Abstract
Heat shock protein 70 (Hsp70) is an evolutionarily well-conserved molecular chaperone involved in several cellular processes such as folding of proteins, modulating protein-protein interactions, and transport of proteins across the membrane. Binding partners of Hsp70 (known as “clients”) are identified on an individual basis as researchers discover their particular protein of interest binds to Hsp70. A full complement of Hsp70 interactors under multiple stress conditions remains to be determined. A promising approach to characterizing the Hsp70 “interactome” is the use of protein epitope tagging and then affinity purification followed by mass spectrometry (AP-MS/MS). AP-MS analysis is a widely used method to decipher protein-protein interaction networks and identifying protein functions. Conventionally, the proteins are overexpressed ectopically which interferes with protein complex stoichiometry, skewing AP-MS/MS data. In an attempt to solve this issue, we used CRISPR/Cas9-mediated gene editing to integrate a tandem-affinity (TAP) epitope tag into the genomic locus of HSC70. This system offers several benefits over existing expression systems including native expression, no requirement for selection, and homogeneity between cells. This cell line, freely available to chaperone researchers, will aid in small and large-scale protein interaction studies as well as the study of biochemical activities and structure-function relationships of the Hsc70 protein.
Electronic supplementary material
The online version of this article (10.1007/s12192-017-0845-2) contains supplementary material, which is available to authorized users.
Keywords: Heat shock protein, HSC70, CRISPR/Cas9, Genome-editing
Introduction
The highly conserved molecular chaperone 70 kDa heat shock protein family (Hsp70s) are key players in protein homeostasis not only during stress, but also in optimal growth conditions. Members of the Hsp70 family are involved in folding of newly synthesized and misfolded proteins, solubilization of protein aggregates, degradation via the proteasome and autophagy pathways, transport of proteins through membranes, and assembly and disassembly of protein complexes (Hartl et al. 2011). The structure of Hsp70 is comprised of highly conserved amino acid sequences and domains among the different family groups. These domains include the following: N-terminal ATPase domain, which is a 44-KDa structure that is involved in the binding of Hsp70 to client proteins and in the hydrolysis of ATP, mid region containing protease sensitive sites, substrate binding domain, weighing about 28-KDa and known to bind to substrates such as polypeptides, and a C-terminal region containing leucine rich EEVD motif, essential for co-chaperone binding and is missing in ER-specific/Grp78 (Mayer and Bukau 2005). There are 17 different isoforms of Hsp70 family that have been identified but their functions are still unclear. These isoforms can be classified into two broad categories. The two most common isoforms are Hsp70, a stress-inducible form and the constitutively expressed Hsc70 that provides major housekeeping functions, and essential cell viability (Boorstein et al. 1994; Kelley 1999; Rohde et al. 2005).
Co-chaperones of Hsp70 associate with the N-terminus and mediate both client protein binding activities often via stimulation of ATP-binding and hydrolysis. There are suites of co-chaperones present at any one time in chaperone-client complexes that include J-domain, BCL2-associated athanogene (Bag), Hsp70 interacting proteins (Hip), Hsp70/Hsp90 organizing proteins (Hop), CHIP (Carboxyl-terminus of Hsp70 Interacting Protein), and Nucleotide Exchange Factors (NEFs). J-domain proteins assist in the targeting of the client protein to their substrate-binding cavity (Kelley 1999) whereas Bag proteins promote substrate release by binding to Hsp70 chaperone, thus having an inhibitory role (Takayama and Reed 2001).
Characterizing the interactome of a target protein by AP-MS offers a powerful approach to understanding its role in the cell (Aebersold and Mann 2003; Gavin et al. 2002; Ho et al. 2002; Kocher and Superti-Furga 2007). This holds especially true for chaperones whose interactomes are typically very large yet specific (Taipale et al. 2012; Zhao et al. 2005). In addition, chaperone interactions are dynamic, changing upon stress, and post-translational modification (Truman et al. 2012; Truman et al. 2015; Woodford et al. 2016b). The study of Hsp70 complexes has been greatly aided by the generation of isoform-specific monoclonal antibodies for Hsc70 and Hsp72 (Shiota et al. 2010; Tanaka et al. 2011). These antibodies have been used to immunoprecipitate and analyze chaperone complexes via high-resolution mass spectrometry. This methodology has been particularly effective in detecting low-abundance interactors of Hsp70 in a variety of conditions, allowing purification of complexes at native stoichiometry (Tanaka et al. 2014; Tanaka et al. 2016). Alternative strategies for chaperone interactome analysis have employed the use of an epitope-tagged bait protein in cell lines from a transient, CMV-driven expression plasmid. Given that the HIS epitope and associated affinity reagents often used for such experiments are unaffected by denaturing reagents such as urea or guanidine HCl, purification of chaperone complexes can be achieved under more stringent conditions. This has been employed with substantial success in a number of Hsp70 and Hsp90 interactome studies (Dunn et al. 2015; Truman et al. 2012; Truman et al. 2015; Woodford et al. 2016b).
Programmable sequence specific nucleases such as CRISPR/Cas9 facilitate precise editing of endogenous genomic loci. CRISPR/Cas9 is used to generate cell lines with tailored modifications such as gene knockouts, point mutation, and knock-in of exogenous DNA (Ran et al. 2013). A particularly useful application of CRISPR/Cas9 is the endogenous epitope tagging of genes at their genomic loci. Several DNA repair and chromatin-modifying proteins have been tagged and purified using this methodology (Dalvai et al. 2015).
In this study, we have optimized the CRISPR/Cas9 system to epitope tag HSC70 at its N-terminus with a tandem affinity tag (HIS6-FLAG) using a single stranded oligonucleotide template (ssODN) with substantially smaller homology arms than is traditionally used. The resulting cell line (HEK293THIS-FLAG-Hsc70) expresses tagged Hsc70 from its native promoter offering several benefits for researchers interested in the systematic and unbiased mapping of Hsc70 interactions.
Materials and methods
Design of HSC70 sgRNA and generation of the HSC70-sgRNA-Cas9 plasmid
An HSC70-targeting sgRNA was generated using the CRISPOR algorithm (http://crispor.tefor.net/). We identified a suitable sgRNA sequence (TTTTCAGCAACCATGTCCAA) based on two criteria-minimal off-targeting and proximity to 5′ end of HSC70. The HSC70-targeting sgRNA was cloned via BbsI into pX458 plasmid (Ran et al. 2013) that allows simultaneous expression of chosen sgRNA, Cas9, and a GFP marker.
Cell culture and transfection
HEK293T cells were obtained from the ATCC and maintained at 37 °C under 5% CO2 in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), penicillin-streptomycin (Gibco), and GlutaMAX (Gibco). Cells were transfected in six-well plates at 70% confluency using 5 μg of HSC70 gRNA-pX458 or pX458 control using Lipofectamine 3000 (Life Technologies). Twenty-four hours after DNA transfection, cells were washed one time with PBS (Gibco) and media was changed. After 72 h, the cells were prepared for FACS Cell Sorting.
FACS cell sorting
Cells were washed with PBS, trypsinized, and then resuspended in PBS and 1% BSA solution and kept in ice prior to sorting. Cells positive for GFP (and therefore Cas9) expression were sorted by flow cytometry. 2 × 104 cells were sorted in one well of a six-well plate in DMEM using a BD FACS ARIA II flow cytometer and accompanying software (BD Biosciences).
Limiting dilution
Cells were allowed to reach 70% confluency after sorting and harvested by trypsinizing. Cells were resuspended in a 10-ml media and counted using Cell Counter. The cells were diluted to 103 cells/ml and were diluted to 1 cell/well. The diluted cells were allowed to expand in a 12-well plate. The wells containing single cell colonies were selected. When the colonies reached 70% confluency, the cells were harvested for further analysis.
SURVEYOR assay
The SURVEYOR assay was performed according to the manufacturer’s instructions (Integrated DNA Technologies). Genomic DNA was isolated using QuickExtract DNA Extraction Solution (Epicenter) according to manufacturer’s protocol. Briefly, pelleted cells were resuspended in QuickExtract solution and incubated at 65 °C for 15 min, 68 °C for 15 min, and 98 °C for 10 min. The genomic region flanking the CRISPR target site for each gene was PCR amplified using Forward primer (5′-GTGCAGCCTCCACACAGGCCTGTTG-3′) and Reverse primer (5′-GGTTCGGTTTCCCTGATCATTGGC-3′). PCR product was purified using PCR Purification kit. PCR products were then mixed with 2 μl 10X Dream TaqDNA Polymerase PCR buffer (Thermo) to a final volume of 20 μl and subjected to a re-annealing process to enable heteroduplex formation: 95 °C for 10 min, 95 °C to 85 °C ramping at − 2 °C/s, 85 °C to 25 °C at − 0.25 °C/s, and 25 °C hold for 1 min. After re-annealing, products were treated with SURVEYOR nuclease and SURVEYOR enhancer S following the manufacturer’s recommended protocol and analyzed on 10% TBE Gel (Invitrogen) and imaged using Gel Doc imaging system (Bio-rad).
In-out PCR of HIS6-FLAG-HSC70 genomic region
Genomic DNA from the clones was purified using QuickExtract DNA Extraction Solution (Epicenter). PCR amplification for SURVEYOR assay was performed by an initial amplification using Forward primer 5′-GTGCAGCCTCCACACAGGCCTGTTG-3′, and Reverse primer 5′-CTCCTCACGTTTCATAAACTTTTGTGC-3′ was done with a denaturation step at 98 °C for 10 min, followed by 34 cycles of denaturation at 98 °C for 1 min, primer annealing at 64 °C for 30 s, and primer extension at 72 °C for 45 s. Upon completion of the cycling steps, a final extension at 72 °C for 5 min was done and then the reaction was stored at 4 °C. PCR was carried out using a Bio-Rad PCR machine. The In-out PCR was done on genomic DNA isolated from the clones using Forward primer 5′-GACTACAAGGACGACGATGACAAAGGTTC-3′ and Reverse primer 5′-CTTAACCCTGAGCTGAGCCCCATCTGTTC-3′ using the same PCR program as above.
Sequencing of CRISPR-edited region
A 1 kb region of DNA containing 5′ sequence of HSC70 gene along with gRNA-binding site and HIS-FLAG epitope tag was amplified via PCR and cloned into pGEX-6P-1 for sequencing. To allow for multiple integration events, multiple clones were sequenced via multiple primers in forward and reverse orientation.
Immunoprecipitation of Hsc70 complexes
Total cell extract was prepared from the individual clones using M-PER (Thermo) containing EDTA-free protease and phosphatase inhibitor cocktail (Thermo) according to the manufacturer’s recommended protocol. Protein was quantitated using the Bradford Assay. His-tagged proteins were purified as follows: 200 μg of cell lysate was incubated with 30 μl of His-Tag Dynabeads (Invitrogen) with gentle agitation for 20 min at 4 °C. Dynabeads were collected by magnet then washed five times with 500-μl-Binding/Wash buffer. After final wash, buffer was aspirated and beads were incubated with 100-μl Elution buffer (300 mM imidazole, 50 mM Na-phosphate pH 8.0, 300 mM NaCl, 0.01% Tween-20) for 20 min, then beads were collected via magnet. The supernatant containing purified Hsc70 complex was transferred to a fresh tube, 25 μl of 5× SDS-PAGE sample buffer was added and the sample was denatured for 5 min at 95 °C. Twenty microliter of sample was analyzed by SDS-PAGE and processed for conventional Western blot analysis. FLAG-tag proteins were purified as follows: 200 μg of cell lysate was incubated with 30-μl Anti-FLAG® M2 Magnetic Beads (Sigma) overnight on a rotator at 4 °C. FLAG beads were collected by magnet then washed five times with 500 μl 1× TBS. After the final wash, buffer was aspirated and beads were incubated with 100-μl Elution buffer (TBS supplemented with 10 μg/ml FLAG peptide) for 20 min. The supernatant containing purified FLAG-Hsc70 complex was transferred to a fresh tube, 25 μl of 5× SDS-PAGE sample buffer was added, and the sample was denatured for 5 min at 95 °C. Twenty microliter of sample was analyzed by SDS-PAGE and processed for conventional Western blot analysis with HSP110 (StressMarq, SPC-195), HDJ2 (Thermo, MA512748), HSP27 (Thermo, MA3015), FLAG (Sigma, F3165), and HIS (Qiagen, 34,670) antibodies.
Immunoblotting
Total cell extracts were prepared from the single-cell clones using Mammalian Protein Extract Reagent (Thermo). Samples were loaded on 8–12% Bis-Tris Gel (Invitrogen) and ran at 200 V for approximately 60 min. Gels were transferred onto Nitrocellulose membrane by transfer at 500 mA for 60 min. Membranes were blocked for 1 h with TBS-Tween and 1% BSA and probed with HIS, FLAG, and α-Tubulin primary antibodies overnight in blocking solution overnight at 4 °C. Membranes were incubated with α-mouse and α-rabbit secondary antibodies (GE Healthcare) for 1 h at room temperature in blocking buffer. The blots were imaged using MP Gel Doc imaging system (Bio-rad).
Luminespib treatment of cells
Wildtype and CRISPR Clones were seeded in a six-well plate and treated with 50-nM Luminespib/AUY922 (LC Laboratories N-5300) for 24 h. Protein was extracted, run on SDS-PAGE gels, and Western blotted with antibodies to either HSP72 (Enzo, C92F3A-5), HIS, or α-tubulin.
Results
Designing a tandem affinity tag for endogenous tagging of Hsc70
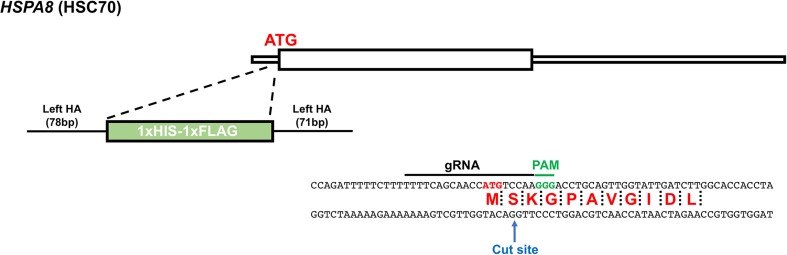
An ideal Tandem Affinity Purification (TAP) tag aids in the recovery of a fusion protein and associated complexes with minimal background contaminants. We chose a TAP tag that comprised of two highly utilized epitope tags, hexahistidine (HIS6) FLAG (DYKDDDK). Both of these tags are relatively small in size, having minimal impact on protein function. Tagging of Hsp70s in model organisms such as budding yeast with HIS6 and FLAG does not impair essential chaperone function (Truman et al. 2012). Both of these tags can be utilized to isolate highly purified native protein complexes (Rigaut et al. 1999). Given that epitope tagging of Hsp70s on the C-terminus impairs client binding, we chose to epitope tag Hsc70 on its N-terminus (Shaner et al. 2005). CRISPR-mediated epitope tagging requires expression of three components in the cell: (1) the Cas9 enzyme which creates a DNA double strand break, (2) a gRNA that binds Cas9 and targets it to the desired location, and (3) a repair template containing both the epitope tag and regions of homology to the location of insertion. The majority of CRISPR knock-in studies have utilized repair templates between 250 and 1000 bp (Dalvai et al. 2015; He et al. 2016), amplifying expense and technical difficulties. For this study, we decided to examine the feasibility of endogenously tagging Hsc70 using a repair template with exceptionally small overhangs less than 100 bp each side. Using the maximum size of IDT’s ultramers (200 bp), we designed an ssODN repair template that contained the sequence for the Start codon, HHHHHH (His6 tag), DYKDDDDK (Flag tag), and GG linker. In addition, the ssODN contained a 5′ homology arm of 78 bp and a 3′ homology arm of 71 bp (Fig. 1). To prevent continued Cas9 digestion of the 5′ of the HSC70 gene post-CRISPR-mediated epitope tagging, a silent mutation was incorporated into Serine 2 of the Hsc70 gene (TCC to TCA).
Fig. 1.
Design of a tandem HIS-FLAG epitope suitable for endogenous tagging of Hsc70. Schematic of the HSC70 locus, Cas9 targeting site, and donor construct used to insert the HIS-FLAG tag. Annotated are the positions of the stop codon (TAG), the Protospacer Adjacent Motif (PAM) that specifies the cleavage site, and homology arms left and right (Left-HA, Right-HA)
Generation of HSC70-sgRNA-pX458
CRISPR-mediated genome requires expression of the Cas9 nuclease and a guide RNA (gRNA) that targets Cas9 to a required region. A suite of vectors have been created for targeted gene deletion and knock-in, but pX458 (pSpCas9(BB)-2A-GFP, (Ran et al. 2013)) was chosen for our study based on its ability to co-express Cas9, a GFP marker and specific gRNA from the U6 promoter. Using the CRISPOR algorithm (http://crispor.tefor.net/), we designed a gRNA (TTTTTCAGCAACCATGTCCA) that had a specificity score of 67/100 and produces no off-targets even with single-base mismatches present next to the genomic PAM sequence.
HSC70-sgRNA-pX458 induces DSBs at the 5′ region of the HSC70 gene
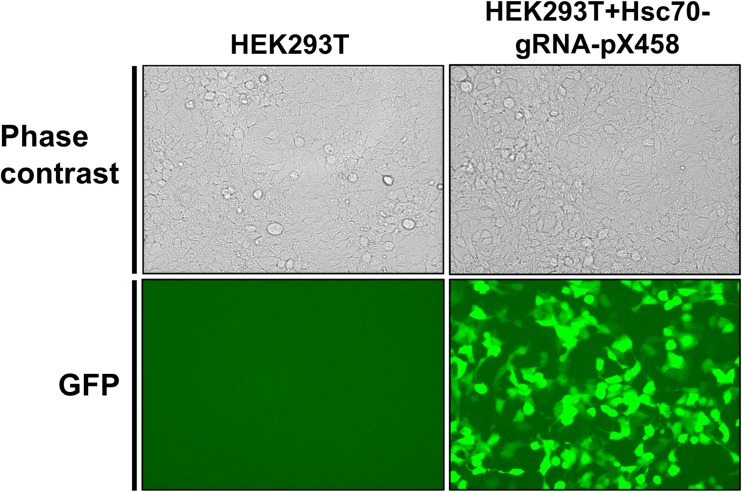
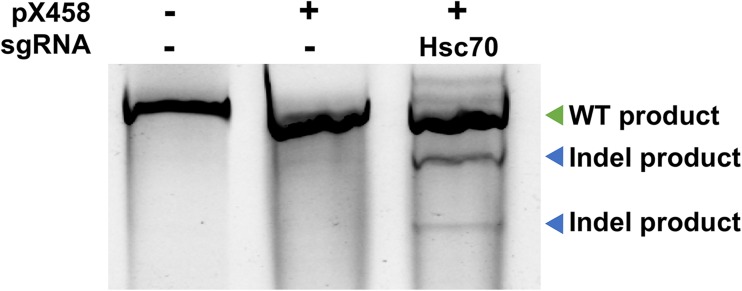
HEK293T cells were chosen to express our CRISPR construct given their high-average transfection efficiency, ease of culturing and their tolerance for limiting dilution, and genome editing experiments (de Los Milagros Bassani Molinas et al. 2014; Yang et al. 2014). Moreover, the HEK cell line is designated as a tier 3 cell line by the Encyclopedia of DNA Elements (ENCODE) project meaning; a multitude of genomic data is available for this cell line (Consortium 2012). We transfected the HEK293T cell line with HSC70-sgRNA-pX458 and checked expression of Cas9 by monitoring of GFP expression in the cells using microscopy. Approximately 70% of cells expressed GFP, expected given the standard rate of transfection of the HEK293T cell line (Fig. 2). To isolate cells that had been edited by Cas9, we sorted the GFP-positive cells using FACS in a petri dish 72 h post-transfection. To examine the ability of HSC70-sgRNA-pX458 to promote cleavage at the HSC70 gene, we isolated the pooled DNA from sorted cells and subjected them to the SURVEYOR nuclease assay. Genome editing was observed in cells transfected with HSC70-sgRNA-pX458 but not with untransfected cells or cells transfected pX458 plasmid lacking a targeting gRNA (Fig. 3).
Fig. 2.
Expression of Cas9-2A-GFP in HEK293T cells transfected Hsc70-gRNA-pX458. Phase contrast and GFP fluorescence images of HEK293T cell line and HEK 293 T cell line transfected with Hsc70-gRNA-pX458
Fig. 3.
Determining CRISPR-mediated cleavage using SURVEYOR assay. DNA was extracted from unsorted untransfected HEK293T cells (control), transfected with either Cas9 expressing plasmid with no guide RNA, or a plasmid expressing both Cas9 and HSC70-targeting gRNA. CRISPR-mediated genome editing was assessed by SURVEYOR assay. Correctly sized Indel products obtained from the SURVEYOR assay are annotated (Blue arrow)
HSC70-sgRNA-pX458 allows creation of cell lines expressing epitope tagged Hsc70 at native levels
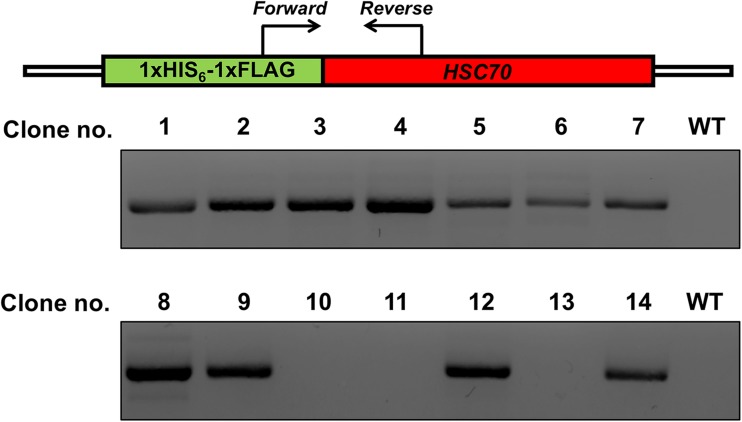
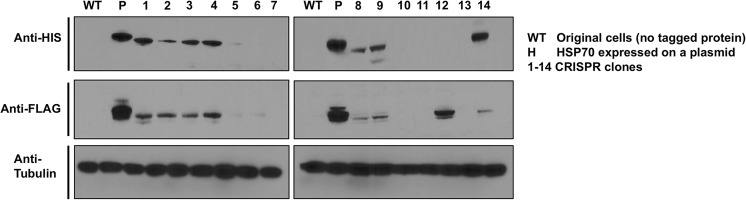
Based on the success of HSC70-sgRNA-pX458 to create DSBs at the specified genomic region, we decided to attempt creation of cells that would express HIS6-FLAG Hsc70 at native promoter levels. As before, we transfected HEK293T cells with HSC70-sgRNA-pX458, but added our tailored ssODN expressing the tandem HIS-FLAG tag along with homology arms to the HSC70 gene. GFP+ cells were sorted via FACS and were cloned by limiting dilution. Single cell clones were picked after 10 days and expanded. We checked the integration of a single HIS6-FLAG tag at the N-terminus of HSC70 in each of the isolated clones via PCR amplification of the genome using the primers flanking the epitope tag HSC70. Presence of the HIS6-FLAG tag was observed in 11/14 (79%) clones (Fig. 4). After limited dilution, single cells were identified in 12-well plate format and expanded.
Fig. 4.
Schematic and results of a PCR-based assay (in-out PCR). PCR-based assay used to detect targeted integration of the tag sequence in single-cell-derived HEK293T clones obtained by limiting dilution following CRISPR/Cas9-driven gene targeting. Primers are located outside of the homology arms and are designed to yield a PCR product if the tag is inserted. WT indicates untagged cells and 1–14 are single-cell-derived clones
Expression of HIS6-FLAG-Hsc70 protein from native promoter in HEK293T cells
Given the myriad of factors that controls protein expression, correct integration of the HIS6-FLAG tag at the genomic level by no means guaranteed correct expression of the fusion protein. To confirm the expression of HIS6-FLAG-Hsc70, we analyzed our individual clones via Western blotting of total lysate using HIS and FLAG (α-Tubulin was used as a loading control). Seven out of 14 clones (50%, clones 1–4, 8, 9, and 14) correct expression of HIS6-FLAG-tagged Hsc70 protein while one clone (clone 12) lacked a detectable HIS tag despite showing cross-reaction with the FLAG antibody.
Standard mammalian plasmids drive constitutive protein expression via promoters such as CMV. To examine how our native promoter controlled HIS6-FLAG Hsc70 compared with traditional methods, we also ran a control of cell lysate from cells transiently expressing a CMV-driven HIS6-FLAG-Hsc70 (Fig. 5). CMV-driven Hsc70 protein levels were substantially higher than those detected for the CRISPR clones (Figs. 5, 7). The DNA region containing HSC70 and HIS-FLAG tag was amplified from multiple clones and sequenced. The sequence contained the HIS-FLAG tag in frame with HSC70. No mutations were detected, except the silent mutation engineered to prevent repeat gRNA binding (Sequence attached as supplemental File S1).
Fig. 5.
Western Blot analysis of single-cell-derived HEK293T clones. Western Blots showing HIS-tag (upper panel) and FLAG-tag protein expression in single-cell-derived HEK293T clones obtained by limited dilution following CRISPR/Cas9-driven gene targeting using HIS and FLAG antibody, α-tubulin is used as a loading control. WT indicates untagged cells, P indicates Hsc70 expressed on a plasmid, and 1–14 are single-cell-derived clones
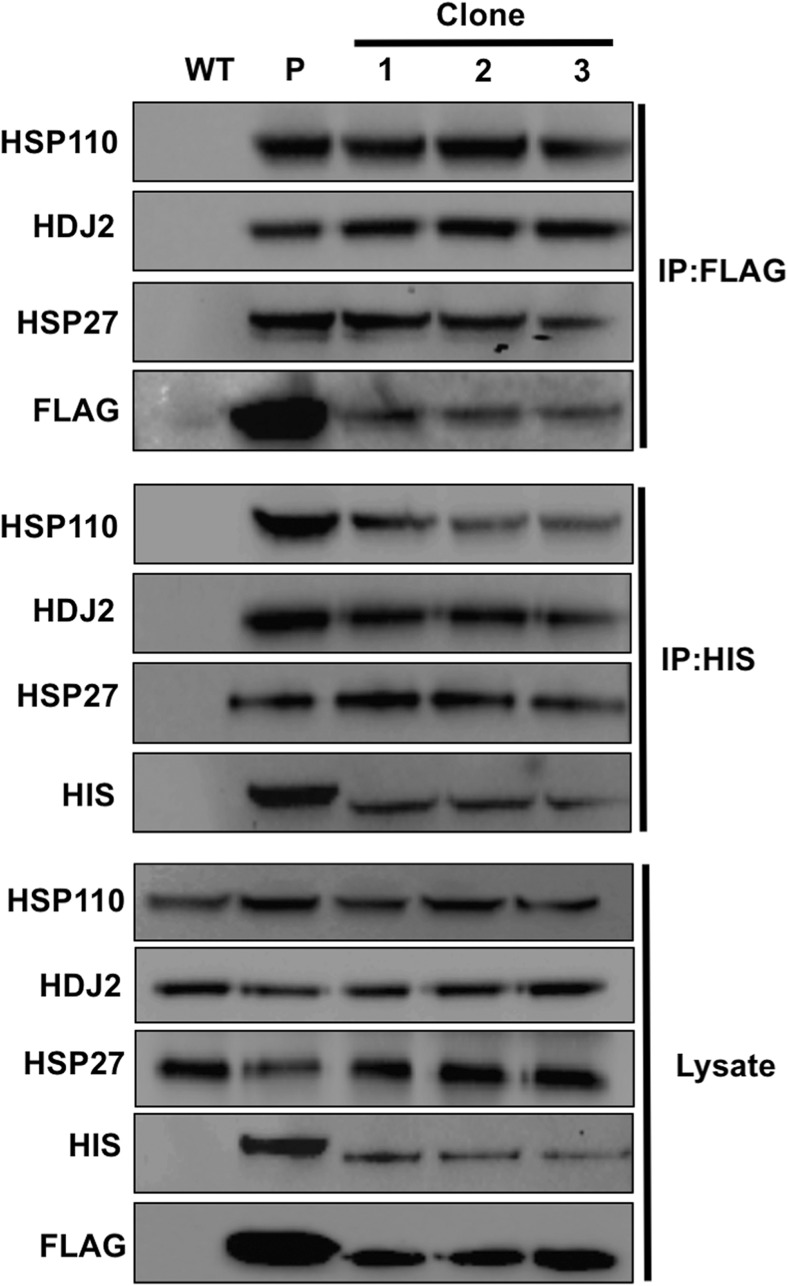
Fig. 7.
Validation of known Hsc70 interactions in HEK293THIS-FLAG-Hsc70 cells. Protein extracts were obtained from WT and three separate HEK293THIS-FLAG-Hsc70 clones. Hsc70 complexes were purified using either HIS or FLAG magnetic beads. Hsc70 complexes were resolved by SDS-PAGE and analyzed by Western Blot using antibodies to co-chaperone proteins HSP27, HDJ2, and HSP110
CRISPR-mediated tagging is isoform specific
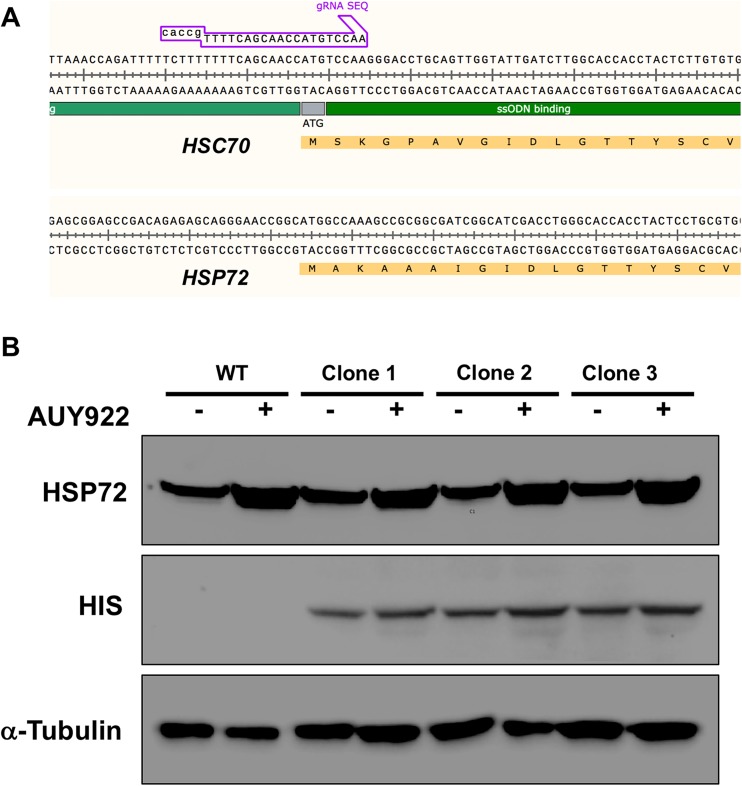
Several different isoforms of Hsp70 exist in cells, the most common being Hsc70 and Hsp72. To assess the probability of unwanted tagging of Hsp72 with our methodology, we compared the amino acid and genomic sequences of HSC70 and HSP72 genes. Interestingly, we identified low homology between the two genes, suggesting minimal probability that either the HSC70 gRNA or HSC70 ssODN could bind to the equivalent region of the HSP72 gene (Fig. 6a). To confirm this, we relied on (1) specific antibodies that selectively detect Hsp72 protein and (2) that Hsp72 can be induced by several stresses/small molecules that leave Hsc70 protein unaffected. We grew WT and three CRISPR clones in untreated media or media containing 50 mM AUY922/Luminespib. Luminespib is a well-characterized small molecule inhibitor of Hsp90 and due to indirect effects on the HSF1 transcription factor triggers increased expression of Hsp72 (Taniguchi et al. 2014). While Hsp72 levels were induced upon Luminespib treatment, levels of HIS protein remained constant. In addition, the molecular weight of Hsp72 protein observed in the CRISPR clones corresponded to that seen in WT cells. These data suggest that as expected, the epitope tagging seen was only present on the Hsc70 protein and not on the Hsc72 isoform (Fig. 6b).
Fig. 6.
CRISPR-mediated tagging of Hsc70 is isoform specific. a Amino acid and genomic DNA sequences of HSC70 and HSP72. The HSC70 gRNA binding site is absent in the HSP72 gene. b Luminespib induces Hsp72 levels but not HIS-Hsc70 levels. WT indicates untagged cells; clones 1–3 are three separate HEK293THIS-FLAG-Hsc70 clones. α-tubulin is used as a loading control in this instance
Recapitulation of well-characterized Hsc70 interactions in HEK293THIS-FLAG-Hsc70
Hsc70 interacts with a suite of co-chaperone proteins that activate Hsc70 through stimulation of the N-terminal ATPase domain (Radons 2016). Given that a primary use of this cell line is analyze Hsc70 complexes at native stoichiometry, we examined whether known interactions of Hsc70 were maintained in HEK293THIS-FLAG-Hsc70 cells. We purified Hsc70 complexes from control cells that had been transiently transfected with CMV-HIS6-FLAG-Hsc70 and from three independent CRISPR clones expressing HIS6-FLAG-Hsc70 from the native HSC70 promoter using either FLAG affinity beads or IMAC. Confirming that the CRISPR-driven Hsc70 epitope tag functioned as intended, Hsc70 interactions with Hsp110, Hdj2, and Hsp27 were detected in HEK293THIS-FLAG-Hsc70 cells but not in cells lacking epitope-tagged Hsc70 (Fig. 7).
Discussion
Although Hsp70 has been studied for several decades, much of this work has focused on the biochemical and biophysical properties of the molecule (Liu et al. 2017; Mayer 2013). Several studies have attempted to obtain a true “systems view” of the Hsp70 interaction network using yeast two-hybrid and affinity-purification followed by mass spectrometry (AP-MS) (Gong et al. 2009; Truman et al. 2012). These attempts have been made more challenging by the fact that the Hsp70 interactome is highly dynamic, changing in response to cellular stress, post-translational modifications, and disease state of the cell (Truman et al. 2012; Truman et al. 2015). Recent work has suggested that the “chaperome” of a cell may be an important trigger and/or response to disease (Rodina et al. 2016). Although drug-based affinity reagents for purifying and identifying Hsp70 complexes have been developed, these suffer from several drawbacks. Cost of generating the affinity reagent is high, and the specificity of the drug may result in purification of unwanted Hsp70 isoform complexes. It has yet to be determined whether post-translational modification on Hsp70 alter drug binding in the same way they do for the related chaperone Hsp90 (Walton-Diaz et al. 2013; Woodford et al. 2016a).
Conventional approaches to expressing epitope-tagged proteins in mammalian cells for affinity purification-mass spectrometry typically fall into two categories; transient transfection with a plasmid expression, the fusion protein of interest (typically from a constitutive promoter such as CMV), or stable transfection where cells are transfected with an expression plasmid and then maintained for several weeks to months on selectable media to induce integration into the cell’s genome. Endogenous epitope tagging of proteins offers significant advantages over conventional approaches used to study protein complexes in the mammalian cells. Tagging of genes at their natural chromosomal locations retains native promoter control of the target protein. This is important because overexpressing (or underexpressing) a target gene may change the stoichiometry of its protein-protein or protein-DNA interactions, creating artifacts in interactomic data. Many genes are finely regulated at the transcriptional level, changing based on factors such as cell cycle stage and stress. Native promoter control allows observation of native protein expression under any cellular condition.
In this study, we set out to create a mammalian cell line expressing tap-tagged Hsc70 from its endogenous promoter using CRISPR. We managed to achieve this, utilizing only a CAS9-Hsc70gRNA targeting plasmid and a small ssODN containing a tandem HIS-FLAG tag and overhangs homologous to the genome. A relatively high success rate was achieved, with 50% of clones expressing HIS6-FLAG-Hsc70. This is encouraging given the homology arms of the ssODN were only 71 and 78 bp, respectively.
The HEK293THIS-FLAG-Hsc70 cells produced in this study are stable and thus are suitable for experiments lasting longer than 72 h after which expression from transient transfection would decline. Although traditional stable transfection methodologies mitigate this issue, this technology suffers from lack of control over copy number of and location of integration events, with causing varied expression between cells in a growing population. Our CRISPR cells were expanded from a single clone producing homogeneity of expression. In addition, given that the CRISPR integration method does not use any selectable resistance markers, cells can be grown in drug-free media conditions. A common concern of epitope tagging is potential disruption of protein function and interactions. While HIS and FLAG-tagged Hsc70 constructs have been utilized in many previous studies, it was hard to assess functionality given that the native Hsc70 was still expressed alongside the exogenous protein. All interactions of well-established Hsc70 binding proteins tested in this study were maintained in HEK293THIS-FLAG-Hsc70 cells. In addition, no growth defects were observed for these cells compared to standard HEK293T cells, attesting to the functionality of these constructs when expressed as the sole Hsc70 in the cell. Advantages of the tandem HIS6-FLAG tag include flexibility in choice of affinity reagents that can be used and ability to significantly reduce non-specific complex binding through tandem purification protocols (typically IMAC purification followed by FLAG purification).
In conclusion, we have developed a protocol for generating epitope-tagged chaperones from their native promoter using only minimal reagents. The cell line generated in this study (HEK293THIS-FLAG-Hsc70 cells) will be a freely available and useful reagent for chaperone researchers, especially in the quest to understand the complexities of the Hsc70 interactome.
Electronic supplementary material
(SEQ 1 kb).
Acknowledgements
The authors thank F. Zhang for providing materials used in this study and M. Mollapour for helpful comments. We thank C. Richardson and D. Dreau for their technical assistance. This project was supported by NCI R15CA208773.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12192-017-0845-2) contains supplementary material, which is available to authorized users.
References
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvai M, et al. A scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell Rep. 2015;13:621–633. doi: 10.1016/j.celrep.2015.09.009. [DOI] [PubMed] [Google Scholar]
- de Los Milagros Bassani Molinas M, Beer C, Hesse F, Wirth M, Wagner R. Optimizing the transient transfection process of HEK-293 suspension cells for protein production by nucleotide ratio monitoring. Cytotechnology. 2014;66:493–514. doi: 10.1007/s10616-013-9601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn DM, et al. c-Abl mediated tyrosine phosphorylation of Aha1 activates its co-chaperone function in cancer cells. Cell Rep. 2015;12:1006–1018. doi: 10.1016/j.celrep.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- He X, et al. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Kelley WL. Molecular chaperones: how J domains turn on Hsp70s. Curr Biol. 1999;9:R305–R308. doi: 10.1016/S0960-9822(99)80185-7. [DOI] [PubMed] [Google Scholar]
- Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- Liu Q, Li H, Yang Y, Tian X, Su J, Zhou L, Liu Q. A disulfide-bonded DnaK dimer is maintained in an ATP-bound state. Cell Stress Chaperones. 2017;22:201–212. doi: 10.1007/s12192-016-0752-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rodina A, et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature. 2016;538:397–401. doi: 10.1038/nature19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–582. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- Shiota M, et al. Generation of a rat monoclonal antibody specific for heat shock cognate protein 70. Hybridoma (Larchmt) 2010;29:453–456. doi: 10.1089/hyb.2010.0024. [DOI] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–E241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- Tanaka M, et al. Generation of a rat monoclonal antibody specific for hsp72. Hybridoma (Larchmt) 2011;30:397–400. doi: 10.1089/hyb.2011.0015. [DOI] [PubMed] [Google Scholar]
- Tanaka M, et al. Hsc70 contributes to cancer cell survival by preventing Rab1A degradation under stress conditions. PLoS One. 2014;9:e96785. doi: 10.1371/journal.pone.0096785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, et al. Identification of low-abundance proteins in serum via the isolation of HSP72 complexes. J Proteomics. 2016;136:214–221. doi: 10.1016/j.jprot.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, et al. Heat shock protein 90 inhibitor NVP-AUY922 exerts potent activity against adult T-cell leukemia-lymphoma cells. Cancer Sci. 2014;105:1601–1608. doi: 10.1111/cas.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW, et al. CDK-dependent Hsp70 phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell. 2012;151:1308–1318. doi: 10.1016/j.cell.2012.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW, et al. Quantitative proteomics of the yeast Hsp70/Hsp90 interactomes during DNA damage reveal chaperone-dependent regulation of ribonucleotide reductase. J Proteomics. 2015;112:285–300. doi: 10.1016/j.jprot.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton-Diaz A, Khan S, Bourboulia D, Trepel JB, Neckers L, Mollapour M. Contributions of co-chaperones and post-translational modifications towards Hsp90 drug sensitivity. Future Med Chem. 2013;5:1059–1071. doi: 10.4155/fmc.13.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford MR, Dunn D, Miller JB, Jamal S, Neckers L, Mollapour M. Impact of posttranslational modifications on the anticancer activity of Hsp90 inhibitors. Adv Cancer Res. 2016;129:31–50. doi: 10.1016/bs.acr.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Woodford MR, et al. Mps1 mediated phosphorylation of Hsp90 confers renal cell carcinoma sensitivity and selectivity to Hsp90 inhibitors. Cell Rep. 2016;14:872–884. doi: 10.1016/j.celrep.2015.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Yang JL, Byrne S, Pan J, Church GM. CRISPR/Cas9-directed genome editing of cultured cells. Curr Protoc Mol Biol. 2014;107:31.31.31–31.31.17. doi: 10.1002/0471142727.mb3101s107. [DOI] [PubMed] [Google Scholar]
- Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(SEQ 1 kb).