation and apoptosis inhibition (Calderwood et al. 2006). HSPs have also been involved in resistance to chemotherapy and radiotherapy (Wu et al. 2017).
HSPs were traditionally classified according to their molecular weight, but to avoid confusions because of the expanding number of members in the families, the nomenclature system recommended by the Human Genome Organization (HUGO) Gene Nomenclature Committee was adopted for all HSPs. It includes HSPH (HSP110), HSPC (HSP90), HSPA (HSP70), DNAJ (HSP40), and HSPB (small HSP) as well as HSPD/E (HSP60/HSP10) and CCT (TRiC) for the human chaperonin families (Kampinga et al. 2009). The expression of HSPs is transcriptionally regulated by heat shock factors (HSFs), which can bind to sequences located upstream of HSP genes called heat shock elements (HSEs) to induce gene expression (Akerfelt et al. 2010).
As mentioned previously, several stressful conditions have been shown to induce the expression of HSPs. Hyperthermia is one of the most exhaustively studied agents. It causes DNA, protein and membrane damage, interferes with cell cycle, DNA, and protein synthesis and may result in cell death (Roti Roti 2008). Hyperthermia is known to cause an increase in the protein/DNA ratio, mainly of non-histone proteins, in the nuclei of heat shocked CHO and HeLa cells. This protein aggregation is reflected in a relative increase of the nuclear protein mass (Tomasovic et al. 1978; Roti Roti et al. 1979; Roti Roti and Wilson 1984). The protein/DNA ratio increase seems to be time/temperature dependent (Roti Roti et al. 1979; Laszlo et al. 1992).
In parallel, several research groups have extensively described the phenomenon of thermotolerance and thermoresistance. A transient and moderate elevation of the temperature (heat shock (HS)) can induce the synthesis of HSPs and the resistance against other damaging agents, such as ionizing radiation (Plesset et al. 1987; Mitchel and Morrison 1982a, b, 1986). Furthermore, in 1991, it was shown that HSPA is involved in the protection of the cells against a lethal heat treatment and may be responsible for the phenomenon of thermotolerance (Angelidis et al. 1991). In this context, a direct role of inducible DNA repair has been proposed based on the observation of a decrease of the mutation rate induced by MNNG (N-methyl-N′-nitro-N-nitroso-guanidine) and MNU (N-methyl-N-nitrosourea) after a HS (Mitchel and Morrison 1987). Moreover, the heat shock blocked the increase in mutation frequency induced by elevated temperatures in Saccharomyces cerevisiae, which suggests that the enzymes involved in repair of premutational damage may be more resistant to denaturation (Nunes et al. 1993).
The nuclear matrix mass increase induced by HS was defined as “heat-induced excess nuclear proteins” (HIENP) that were involved in heat-induced cell killing, inhibition of DNA synthesis, and inhibition of DNA repair in irradiated cells (Roti Roti and Painter 1982; Bodell et al. 1984; Warters et al. 1987). HIENP were later characterized in HeLa cells, and the HSP70 family members were identified as the major components of the excess of nuclear proteins, excluding cytoskeletal elements (Laszlo et al. 1992). Nuclear proteins with molecular weights of 130, 95, 75, 58, 53, 48, 46, 37, 28, and 26 kDa were identified, among others. The HIENP response was possibly due to the increased binding of soluble nuclear proteins and the migration of cytoplasmic proteins into the nucleus (Laszlo et al. 1992).
HSPs also play important roles in protecting cells against damage, such as oxidative injury (Donati et al. 1990). For instance, HSP70 translocates to the nucleus after treatment of WISH cells (derived from human amniotic tissue) with hydrogen peroxide. The timing of the transient translocation of HSP70 into the nucleus coincides with the highest 8-hydroxy-2′-deoxyguanosine (8-OH-dG) levels in DNA. Similar results were found after adriamycin treatment, a chemotherapeutic drug that induces oxidative DNA damage. Hence, HSP70 may contribute to the nuclear transport of proteins to either repair DNA damage or to protect DNA from further damage (Abe et al. 1995).
Several studies have documented the cytoprotective role of HSPs and investigated the mechanisms involved. DNA is subject to several endogenous and exogenous agents that can lead to a variety of DNA lesions, but nevertheless, complex DNA repair machineries have evolved in human and other eukaryotic cells to maintain genome stability. UV and γ-ray-induced Hsp70 may bind to p53, suggesting that it has a regulating activity on cell cycle, DNA repair processes, and apoptosis (Matsumoto et al. 1995). HSPs, particularly Hsp70, also seem to be involved in radioresistance which is associated with the induction of radiation-induced DNA repair (Park et al. 2000). Furthermore, HSPs may have functional roles in BER (base excision repair), as the BER enzymes uracil DNA glycosylase (UDG) and human AP endonuclease (HAP1) have been found to be associated with Hsp70 and Hsp27 in HeLa cells (Mendez et al. 2000). These findings have provided a basis for examining the roles of HSPs in each DNA repair mechanism. In light of these new findings, HSPs have emerged as modulators of certain DNA repair pathways, an exciting field of research aiming at identifying novel candidate molecules as therapeutic targets for cancer therapy. Since most anticancer therapies exert their cytotoxic activities by disrupting normal DNA structure and function, proper DNA repair mechanisms become crucial for genome maintenance. An adequate DNA damage response is therefore critical to ensure an optimal therapeutic efficacy.
In this review, we will discuss the complex set of HSP interactions with components of DNA repair mechanisms and their potential interplay in these pathways, focusing on the implication in cancer treatment.
DNA damage repair and heat shock proteins
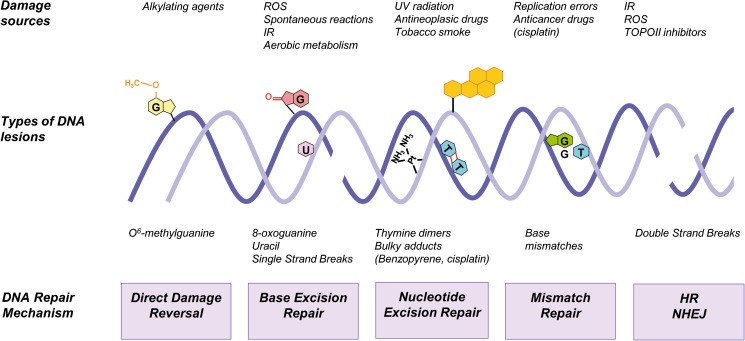
Cellular DNA constantly suffers numerous lesions caused by exogenous factors, such as ultraviolet light (UV), ionizing radiation (IR), and genotoxic chemicals (Barnes and Lindhal 2004), or by endogenous factors produced by normal cellular metabolism such as reactive oxygen species (ROS) (Tubbs and Nussenzweig 2017). These agents can cause different types of DNA damage, including single-stranded breaks (SSBs), double-stranded breaks (DSBs), single base modifications, and cross-links. If these errors are not properly corrected, the main consequences will be cell cycle arrest, cell death, or dangerous mutations with increased oncogenesis risk (Roos et al. 2016; Jeggo et al. 2016). However, mammalian cells bear a complex network of signaling pathways and DNA repair mechanisms to protect genome stability. The term DNA damage response (DDR) comprises complex pathways involving the detection of the damage, activation of signaling networks and cell cycle checkpoint, and DNA repair or induction of cell death (Ciccia and Elledge 2010). Five DNA repair mechanisms are usually distinguished: (a) direct DNA damage reversal, (b) BER, (c) mismatch repair (MMR), (d) nucleotide excision repair (NER), and (e) homologous recombination (HR) and non-homologous end joining (NHEJ) (Fig. 1). DNA repair pathways were originally restricted to the nuclear compartment. Ample evidences indicate that mitochondria possess a number of DNA repair factors and mechanisms shared with the nuclear processes (Boesch et al. 2011). BER is the predominant DNA repair pathway in mitochondria, but it lacks effective MMR and completely lacks NER and classical NHEJ. The mitochondria possess efficient microhomology-mediated end joining activity (MMEJ) (Vasileiou et al. 2017). However, the mechanisms that maintain genome stability within the nucleolus are still poorly understood and the findings are inconclusive. Here, we provide a brief overview of the DDR and the major DNA repair mechanisms.
Fig. 1.
Sources of DNA damage and repair mechanisms. Endogenous and exogenous agents constantly impact on DNA. They may cause many different forms of DNA damage. The scheme shows the five major DNA repair mechanisms operating in the nucleus of mammalian cells capable of removing a wide range of DNA lesions: direct damage reversal, base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), homologous recombination (HR), and non-homologous end joining (NHEJ). The BER system may also be found in the mitochondria. ROS reactive oxygen species, IR ionizing radiation, TOPOII topoisomerase II
DNA damage response signaling pathway (DDR)
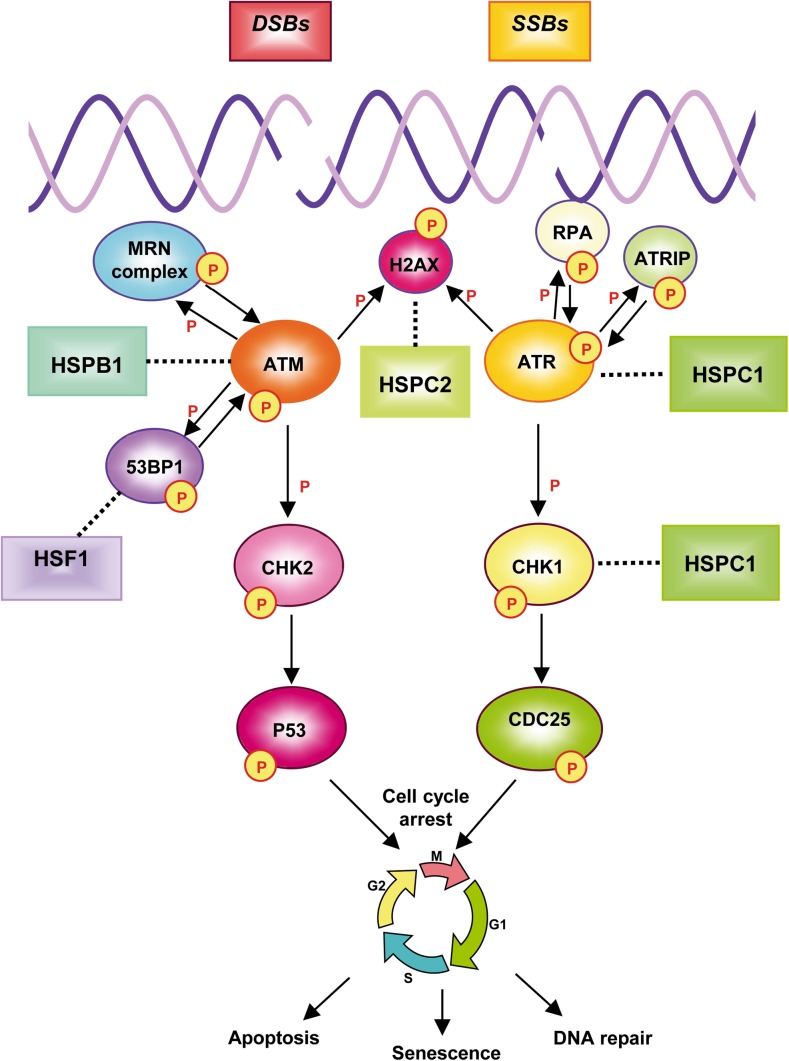
The DDR comprises coordinated pathways from DNA damage detection to cell cycle arrest, DNA repair, or even to apoptosis (Jackson and Bartek 2009). The phosphatidylinositol 3-kinase (PI3K)-like kinase (PIKK) family members ATM (ataxia telangiectasia mutated), ATR (ataxia telangiectasia and Rad3-related protein), and DNA-PK (DNA-dependent protein kinase) are considered the main regulators of the DDR. ATM and DNA-PK are critical for the signaling of DSBs, whereas ATR is mainly involved in the response to DNA SSBs and stalled replication forks (Bohgaki et al. 2010). ATM regulates the cell cycle through the phosphorylation of its downstream factors, CHK2 and p53, whereas ATR kinase phosphorylates CHK1 and p53. HSPs have been associated with proteins involved in the DDR signaling pathway. A comprehensive description of the DDR can be found from Jones (2012). Figure 2 reviews the relationship between the HSPs and the DDR cascade.
Fig. 2.
HSPs and the DNA damage response (DDR). Complex processes and signaling pathways take place in the cell in response to DNA single-strand breaks (SSBs) and double strand breaks (DSBs). SSBs are detected by RPA and DSBs by the MRN complex (MRE11-RAD50-NBS1). RPA and MRN mediate the recruitment of ATM and ATR, respectively. ATR is bound by the ATR-interacting protein (ATRIP), which interacts with RPA. Both ATM and ATR can phosphorylate (P) the histone variant H2AX on Ser139 (γH2AX) in the damaged DNA region. p53-binding protein 1 (53BP1) participates in the nuclear foci organization, promotes ATM activation, and facilitates the phosphorylation of specific substrates by ATM. The DNA damage signaling cascade continues with the phosphorylation of the cell cycle checkpoint kinases CHK1 and CHK2, which activate the downstream effectors: the tumor suppressor protein p53 or the CDC25 protein phosphatase. As a result, cell cycle progression is interrupted to allow DNA repair, senescence, or, in cases where DNA damage is too severe, apoptosis. The solid arrows show the DDR activation pathway. The dotted lines indicate the relationship between DDR and HSPs
The kinase CHK1 was the first DNA repair factor that had been proposed as an HSPC1 client protein. The interaction between CHK1 and HSPC1 was demonstrated by immunoprecipitation. The HSPC inhibitor 17-allylamino-17-demethoxygeldanamycin (17-AAG) induces the depletion of CHK1 and sensitizes cancer cells to gemcitabine (Arlander et al. 2003). Moreover, the inhibition of HSPC1 triggers CHK1 degradation by the proteasome pathway in multiple cell lines, demonstrating the importance of HSPC1 in stabilizing CHK1 (Nomura et al. 2005). As a general feature, the inhibition of HSPC1 usually causes destabilization and proteasomal degradation of its molecular partners. The Hsp90 (HSPC1) inhibitors geldanamycin (GA) and its derivative 17-AAG bind to the NH2-terminal ATP-binding domain of HSPC1 and lock it into an ATP-bound conformation (Blagg and Kerr 2006). HSPC inhibitors have been a helpful tool to support the discovery of new client proteins.
HSPB1, another HSP, is linked with ATM. IR leads to increased HSPB1 phosphorylation on Ser 78, while ATM inhibition prevents the post-translational changes (Cosentino et al. 2011). HSPB1 colocalizes with ATM in untreated and irradiated human head and neck squamous cell carcinoma cell lines. The inhibition of HSPB1 impairs ATM and CHK2 phosphorylation after radiation, increases γH2AX foci formation, and, consequently, attenuates DSB repair. In addition, the inhibition of ATM does not affect radiosensitization in shNT and shHSP27 cell lines, suggesting that the effect of HSPB1 inhibition is ATM dependent and implicating an impaired activation of ATM and, consequently, retarded DNA repair (Guttmann et al. 2013). It is important to clarify that H2AX is a member of the histone protein H2A family and that its phosphorylation on Ser139 is an early event in the response to DSBs, which is referred to as γH2AX because it was first observed in cells exposed to γ-rays (Kuo and Yang 2008).
The DNA damage sensor protein ATR has also been reported to be a client protein of HSPC1. The interaction between ATR and HSPC1 is disrupted by the presence of the HSPC1 inhibitor AUY922. The inhibitor also decreases ATR, phospo-ATR (pATRSer428), and CHK1 protein levels and increases γH2AX foci. Following radiation exposure, the HSPC1 inhibitor reduces the recruitment of ATR, CHK1, and 53BP1 to the DNA damage sites without affecting the recruitment of ATM. Finally, cells pretreated with AUY922 and collected at different time points after radiation exhibit longer comet tail moments than irradiated cells, suggesting that the inhibition of HSPC1 not only attenuates the accumulation of repair factors but also diminishes the DNA repair response (Ha et al. 2011).
Phosphorylated HSPC2 (HSP90α) has recently been proposed as a surrogate for γH2AX. HSPC2 phosphorylates on Thr7 and accumulates at radiation-induced DNA damage sites, forming repair foci (Quanz et al. 2012). Elevated levels of phosphorylated HSPC2 correlate with γH2AX in response to DNA damage across different cell lines. Interestingly, γH2AX shows a diffuse pattern about 3 h after irradiation in HSPC2-downregulated cells, whereas characteristic γH2AX foci remain in cells expressing HSPC2, thus indicating that HSPC2 is required for the maintenance of γH2AX foci and an efficient repair of the radiation-induced DNA damage. Basal levels of phosphorylated HSPC2 correlate with γH2AX expression in several human tumors, suggesting that HSPC2 phosphorylation may be a useful biomarker of DNA damage in tumor cells (Quanz et al. 2012). Recent studies revealed that ATM phosphorylates only nuclear HSPC2 in irradiated mouse embryonic fibroblasts (MEF). Moreover, HSPC2 knockdown sensitizes cell lines to IR and low levels of γH2AX express in HSPC2 null MEF cells, indicating that HSPC2 plays a role in radiation-induced DNA damage response. In summary, ATM is the kinase responsible for nuclear HSPC2 phosphorylation and, as previously proposed, HSPC2 is associated with γH2AX foci formation (Elaimy et al. 2016).
53BP1 (p53-binding protein 1) is an important regulator of the cellular response to DSBs that acts as a molecular scaffold in the recruitment of additional DSB responsive proteins to damaged chromatin, which promotes end joining of distal DNA ends. 53BP1 amplifies the ATM-dependent signaling pathway activation in response to low levels of DNA damage (Panier and Boulton 2014). Irradiation of rat fibroblast cells lacking HSF1 leads to the loss of 53BP1 foci formation at sites of DNA damage and a decrease in DSBs repair capacity, suggesting that the HSF1 activity is essential for its role in DNA damage repair (Li and Martinez 2011). Despite this promising result, the relationship between HSF1 and 53BP1 has been poorly studied.
Direct DNA damage reversal
In humans, mismatches and alkylated bases in DNA can be recognized and repaired by direct DNA damage reversal. DNA alkylation damage may arise from exposure to external agents, such as N-methyl-N′-nitro-N-nitroso-guanidine (MNNG), or from endogenous sources. Some common examples of these lesions are O6-methylguanine (O6-meG), 7-methylguanine (7-meG), and 3-methyladenine (3-meA), which can be recognized by specialized alkyltransferases, such as O6-meG-DNA methyltransferase (MGMT) (Tubbs and Nussenzweig 2017).
The expression of the repair enzyme MGMT is elevated in human tumors, but only 15–20% of the established cell lines appear to have MGMT deficiencies as a result of epigenetic gene silencing associated with promoter methylation (Bhakat and Mitra 2003). More than a decade ago, human MGMT emerged as target for improving cancer chemotherapy (Gerson 2004). MGMT strongly associates with HSPC2 (Hsp90alpha) and HSPC3 (Hsp90beta) in extracts of HT29 human colon cancer cells. These results obtained by tandem mass spectrometry provide unequivocal evidence that the HSPC family members are interacting partners of MGMT. No functional experiments have been, however, performed to clarify whether HSPC2 and HSPC3 play a role in direct DNA damage reversal. Given that these HSPs are inducible members of the HSPC family, they are probably accomplishing other functions besides their chaperone role (Niture et al. 2005).
Base excision repair
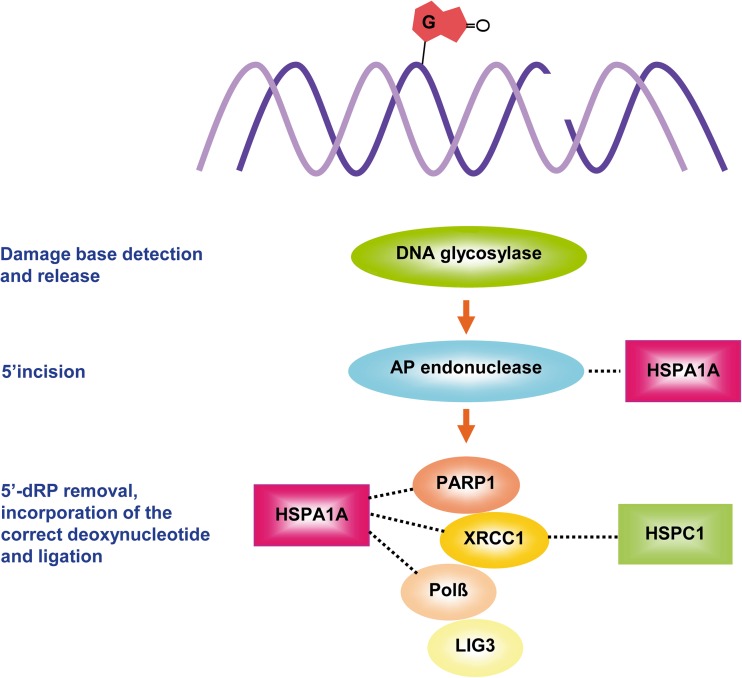
BER is the main mechanism for preventing mutagenesis. It removes different types of common DNA lesions from endogenous sources, such as chemical alterations of bases, including oxidations produced by ROS (8-hydroxyguanine), alkylations (O6-methylguanine), deaminations (guanine to xanthine), and hydroxylations (thymine glycol, cytosine glycol) (Dizdaroglu 2005). BER may also recognize single-strand breaks generated by ROS or by IR (Okano et al. 2003). Two BER subpathways have been characterized: the short and the long patch (Brenerman et al. 2014). Figure 3 summarizes the cascade of events that occurs in BER and HSP participation.
Fig. 3.
Participation of HSPs in the main steps of the base excision repair pathway. In human cells, BER mechanisms are achieved by two subpathways, short or long patch. The figure shows the main steps of the short-patch BER, which usually removes specific base damages. The recognition is initiated by specific DNA glycosylases (partially overlapping between them), which flip out the damaged base to form an abasic apurinic/apyrimidinic (AP) site in the DNA. An AP endonuclease (APE1 in mammalian cells) cleaves the 5′ phosphodiester bond, generating 3′OH and 5′dRP terminus, then produces the excision of the 3′ abasic fragment to form a gap that is filled by DNA polymerase β (Polβ). Finally, DNA ligase 3 (LIG3) performs the ligation step in association with XRCC1 (X-ray repair cross-complementing protein 1). Single-strand breaks, generated by ionizing radiation or OH radical, may also be corrected by the short-patch BER with the participation of the PARP1/XRCC1 complex. The figure also shows the involvement of HSPA1A and HSPC1 in BER steps
Over the past two decades, an increasing number of studies have revealed that the cell stress response is involved in DNA repair and, more specifically, in the BER system. HSPA1A binds to the human apurinic/apyrimidinic endonuclease 1 (HAP1 or APE1) and enhances its specific endonuclease activity 10- to 100-fold (Kenny et al. 2001). HSPA1A also stimulates repair by Polβ 5- to 10-fold through its 43- to 48-kDa N-terminal adenosine triphosphate (ATP-binding portion) domain (Mendez et al. 2000, 2003). The downregulation of HSPA1A by siRNA interferes with the glycosylase activity and BER after ionizing radiation in THP1 leukemic cells, while cell treatment with recombinant HSPA1A stimulates the rate of abasic site repair (Bases 2006). In addition, heat shock treatment of HeLa cells leads to a translocation of HSPA1A to the nuclei/nucleoli. HSPA1A associates with PARP-1 and/or XRCC1 at BRCT domains (BRCA1 C-terminus), protecting both proteins and allowing DNA SSB repair (Kotoglou et al. 2009).
HSPC is also involved in the BER system, but the underlying mechanisms have been less studied than in HSPA1A. The treatment of human lung cancer cell lines with the HSPC inhibitor 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin (17-DMAG) before radiation leads to an impaired DNA damage repair, measured by alkaline comet assay. The pretreatment with 17-DMAG suppresses the enzymatic activation of APE1 and DNA Polβ without affecting their protein expression levels. A decreased expression of phospho-ATM following HSPC inhibitor and radiation can, however, be observed (Koll et al. 2008).
HSPC1 interacts with XRCC1 unbound to Polβ in human glioblastoma cell lines and to protect XRCC1 from degradation mediated by E3 ubiquitin ligase CHIP. In addition, cisplatin and IR induce the phospho-HSPC1/XRCC1 heterodimer, especially in proliferating cells. The heterodimer may thus facilitate the role of XRCC1 in another DNA repair pathway independent of Polβ and BER. In summary, HSPC1 could contribute to the regulation of the DNA repair pathway choice through the stabilization and interaction with XRCC1 (Fang et al. 2014).
Mismatch repair
The mismatch repair system constitutes a post-replicative machinery that corrects base-base mismatches and IDLs (insertion/deletion loops) mainly generated during DNA replication. Mismatches may, however, also be caused by base oxidation or alkylation. MMR is highly conserved through evolution and occurs from bacteria and yeast to mammals. DNA mismatches that are not corrected can lead to genomic instability, especially at repeated sequence motifs known as microsatellites (Peltomaki 2003). Microsatellite instability (MSI) is frequently found in MMR deficient tumors (colon, endometrium, and other organs) linked to the Lynch syndrome and other sporadic cancers. In human cells, the MMR system is accomplished through the interaction of protein heterodimers (Jiricny 2006). A brief description of the MMR system and its linkage with HSPs is included in Fig. 4.
Fig. 4.
HSPs and the mismatch repair system (MMR). The heterodimer composed by the proteins MSH2 and MSH6 (referred to as MutSα, E. coli MutS homolog) recognizes base-base mismatches and IDLs of one or two extrahelical nucleotides. The repair of larger IDLs is initiated by MutSβ, a heterodimer of MSH2 and MSH3 (not shown). MutLα, a heterodimer of MLH1 and PMS2, is also a core complex. The MutSα-MutLα complex remains bound to the mismatch and initiates the repair reaction in coordination with other factors: PCNA (proliferating cell nuclear antigen), RFC (replication factor C), RPA (replication protein A), EXO1 (exonuclease 1), DNA polymerase δ and/or DNA polymerase ε, and DNA ligase I. Dotted lines indicate the relationship between HSPs and MMR components
Heat shock before cisplatin or doxorubicin treatment induces nuclear accumulation of HSPB1 and HSPA1A in cultured peripheral blood lymphocytes (PBL) from healthy individuals, which is associated with an elevated DNA repair capacity and high expression of MLH1 and MSH2 proteins (Nadin et al. 2003). Cancer patients with complete clinical response to cisplatin-based chemotherapy show increased MLH1 and MSH2 expression in PBL following in vitro exposure to the drug (Nadin et al. 2006). In vitro exposure to hyperthermia and cisplatin of PBL from cancer patients increases nuclear expression of MLH1, MSH2, and HSPA1A. Moreover, a higher N/C (nuclear/cytoplasmic) HSPB1 ratio correlates with elevated disease-free survival and overall survival, while high expression of MSH2 is associated with increased overall survival (Nadin et al. 2007). HSPB1 and HSPA1A colocalize with the MMR proteins MLH1 and MSH2 in PBL from healthy persons exposed to hyperthermia 24 h before cisplatin or doxorubicin (Nadin et al. 2007). Coimmunoprecipitation reveals that there is an interaction between HSPB1 and MSH2 in glioma cells exposed to temozolomide. Nuclear colocalization of these proteins occurs at basal levels in human glioblastoma cells; it significantly increases after drug administration (Castro et al. 2015). Furthermore, MLH1 and MSH2 interact with HSPB1 and HSPA1A in human colon cancer cells. The nuclear association of these proteins increases after cisplatin exposure, but the role of HSPA1A and HSPB1 in the MMR system remains unclear (Sottile et al. 2015).
Radicicol, the specific inhibitor for HSPC1 that is structurally unrelated to geldanamycin, increases sensitivity to cisplatin up to 1.6-fold in MLH1-proficient compared to MLH1-deficient colon cancer cells, suggesting a functional relationship between HSPC1 and MLH1. HSPC1 might affect the function of MLH1, leading to the counter-regulation of cytotoxic pathways initiated by the MMR system after cisplatin-induced DNA damage (Fedier et al. 2005).
Increased expression of MSH2 in human lung cancer cell lines exposed to the anticancer agent pemetrexed has been reported. Drug treatment in combination with the HSPC1 inhibitor 17-AAG leads to a reduction of MSH2 levels and enhances the cytotoxic effect of pemetrexed. These results suggest a possible role of HSPC1 in stabilizing MSH2 protein and a potential utility of HSPC1 as a tool for the treatment of pemetrexed resistance in lung cancer (Tung et al. 2014).
Nucleotide excision repair
The NER system recognizes DNA helix-distorting lesions, which are mainly caused by UV light (cyclobutane pyrimidine dimers (CPDs)), polycyclic aromatic hydrocarbons, and DNA cross-linking agents such as platinum anticancer drugs (Nouspikel 2009). NER is accomplished by global genome NER (GG-NER) for distorting damages of the entire genome or by transcription-coupled NER (TC-NER), which specifically targets lesions that block transcription (Marteijn et al. 2014). Figure 5 summarizes the involvement of HSPs in NER subpathways.
Fig. 5.
HSPs and nucleotide excision repair (NER). NER can be divided in two subpathways, GG-NER and TC-NER. GG-NER is mediated by the XPC-HR23B-Centrine 2 heterodimer, which senses distortions. XPC (xeroderma pigmentosum C) is a DNA binding protein with high affinity for damaged DNA. HR23B (human homolog of the yeast protein Rad23) and Centrine 2 stabilize the complex. Additional factors then interact to form a multiprotein complex composed of XPA, RPA, transcription factor IIH (TFIIH), XPG, and XPF. RPA and TFIIH facilitate the unwinding of the double helix. RPA also binds to single-strand DNA. In the next step, the damaged strand is excised by two endonucleases. XPG cuts at the 3′ side, and XPF/ERCC1 (excision repair cross-complementing 1) cuts at the 5′ side, generating an incision 15 to 24 nucleotides away. The resulting gap is filled by DNA polymerase δ and ε, RFC and PCNA, and sealed by DNA ligase 1 or ligase 3-XRCC1. TC-NER removes lesions from the transcribed strand of active genes. GG-NER and TC-NER differ in the recognition step. RNA polymerase II stalls at damages sites. CSA and CSB proteins (Cockayne syndrome A and B, respectively) interact and cooperate with RNA polymerase II and with XPG. All downstream steps are common to both pathways. The implications of HSPB1 and HSPA1A are highlighted in the figure
First reports about the participation of HSPs in NER showed that the small heat shock protein HSPB1 is involved in the excision of the DNA damage induced by UV light. In UVC-sensitive RSa cells (established from human embryo-derived fibroblastic cells) with upregulation of HSPB1, the removal capacity of the two major types of UVC-damaged DNA (CPD and 6,4-photoproducts) is moderately elevated compared to that in control cells. HSPB1 downregulation suppresses the DNA damage repair process, which suggests that this protein is involved in the repair of UVC-induced DNA damage through the NER system (Wano et al. 2004). HSPB1 has also been shown to participate in this process by maintaining NER repair proteins in their properly folded state in Escherichia coli (Zou et al. 1998).
Another HSP implicated in NER is HSPA1A. Overexpression of HSPA1A in human lung adenocarcinoma cells may protect them from accumulating DNA damage caused by UVC irradiation (Niu et al. 2006). HSPA1A also seems to be involved in benzo[a]pyrene repair process, as HSPA1A expression is inversely correlated to residual DNA damage (Xiao et al. 2002; Gao et al. 2004). Exposure of human bronchial epithelia cells to different concentrations of benzo[a]pyrene for 24 h leads to an increased colocalization of HSPA1A with XPA and XPG in the nuclei (Yang et al. 2009).
Human bronchial epithelia cells transfected with plasmids carrying HSPA1A show a higher DNA repair capacity to remove benzo[a]pyrene diol epoxide (BPDE)-DNA adducts, whereas downregulation of HSPA1A inhibits DNA repair. The latter results provide evidence that HSPA1A has the potential to modulate the repair of BPDE-DNA adducts via the NER pathway (Duan et al. 2014).
Double-strand break repair: non-homologous end joining/homologous recombination
Double strand breaks (DSBs) constitute one of the most dangerous DNA lesions. They can result from ionizing radiation, X-rays, ROS, chemicals (topoisomerase inhibitors used in chemotherapy), or by physical stress when chromosomes are pulled to opposite poles during mitosis. There are two pathways for repairing DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR). DNA repair by HR is an error-free process restricted to late S or G2 phases, whereas the error-prone NHEJ occurs throughout the cell cycle (Ceccaldi et al. 2016a). Figure 6 shows the main events of the two pathways and the intervention of HSPs.
Fig. 6.
HSPs and double strand break repair. Double strand breaks (DSB) are repaired by two major mechanisms: homologous recombination repair (HR) and non-homologous end joining (NHEJ). HR operates after replication, when a second identical DNA copy is available. The pathway begins with the recruitment of the MRN complex (MRE11-NBS1-RAD50) to the DSB. ATM, ATR, and the MRN complex act as damage sensors. MRE11 has DNA exonuclease and DNA unwinding activity. RAD50 contains motifs for nucleotide binding. BRCA1 interacts with p53, BRCA2, RAD51, MRN complex, p21, and cyclin B to form multisubunit complexes. hSNM1B/Apollo is a DNA 5′ exonuclease with a preference for single-stranded substrates. The resection of 5′ DNA on either side of the DSB is accomplished by a BRCA1-dependent process, resulting in the exposure of two regions of single-stranded DNA (ssDNA). BRCA2 localizes the DNA recombinase RAD51 to the exposed ssDNA regions. RAD51 forms a nucleoprotein filament that can invade the DNA double helix and pair with undamaged homologous sequences. DNA polymerases δ/ε use the homologous DNA sequence as a template and synthesize new DNA. After DNA synthesis occurs, recombination between chromatids can be resolved by endonucleases and the nicks sealed by DNA ligase 1. Meanwhile, the NHEJ system ligates two broken ends without a sequence homology and when DSB is affecting a short region (1 to 6 bp). The first step consists in the DNA binding of the Ku70/Ku80 heterodimer, protecting the DNA from exonuclease digestion. The Ku heterodimer then binds to the catalytic subunit of DNA-PK (DNA-dependent protein kinase), activating the enzyme, and Artemis stabilizes the ends. After juxtaposition of the two DNA ends, DNA-PK autophophorylates and DNA ligation is performed by the DNA ligase 4 (LIG4)-XRCC4. When end processing is required, DNA polymerase μ (Pol μ) and DNA polymerase λ (Pol λ) can fill in 5′-single-strand extensions, which are sealed by the LIG4-XRCC4 complex. The participation of HSPs is indicated with dotted lines
Several proteins involved in DSB repair, including components of the MRE11/RAD50/NBN (MRN) complex, RAD51, BRCA1, BRCA2, CHK1, DNA-PKcs, and members of the Fanconi anemia (FA) pathway, have been identified as HSPs clients. We will describe the cooperation of HSPs in NHEJ and HR repair pathways separately.
The first component of the NHEJ machinery related to HSPs is the DNA-dependent protein kinase (DNA-PK), a member of the phosphatidylinositol 3-kinase-related (PIKK) family of protein kinases. It is involved in the ligation of the broken ends of DSBs (Lieber 2010). DNA-PK phosphorylates Hsp90α (HSPC2) at N-terminal threonines 5 and 7 (Lees-Miller and Anderson 1989). Briefly, the HSP90 family is composed of five members, HSPC1, HSPC2, HSPC3, HSPC4, and HSPC5, plus the so-called new member Hsp89-alpha-deltaN (HSP90N) (Kampinga et al. 2009). The heat shock transcription factor HSF1 stimulates the activity of purified DNA-PK in an in vitro reaction (Peterson et al. 1995). HSF1 binds to each component of DNA-PK in vitro through its regulatory domain. This factor may therefore participate in cellular survival and recovery from heat shock, rather than promoting the activation of the heat shock genes (Huang et al. 1997). HSPC2, the inducible form of HSPC1, is a substrate and coactivator of DNA-PK. The latter can phosphorylate HSPC2 on Thr5 and Thr7 during apoptosis, and phosphorylated HSPC2 and DNA-PK colocalize in the apoptotic ring. Observations in HCT116, HeLa, and Jurkat cells exposed to different apoptotic agents revealed that HSPC2 is a substrate and chaperone of DNA-PK in the apoptotic response (Solier et al. 2012). It is important to mention that the “apoptotic ring” is characterized by the phosphorylation of histone H2AX on Ser139 (γ-H2AX) by DNA-PK, but lacks MDC1 and 53BP1 as the characteristic nuclear foci. Treatment with 17-DMAG (HSPC inhibitor) alone or in combination with temozolomide (TMZ) previous radiation exposure significantly increases the γH2AX foci formation rate, delayed DNA repair, and attenuated DNA-PK phosphorylation (Thr2609) in glioma cell lines (Choi et al. 2014).
The role of HSPB1 in DSB repair pathways has been scarcely studied. It can interact with Ku80 after recovery of Zeocin™ treatment. The Ku70/Ku80 heterodimer, a central component of the NHEJ pathway, forms a ring through which the DSB threads. siHSPB1-transfected cells have a lower NHEJ-mediated-DSB repair efficiency (measured by in vitro NHEJ assay) and accumulate less H2AX foci than si-control cells. These results strongly suggest that HSPB1 could negatively regulate NHEJ repair by interacting with KU80 and thus preventing the binding of this protein with DNA-PK (Katsogiannou et al. 2014).
Among the proteins involved in the HR repair mechanism, the MRE11-RAD50-NBS1 (MRN) complex, a DSB sensor and coactivator of DSB-induced signaling, was the first multiprotein complex reported to bind HSPC1 in human pancreatic carcinoma cell lines. Exposure to 17-DMAG reduces the ability of the MRN complex to form nuclear foci after irradiation and reduces the interaction between NBS1 and ATM (Dote et al. 2006). The tumor suppressor BRCA2 assists RAD51 loading on ssDNA during HR repair. BRCA2 directly interacts with HSPC1, thus BRCA2 acts as a client protein for HSPC1 (Noguchi et al. 2006). The presence of the HSPC1 inhibitor 17-DMAG causes the degradation of BRCA2 and behavioral alteration of RAD51, delaying the IR-induced BRCA2-mediated RAD51 foci formation (Yao et al. 2007).
Different HSPC inhibitors, such as NVP-AUY922, NVP-BEP800, and 17-DMAG, alone or in combination with IR lead to a radiosensitization of tumor cell lines. HSPC1 inhibition increases IR-induced DSBs and causes a delay of the DNA damage repair (Stingl et al. 2010). The interaction between BRCA1 and HSPC1 in breast cancer cells increases after radiation (Stecklein et al. 2012), but the presence of the inhibitor 17-AAG abolishes this interaction. Treatment with other HSPC inhibitors, such as radicicol and novobiocin, leads to a loss of BRCA1 due to its proteasomal degradation. In addition, the 17-AAG pretreatment increases γH2AX foci formation by irradiation and affects BRCA1 recruitment to ionizing radiation induced foci (IRIF). BRCA1 depletion by 17-AAG impairs DNA DSBs repair, suggesting that HSPC1 is required for HR and NHEJ mechanisms (Stecklein et al. 2012). The tumor suppressor BRCA1 hyperphosphorylates in response to DNA damage and regulates MRN complex functions. Tumors overexpressing BRCA1 are resistant to conventional antineoplastic treatments. For this reason, the suppression of BRCA1-dependent HR repair through HSPC inhibition currently represents a promising anticancer strategy that restores the sensitivity of cancer cells to antitumor agents (Shrestha et al. 2016).
The Hsp90 inhibitor NVP-AUY922 induces radiosensitization both in vitro and in vivo. The number of γH2AX foci increases 4 and 24 h after the combined treatment in comparison with radiation alone, which demonstrates an impairment of DSB repair. The latter result correlated with a delay of RAD51 foci formation across all studied cell lines (Zaidi et al. 2012). Exposure of lung carcinoma cell lines to 17-AAG causes RAD51 degradation through the proteasome pathway and decreases the DNA repair capacity, indicating that RAD51 is a client protein of HSPC1. The transfection of RAD51 expression vector restores, however, the protein levels and the DNA repair function, indicating that 17-AAG cytotoxicity may be due to a diminished DNA repair capacity as consequence of RAD51 downregulation (Ko et al. 2012). The use of PU-H71, a novel HSPC inhibitor, increases the persistence of γH2AX foci, while it reduces RAD51 foci and phosphorylates DNA-PKcs expression in irradiated human lung cancer cells. PU-H71 also sensitizes cancer cells to irradiation by inhibiting HR and NHEJ repair mechanisms (Lee et al. 2016).
With regard to HSPA, HSPA1A protects C3H10T1/2 cells (mouse embryo fibroblasts) from γ-induced DNA damage (Calini et al. 2003). The human SNM1B (hSNM1B) protein, also known as Apollo, has intrinsic 5′ to 3′ exonuclease activity and plays a significant role in DNA interstrand cross-links and IR-induced DNA damage repair (Schmiester and Demuth 2017). Apollo interacts with the MRN complex, which is involved in the initial processing of DSBs, and with FANCD2, a member of the FA pathway (Bae et al. 2008). Two members of the HSPA protein family, HSPA8 and HSPA1A, copurify with Apollo. This interaction is mediated through the C-terminal substrate-binding domain of HSPA1A (Anders et al. 2009).
Finally, HSPC1 might regulate the FA pathway, specifically, the cytoplasmic fraction of FANCA interacts with HSPC1. Treatment of different cell lines with 17-AAG induces rapid proteasomal degradation and cytoplasmic retention of FANCA. These data suggest that HSPC1 is critical for the stabilization and nuclear import of cytoplasmic FANCA, whereas inhibition of HSPC1 induces an impaired activation of the FA DNA repair pathway (Oda et al. 2007). A detailed description of the FA pathway and cross-links repair can be found from Ceccaldi et al. (2016b).
Concluding remarks
The antiapoptotic role of HSPs in living cells is well documented (Beere 2004). In this review, we have shown that HSPs promote survival through their multiple interactions with DNA repair mechanisms, besides their antiapoptotic role. Due to this peculiarity, HSPs have gained importance in targeted molecular therapies, a powerful approach in cancer treatment.
Despite the fact that to date HSPs are not considered crucial for the DNA repair reaction, the chaperones are required to stabilize core components of DNA repair mechanisms. Altered expression levels of HSPs could lead to impaired detection as well as delayed untimely repair of damaged DNA. HSPs inhibitors are currently being used to repress DNA repair pathways and to potentiate the cytotoxicity of DNA-damaging anticancer drugs. For example, antagonists of the mitochondrial HSP, TRAP1, involved in protection against oxidant-induced DNA damage and apoptosis, have shown promising results as molecular targets in the treatment of ovarian cancer. TRAP1 antagonists induce the collapse of the mitochondrial function and selective tumor cell death (Landriscina et al. 2010). The combination of 17-AAG with the PARP inhibitor olaparib can sensitize human glioma cells to IR (Dungey et al. 2009). PARP inhibitors increase the DSBs generation in cells. Given that HR repair components are client proteins of HSPC, the HR suppression through HSPC inhibitors supports their combination with olaparib as therapeutic strategy (Eppink et al. 2012). The administration of AUY922 in combination with concurrent cisplatin radiotherapy has a synergistic effect in mutant p53 head and neck carcinoma cells. Downregulation of RAD51 and BRCA1 expression leads to disruption of DDR signaling and DSB repair by HR (McLaughlin et al. 2017). Given the complexity of the HSPs’ associations, further studies involving DNA-damaging anticancer agents in combination with HSPs inhibitors may become an interesting challenge to develop new strategies for cancer therapy.
Compliance with ethical standards
Financial & competing interest disclosure
This review was supported by grants from Alberto J. Roemmers Foundation (Dr. SB Nadin) and the National Agency for Scientific and Technological Promotion of Argentina (PICT 2015-1171, Préstamo BID, Dr. SB Nadin). The authors have no financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
References
- Abe T, et al. Possible correlation between DNA damage induced by hydrogen peroxide and translocation of heat shock 70 protein into the nucleus. Biochem Biophys Res Commun. 1995;206:548–555. doi: 10.1006/bbrc.1995.1078. [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders M, Mattow J, Digweed M, Demuth I. Evidence for hSNM1B/Apollo functioning in the HSP70 mediated DNA damage response. Cell Cycle. 2009;8:1725–1732. doi: 10.4161/cc.8.11.8605. [DOI] [PubMed] [Google Scholar]
- Angelidis CE, Lazaridis I, Pagoulatos GN. Constitutive expression of heat-shock protein 70 in mammalian cells confers thermoresistance. Eur J Biochem. 1991;199:35–39. doi: 10.1111/j.1432-1033.1991.tb16088.x. [DOI] [PubMed] [Google Scholar]
- Arlander SJ, et al. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. J Biol Chem. 2003;278:52572–52577. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- Bae JB, et al. Snm1B/Apollo mediates replication fork collapse and S Phase checkpoint activation in response to DNA interstrand cross-links. Oncogene. 2008;27:5045–5056. doi: 10.1038/onc.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Lindhal T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Bases R. Heat shock protein 70 enhanced deoxyribonucleic acid base excision repair in human leukemic cells after ionizing radiation. Cell Stress Chaperones. 2006;11:240–249. doi: 10.1379/CSC-185R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beere HM. The stress of dying: the role of heat shock proteins in the regulation of apoptosis. J Cell Sci. 2004;117:2641–2651. doi: 10.1242/jcs.01284. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Mitra S. CpG methylation-dependent repression of the human O6-methylguanine-DNA methyltransferase gene linked to chromatin structure alteration. Carcinogenesis. 2003;24:1337–1345. doi: 10.1093/carcin/bgg086. [DOI] [PubMed] [Google Scholar]
- Blagg BS, Kerr TD. Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med Res Rev. 2006;26:310–338. doi: 10.1002/med.20052. [DOI] [PubMed] [Google Scholar]
- Bodell WJ, Cleaver JE, Roti Roti JL. Inhibition by hyperthermia of repair synthesis and chromatin reassembly of ultraviolet-induced damage to DNA. Radiat Res. 1984;100:87–95. [PubMed] [Google Scholar]
- Boesch P, et al. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta. 2011;1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Bohgaki T, Bohgaki M, Hakem R. DNA double-strand break signaling and human disorders. Genome Integr. 2010;1:15. doi: 10.1186/2041-9414-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenerman BM, Illuzzi JL, Wilson DM., 3rd Base excision repair capacity in informing healthspan. Carcinogenesis. 2014;35:2643–2652. doi: 10.1093/carcin/bgu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Calini V, Urani C, Camatini M. Overexpression of HSP70 is induced by ionizing radiation in C3H 10T1/2 cells and protects from DNA damage. Toxicol in Vitro. 2003;17:561–566. doi: 10.1016/s0887-2333(03)00116-4. [DOI] [PubMed] [Google Scholar]
- Castro GN, et al. Effects of temozolomide (TMZ) on the expression and interaction of heat shock proteins (HSPs) and DNA repair proteins in human malignant glioma cells. Cell Stress Chaperones. 2015;20:253–265. doi: 10.1007/s12192-014-0537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D'Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D'Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- Choi EJ, et al. Enhanced cytotoxic effect of radiation and temozolomide in malignant glioma cells: targeting PI3K-AKT-mTOR signaling, HSP90 and histone deacetylases. BMC Cancer. 2014;14:17. doi: 10.1186/1471-2407-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res. 2005;591:45–49. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Donati YR, Slosman DO, Polla BS. Oxidative injury and the heat shock response. Biochem Pharmacol. 1990;40:2571–2577. doi: 10.1016/0006-2952(90)90573-4. [DOI] [PubMed] [Google Scholar]
- Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66:9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- Duan Y, et al. HspA1A facilitates DNA repair in human bronchial epithelial cells exposed to Benzo[a]pyrene and interacts with casein kinase 2. Cell Stress Chaperones. 2014;19:271–279. doi: 10.1007/s12192-013-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungey FA, Caldecott KW, Chalmers AJ. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol Cancer Ther. 2009;8:2243–2254. doi: 10.1158/1535-7163.MCT-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elaimy AL, et al. ATM is the primary kinase responsible for phosphorylation of Hsp90α after ionizing radiation. Oncotarget. 2016;7:82450–82457. doi: 10.18632/oncotarget.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppink B, Krawczyk PM, Stap J, Kanaar R. Hyperthermia-induced DNA repair deficiency suggests novel therapeutic anti-cancer strategies. Int J Hyperth. 2012;28:509–517. doi: 10.3109/02656736.2012.695427. [DOI] [PubMed] [Google Scholar]
- Fang Q, et al. HSP90 regulates DNA repair via the interaction between XRCC1 and DNA polymerase β. Nat Commun. 2014;5:5513. doi: 10.1038/ncomms6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedier A, Stuedli A, Fink D. Presence of MLH1 protein aggravates the potential of the HSP90 inhibitor radicicol to sensitize tumor cells to cisplatin. Int J Oncol. 2005;27:1697–1705. [PubMed] [Google Scholar]
- Gao YJ, et al. In vitro study on role of Hsp70 expression in DNA damage of human embryonic lung cells exposed to benzo[a]pyrene. Biomed Environ Sci. 2004;17:144–152. [PubMed] [Google Scholar]
- Gerson SL. MGMT: its role in cancer etiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- Guttmann DM, et al. Inhibition of Hsp27 radiosensitizes head-and-neck cancer by modulating deoxyribonucleic acid repair. Int J Radiat Oncol Biol Phys. 2013;87:168–175. doi: 10.1016/j.ijrobp.2013.05.028. [DOI] [PubMed] [Google Scholar]
- Ha K, et al. Hsp90 inhibitor-mediated disruption of chaperone association of ATR with hsp90 sensitizes cancer cells to DNA damage. Mol Cancer Ther. 2011;10:1194–1206. doi: 10.1158/1535-7163.MCT-11-0094. [DOI] [PubMed] [Google Scholar]
- Huang J, Nueda A, Yoo S, Dynan WS. Heat shock transcription factor 1 binds selectively in vitro to Ku protein and the catalytic subunit of the DNA-dependent protein kinase. J Biol Chem. 1997;272:26009–26016. doi: 10.1074/jbc.272.41.26009. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 2016;16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Jones B. DNA damage response: rapid response. Nat Rev Mol Cell Biol. 2012;13:602. [Google Scholar]
- Kampinga HH, et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsogiannou M, et al. The functional landscape of Hsp27 reveals new cellular processes such as DNA repair and alternative splicing and proposes novel anticancer targets. Mol Cell Proteomics. 2014;13:3585–3601. doi: 10.1074/mcp.M114.041228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny MK, et al. Heat shock protein 70 binds to human apurinic/apyrimidinic endonuclease and stimulates endonuclease activity at abasic sites. J Biol Chem. 2001;276:9532–9536. doi: 10.1074/jbc.M009297200. [DOI] [PubMed] [Google Scholar]
- Ko JC, et al. HSP90 inhibition induces cytotoxicity via down-regulation of Rad51 expression and DNA repair capacity in non-small cell lung cancer cells. Regul Toxicol Pharmacol. 2012;64:415–424. doi: 10.1016/j.yrtph.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Koll TT, et al. HSP90 inhibitor, DMAG, synergizes with radiation of lung cancer cells by interfering with base excision and ATM-mediated DNA repair. Mol Cancer Ther. 2008;7:1985–1992. doi: 10.1158/1535-7163.MCT-07-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoglou P, et al. HSP70 translocates to the nuclei and nucleoli, binds to XRCC1 and PARP-1, and protects HeLa cells from single-strand DNA breaks. Cell Stress Chaperones. 2009;14:391–406. doi: 10.1007/s12192-008-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo LJ, Yang LX. Gamma-H2AX—a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- Landriscina M, Amoroso MR, Piscazzi A, Esposito F. Heat shock proteins, cell survival and drug resistance: the mitochondrial chaperone TRAP1, a potential novel target for ovarian cancer therapy. Gynecol Oncol. 2010;117:177–182. doi: 10.1016/j.ygyno.2009.10.078. [DOI] [PubMed] [Google Scholar]
- Laszlo A, Wright W, Roti Roti JL. Initial characterization of heat-induced excess nuclear proteins in HeLa cells. J Cell Physiol. 1992;151:519–532. doi: 10.1002/jcp.1041510311. [DOI] [PubMed] [Google Scholar]
- Lee Y, et al. The purine scaffold Hsp90 inhibitor PU-H71 sensitizes cancer cells to heavy ion radiation by inhibiting DNA repair by homologous recombination and non-homologous end joining. Radiother Oncol. 2016;121:162–168. doi: 10.1016/j.radonc.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller SP, Anderson CW. The human double-stranded DNA-activated protein kinase phosphorylates the 90-kDa heat-shock protein, hsp90 alpha at two NH2-terminal threonine residues. J Biol Chem. 1989;264:17275–17280. [PubMed] [Google Scholar]
- Li Q, Martinez JD. Loss of HSF1 results in defective radiation-induced G(2) arrest and DNA repair. Radiat Res. 2011;176:17–24. doi: 10.1667/rr2393.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macario AJ, Conway de Macario E. Molecular chaperones: multiple functions, pathologies, and potential applications. Front Biosci. 2007;1:2588–2600. doi: 10.2741/2257. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Wang X, Ohnishi T. Binding between wild type p53 and hsp72 accumulated after UV and γ-ray irradiation. Cancer Lett. 1995;92:127–133. doi: 10.1016/0304-3835(95)03769-s. [DOI] [PubMed] [Google Scholar]
- McLaughlin M, et al. HSP90 inhibition sensitizes head and neck cancer to platin-based chemoradiotherapy by modulation of the DNA damage response resulting in chromosomal fragmentation. BMC Cancer. 2017;17:86. doi: 10.1186/s12885-017-3084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez F, et al. Heat-shock proteins associated with base excision repair enzymes in HeLa cells. Radiat Res. 2000;153:186–195. doi: 10.1667/0033-7587(2000)153[0186:hspawb]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Mendez F, Kozin E, Bases R. Heat shock protein 70 stimulation of the deoxyribonucleic acid base excision repair enzyme polymerase β. Cell Stress Chaperones. 2003;8:153–161. doi: 10.1379/1466-1268(2003)008<0153:hspsot>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel RE, Morrison DP. Heat-shock induction of ionizing radiation resistance in Saccharomyces cerevisiae, and correlation with stationary growth phase. Radiat Res. 1982;90:284–291. [PubMed] [Google Scholar]
- Mitchel RE, Morrison DP. Heat-shock induction of ionizing radiation resistance in Saccharomyces cerevisiae. Transient changes in growth cycle distribution and recombinational ability. Radiat Res. 1982;92:182–187. [PubMed] [Google Scholar]
- Mitchel RE, Morrison DP. Inducible error-prone repair in yeast. Suppression by heat shock. Mutat Res. 1986;159:31–39. doi: 10.1016/0027-5107(86)90109-0. [DOI] [PubMed] [Google Scholar]
- Mitchel RE, Morrison DP. Inducible DNA-repair systems in yeast: competition for lesions. Mutat Res. 1987;183:149–159. doi: 10.1016/0167-8817(87)90057-5. [DOI] [PubMed] [Google Scholar]
- Nadin SB, Vargas-Roig LM, Cuello-Carrión FD, Ciocca DR. Deoxyribonucleic acid damage induced by doxorubicin in peripheral blood mononuclear cells: possible roles for the stress response and the deoxyribonucleic acid repair process. Cell Stress Chaperones. 2003;8:361–372. doi: 10.1379/1466-1268(2003)008<0361:dadibd>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin SB, et al. DNA damage and repair in peripheral blood lymphocytes from healthy individuals and cancer patients: a pilot study on the implications in the clinical response to chemotherapy. Cancer Lett. 2006;239:84–97. doi: 10.1016/j.canlet.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Nadin SB, et al. Hsp27, Hsp70 and mismatch repair proteins hMLH1 and hMSH2 expression in peripheral blood lymphocytes from healthy subjects and cancer patients. Cancer Lett. 2007;252:131–146. doi: 10.1016/j.canlet.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Niture SK, et al. Proteomic analysis of human O6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry. Biochem Biophys Res Commun. 2005;337:1176–1184. doi: 10.1016/j.bbrc.2005.09.177. [DOI] [PubMed] [Google Scholar]
- Niu P, et al. Overexpressed heat shock protein 70 protects cells against DNA damage caused by ultraviolet C in a dose-dependent manner. Cell Stress Chaperones. 2006;11:162–169. doi: 10.1379/CSC-175R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M, et al. Inhibition of homologous recombination repair in irradiated tumor cells pretreated with Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Biochem Biophys Res Commun. 2006;351:658–663. doi: 10.1016/j.bbrc.2006.10.094. [DOI] [PubMed] [Google Scholar]
- Nomura M, Nomura N, Yamashita J. Geldanamycin-induced degradation of Chk1 is mediated by proteasome. Biochem Biophys Res Commun. 2005;335:900–905. doi: 10.1016/j.bbrc.2005.07.160. [DOI] [PubMed] [Google Scholar]
- Nouspikel T. DNA repair in mammalian cells: nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:965–967. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes E, Candreva EC, Keszenman D, Salvo VA. The mutagenic effect of elevated temperatures in yeast is blocked by a previous heat shock. Mutat Res. 1993;289:165–170. doi: 10.1016/0027-5107(93)90066-o. [DOI] [PubMed] [Google Scholar]
- Oda T, Hayano T, Miyaso H, Takahashi N, Yamashita T. Hsp90 regulates the Fanconi anemia DNA damage response pathway. Blood. 2007;109:5016–5026. doi: 10.1182/blood-2006-08-038638. [DOI] [PubMed] [Google Scholar]
- Okano S, et al. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Park SH, et al. Inducible heat-shock protein 70 is involved in the radioadaptive response. Radiat Res. 2000;153:318–326. doi: 10.1667/0033-7587(2000)153[0318:ihspii]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- Peterson SR, et al. Stimulation of the DNA-dependent protein kinase by RNA polymerase II transcriptional activator proteins. J Biol Chem. 1995;270:1449–1454. doi: 10.1074/jbc.270.3.1449. [DOI] [PubMed] [Google Scholar]
- Plesset J, Ludwig JR, Cox BS, McLaughlin CS. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J Bacteriol. 1987;169:779–784. doi: 10.1128/jb.169.2.779-784.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quanz M, et al. Heat shock protein 90α (Hsp90α) is phosphorylated in response to DNA damage and accumulates in repair foci. J Biol Chem. 2012;287:8803–8815. doi: 10.1074/jbc.M111.320887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa FM, Vonborstel RC. Chromosome puffs in Drosophila induced by ribonuclease. Science. 1964;145:513–514. doi: 10.1126/science.145.3631.513. [DOI] [PubMed] [Google Scholar]
- Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- Roti Roti JL. Cellular responses to hyperthermia (40–46 degrees C): cell killing and molecular events. Int J Hyperth. 2008;24:3–15. doi: 10.1080/02656730701769841. [DOI] [PubMed] [Google Scholar]
- Roti Roti JL, Painter RB. Effects of hyperthermia on the sedimentation of nucleoids from HeLa cells in sucrose gradients. Radiat Res. 1982;89:166–175. [PubMed] [Google Scholar]
- Roti Roti JL, Wilson CF. The effects of alcohols, procaine and hyperthermia on the protein content of nuclei and chromatin. Int J Radiat Biol Relat Stud Phys Chem Med. 1984;46:25–33. doi: 10.1080/09553008414551031. [DOI] [PubMed] [Google Scholar]
- Roti Roti JL, Henle KJ, Winward RT. The kinetics of increase in chromatin protein content in heated cells: a possible role in cell killing. Radiat Res. 1979;78:522–531. [PubMed] [Google Scholar]
- Schmiester M, Demuth I. SNM1B/Apollo in the DNA damage response and telomere maintenance. Oncotarget. 2017;8:48398–48409. doi: 10.18632/oncotarget.16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha L, Bolaender A, Patel HJ, Taldone T. Heat shock protein (HSP) drug discovery and development: targeting heat shock proteins in disease. Curr Top Med Chem. 2016;16:2753–2764. doi: 10.2174/1568026616666160413141911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solier S, et al. Heat shock protein 90α (HSP90α), a substrate and chaperone of DNA-PK necessary for the apoptotic response. Proc Natl Acad Sci U S A. 2012;109:12866–12872. doi: 10.1073/pnas.1203617109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile ML, et al. Hyperthermia effects on Hsp27 and Hsp72 associations with mismatch repair (MMR) proteins and cisplatin toxicity in MMR-deficient/proficient colon cancer cell lines. Int J Hyperth. 2015;31:464–475. doi: 10.3109/02656736.2015.1026848. [DOI] [PubMed] [Google Scholar]
- Stecklein SR, et al. BRCA1 and HSP90 cooperate in homologous and non-homologous DNA double-strand-break repair and G2/M checkpoint activation. Proc Natl Acad Sci U S A. 2012;109:13650–13655. doi: 10.1073/pnas.1203326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl L, et al. Novel HSP90 inhibitors, NVP-AUY922 and NVP-BEP800, radiosensitise tumour cells through cell-cycle impairment, increased DNA damage and repair protraction. Br J Cancer. 2010;102:1578–1591. doi: 10.1038/sj.bjc.6605683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasovic SP, Turner GN, Dewey WC. Effect of hyperthermia on nonhistone proteins isolated with DNA. Radiat Res. 1978;73:535–552. [PubMed] [Google Scholar]
- Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CL, et al. Down-regulation of MSH2 expression by an Hsp90 inhibitor enhances pemetrexed-induced cytotoxicity in human non-small-cell lung cancer cells. Exp Cell Res. 2014;322:345–354. doi: 10.1016/j.yexcr.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Vasileiou PVS, Mourouzis I, Pantos C. Principal aspects regarding the maintenance of mammalian mitochondrial genome integrity. Int J Mol Sci. 2017;18:1821. doi: 10.3390/ijms18081821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wano C, et al. Protective role of HSP27 against UVC-induced cell death in human cells. Exp Cell Res. 2004;298:584–592. doi: 10.1016/j.yexcr.2004.04.048. [DOI] [PubMed] [Google Scholar]
- Warters RL, Lyons BW, Axtell-Bartlett J. Inhibition of repair of radiation-induced DNA damage by thermal shock in Chinese hamster ovary cells. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;51:505–517. doi: 10.1080/09553008714550981. [DOI] [PubMed] [Google Scholar]
- Wu J, et al. Heat shock proteins and cancer. Trends Pharmacol Sci. 2017;38:226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- Xiao C, et al. Association of HSP70 and genotoxic damage in lymphocytes of workers exposed to coke-oven emission. Cell Stress Chaperones. 2002;7:396–402. doi: 10.1379/1466-1268(2002)007<0396:aohagd>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. Correlations and co-localizations of Hsp70 with XPA, XPG in human bronchial epithelia cells exposed to benzo[a]pyrene. Toxicology. 2009;2009(265):10–14. doi: 10.1016/j.tox.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Yao Q, Weigel B, Kersey J. Synergism between etoposide and 17-AAG in leukemia cells: critical roles for Hsp90, FLT3, topoisomerase II, Chk1, and Rad51. Clin Cancer Res. 2007;13:1591–1600. doi: 10.1158/1078-0432.CCR-06-1750. [DOI] [PubMed] [Google Scholar]
- Zaidi S, et al. The HSP90 inhibitor NVP-AUY922 radiosensitizes by abrogation of homologous recombination resulting in mitotic entry with unresolved DNA damage. PLoS One. 2012;7:e35436. doi: 10.1371/journal.pone.0035436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Crowley DJ, Van Houten B. Involvement of molecular chaperonins in nucleotide excision repair. J Biol Chem. 1998;273:12887–12892. doi: 10.1074/jbc.273.21.12887. [DOI] [PubMed] [Google Scholar]