Abstract
Abiotic stresses like drought, salinity, high and low temperature, and submergence are major factors that limit the crop productivity. Hence, identification of genes associated with stress response in crops is a prerequisite for improving their tolerance to adverse environmental conditions. In an earlier study, we had identified a drought-inducible gene, vesicle-associated membrane protein-associated protein (TaVAP), in developing grains of wheat. In this study, we demonstrate that TaVAP is able to complement yeast and Arabidopsis mutants, which are impaired in their respective orthologs, signifying functional conservation. Constitutive expression of TaVAP in Arabidopsis imparted tolerance to water stress conditions without any apparent yield penalty. Enhanced tolerance to water stress was associated with maintenance of higher relative water content, photosynthetic efficiency, and antioxidant activities. Compared to wild type, the TaVAP-overexpressing plants showed enhanced lateral root proliferation that was attributed to higher endogenous levels of IAA. These studies are the first to demonstrate that TaVAP plays a critical role in growth and development in plants, and is a potential candidate for improving the abiotic stress tolerance in crop plants.
Electronic supplementary material
The online version of this article (10.1007/s12192-017-0854-1) contains supplementary material, which is available to authorized users.
Keywords: Abiotic stress, TaVAP, Triticum aestivum, Lateral roots, Auxin
Introduction
Plants respond to imposition of abiotic stress by modulating various physiological, biochemical, and molecular processes (Shinozaki and Dennis 2003; Wang et al. 2003; Shi and Chan 2014; Mickelbart et al. 2015). Although considered a housekeeping process, vesicle trafficking also plays an important role in regulation of stress response (Levine et al. 2001; Mazel et al. 2004). In fact, vesicle transport is important for maintaining homeostasis, cell growth and development, and polarity (Blatt and Thiel 2003). The membrane vesicle transport machinery includes phospholipids and integral membrane proteins, such as vesicle-associated membrane proteins (VAMPs), the latter being the major constituent of soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNARE) complexes. SNARE complexes are responsible for fusion of vesicles with the target membranes (Duman and Forte 2003; Jena 2011; Woodman 1997).
A protein interacting with VAMP was first identified in mollusk Aplysia californica and designated as VAMP-associated protein-33 (VAP-33) (Skehel et al. 1995). The first ortholog of VAP-33 in plants, VAP-27, which exhibited high homology to the inositol regulator SCS2 of yeast (Kagiwada et al. 1998), was characterized from Nicotiana tabacum (Laurent et al. 2000). The VAPs are conserved proteins, with homologs reported in all eukaryotes (Weir et al. 1998; Nishimura et al. 1999; Lipka et al. 2007). The VAPs are characterized by the presence of a motile sperm protein (MSP-VAP) domain, a linker that contains a coiled-coil (cc) domain, and a carboxyl-terminal trans-membrane domain containing a GXXXG dimerization motif (Russ and Engalman 2000). VAP homologs have very prominent VAP consensus sequence (VCS), a 16-residue motif that is highly conserved in VAPs but not in MSP1. The orthologs of VAP in S. cerevisiae (SCS2) and other fungi have a similar domain structure but lack coiled-coil domain in the linker and the GXXXG motif.
Studies on A. californica ascribed VAP-33 a role in exocytosis of neurotransmitters, providing the first insight into the functioning of these proteins (Skehel et al. 1995). Later studies showed that these proteins are also involved in several other fundamental cellular processes like microtubule organization, lipid metabolism, inositol auxotrophy, and unfolded protein response (Hosaka et al. 1992; Skehel et al. 2000; Lev et al. 2008; Rutkowski and Kaufman 2004; Ron and Walter 2007; Petersen et al. 2009; Saravanan et al. 2009). Recent studies have demonstrated that Arabidopsis VAP-27 genes are essential for normal growth of plants (Wang et al. 2016). The implication(s) of these genes in stress response of plants is still a matter of conjecture. Our earlier studies on the cloning and characterization of a gene encoding wheat VAP (TaVAP) demonstrated that expression of this gene in contrasting cultivars of wheat (drought tolerant and susceptible) was modulated differentially in response to water stress (Singh et al. 2007), indicating its likely role in stress adaptation. To further unravel the physiological implications of this gene in abiotic stress response of plants, we carried out functional analysis of TaVAP by developing transgenics of Arabidopsis. These studies are the first to demonstrate that overexpression of TaVAP imparts tolerance to different abiotic stress conditions in transgenic Arabidopsis plants without any apparent yield penalty under control conditions, and it appears to be a potential candidate for enhancing stress tolerance in crop plants.
Experimental procedures
Yeast culture media and strains
Yeast extract-peptone-dextrose (YPD) and yeast minimal media for culturing yeast (Saccharomyces cerevisiae) cells were used as described by Kaiser et al. (1994) and Klig et al. (1985), respectively. The strains of yeast used in this study were YER120W (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the parental strain, BY4741 (GE Healthcare Dharmacon Inc., USA).
Plant material and growth conditions
For studies at seedling stages, the plants of wheat (Triticum aestivum) cv. PBW343 were grown hydroponically, whereas observations at later stages of development were recorded using plants raised in soil in clay pots. The plants were grown in the culture room at 22 ± 1 °C, maintaining 16:8 h light and dark regime with light intensity of 100–125 μmol m−2 s−1. Heat stress (HS) was imposed by transferring 10-day-old seedlings to 37 °C for 2 h, followed by 42 °C for 2 h in the growth chamber (Conviron, Canada). Seedlings were exposed to cold stress (CS) by incubating at 4 °C for 24 h. Salt and osmotic stress (OS) were imposed by exposing the seedlings to 150 mM NaCl and 2% PEG-6000 solutions, respectively, for 24 h. For hormone treatments, the seedlings were treated with 10 μM abscisic acid (ABA) (Sigma-Aldrich Co., USA), brassinosteroid (BR) (1 μM epibrassinolide), and 50 μM salicylic acid (SA) for 4 h. Following the treatments, the tissues were frozen in liquid N2 and stored at − 80 °C until RNA isolation. Arabidopsis thaliana ecotype Columbia (strain Col-0) was used in the present study. Arabidopsis plants were grown on Murashige and Skoog (MS) medium in the Petri plates for studies with seedlings, whereas the plants grown in Soilrite (Vermiculite:Perlite:Sphagnum moss; 1:1:1) were used for observations at later stages of development.
Expression analysis of TaVAP
Total RNA from different tissues of wheat and Arabidopsis plants was isolated using Trizol Reagent (Invitrogen, USA) as per the manufacturer’s instructions. The primers for real-time PCR analysis of TaVAP were designed using the Primer Express 2.0 (PE Applied Biosystems, USA) (Table S1). The quality and quantity of the extracted RNA was checked on Nanovue® Spectrophotometer (GE healthcare, UK) by determining the ratio of A260/280 and A260/230, which were between 1.9–2.1 and 2–2.3, respectively. Integrity of the RNA was checked by denaturing 1.5% agarose gel electrophoresis. At least two different RNA isolations were used for cDNA syntheses, and each cDNA was analyzed in triplicate. First strand cDNA synthesis reaction was set up as per manufacturer’s instructions using High capacity cDNA Reverse transcription Kit (Applied Biosystems, USA). Two micrograms of RNA was taken in a 40-μl total reaction volume. Real-time PCR analysis was carried out using 2× Roche SYBR Green I master mix on a Roche Light Cycler 480 II Instrument (Roche, Switzerland). The 18-μl reaction volume, in three technical replicates, consisted of 6.25 μl PCR grade water, 9 μl 2× Roche SYBR Green I master mix, 1 μl cDNA, and 1.75 μl of 5 μM of forward and reverse primers. The conditions used for real-time PCR cycles were pre-incubation for 10 min at 95 °C, followed by 45 cycles of 10 s at 95 °C, 20 s at 60 °C, and 10 s at 72 °C, followed by cooling at 40 °C for 30 s. The Ct values obtained were processed further to calculate the relative mRNA levels for different genes by ΔΔCt method. The relative mRNA levels of the TaVAP in different RNA samples were normalized using three internal standards, i.e., actin (ACTIN), ribosylation factor (ADP), and RNase L inhibitor (RLI) protein (Gimenez et al. 2011) for wheat, whereas actin (ACTIN2), paired amphipathic helix repeat containing protein (PAH2), and zinc finger family protein (ZFP) were used for A. thaliana (Czechowski et al. 2005). The data obtained, presented as the mean ± S.E, were subjected to t test and one way ANOVA to determine the significance at p ≤ 0.01.
Induction of TaVAP and its immunoblotting
For raising polyclonal antibodies, the cDNA encoding full length open reading frame (ORF) of TaVAP sequence was amplified from pDrive::TaVAP (Singh et al. 2007) with suitable primers (Table S1), and cloned into EcoRI and NheI sites in pET28a(+) vector (Roche Applied Science, Switzerland). The recombinant plasmid pET28a::TaVAP was transformed into E. coli strain BL21-CodonPlus(DE3)-RIL for expression. The recombinant TaVAP protein was induced with 0.4 mM IPTG with overnight shaking at ~ 7 rcf at 18 °C. IPTG was added when A600 of the culture reached 0.5. The induced cells were pelleted by centrifugation at ~ 20,000 rcf for 2 min at 4 °C. The pellet was suspended in the lysis buffer [20 mM Tris buffer (pH 8.0), 10 mM ß-mercaptoethanol, 1 mM PMSF], followed by sonication (6 short bursts of 10 s each followed by intervals of 30 s at 4 °C). The supernatant was collected after centrifugation at ~ 20,000 rcf for 20 min at 4 °C. The induction of recombinant protein was confirmed by 12% SDS-polyacrylamide gel electrophoresis. The induced protein was purified using Ni-NTA column (Sigma-Aldrich Co., USA) and used for raising polyclonal antibodies in rabbit (outsourced to Bangalore Genei, Karnataka, India). Immunoblotting studies were carried out as described earlier (Chauhan et al. 2012).
Complementation of the yeast mutant
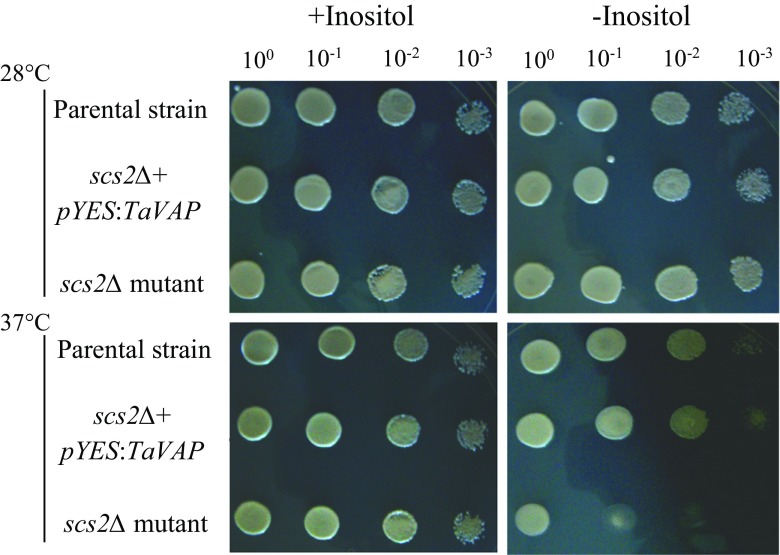
The open reading frame of TaVAP, cloned earlier in our laboratory (Singh et al. 2007), was amplified using gene-specific primers (Table S1). The amplified fragment was cloned into pDrive vector by TA cloning, and after digestion with EcoRI and HindIII enzymes, ligated into the pYES 2.0 vector. Following confirmation by restriction digestion and sequencing, the recombinant pYES-TaVAP construct was transformed into the S. cerevisiae Scs2∆ mutant strain using lithium acetate method as described in yeast protocols handbook (Clontech Laboratories Inc., USA). The parental yeast strain BY4741 was used as a positive control. Since Scs2∆ mutant cells show inositol auxotrophy at higher temperature (Kagiwada et al. 1998), complementation assays in the absence and presence of inositol (100 μM) were performed at 28 and 37 °C.
Overexpression of TaVAP in A. thaliana
The AtVAP (AT4G00170) gene in Arabidopsis is homologous to TaVAP and shares ca. 57% homology at protein level (Table S2). Seeds of the Atvap mutant [Columbia (Col-0) background; SALK_119355], used in this study, were procured from the ABRC (Arabidopsis Biological Resource Center; http://www.arabidopsis.org/). The Atvap mutant contains T-DNA insertion in the first exon of the AtVAP gene. The PCR-amplified TaVAP amplicon (Table S1) was cloned into XbaI and BamHI sites of the plant binary vector PBI121 to generate PBI121–35S pro::TaVAP construct. The recombinant PBI121–35S pro::TaVAP was transformed into the Agrobacterium tumefaciens strain Agl1 which was used for transformation of Arabidopsis plants (strain Col-0) by floral dip method (Clough and Bent 1998). T0 transgenic plants were selected on MS medium containing kanamycin (50 mg/ml). The plants selected on antibiotic medium were transferred and allowed to grow in pots with Soilrite, and selfed in successive generations to obtain homozygous T4 lines that showed 100% germination on medium containing kanamycin. Transgenic and wild type Col-0 seedlings were grown in Soilrite-filled pots in a culture room maintained at 22 ± 1 °C with daily photoperiodic cycles of 16 h light and 8 h dark. The effect of hormones on the growth of Atvap mutants was studied by treating the 10- and 15-day-old seedlings grown in MS medium and Soilrite, respectively, with gibberellic acid (GA), auxin (indole-3-acetic acid; IAA), and ethylene (1-aminocyclopropane-1-carboxylic acid; ACC) at concentrations of 0.5, 0.1, and 1 μM. The effect of these growth regulators on seedlings grown in MS medium was recorded until the plants reached 20-day-old stage, as compared to 30-day-old stage for the plants raised in Soilrite.
Seed germination assays
Seeds of wild type (WT) and transgenic Arabidopsis lines (TaV1, TaV2, TaV3, TaV4) were surfaced-sterilized using 2% sodium hypochlorite and Triton X-100 mix, washed thoroughly (4–5 times) with sterilized deionized water, and plated on half-strength MS medium (pH 5.8) containing 2% sucrose and 0.8% agar (supplemented with various concentrations of NaCl and PEG, when required). Plates were kept for seed stratification at 4 °C in the dark for 72 h, and transferred to a culture room at 22 ± 1 °C under long-day (16 h light/8 h dark) conditions. Germination, as evident by radicle emergence, was recorded after 3 days of plating of seeds.
Abiotic stress assays
After some initial standardization, taking into consideration the number of rosette leaves and their size, the 15-day-old plants (harboring 8 leaves) were selected for stress imposition (Boyes et al. 2001). For survival assays, 15-day-old plants of the mutant, WT, and transgenic lines (TaV1), grown in pots, were subjected to drought stress by withholding water supply for 14 days, followed by rewatering for 7 days. Since 14 days of drought was lethal to the WT plants, therefore, further studies were carried after 10 days of water stress followed by 7 days of re-watering. Relative water content (RWC) of the leaves of 15-day-old plants was calculated according to Barrs and Weatherley (1962). Water potential was measured using a dewpoint hygrometer (WP4, Decagon Devices, Inc., Pullman, WA, USA). Photosystem II (PSII) activity was measured according to Krause and Weiss (1991). Measurements of modulated chlorophyll fluorescence emission from the upper surface of the leaf were made using a pulse amplitude modulation fluorometer (Junior-PAM chlorophyll fluorometer, H. Walz, Eifeltrich, Germany). Leaves were dark adapted for 30 min before measuring the induction of fluorescence, and maximum photochemical efficiency (Fv/Fm) was recorded.
Biochemical estimations
Chlorophyll and carotenoids were estimated from fresh leaves of mutant, WT, and transgenic plants by non-maceration method according to Hiscox and Israelstam (1979). The proline content of the dry leaves was determined as described earlier (Bates et al. 1973). Malondialdehyde (MDA) content of the dried leaves, which is used as a measure of lipid peroxidation in the tissue, was estimated as a total reactive substance of 2-thiobarbituric acid (TBA) (Heath and Packer 1968). The MDA concentration was calculated using an extinction coefficient of 155 mM−1 cm−1 with the formula: MDA (mmol g−1 DW) = [(A532–A600)/155] × 103 × dilution factor. Total soluble sugars in the dry leaves were determined according to the protocol developed by Dubois et al. (1956) using different concentrations of glucose as standard.
Measurement of the primary and lateral root length
For evaluating the rooting pattern, 5-day-old seedlings of mutant, WT, and TaVAP-overexpressing (TaVAP OE) were transferred to the fresh MS plates and grown vertically for 10 days. Photographs were clicked with the digital camera and analyzed using ImageJ software (http://rsbweb.nih.gov/ij/download.html). The length of the primary and lateral roots was measured using free hand tool of ImageJ. The total length of lateral roots of 10 plants each in three replicates was calculated, and the mean for each line was used as an index to measure the lateral root growth.
Estimation of IAA
The homogenates of 100 mg leaf samples from the mutant, WT, and TaVAP OE plants each were extracted with 80% methanol after homogenizing in liquid N2 (Merck, HPLC grade), followed by overnight incubation at 4 °C in the dark (Nayar et al. 2013). The methanol fractions were passed twice through C18 Sep-Pak cartridge (Waters Ass., Milford, MA). The eluates were evaporated completely in vacuum at 40 °C and redissolved in 200 μl absolute methanol. A 5-μl aliquot of the crude extract was subjected to ultra-high performance chromatography (Waters, USA) with the following parameters: C18 column (Waters BEH C18 2.1 × 50 mm, particle size 1.7 μm), solvents 5 mM ammonium formate (pH 4.0), and methanol, flow rate of 0.25 ml min−1, temperature: 40 °C, and total run time: 10 min. The chromatograms of the mutant, WT, and transgenic plants were compared to the standard indole-3-acetic acid (Sigma-Aldrich Co., USA) profile.
Microarray hybridization and data analysis
For microarray analysis, 5 μg aliquot of high-quality total RNA isolated from the 15-day-old seedlings of mutant, WT, and transgenic plants grown under normal growth conditions was used for processing using Affymetrix gene chips. The analysis was carried out employing one-cycle target labeling and control reagents (Affymetrix, Santa Clara, CA, USA) and Arabidopsis genome arrays (Affymetrix: ATH1-121501) as per manufacturer’s instructions (Jain et al. 2006). For data analysis, the image (cel) files were imported into Gene Spring software (Stratagene, La Jolla, CA, USA). Normalization of the data was performed by Gene Chip robust multiarray analysis (GCRMA) algorithm. The data were log transformed, and average log signal intensity values of two biological replicates were computed for each of the three samples and subjected to further analysis. The genes that were up- or downregulated ≥ twofold with a corrected P value ≤ 0.05 in transgenic compared to WT, and mutant compared to WT, were identified. Data was submitted to the online NCBI GEO, with accession no. GSE86848.
Assay of auxin-inducible GUS activity
To gain insight into auxin homeostasis in the TaVAP OE lines, the transgenic plants were crossed with the Arabidopsis plants harboring DR5:GUS. F1 generation was allowed to self and the F2 plants, that showed beta-glucuronidase (GUS) activity in the roots (35S::TaVAP X DR5::GUS), were selected for the present study. The reporter gene activity in the 3-day-old seedlings was visualized histochemically according to Jefferson et al. (1987). For analyzing the inhibitory effects, the 5-day-old plants grown on MS medium were transferred to medium containing 1-N-naphthylphthalamic acid (NPA; 1, 5, 10 μM) and used for histochemical GUS staining. Briefly, the tissue samples were incubated overnight at 37 °C in histochemical assay buffer [0.1 M sodium phosphate buffer (pH 7.0); 50 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100, 1 mg ml−1 X-gluc (Sigma-Aldrich Co., USA)]. Excess of staining and cholorophyll pigmentation from the plants was removed by washing with absolute alcohol:acetone (3:1), followed by observations under stereozoom microscope (Olympus, UK).
Results
Our earlier studies demonstrated that the transcript abundance of TaVAP was modulated differentially by water stress in the leaves and grains of drought tolerant and susceptible cultivars of wheat, thus, suggesting a possible role of this gene in drought stress response (Singh et al. 2007). In the present study, work has been extended further to elucidate the function of TaVAP by overexpression in Arabidopsis and comparing its phenotype with the wild type and a mutant of its orthologous gene in Arabidopsis. Although we do intend to do its functional validation eventually in wheat but raising transgenic in wheat still poses a formidable challenge.
Expression analysis of TaVAP
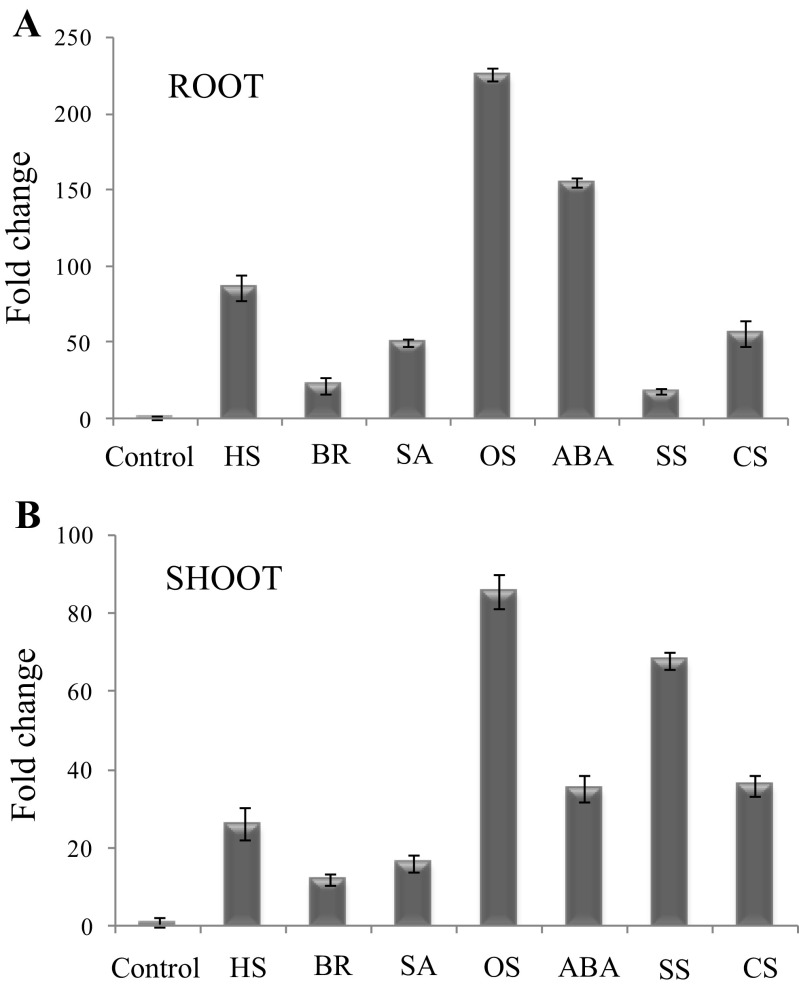
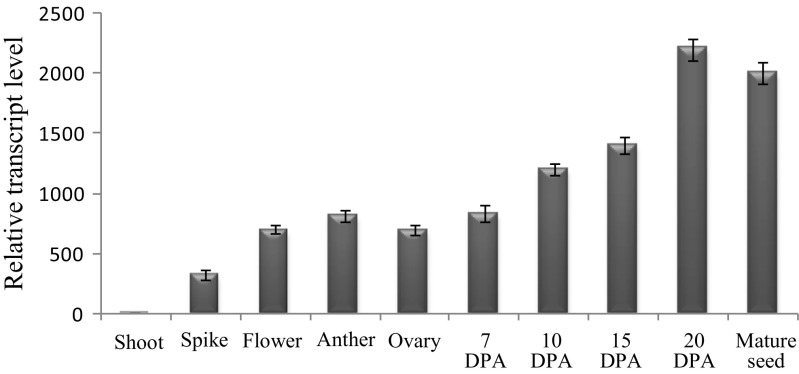
To understand the role of TaVAP in stress tolerance, expression of this gene in the roots and shoots of 10-day-old wheat seedlings was studied in response to different stress conditions and on treatment with growth regulators, BR, and ABA. Compared to the control plants, exposure to different treatments resulted in enhanced expression of TaVAP in both root and shoot, with maximum increase observed in response to osmotic stress. The effect of salt stress on the expression of TaVAP was more pronounced in the shoots, whereas expression of this gene in response to ABA was higher in the roots (Fig. 1a, b). These observations indicate that regulation of TaVAP under stress conditions is tissue-dependent, and it may be involved in adaptation of plants against multiple stress conditions. Detailed studies are, however, required to understand the role of ABA in stress-induced expression of TaVAP. Further, compared to other floral tissues, the expression of TaVAP increased consistently during grain development, with maximum transcript accumulation observed at the later stages (20 DPA) of grain maturation (Fig. 2). These observations are further supported by the cloning of the 2 kb region, upstream to the TaVAP, revealed the presence of several putative cis regulatory sequences that may be responsible for stress-inducibility of this gene (Fig. S1A).
Fig. 1.
Effect of different hormones and abiotic stress treatments on expression of TaVAP gene in the roots and shoots of 10-day-old seedlings of T. aestivum. The changes in transcript were studied by real-time PCR. The relative mRNA levels of TaVAP were computed with respect to the internal standards viz.; actin (ACTIN), ribosylation factor (ADP), and RNase L inhibitor (RLI). Control plants were maintained at 22 ± 1 °C and irrigated with autoclaved double distilled water. The data represent the mean ± S.E. of three independent experiments. The data presented represent fold change with respect to the control, whose value is 1. HS heat stress, BR brassinosteroid, SA salicylic acid, OS osmotic stress, ABA abscisic acid, SS salt stress, CS cold stress
Fig. 2.
Real-time PCR expression analysis of TaVAP at different stages of seed development in T. aestivum. The relative mRNA levels of TaVAP were computed with respect to the internal standards ACTIN, RIBOSYLATION FACTOR, and RNASE L INHIBITOR. The data represent the mean ± S.E. of three independent experiments
Complementation of mutants of yeast and A. thaliana
TaVAP shares 28.7 and 56.6% identity with its yeast and Arabidopsis orthologs, respectively (Table S2). Therefore, functional analysis of TaVAP was carried out by transforming the yeast strain and Arabidopsis plants, which were mutated in their respective orthologous genes. The yeast cells having mutation (Scs2∆) in the ortholog of TaVAP are unable to grow at 37 °C in the absence of inositol (Kagiwada et al. 1998). Transformation with TaVAP gene enabled the Scs2∆ mutant cells to grow at 37 °C even in the absence of inositol (Fig. 3), indicating functional complementation. The T-DNA lines of Arabidopsis, mutated in AtVAP (AT4G00170), were obtained from ABRC. Relative to WT (Col-0) plants (53.6 ± 0.98 cm), the T-DNA mutants exhibited significant decrease in height (32.77 ± 0.25 cm) (Table 1; Fig. S1B), implying that TaVAP is playing an important role in the growth of Arabidopsis. The mutant plants, however, did not show any impairment in the reproductive process and were able to set viable seeds. The mutant Arabidopsis plants transformed with TaVAP grew to a height similar to that of WT (Fig. S1B). Morphometric analysis revealed that there was no significant difference in the number of nodes/internodes of the mutant and WT plants (Table 1), implying that the dwarf character is primarily due to decrease in the length of internodes.
Fig. 3.
Complementation of yeast scs2Δ mutant with TaVAP. Inositol auxotrophy of yeast scs2Δ mutant strain, scs2Δ mutant cells transformed with pYES2.0:TaVAP, and parental strain was determined by streaking the cells on either inositol-containing (INO+) or inositol-free (INO-) minimal synthetic dropout (SD) medium followed by incubation at 28 °C for 72 h or at 37 °C for 96 h. SD medium was added with galactose (2%) and raffinose (1%)
Table 1.
Morphometric analysis of the mutant, wild type (WT), and transgenic lines overexpressing TaVAP (TaV1, TaV2), under control conditions. Data represent the mean ± S.E of ten different plants
| Morphological characters | Mutant | Wild type | TaV1 | TaV2 |
|---|---|---|---|---|
| Plant heighta (cm) | 32.77 ± 0.25 | 53.6 ± 0.98 | 54.6 ± 1.25 | 52.63 ± 1.45 |
| Number of nodes | 27.20 ± 1.51 | 26.12 ± 1.70 | 26.90 ± 3.21 | 27.02 ± 1.05 |
| Leaf lengthc (cm) | 2.69 ± 1.27 | 2.53 ± 0.15 | 2.85 ± 0.07 | 2.48 ± 0.14 |
| Leaf widthc (cm) | 1.20 ± 0.20 | 1.15 ± 0.06 | 1.19 ± 0.03 | 1.13 ± 0.08 |
| Rosette diameterb (cm) | 5.7 ± 0.23 | 5.23 ± 0.11 | 5.07 ± 0.19 | 4.96 ± 0.09 |
| Rosette leaf numberb | 12.02 ± 0.34 | 11.2 ± .56 | 12.1 ± 0.48 | 11.5 ± .98 |
| Silique lengthd (cm) | 1.45 ± 0.12 | 1.65 ± 0.02 | 1.59 ± 0.06 | 1.53 ± 0.08 |
| No. of seeds/silique | 46 ± 0.24 | 48 ± 1.23 | 52 ± 1.53 | 53 ± 1.94 |
| Bolting timee (days) | 28 ± 0.2 | 29.50 ± 1.6 | 28.76 ± 0.9 | 30 ± 1.2 |
aMaximum height attained by the plant during its course of growth
bMaximum height attained by the plant at the time of bolting
cMaximum height attained by the largest leaf of the plant at the time of bolting
dMaximum height attained by the length of the second silique on the primary inflorescence
eMaximum height attained by the plant days after stratification
Role of TaVAP in abiotic stress tolerance
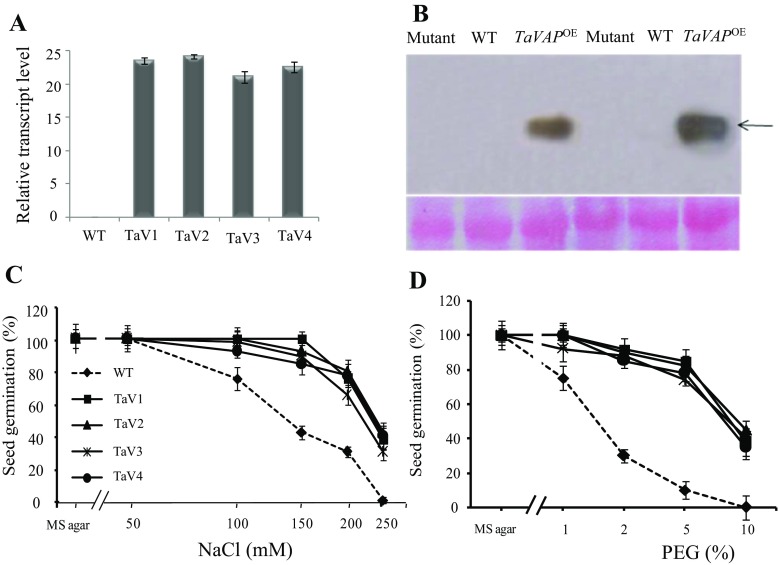
Four independent transgenic lines of A. thaliana overexpressing wheat TaVAP (driven by CaMV35S promoter) were raised and carried over to homozygous T4 generation for studying the role of this gene in abiotic stress tolerance. The transcript levels of TaVAP were high in all these four transgenic lines (Fig. 4a). One of the transgenic lines (TaV1), selected for detailed analysis, was also analyzed for translation of the protein by immunoblotting studies using TaVAP-specific polyclonal antibodies. Immunoblotting studies revealed the presence of a specific band corresponding to ca. 30 kDa (Fig. 4b) in the transgenic plants only; the mutant and the WT Arabidopsis plants did not show the presence of this protein. These results, therefore, demonstrate that the transgenic line accumulated TaVAP protein at detectable levels. The TaVAP OE plants were morphologically similar to WT and no significant difference in seed yield and the yield components was observed between WT and the two transgenic lines evaluated under control conditions (Table 2).
Fig. 4.
a Real-time PCR expression analysis of TaVAP gene in wild type (WT) and four independent transgenic lines (TaV1, TaV2, TaV3, TaV4) of A. thaliana. ACTIN was used as an internal control. b Immunoblotting of total soluble proteins extracted from the rosette leaves of 10-day-old seedlings of mutant, WT, and transgenic (TaV1) lines grown under control and stress conditions was carried out using anti-TaVAP polyclonal antibodies. Seed germination of the WT and transgenic lines was studied on MS basal medium containing different concentrations of NaCl (c), and polyethylene glycol (PEG-6000) (d). Inhibition of seed germination was recorded as non-emergence of the radical after 3 days of plating. Each experiment was conducted in three replicates containing 100 seeds each. Values are mean ± S.E. Wild type and TaVAP OE lines were represented as dotted and solid lines, respectively
Table 2.
Effect of water stress on yield and its different components in the wild type (WT) and transgenic lines (TaV1 and TaV2). Fifteen-day-old plants were exposed to water stress for 6 days followed by regular watering. Data represent the mean ± S.E of ten replicates comprising of three plants each
| Yield components | Control | 6 DAS | ||||
|---|---|---|---|---|---|---|
| WT | TaV1 | TaV2 | WT | TaV1 | TaV2 | |
| Siliques per plant | 50 ± 2.3 | 48.9 ± 2.0 | 49 ± 2.7 | 15 ± 1.2 | 21.8 ± 1.3 | 19.1 ± 2.0 |
| Seeds per silique | 53 ± 1.2 | 52 ± 1.6 | 52 ± 1.9 | 9 ± 0.92 | 23.4 ± 1.7 | 20.8 ± 2.7 |
| 100 seed weight (mg) | 20 ± 1.1 | 19.5 ± 1.0 | 19.9 ± 0.95 | 1.6 ± 0.12 | 1.8 ± 0.07 | 1.8 ± 0.19 |
| Seed yield per plant (mg) | 53 ± 1.4 | 49.6 ± 2.1 | 50.7 ± 2.5 | 2.1 ± 0.15 | 9.4 ± 0.09 | 7.1 ± 0.7 |
DAS days after stress
The role of TaVAP in conferring abiotic stress tolerance in plants was studied by evaluating the germination of WT and transgenic seeds under salt and osmotic stress. Germination of WT seeds was significantly decreased at 100 mM NaCl, whereas germination of seeds of the four different transgenic lines tested was more or less unaffected up to the level of 200 mM NaCl (Fig. 4c). Although germination of the WT seeds was completely inhibited at 250 mM NaCl, the transgenic lines exhibited 40–60% germination at this salt concentration also (Fig. 4c). These results clearly demonstrate that overexpression of TaVAP enhanced the ability of transgenic seeds to tolerate potentially inhibitory levels of NaCl. Ectopic expression of TaVAP also conferred tolerance against osmotic stress in the transgenic seeds. This was evident since as compared to WT seeds, which failed to germinate in the presence of 10% PEG, the transgenic seeds exhibited 60–70% germination (Fig. 4d). Since ABA is a crucial hormone which regulates the response of plants to different abiotic stress conditions (Yamaguchi-Shinozaki and Shinozaki 2006; Shinozaki and Yamaguchi-Shinozaki 2007), therefore, we also studied the effect of ABA on germination of seeds of WT and different transgenic lines. Germination of transgenic seeds at 5 μM ABA ranged between 60 and 80%, whereas the germination of WT was completely inhibited (Fig. S2). These results, therefore, imply that overexpression of TaVAP resulted in reduced sensitivity toward ABA. Further studies including estimation of endogenous ABA and its turnover are required to understand the precise physiological and molecular implications of these observations.
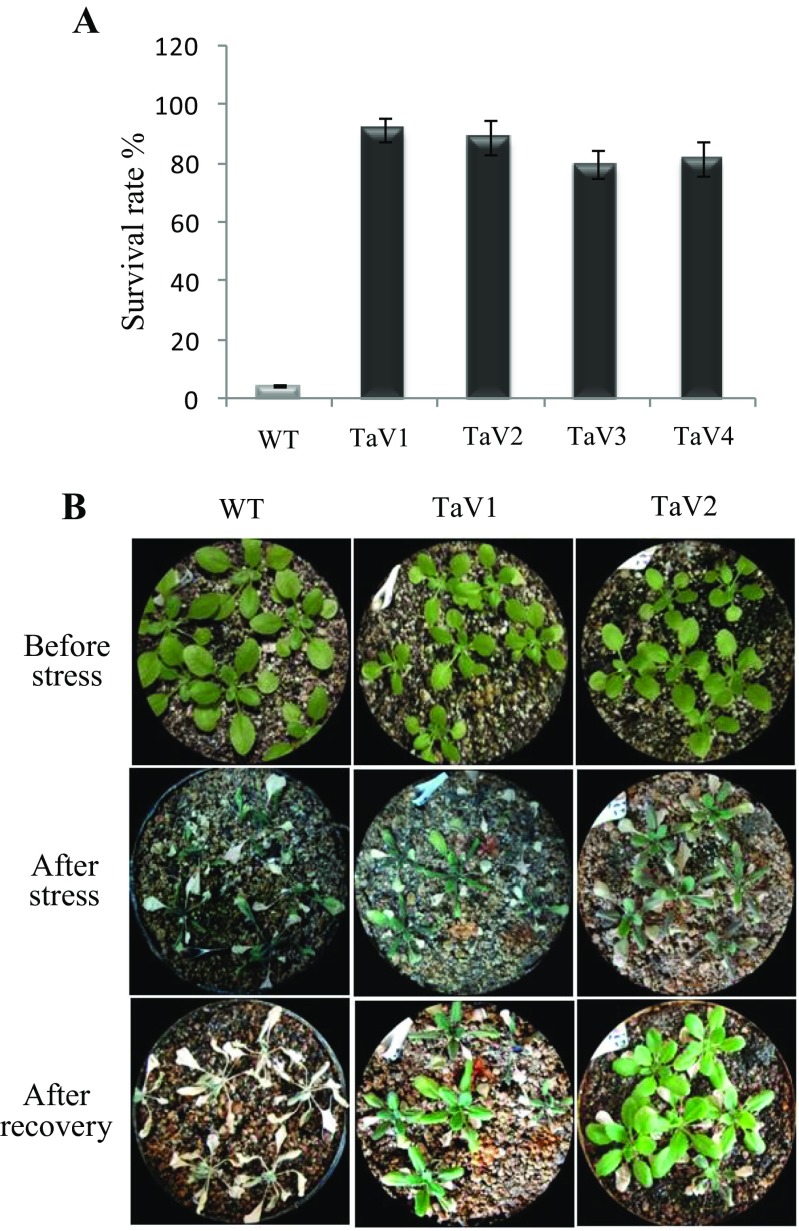
The expression of TaVAP in wheat seedlings was enhanced maximally in response to osmotic stress (Fig. 1), and since water deficit is one of the major factors that limits crop productivity (Boyer 1982), therefore, physiological, biochemical, and molecular basis of TaVAP-induced tolerance in the transgenic plants was studied in response to drought stress. Exposure of 15-day-old plants to drought stress for 14 days, followed by rewatering for 7 days, resulted in only 5–7% survival in the WT plants, as compared to 80–85% survival for the TaVAP OE transgenic lines (Fig. 5). The effect of water stress was also studied on seed yield and its components by exposing the 15-day-old plants to 6 days of stress. Imposition of drought stress although caused a significant decrease in seed yield of both WT and transgenic plants, the extent of decrease was significantly lesser in the latter, which showed three- to fourfold higher seed yield per plant (Table 2). Higher seed yield in the transgenic under stress conditions could be attributed to maintenance of higher number of pods per plant and seeds per pod.
Fig. 5.
a Effect of drought stress on survival of wild type (WT) and transgenic (TaV1, TaV2, TaV3, TaV4) plants. The 15-day-old plants were subjected to drought stress by withholding water supply for 14 days followed by rewatering for 7 days before recording their survival. b Pictorial representation of WT and transgenic plants (TaV1 and TaV2) after stress followed by recovery
Physiological basis of TaVAP-induced stress tolerance
Relative water content and osmolytes accumulation
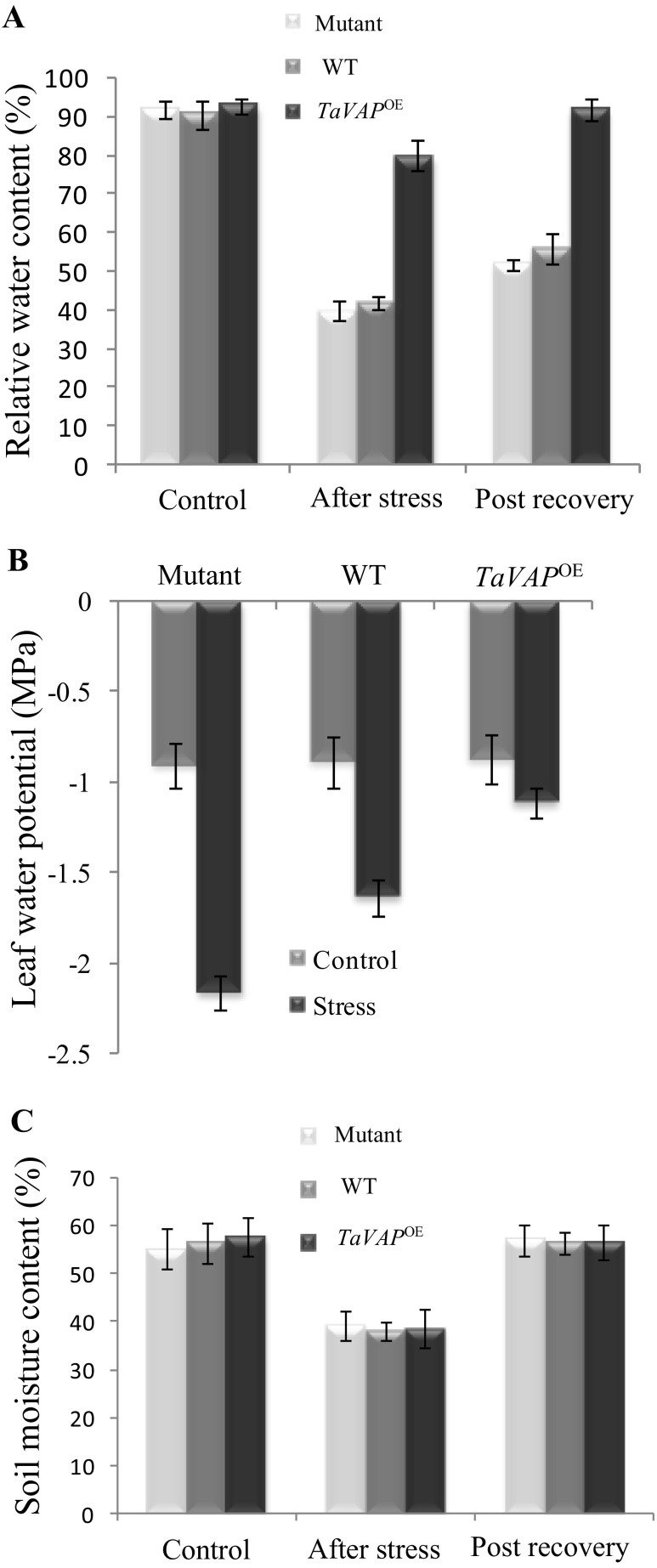
Changes in water relations of plants play a critical role in the ability of plants to cope up with drought stress. Maintenance of higher RWC and osmotic adjustment have been used as indicators of stress tolerance (Dacosta and Huang 2006; Huda et al. 2013; Zhang et al. 2010). We estimated the drought-induced changes in leaf RWC and water potential of the upper two pairs of younger leaves. RWC and water potential of mutant, WT, and TaVAP OE leaves did not show any significant difference under irrigated control conditions and was maintained between 90 to 95% and − 0.87 to − 0.90 MPa, respectively (Fig. 6a, b). Imposition of drought stress for 10 days, however, resulted in a dramatic decrease in RWC in the mutant (40%) and WT (42%) plants as compared to irrigated controls. In comparison, the RWC of transgenic leaves was maintained at significantly higher levels (ca. 80%) even under water deficit conditions. Moreover, relative to the mutant and WT leaves, which exhibited only marginal recovery in RWC after rewatering for 7 days, the RWC of transgenic leaves showed complete recovery and attained the pre-stress levels. Compared to irrigated control, the leaf water potential also registered a significant decline on imposition of water stress, with the decrease being greater in the mutant (from − 0.9 to − 2.2 MPa) and WT (from − 0.88 to − 1.65 MPa) as compared to the transgenic plants (from − 0.87 to − 1.15 MPa). The soil water content for the mutant, WT, and transgenic lines was similar under control conditions (Fig. 6c). The soil moisture content decreased significantly after 10 days of drought stress (from 55 to 60% at the beginning of treatment to approximately 30%) for all the genotypes. Although soil water potential was not estimated, these observations indicate that mutant, WT, and transgenic plants might have been exposed to similar levels of stress at the time of sampling (Fig. 6c). Detailed studies on soil and plant water relations are, however, required to determine the contribution of osmotic adjustment to the improved performance of transgenic plants under stress.
Fig. 6.
Changes in leaf relative water content (a), leaf water potential (b), and soil moisture content (c) of 15-day-old mutant, wild type (WT), and transgenic (TaVAPOE) plants exposed to drought stress by withholding water for 10 days followed by rewatering for 7 days. Data represent the mean of three biological replicates ±S.E
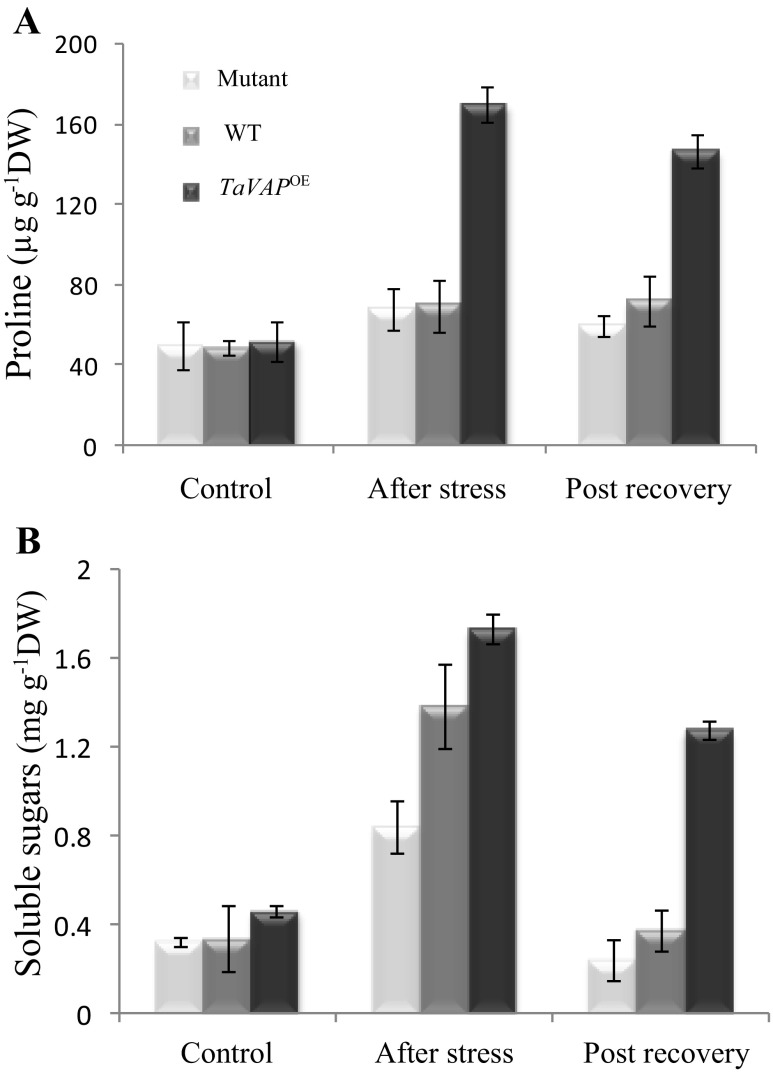
Root proliferation and accumulation of osmolytes, such as proline and soluble sugars, contribute to maintenance of higher RWC in the plants under drought stress (Ingram and Bartels 1996; Karaba et al. 2007). The exposure to drought stress resulted in more than twofold increase in the proline content of transgenic leaves, as compared to a non-significant enhancement in the mutant and WT leaves (Fig. 7a). On the contrary, the water stress-induced increase in soluble sugar content was significant in the leaves of mutant, WT, and transgenic plants, with the TaVAP OE plants exhibiting highest levels and mutant plants the lowest (Fig. 7b). Compared to mutant and WT plants, the levels of sugar and proline in the TaVAP OE leaves were significantly higher even in the post-recoveryphase. However, the soluble sugar content of mutant and WT plants declined to pre-stress levels after recovery.
Fig. 7.
Changes in proline (a) and soluble sugars (b) in the leaves of 15-day-old mutant, wild type (WT), and transgenic (TaVAP OE) plants exposed to drought stress for 10 days, followed by rewatering for 7 days. Data represent the mean ± S.E. of three biological replicates
Photosynthetic efficiency and pigment accumulation
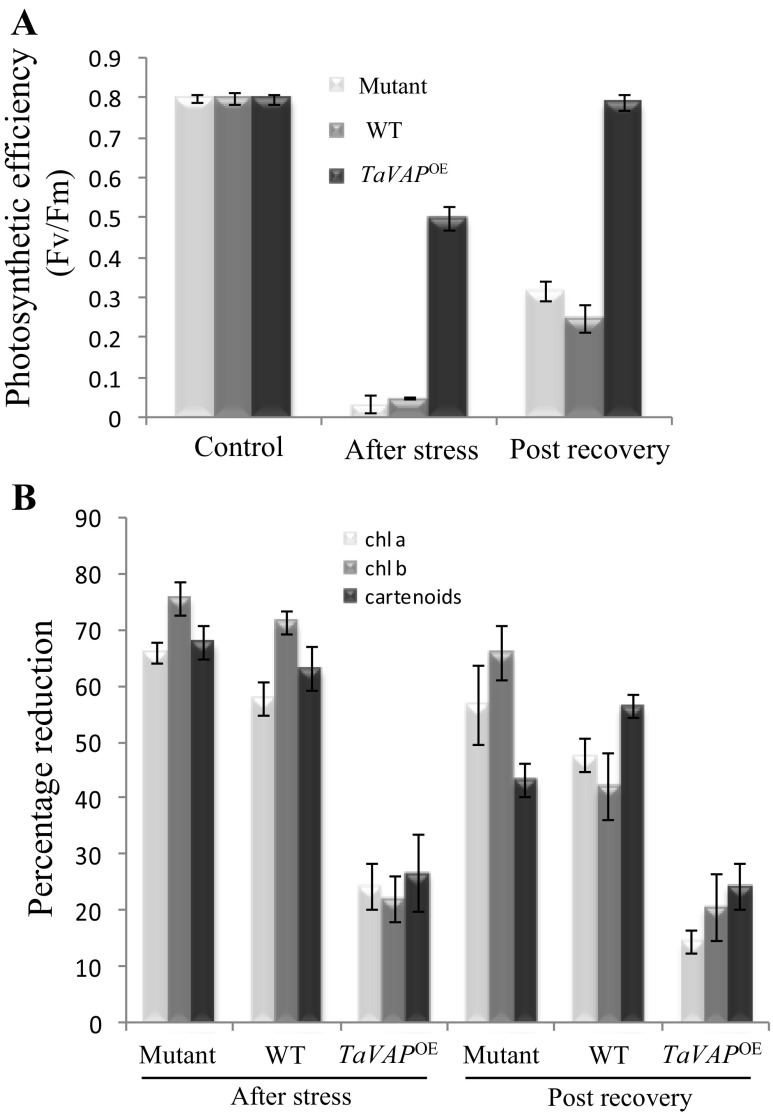
Maintenance of higher photosynthetic rates under stress is considered a crucial factor that contributes to stress tolerance (Ashraf and Harris 2013). Photosynthetic efficiency (Fv/Fm value) is used as a measure of photooxidative damage to PSII, and decrease in its value indicates damage to PSII (Maxwell and Johnson 2000). No significant difference in the Fv/Fm values of mutant, WT, and TaVAP OE plants was observed under control conditions (Fig. 8a), implying that loss or gain of VAP gene had no apparent effect on the photosynthetic efficiency. Imposition of drought stress, however, resulted in a drastic decrease (90–95%) in the Fv/Fm values of mutant and WT plants relative to their irrigated controls, whereas the transgenic plants showed only about 40% decline (Fig. 8a). Relieving of stress resulted in almost complete recovery in the photosynthetic efficiency of the TaVAP OE plants, as compared to only about 40 and 31% recovery observed in WT and mutant plants, respectively.
Fig. 8.
Effect of water deficit conditions on photosynthetic efficiency (Fv/Fm) (a) and photosynthetic pigments (b) in the rosette leaves of 15-day-old plants of mutant, wild type (WT), and transgenic (TaVAP OE) lines. Plants were exposed to drought stress by withholding water for 10 days followed by rewatering for 7 days. Data represent the mean of three biological replicates ±S.E
The photosynthetic machinery is composed of both plastidial and nuclear gene products which are necessary for chloroplast biogenesis. As chlorophyll a, chlorophyll b, and carotenoids are the primary components involved in the photosynthetic machinery; therefore, effect of water stress was also studied on accumulation of these pigments so as to understand the basis of enhanced photosynthetic efficiency of the TaVAP OE plants. Relative to the irrigated controls, the stress-induced reduction in chlorophylls a and b, and carotenoids at the end of stress period ranged between 60 and 75% in the mutant and WT, as compared to only 20–25% in the TaVAP OE plants (Fig. 8b). Relieving of water stress resulted in a marginal increase in the pigment content of all the three types of plants, but transgenic leaves showed higher pigment content during this phase also.
Changes in antioxidative activities
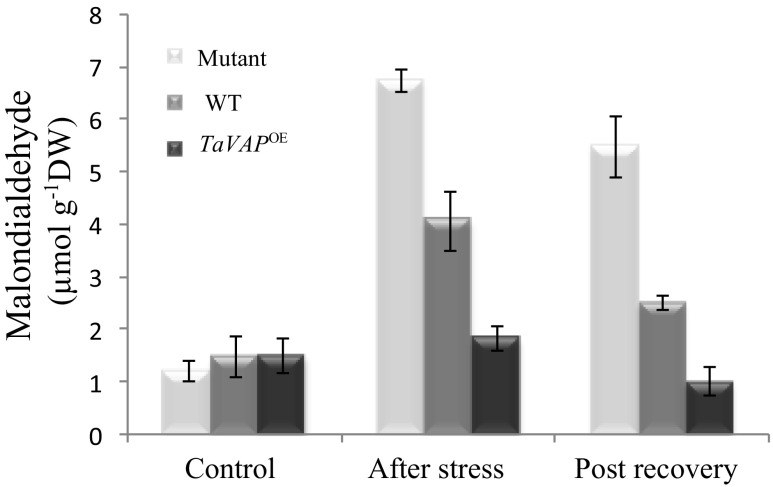
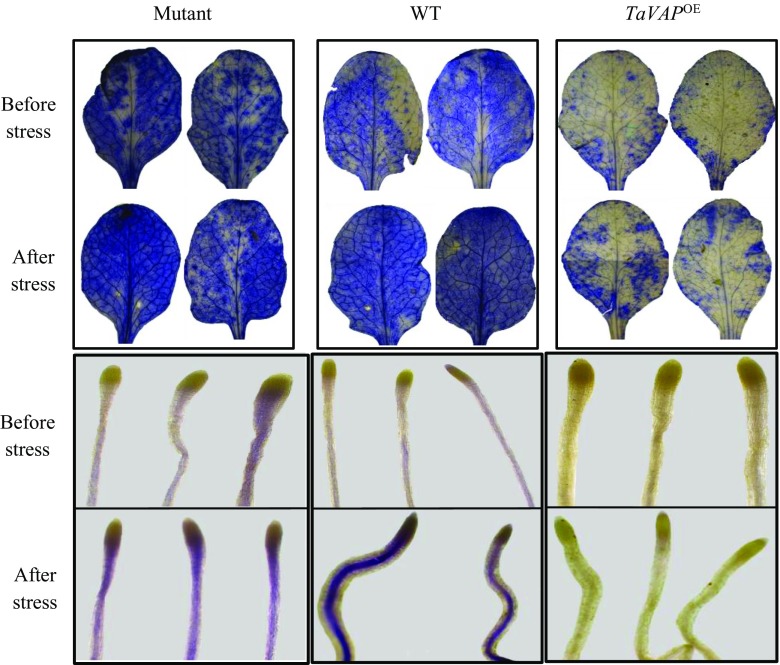
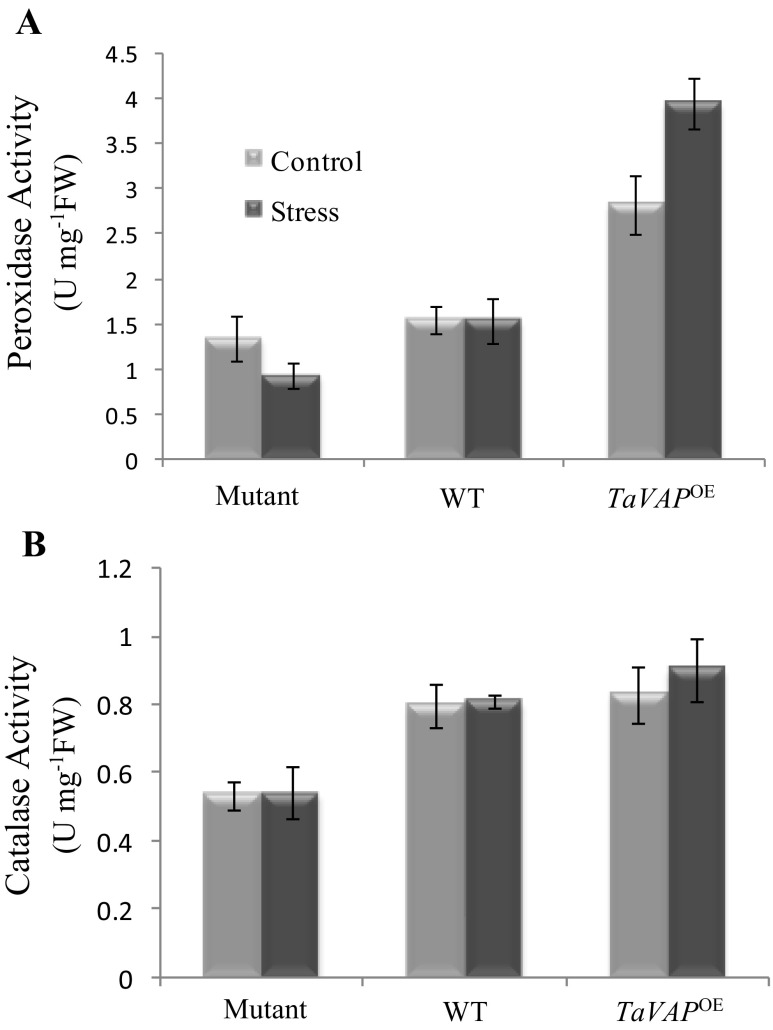
MDA, produced as a result of peroxidation of membrane lipids, is used as an indicator of stress-induced oxidative damage at the cellular level (Jain et al. 2001). The MDA levels in the leaves of the mutant, WT, and TaVAP OE plants were similar under control conditions (Fig. 9). Imposition of drought stress, however, resulted in about seven- and threefold increase in the MDA content of mutant and WT leaves, respectively, which was maintained at significantly higher levels even after recovery (Fig. 9). However, imposition of water stress had no significant effect on MDA levels of the transgenic leaves. Further validation for the effect of TaVAP-overexpression on free radical production was carried out by in situ detection of superoxides in the roots and leaves of 10-day-old seedlings. Compared to the mutant and WT, the intensity of superoxide staining in the roots and leaves of transgenic plants was considerably lesser under both control and water stress conditions. These observations indicate that ectopic expression of TaVAP resulted in a decrease in generation of reactive oxygen species (ROS) (Fig. 10). Our studies revealed that as compared to mutant plants, the peroxidase and catalase activities were significantly higher in the transgenic leaves under both control and water deficit conditions (Fig. 11a, b). Overexpression of TaVAP also resulted in significantly higher peroxidase activity than WT under both irrigated and stressed conditions. Significant increase in peroxidase activity in response to water stress was observed only in the TaVAP OE plants. Catalase activity was not affected appreciably by drought stress in any of the genotypes (Fig. 11b).
Fig. 9.
Drought-induced changes in malondialdehyde content of the rosette leaves of 15-day-old mutant, wild type (WT), and transgenic (TaVAP OE) plants subjected to water stress for 10 days followed by recovery for 7 days. Data represent the mean ± S.E. of three biological replicates
Fig. 10.
In situ detection of superoxides by nitroblue tetrazolium staining in the rosette leaves and roots of 15-day-old mutant, wild type (WT), and trangenic (TaVAP OE) plants before and after 10-day drought stress. The results are representative of ten independent biological replicates
Fig. 11.
Estimation of peroxidase and catalase activities in the rosette leaves of 10-day-old mutant, wild type (WT), and transgenic (TaVAP OE) plants under control and drought stress conditions. Water stress was imposed by withholding the water supply for 10 days. Experiments were repeated twice with three replicates each. Values are the mean ± S.E
Changes in endogenous auxin levels and rooting pattern
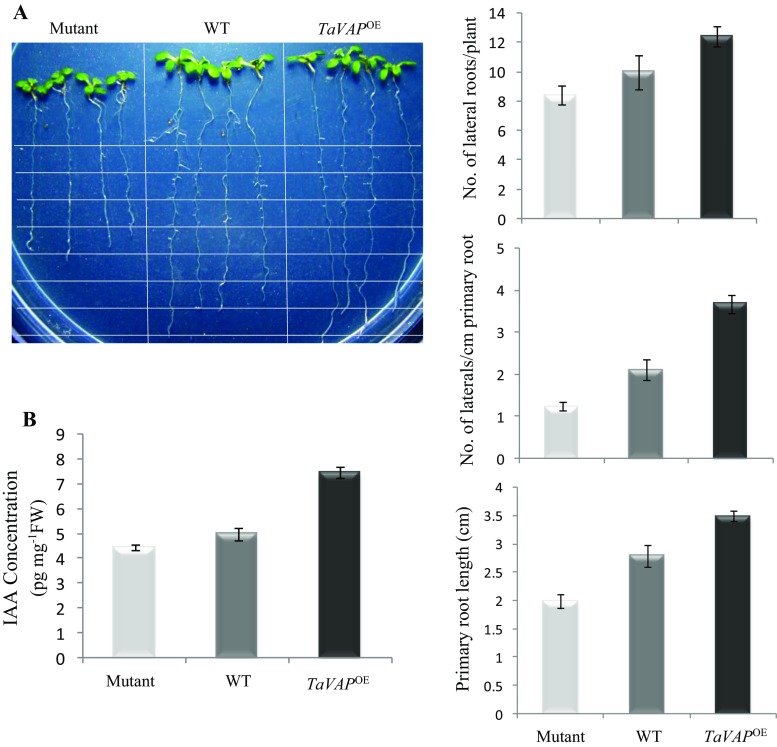
Although the stress-induced increase in proline and soluble sugars was higher in the TaVAP OE plants (Fig. 7a, b), the decrease in water potential of the transgenic lines was substantially lower than the mutant and the WT plants (Fig. 6b). Moreover, the transgenic leaves maintained significantly higher water content relative to the mutant and WT plants under stress conditions (Fig. 6a). These observations imply that ectopic expression of TaVAP resulted in retention of higher moisture content under drought stress conditions and, thus, avoided stress. As enhanced lateral rooting is one of the major determinants in increasing the water uptake of plants, particularly under stress conditions (Comas et al. 2013), we carried out comparative analysis of root morphology of the mutant, WT, and transgenic plants. The data presented in Fig. 12a clearly show that overexpression of TaVAP caused a significant increase in not only the length of primary roots but also the number and density of lateral roots in the transgenic plants. On the contrary, the disruption of this gene in the mutant plants caused a decrease in these parameters.
Fig. 12.
Changes in the phenotype of roots (a) and estimation of indole-3-acetic acid (IAA) (b) in the 10-day-old seedlings of mutant, wild type (WT), and the transgenic (TaVAP OE) plants grown on MS medium. Bar graphs show the primary root length and number of lateral roots per plant and their density. Endogenous levels of IAA were estimated under control conditions using IAA standard as reference, and data represent the mean (± S.E.) of four to five independent replicates
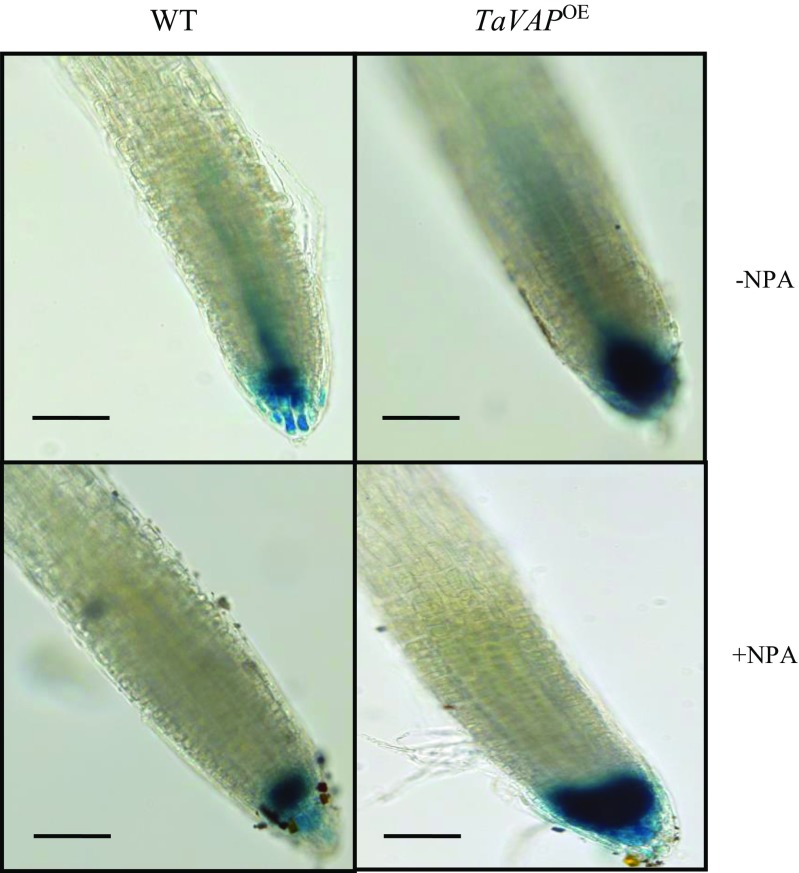
Since auxin is reported to initiate the lateral root primodia (Laskowski et al. 1995), we estimated the auxin content in the 10-day-old seedlings grown on MS medium for elucidating the physiological basis of differential lateral root formation in the Atvap mutant, WT, and TaVAP OE plants. Highest level of IAA was observed in the transgenic plants (7.5 pg mg−1 FW) as compared to WT (5 pg mg−1 FW) and mutants (4.2 pg mg−1 FW) (Fig. 12b). These values are comparable with the IAA content in Arabidopsis plants reported in other studies (Li et al. 2015). It, thus, appears that the increase in lateral roots in transgenic plants might be due to enhanced levels of IAA. Further validation for the role of auxin in lateral root formation of transgenic plants was carried out by crossing TaVAP OE plants with the Col-0 plants harboring GUS gene fused to auxin-responsive promoter DR5 (Ulmasov et al. 1997). Comparative analysis revealed that as compared to WT, the expression of GUS was higher in the zone of elongation and tip region of transgenic roots, implying greater levels of IAA in these cells, which also corresponded with an increase in the number of lateral root primodia (Fig. S3). The role of auxin in proliferation of roots in the TaVAP OE plants was further confirmed by exogenous application of N-1-naphthylphthalamic acid (NPA), which blocks the delivery of IAA to the zone of lateral root initiation by inhibiting both acropetal and basipetal polar auxin transport (Rashotte et al. 2000). It was observed that GUS activity in the roots of transgenic and WT plants exposed to NPA was localized only to the tip region, and the zone of elongation exhibited negligible staining (Fig. 13).
Fig. 13.
The GUS staining signifying accumulation of indole-3-acetic acid (IAA) in the root apex of the 5-day-old wild type (WT) (DR5::GUS) and transgenic (35S::TaVAPXDR5::GUS) plants grown on MS medium in the presence and absence of inhibitor (1-N-naphthylphthalamic acid; NPA) of basipetal transport of IAA
Microarray analysis
To understand the molecular basis of TaVAP-induced changes, we studied the changes in gene expression in WT, mutant, and transgenic lines by microarray analysis. Compared to WT, overexpression of TaVAP resulted in enhanced transcript levels of 58 genes in the transgenic plants that also showed downregulation in the mutant plants (Table S3). These data revealed approximately tenfold decrease in expression of one of the profilin genes, PFN3, in the mutant plants, whereas the TaVAP OE lines depicted fivefold increase (Table S3). Several genes (31) such as NITRILASE1, 2, 3 (AT3G44320), CYTOCHROME P450 MONOOXYGENASE 83B1 (CYP83B1; AT4G31500), and CYTOCHROME P450, FAMILY 79, SUBFAMILY B, and POLYPEPTIDE 2, 3 (CYP79B2, CYP79B3; AT4G39950, AT2G22330), which are implicated in auxin pathway (Zhao 2013), exhibited upregulation by two- to tenfold in the TaVAP OE plants as compared to WT (Table S3), suggesting their role in enhanced accumulation of auxin in the transgenic plants. Although tryptophan-dependent pathway is the major route for auxin biosynthesis in plants, auxin synthesis is also regulated through crosstalk of ethylene and auxin pathways (Stepanova et al. 2005). Methionine is converted to ethylene by SAM synthetase, ACC synthase, and ACC oxidase (Stepanova et al. 2005). EIN3, a receptor of ethylene, triggers the transcription of different ethylene response factors (ERFs). One of the ERFs, ANTHRANILATE SYNTHASE BETA SUBUNIT (ASB1/WEI7), is also involved in auxin biosynthesis. Compared to WT plants, significant increase in transcript levels of the genes encoding SAM synthetase (threefold), ACC synthase (fourfold), EIN3 (3.64-fold), and ASB1 (4.2-fold) was observed in transgenic, implying that these pathways may also be contributing to enhanced levels of auxin in the TaVAP OE plants.
In addition to biosynthesis, the spatio-temporal distribution of auxin is also essential for plant development (Vanneste and Friml 2009). The auxin influx carriers such as AUXIN-RESISTANT 1/LIKE AUX1 (AUX1/LAX), and the auxin efflux carriers, PIN-FORMED (PIN), and ATP-binding cassette (ABC) transporters, have been implicated in auxin transport (McSteen 2010). The PIN family of efflux carriers consists of eight different proteins in Arabidopsis (Krecek et al. 2009). One of the members of this family, AtPIN4, in Arabidopsis was shown to generate a sink for auxin below the quiescent center of the root meristem that is essential for auxin distribution and patterning (Friml et al. 2002). Relative to WT, we observed significant upregulation (2.2-fold) of AtPIN4 (AT2G01420) in the TaVAP OE plants, indicating its likely contribution to higher accumulation of auxin in the basipetal region. Expression analysis of few representative genes viz. PIN4, CYP79B2, EIN3, SAM1, ASB1, ACS8, NIT1, and NIT3 that are implicated in auxin response, by real-time PCR also validated the findings of microarray analysis (Fig. S4). Further, enhancement in transcript levels in the transgenic plants was also observed for genes encoding antioxidative enzymes such as catalase 2 (CAT2), catalase 3 (CAT3), ascorbate peroxidase 1 (APX1), ascorbate peroxidase 3 (APX3), and peroxidase 42 (PRXR1) which is consistent with the higher antioxidative activity in the TaVAP OE plants relative to the mutant and WT plants. Taken together, these observations suggest that the TaVAP overexpression causes pleiotropic effects in Arabidopsis transgenic and many different pathways converge to impart tolerance to abiotic stress.
Discussion
The VAPs are type II integral ER membrane-bound proteins tethered to the membranes and have been implicated in different processes such as membrane trafficking (Kagiwada et al. 1998; Nishimura et al. 1999; Skehel et al. 1995, 2000), lipid transport and metabolism (Nikawa et al. 1995; Kagiwada and Zen 2003; Kawano et al. 2006), and unfolded protein response (Brickner and Walter 2004; Kanekura et al. 2006). However, information on the role of these proteins in growth and development of plants is scanty (Wang et al. 2016). Further, the implications of these genes in abiotic stress adaptation of plants have also not been explored yet. In the present study, we have investigated the role of TaVAP in stress tolerance of plants by overexpressing in Arabidopsis. The expression of TaVAP in wheat seedlings was enhanced in response to ABA and different abiotic stress conditions (Fig. 1), suggesting that TaVAP may be playing an important role in modulating the response of plants to multiple stresses.
Complementation studies using yeast and Arabidopsis mutants were carried out to gain insight into the role of TaVAP in the cell. The TaVAP gene was able to complement yeast and Arabidopsis mutants, suggesting functional conservation of this gene during the course of evolution. GA has been implicated as a key regulator of cell elongation and contributes to plant height (de Lucas et al. 2008). Dwarf mutants such as spindly and ga1-3 in Arabidopsis (Jacobsen and Olszewski 1993; Silverstone et al. 1998), le in pea (Duckett and Lloyd 1994), sin in barley (Ross et al. 1997), and d18 and d35 in rice (Murakami 1995; Futsuhara and Kikuchi 1995) were rescued by exogenous application of GA. However, the dwarf character of Atvap mutants was not affected by exogenous GA, auxin, or ethylene (data not shown). GA insensitivity was also observed for dwarf mutants that were impaired in genes such as profilins (PFNs) in Arabidopsis (Ramachandran et al. 2000), PROBENAZOLE-INDUCIBLEPROTEIN (PBZ1) (Tanaka et al. 2006), and Dwarf1 in rice (Ashikari et al. 1999). It is, thus, likely that AtVAP may not be regulating the height of plants through GA signaling.
Microarray analysis revealed dramatic reduction (tenfold) in expression of PFN3 in the mutant lines as compared to fivefold enhancement in the TaVAP OE plants (Table S3). Four different genes PFN1, PFN2, PFN3, and PFN4, which encode profilins, have been identified in Arabidopsis (Christensen et al. 1996). Profilins are ubiquitous actin-binding proteins that are present in all eukaryotic cells (Valenta et al. 1991), and have been implicated in polymerization and sequestration of actin (Radauer et al. 2006; Lopez et al. 2007). Suppression of PFN gene in tomato and Arabidopsis plants was also reported to result in growth defects, suggesting their possible roles in the plant development (Ramachandran et al. 2000; McKinney et al. 2001). Although involvement of other genes cannot be ruled out, we speculate that the decrease in height of Atvap mutants may be mediated through suppression of PFN3 expression which may be leading to impairment of actin organization. Role of TaVAP in organization of actin network is also supported by recent studies, which demonstrated that Arabidopsis orthologues VAP27-1 and VAP27-3 (48.8 and 29% identical, respectively, to TaVAP), interact with an actin-binding protein, NET3C. The role of AtVAP-mediated regulation of PFN3 in controlling plant height is further supported by the fact that expression of genes such as PBZ1 (Tanaka et al. 2006) and Dwarf1 (Ashikari et al. 1999), which are also reported to modulate plant height, was not affected significantly in the Atvap mutants. To our knowledge, this is the first study to provide evidence that VAP gene is important for normal growth and development in plants. Elucidation of actin network and its organization in Atvap mutants and TaVAP OE transgenic will help in unraveling the cellular basis of TaVAP-induced changes.
Overexpression of TaVAP had no apparent adverse effect on the transgenic plants (Table 1; Fig. S1B), which is contrary to the recent study that reported defects in root, pollen, and seed development on suppression or expression of VAP27-1 and VAP27-3 in Arabidopsis (Wang et al. 2016). The differential effects of TaVAP, and VAP27-1 and VAP27-3 genes in the transgenic plants suggest specific functions of these genes that need to be elucidated further. Compared to WT, the transgenic plants depicted greater survival and maintained higher seed yield under drought (Fig. 5, Table 2), providing evidence for the role of TaVAP in stress adaptation. The lack of yield penalty in the TaVAP OE plants, as also observed in transgenic overexpressing genes viz., EDT1/HDG11 (Yu et al. 2013), OsCDPK7 (Saijo et al. 2000), ZmCBF3 (Xu et al. 2011), and PDH45 (Sanan-Mishra et al. 2005), makes TaVAP a suitable candidate for improving productivity of crop plants under stress.
Physiological analysis revealed that although soil water loss under drought stress was similar for the different genotypes (Fig. 6c), the transgenic plants maintained higher leaf RWC and leaf water potential relative to the WT and mutant lines (Fig. 6a, b). Compared to a negligible change in the WT and mutant lines, a significant increase in the proline content of TaVAP OE leaves under drought and during recovery (Fig. 7a) suggests that post-stress rehydration was able to trigger higher levels of proline accumulation in the transgenic lines. Enhanced accumulation of proline and soluble sugars, and maintenance of higher RWC under drought stress in the TaVAP OE plants, as also reported for transgenic plants overexpressing OsSUV3 (Tuteja et al. 2013), EDT1/HDG11 (Yu et al. 2013), TaWRKY10 (Wang et al. 2013), and hrf1 (Zhang et al. 2011), may be contributing to the maintenance of cell turgor and physiological processes. Despite higher stress-induced increase in soluble sugars and proline, the relatively lesser decrease in water potential of the drought stressed TaVAP OE leaves is in accordance with the previous studies which showed that an increase in mannitol and proline is not always associated with significant decrease in osmotic potential (Kishor et al. 1995; Karakas et al. 1997). Due to lack of data on other aspects of water relations such as leaf osmotic potential, it was not possible for us to determine the role of osmotic adjustment toward improved performance of TaVAP OE plants under water stress. This aspect is one of our major objectives for our future studies, which will be carried out to elucidate the molecular mechanisms of TaVAP-induced tolerance to water stress in transgenic plants.
Overexpression of TaVAP also conferred protection against stress-induced damage to the photosynthetic apparatus, as photosynthetic efficiency and pigment content were significantly higher during drought and recovery phases of the transgenic plants (Fig. 8a). Impairment of photosynthetic machinery under stressful conditions is attributed to generation of ROS, which include 1O2, O2.−, H2O2, and OH (Gill and Tuteja 2010). Compared to a significant increase in WT and mutant leaves under drought, the MDA content of the transgenic plants was unaffected by drought, implying lower levels of ROS in the latter. The peroxidase activity in the TaVAP OE leaves was also higher under both irrigated and stressed conditions (Fig. 11). It is likely that enhanced photosynthetic efficiency of the TaVAP OE plants under stress may be due to decrease in oxidative damage to the photosystem machinery, as reported for plants overexpressing OsSUV3 (Tuteja et al. 2013) and EDT1/HDG11 (Yu et al. 2013).
Enhanced root growth may increase the survival of plants under drought by allowing greater uptake of water. Ectopic expression of TaVAP resulted in significant enhancement in the length, number, and density of lateral roots, which was also associated with higher IAA content in the roots (Fig. 12). Auxin is a key regulator of lateral root development (Laskowski et al. 1995; Moriwaki et al. 2011), and mutants with auxin deficiency, such as alf4 and alf3, showed impaired lateral root formation (Celenza et al. 1995). Polar auxin transport is responsible for the movement of IAA to the root apex, and also for acropetal and basipetal transport to the root-shoot junction (Rashotte et al. 2000). Localization of GUS activity only to the root apex in the wild type and transgenic plants carrying DR5::GUS in response to NPA (Fig. 13) indicated enhanced accumulation of IAA in the root apex, likely due to inhibition of basipetal transport (Casimiro et al. 2001). These results suggest that TaVAP may be playing an important role in regulation of auxin homeostasis. However, for evaluating the contribution of enhanced root proliferation to stress tolerance of TaVAP OE plants, studies under field conditions, where the soil volume is not limited, are also required.
IAA biosynthesis in plants occurs through tryptophan-dependent and ethylene-auxin crosstalk pathways (Stepanova et al. 2005; Zhao 2010). The TaVAP-induced enhancement in IAA levels of the transgenic plants appeared to be due to increase in auxin biosynthetic and transport activities. Evidence for this mechanism is provided by microarray analysis, which revealed that relative to WT, the TaVAP OE plants showed significantly enhanced expression of genes involved in auxin synthesis through both tryptophan-dependent and tryptophan-independent pathways, and also its transport (efflux carrier AtPIN4; 2.2-fold) (Table S3). The induction of these genes may be responsible for higher accumulation of auxin in the basipetal region, and consequently greater proliferation of lateral roots. Since PFN-3 protein is reported to interact with actin and facilitates its polymerization (Theriot and Mitchison 1993; Sohn and Goldschmidt-Clermont 1994), it is speculated that the TaVAP-induced increase in auxin accumulation could also be regulated through changes in actin cytoskeleton, as it modulates the vesicle trafficking of PIN proteins (Jia et al. 2015). Identification of TaVAP-interacting proteins and elucidation of changes in actin network are indeed required to validate this hypothesis.
To conclude, this study is the first to demonstrate that VAP plays an essential role in regulating growth and development in plants, and appears to be functionally conserved during evolution. We have also demonstrated that constitutive expression of TaVAP allows the plants to withstand water stress without any apparent yield penalty under normal growth conditions. Further, the TaVAP-overexpressing plants are able to avoid stress through maintenance of higher levels of leaf RWC, proline content, photosynthetic efficiency, and antioxidative activity. The present study, therefore, provides evidence that TaVAP is a promising candidate for improving abiotic stress tolerance of crop plants.
Electronic supplementary material
(DOC 16438 kb)
Acknowledgements
This work was financially supported by the Department of Biotechnology, Government of India. The infrastructural support provided by the Department of Science and Technology, New Delhi, and the University Grants Commission, New Delhi, to both the institutions (GNDU and UDSC) is gratefully acknowledged.
Author contributions
BS carried out all the experiments and drafted the manuscript. PS, JPK, and PK together designed the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12192-017-0854-1) contains supplementary material, which is available to authorized users.
References
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci U S A. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M, Harris PJC. Photosynthesis under stressful environments: an overview. Photosynthetica. 2013;51:163–190. doi: 10.1007/s11099-013-0021-6. [DOI] [Google Scholar]
- Barrs HD, Weatherley PE. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci. 1962;15:413–428. doi: 10.1071/BI9620413. [DOI] [Google Scholar]
- Bates LS, Waldren RP, Teari D. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- Blatt MR, Thiel G. SNARE components and mechanisms of exocytosis in plants. In: Robinson DG, editor. The Golgi apparatus and the plant secretory pathway. Oxford: Blackwell Publishing, CRC Press; 2003. pp. 208–237. [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/tpc.13.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, Bennett M. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Chauhan H, Khurana N, Nijhavan A, Khurana JP, Khurana P. The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 2012;35:1912–1931. doi: 10.1111/j.1365-3040.2012.02525.x. [DOI] [PubMed] [Google Scholar]
- Christensen HEM, Ramachandran S, Tan CT, Surana U, Dong CH, Chua NH. Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 1996;10:269–279. doi: 10.1046/j.1365-313X.1996.10020269.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Comas LH, Becker SR, Cruz VM, Byrne PF, Dierig DA. Root traits contributing to plant productivity under drought. Front Plant Sci. 2013;4:442. doi: 10.3389/fpls.2013.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaCosta M, Huang B. Osmotic adjustment associated with variation in bentgrass tolerance to drought stress. J Am Soc Hortic Sci. 2006;131:338–344. [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- Duckett CM, Lloyd CW. Gibberellic acid-induced microtubule reorientation in dwarf peas is accompanied by rapid modification of an alpha-tubulin isotype. Plant J. 1994;5:363–372. doi: 10.1111/j.1365-313X.1994.00363.x. [DOI] [Google Scholar]
- Duman JG, Forte JG. What is the role of SNARE proteins in membrane fusion? Am J Physiol Cell Physiol. 2003;285:C237–C249. doi: 10.1152/ajpcell.00091.2003. [DOI] [PubMed] [Google Scholar]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, Palme K. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108:661–673. doi: 10.1016/S0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Futsuhara Y, Kikuchi F (1995) In Science of Rice Plant, Matsuo T, Kumazawa K, Ishihara R, Hirata H, eds, Food and Agriculture Policy Research Center, Tokyo, 3, pp 300–308
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Gimenez MJ, Piston F, Atienza SG. Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta. 2011;233:163–173. doi: 10.1007/s00425-010-1290-y. [DOI] [PubMed] [Google Scholar]
- Heath RL, Packer L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- Hiscox JD, Israelstam GF. A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot. 1979;57:1332–1334. doi: 10.1139/b79-163. [DOI] [Google Scholar]
- Hosaka K, Nikawa J, Kodaki T, Yamashita S. A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J Biochem. 1992;111:352–358. doi: 10.1093/oxfordjournals.jbchem.a123761. [DOI] [PubMed] [Google Scholar]
- Huda KM, Banu MS, Garg B, Tula S, Tuteja R, Tuteja N. OsACA6, a P-type IIB Ca(2)(+) ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing the expression of stress-responsive genes. Plant J. 2013;76:997–1015. doi: 10.1111/tpj.12352. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Mathur G, Koul S, Sarin NB. Ameliorative effects of proline on salt stress-induced lipid peroxidation in cell lines of groundnut (Arachishypogaea L.) Plant Cell Rep. 2001;20:463–468. doi: 10.1007/s002990100353. [DOI] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. Overexpression of putative topoisomerase 6 genes from rice confers stress tolerance in transgenic Arabidopsis plants. FEBS J. 2006;273:5245–5260. doi: 10.1111/j.1742-4658.2006.05518.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena BP. Role of SNAREs in membrane fusion. Adv Exp Med Biol. 2011;713:13–32. doi: 10.1007/978-94-007-0763-4_3. [DOI] [PubMed] [Google Scholar]
- Jia H, Hu Y, Fan T, Li J. Hydrogen sulfide modulates actin-dependent auxin transport via regulating ABPs results in changing of root development in Arabidopsis. Sci Rep. 2015;5:8251. doi: 10.1038/srep08251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S, Zen R. Role of the yeast VAP homolog, Scs2p, in INO1 expression and phospholipid metabolism. J Biochem. 2003;133:515–522. doi: 10.1093/jb/mvg068. [DOI] [PubMed] [Google Scholar]
- Kagiwada S, Hosaka K, Murata M, Nikawa J, Takatsuki A. The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J Bacteriol. 1998;180:1700–1708. doi: 10.1128/jb.180.7.1700-1708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kanekura K, Nishimoto I, Aiso S, Matsuoka M. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8) J Biol Chem. 2006;281:30223–30233. doi: 10.1074/jbc.M605049200. [DOI] [PubMed] [Google Scholar]
- Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martinez N, Krishnan A, Nataraja KN, Udayakumar M, Pereira A. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc Natl Acad Sci U S A. 2007;104:15270–15275. doi: 10.1073/pnas.0707294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas B, Ozias-akins P, Stushnoff C, Suefferheld M, Rieger M. Salinity and drought tolerance of mannitol-accumulating transgenic tobacco. Plant Cell Environ. 1997;20:609–616. doi: 10.1111/j.1365-3040.1997.00132.x. [DOI] [Google Scholar]
- Kawano M, Kumagai K, Nishijima M, Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J Biol Chem. 2006;281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- Kishor P, Hong Z, Miao GH, Hu C, Verma D. Overexpression of [delta]-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klig LS, Homann MJ, Carman GM, Henry SA. Coordinate regulation of phospholipid biosynthesis in Saccharomyces cerevisiae: pleiotropically constitutive opi1 mutant. J Bacteriol. 1985;162:1135–1141. doi: 10.1128/jb.162.3.1135-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause GH, Weiss E. Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:313–349. doi: 10.1146/annurev.pp.42.060191.001525. [DOI] [Google Scholar]
- Krecek P, Skupa P, LibusJ NS, Tejos R, Friml J, Zazimalova E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009;10:249. doi: 10.1186/gb-2009-10-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Laurent F, Labesse G, de Wit P. Molecular cloning and partial characterization of a plant VAP33 homologue with a major sperm protein domain. Biochem Biophys Res Commun. 2000;270:286–292. doi: 10.1006/bbrc.2000.2387. [DOI] [PubMed] [Google Scholar]
- Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Levine A, Belenghi B, Damari-Weisler H, Granot D. Vesicle-associated membrane protein of Arabidopsis suppresses Bax-induced apoptosis in yeast downstream of oxidative burst. J Biol Chem. 2001;276:46284–46289. doi: 10.1074/jbc.M107375200. [DOI] [PubMed] [Google Scholar]
- Li W, Wang F, Wang J, Fan F, Zhu J, Yang J, Liu F, Zhong W. Overexpressing CYP71Z2 enhances resistance to bacterial blight by suppressing auxin biosynthesis in rice. PLoS One. 2015;10:e0119867. doi: 10.1371/journal.pone.0119867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V, Kwon C, Panstruga R. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol. 2007;23:147–174. doi: 10.1146/annurev.cellbio.23.090506.123529. [DOI] [PubMed] [Google Scholar]
- Lopez-Torrejon G, Diaz-Perales A, Rodriguez J, Sanchez-Monge R, Crespo JF, Salcedo G, Pacios LF. An experimental and modeling-based approach to locate IgE epitopes of plant profilin allergens. J Allergy Clin Immunol. 2007;119:1481–1488. doi: 10.1016/j.jaci.2007.02.004. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence-a practical guide. J Exp Bot. 2000;51:659–668. doi: 10.1093/jexbot/51.345.659. [DOI] [PubMed] [Google Scholar]
- Mazel A, Leshem Y, Tiwari BS, Levine A. Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e) Plant Physiol. 2004;134:118–128. doi: 10.1104/pp.103.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Kandasamy MK, Meagher RB. Small changes in the regulation of one Arabidopsis profilin isovariant, PRF1, alter seedling development. Plant Cell. 2001;13:1179–1191. doi: 10.1105/tpc.13.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P. Auxin and monocot development. Cold Spring Harb Perspect Biol. 2010;2:a001479. doi: 10.1101/cshperspect.a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16:237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- Moriwaki T, Miyazawa Y, Kobayashi A, Uchida M, Watanabe C, Fujii N, Takahashi H. Hormonal regulation of lateral root development in Arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiol. 2011;157:1209–1220. doi: 10.1104/pp.111.186270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y (1995) In Science of Rice Plant, eds Matsuo T, Kumazawa K, Ishihara R, Hirata H (Food and Agriculture Policy Research Center, Tokyo) 2, pp 182–189
- Nayar S, Sharma R, Tyagi AK, Kapoor S. Functional delineation of rice MADS29 reveals its role in embryo and endosperm development by affecting hormone homeostasis. J Exp Bot. 2013;64:4239–4253. doi: 10.1093/jxb/ert231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Murakami A, Esumi E, Hosaka K. Cloning and sequence of the SCS2 gene, which can suppress the defect of INO1 expression in an inositol auxotrophic mutant of Saccharomyces cerevisiae. J Biochem. 1995;118:39–45. doi: 10.1093/oxfordjournals.jbchem.a124889. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Hayashi M, Inada H, Tanaka T. Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem Biophys Res Commun. 1999;254:21–26. doi: 10.1006/bbrc.1998.9876. [DOI] [PubMed] [Google Scholar]
- Petersen NH, Joensen J, McKinney LV, Brodersen P, Petersen M, Hofius D, Mundy J. Identification of proteins interacting with Arabidopsis ACD11. J Plant Physiol. 2009;166:661–666. doi: 10.1016/j.jplph.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Radauer C, Willerroider M, Fuchs H, Hoffmann-Sommergruber K, Thalhamer J, Ferreira F, Scheiner O, Breiteneder H. Cross-reactive and species-specific immunoglobulin E epitopes of plant profilins: an experimental and structure-based analysis. Clin Exp Allergy. 2006;36:920–929. doi: 10.1111/j.1365-2222.2006.02521.x. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Christensen HE, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH. Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 2000;124:1637–1647. doi: 10.1104/pp.124.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 2000;122:481–490. doi: 10.1104/pp.122.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Murfet IC, Reid JB. Gibberellin mutants. Physiol Plant. 1997;100:550–560. doi: 10.1111/j.1399-3054.1997.tb03060.x. [DOI] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci U S A. 2005;102:509–514. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan RS, Slabaugh E, Singh VR, Lapidus LJ, Haas T, Brandizzi F. The targeting of the oxysterol-binding protein ORP3a to the endoplasmic reticulum relies on the plant VAP33 homolog PVA12. Plant J. 2009;58:817–830. doi: 10.1111/j.1365-313X.2009.03815.x. [DOI] [PubMed] [Google Scholar]
- Shi H, Chan Z. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J Integr Plant Biol. 2014;56:114–121. doi: 10.1111/jipb.12128. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Dennis ES. Cell signalling and gene regulation: global analyses of signal transduction and gene expression profiles. Curr Opin Plant Biol. 2003;6:405–409. doi: 10.1016/S1369-5266(03)00093-1. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Jain M, Kulshreshtha R, Khurana JP, Kumar S, Singh P. Expression analysis of genes encoding translation initiation factor 3 subunit g (TaeIF3g) and vesicle-associated membrane protein-associated protein (TaVAP) in drought tolerant and susceptible cultivars of wheat. Plant Sci. 2007;173:660–669. doi: 10.1016/j.plantsci.2007.09.004. [DOI] [Google Scholar]
- Skehel PA, Martin KC, Kandel ER, Bartsch D. A VAMP-binding protein from Aplysia required for neurotransmitter release. Science. 1995;269:1580–1583. doi: 10.1126/science.7667638. [DOI] [PubMed] [Google Scholar]
- Skehel PA, Fabian-Fine R, Kandel ER. Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc Natl Acad Sci U S A. 2000;97:1101–1106. doi: 10.1073/pnas.97.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn RH, Goldschmidt-Clermont PJ. Profilin: at the crossroads of signal transduction and the actin cytoskeleton. BioEssays. 1994;16:465–472. doi: 10.1002/bies.950160705. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Matsuoka M, Kitano H, Asano T, Kaku H, Komatsu S. gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack. Plant Cell Environ. 2006;29:619–631. doi: 10.1111/j.1365-3040.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. The three faces of profilin. Cell. 1993;75:835–838. doi: 10.1016/0092-8674(93)90527-W. [DOI] [PubMed] [Google Scholar]
- Tuteja N, Sahoo RK, Garg B, Tuteja R. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64) Plant J. 2013;76:115–127. doi: 10.1111/tpj.12277. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta R, Duchene M, Pettenburger K, Sillaber C, Valent P, Bettelheim P, Breitenbach M, Rumpold H, Kraft D, Scheiner O. Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science. 1991;253:557–560. doi: 10.1126/science.1857985. [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng P, Chen L, Wang X, Ma H, Hu W, Yao N, Feng Y, Chai R, Yang G, He G. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One. 2013;8:e65120. doi: 10.1371/journal.pone.0065120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Richardson C, Hawkins TJ, Sparkes I, Hawes C, Hussey PJ. Plant VAP27 proteins: domain characterization, intracellular localization and role in plant development. New Phytol. 2016;210:1311–1326. doi: 10.1111/nph.13857. [DOI] [PubMed] [Google Scholar]
- Weir ML, Klip A, Trimble WS. Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): a broadly expressed protein that binds to VAMP. Biochem J. 1998;333:247–251. doi: 10.1042/bj3330247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman PG. The roles of NSF, SNAPs and SNAREs during membrane fusion. Biochim Biophys Acta. 1997;1357:155–172. doi: 10.1016/S0167-4889(97)00039-6. [DOI] [PubMed] [Google Scholar]
- Xu M, Li L, Fan Y, Wan J, Wang L. ZmCBF3 overexpression improves tolerance to abiotic stress in transgenic rice (Oryza sativa) without yield penalty. Plant Cell Rep. 2011;30:1949–1957. doi: 10.1007/s00299-011-1103-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162:1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li N, Gao F, Yang A, Zhang J. Overexpression of TsCBF1 gene confers improved drought tolerance in transgenic maize. Mol Breed. 2010;26:455–465. doi: 10.1007/s11032-009-9385-5. [DOI] [Google Scholar]
- Zhang L, Xiao S, Li W, Feng W, Li J, Wu Z, Gao X, Liu F, Shao M. Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J Exp Bot. 2011;62:4229–4238. doi: 10.1093/jxb/err131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2013;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 16438 kb)