Abstract
In a database search for homologs of acyl-coenzyme A oxidases (ACX) in Arabidopsis, we identified a partial genomic sequence encoding an apparently novel member of this gene family. Using this sequence information we then isolated the corresponding full-length cDNA from etiolated Arabidopsis cotyledons and have characterized the encoded recombinant protein. The polypeptide contains 675 amino acids. The 34 residues at the amino terminus have sequence similarity to the peroxisomal targeting signal 2 of glyoxysomal proteins, including the R-[I/Q/L]-X5-HL-XL-X15-22-C consensus sequence, suggesting a possible microsomal localization. Affinity purification of the encoded recombinant protein expressed in Escherichia coli followed by enzymatic assay, showed that this enzyme is active on C8:0- to C14:0-coenzyme A with maximal activity on C12:0-coenzyme A, indicating that it has medium-chain-specific activity. These data indicate that the protein reported here is different from previously characterized classes of ACX1, ACX2, and short-chain ACX (SACX), both in sequence and substrate chain-length specificity profile. We therefore, designate this new gene AtACX3. The temporal and spatial expression patterns of AtACX3 during development and in various tissues were similar to those of the AtSACX and other genes expressed in glyoxysomes. Currently available database information indicates that AtACX3 is present as a single copy gene.
Oilseed plants such as Arabidopsis and Brassica napus breakdown stored oil to form Suc shortly after germination. This process requires multiple enzymes located in several subcellular compartments, including lipid bodies, glyoxysomes (a specialized peroxisome), mitochondria, and the cytosol (Kindl, 1987). In fact, electron micrographs show that these three organelles are closely associated during germination of oilseeds (Bewley and Black, 1994). In glyoxysomes, the β-oxidation pathway breaks down fatty acids to form acetyl-coenzyme A (CoA), which is then used by the glyoxylate cycle to produce succinate.
In plants, the β-oxidation pathway consists of three enzymes: acyl-coenzyme A oxidase (ACX), multifunctional protein (enoyl-CoA hydratase/3-hydroxy acyl-CoA dehydrogenase), and 3-ketoacyl-CoA thiolase (thiolase). The first step of β-oxidation is catalyzed by ACX, a flavoprotein responsible for conversion of acyl-CoA to 2-trans-enoyl-CoA. The corresponding reaction in the mitochondria of animals is catalyzed by acyl-CoA dehydrogenase (Ikeda et al., 1985). The existence of ACXs as a family of enzymes was first demonstrated in maize (Hooks et al., 1996). These authors characterized three distinct ACXs that differed in their size, subunit composition, and substrate specificity (short-, medium-, and long-chain specific). Short- and medium-chain-specific ACXs purified to apparent homogeneity were most active with C6:0- and C10:0-CoA, respectively. The partially purified long-chain ACX was most active on C16:0-CoA, and corresponded to a previously described cucumber glyoxysomal long-chain ACX (Kirsch et al., 1986). Recently, several cDNAs for a number of ACX genes from different plant species have been cloned and characterized, among them are two ACXs from Arabidopsis designated AtACX1 and AtACX2 (Hooks et al., 1999). Biochemical analyses of the encoded polypeptides expressed in Escherichia coli showed that ACX1 is active on medium- and long-chain acyl-CoAs with maximal activity on C14:0-CoA, whereas ACX2 is solely active on long-chain acyl-CoAs with highest activity on C18:0-CoA. These authors have shown that both enzymes are active as dimers, similar to that previously described for the long-chain ACX from pumpkin (Hayashi et al., 1998a). A cDNA clone isolated from drought-stressed barley (Grossi et al., 1995) has high sequence homology with AtACX1, however the enzymatic activity profile of this clone, and hence its functional relatedness to that of AtACX, is yet to be determined. Recently, a putative acyl-CoA dehydrogenase from an Arabidopsis expression sequence tag library was also characterized and found to encode an enzyme with short-chain ACX (SACX) activity that is targeted to the glyoxysome (Hayashi et al., 1999). The biochemical characteristics of the Arabidopsis SACX correspond very closely with those described for the maize SACX and are different from AtACX1 and AtACX2. Based on the current findings, there are no reports on the identification of a cDNA encoding a medium-chain-specific ACX.
As a step toward a better understanding of β-oxidation at the molecular and biochemical levels, isolation and characterization of the missing member(s) of the ACX family is essential. Hence, we have cloned a new cDNA (GenBank accession no. AF207994) encoding an enzyme distinct in amino acid sequence and enzyme activity profile from the previously cloned Arabidopsis ACX1, ACX2, and SACX. We therefore designated the gene encoding this enzyme AtACX3, a medium-chain-specific ACX. Currently, available database information indicates that AtACX3 is present as a single copy gene. The developmental and tissue-specific patterns of expression of AtACX3 were similar to that of other β-oxidation genes.
RESULTS
Isolation and Sequence Analysis of AtACX3
In a BLAST search using plant ACX cDNAs, two new Arabidopsis genomic sequences, a 1,317-bp sequence from GenBank (accession no. B12051) and a 650-bp sequence from Cereon (identification no. athlscreen10009), were identified. These sequences were used to design primers for the amplification of the corresponding cDNA. To our knowledge, there are no reports on a cDNA sequence with substantial similarity to this clone, now designated AtACX3 in accordance with established rules on Arabidopsis gene nomenclature (Meinke and Koorneef, 1997). The AtACX3 cDNA is 2,303-bp long and encodes a predicted polypeptide of 675 amino acids with a molecular mass of approximately 75.6 kD (Fig. 1). The predicted pI is 7.6, similar to that of other ACX proteins. The N-terminal 34 amino acids contained a putative type-II peroxisomal targeting signal (PTS2) (R/K)-(L/V/I)-X5-(H/Q)(L/A) (Subramani, 1996). In recent studies it was shown that the amino-terminal presequences of citrate synthase (CS) and malate dehydrogenase (MDH) from pumpkin contained a functional PTS2, which is cleaved upon import into the peroxisome (Kato et al., 1996a, 1998). Alignment of the N-terminal domains of Arabidopsis ACX3, Arabidopsis ACX2 (Hooks et al., 1999), pumpkin long-chain ACX (Hayashi et al., 1998a), Phalaenopsis sp. ACX (Do and Huang, 1997), pumpkin MDH (Kato et al., 1998), pumpkin CS (Kato et al., 1995), Arabidopsis PED1 (peroxisome defective) (Hayashi et al., 1998b), Arabidopsis PKT2 (peroxisomal thiolase) (Ferreira da Rocha et al., 1996), and pumpkin thiolase (Kato et al., 1996b) shows that these sequences share identical or conserved substitutions with the identified consensus sequence R-(I/Q/L)-X5-HL-X15-22-C (Fig. 2). The AtACX3 predicted targeting signal deviates from the consensus sequence at position 10, where an Ala is substituted for the previously described consensus amino acids Ile, Leu, or Gln, and position 17, where Ile is replaced by Leu. The predicted size of the mature AtACX3 protein upon cleavage of the this putative PTS2 is 72.9 kD, similar to that of the long-chain ACXs from Arabidopsis (72 kD) (Hooks et al., 1999), Phalaenopsis sp. (72 kD) (Do and Huang, 1997), and pumpkin (72.4 kD) (Hayashi et al., 1998a).
Figure 1.
Deduced amino acid sequence of Arabidopsis ACX3. The deduced PTS2 is double underlined. The putative FMN binding motif is marked with a single line. The first amino acid of the recombinant AtACX3 polypeptide cloned into the E. coli expression vector is marked with a white box. Asterisks mark the amino acids common to the two protein signatures of acyl-CoA dehydrogenase (protein signature 1 and protein signature 2, respectively).
Figure 2.
Alignment of the amino-terminal presequence of Arabidopsis ACX3 with other microbody proteins that are synthesized as larger precursors. AtACX3, Arabidopsis ACX3; AtACX2, Arabidopsis ACX2 (Hooks et al., 1999); PumLACX, pumpkin long-chain ACX (Hayashi et al., 1998a); PhaACX, Phalaenopsis sp. ACX (Do and Huang, 1997); PumMDH, pumpkin glyoxysomal MDH (Kato et al., 1998); PumCS, pumpkin glyoxysomal CS (Kato et al., 1995); AtPED1, Arabidopsis thiolase (Hayashi et al., 1998b); AtPKT2, Arabidopsis thiolase (Ferreira da Rocha et al., 1996); PumTHI, pumpkin glyoxysomal thiolase (Kato et al., 1996b). Conserved amino acids, where five of the nine sequences are identical are shown in a black box. The first amino acid, as determined by sequencing of amino-terminal residues of the mature proteins, are shown in a gray box.
The ACX3 polypeptide sequence contains a putative FAD binding motif (Dubourdieu and Fox, 1977), stretching from amino acids 442 to 457 (underlined in Fig. 1), similar to other plant short- and long-chain ACXs (Do and Huang, 1997; Hayashi et al., 1998a, 1999). ACX3 also has high identity with the acyl-CoA dehydrogenase protein signals 1 and 2 (Bairoch et al., 1997) (Fig. 2). Residues 187 to 199 are composed of seven of the nine amino acids that represent the acyl-CoA dehydrogenase protein signature1 (PS1: [G/A/C]-[L/I/V/M]-[S/T]-E-2X-[G/S/A/N]-G-S-D-2X-[G/S/A]), and residues 440 to 460 contain seven out of eight residues that represent the acyl-CoA dehydrogenase protein signature 2 (PS2: [Q/E]-2X-G-[G/S]-x-G-[L/I/V/M/F/Y]-2X-[D/E/N]-x(4)-[K/R]-3X-[D/E]).
The phylogenetic relationships between AtACX3 and other known plant ACXs were determined from their deduced amino acid sequences. The available ACX sequences grouped into four clusters. One group, that includes PumLACOX and PhaACOX shares high sequence identity (75%–80%) with AtACX2 and a second group, which includes AtACX1 and a putative barley ACX cDNA, shares 66% identity. The AtSACX and AtACX3 did not group phylogenetically with either AtACX2 or AtACX1. In fact, the AtACX3 was almost equally distant from the other Arabidopsis ACX sequences, sharing 28%, 23%, and 14% identity with AtACX2, AtACX1, and AtSACX, respectively. These comparisons also indicated that AtACX1 and AtACX2 are 25% identical and that AtSACX shares 14% identity with both AtACX1 and AtACX2.
Expression of AtACX3 Recombinant Protein in E. coli and Determination of Substrate Specificity
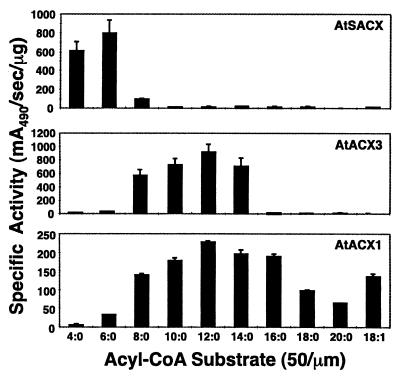
Amplified mature portions of Arabidopsis AtSACX, AtACX1, and AtACX3 cDNAs were cloned into the pQE30 expression system introducing an in-frame His6 tag to the N-terminal end of each expressed protein. SDS-PAGE analysis of the affinity-purified recombinant proteins indicated that each was purified to near homogeneity and had the predicted molecular mass (data not shown). The affinity-purified ACX proteins were assayed with a variety of saturated acyl-CoA substrates ranging from C4:0- to C20:0-CoA, and an unsaturated substrate, C18:1-CoA, all at a concentration of 50 μm (Fig. 3). To directly compare the activity profile of AtACX3 with other reported family members, enzyme assays were performed with affinity purified AtACX1 and AtSACX. AtSACX activity ranged from C4:0- to C8:0-CoA and peaked at C6:0-CoA (Fig. 3), which is identical to that previously described by Hayashi et al. (1999). The activity profile generated with affinity-purified AtACX1 differed from results reported using extracts of E. coli over-expressing the same enzyme (Hooks et al., 1999). Both sets of data showed that AtACX1 is active on a broad range of substrates, C8:0- through C20:0-CoA. However, under our assay conditions, AtACX1 had similar activity levels on C10:0- to C16:0-CoA with slightly higher activity on C12:0-CoA (Fig. 3). Whereas, the AtACX1 activity reported by Hooks et al. (1999) showed a sharper profile with a distinct maximal activity on C14:0-CoA. The discrepancy between these two sets of data may be related to differences in assay conditions and/or the purity of the enzymes examined. To mimic the assay conditions carried out by Hooks et al. (1999), we included a range of bovine serum albumin (BSA) concentrations in these assays (0–400 μg mL−1). The addition of BSA also enabled us to address the potential interference of enzyme activity with long-chain acyl-CoAs caused by micel formation. It was determined that the presence of 100 μg mL−1 BSA only affected ACX activity on substrates longer than C16:0-CoA. In addition, our data indicated that AtACX3 has a novel substrate specificity profile (Fig. 3), in that it is active on medium-chain acyl-CoA substrates ranging from C8:0- to C14:0-CoA, with peak activity with C12:0-CoA and virtually no activity on substrates outside this range. This activity profile was observed repeatedly with a range of substrate concentrations (25–200 μm) in the presence or absence of BSA. As expected, the addition of BSA did not affect AtACX3 activity levels or profile. Furthermore, the specific activity of AtACX3 appeared to be 4-fold higher than that of AtACX1 and is similar to AtSACX under the assay conditions examined. The kinetic parameters of these enzymes were also determined using each of their preferred substrates (Table I). The apparent Km values measured with C12:0-CoA for AtACX1 and AtACX3 were estimated to be 18.9 ± 1.9 and 15.7 ± 2.1 μm, respectively. The apparent Km value measured with C6:0-CoA for AtSACX was 20.3 ± 2.9 μm. The Vmax values for AtACX1, AtACX3, and AtSACX were 0.16 ± 0.01, 1.1 ± 0.07, and 1.2 ± 0.09 mA490 s−1 ng−1, respectively. These data indicate that the higher specific activity of AtACX3 than AtACX1 is the result of this enzyme having a higher intrinsic rate of catalysis.
Figure 3.
Acyl-CoA substrate specificities of affinity-purified recombinant AtSACX, AtACX1, and AtACX3. Specific activities were determined using 50 μm acyl-CoA substrates. AtACX1 activities were determined in the presence of 100 μg mL−1 BSA.
Table I.
Kinetic parameters of ACX enzymes
| Enzyme | Km | Vmax |
|---|---|---|
| mM | mA490 s−1ng−1 | |
| AtACX1 | 18.9 ± 1.9 | 0.16 ± 0.08 |
| AtACX3 | 15.7 ± 2.1 | 1.1 ± 0.07 |
| AtASCX | 20.3 ± 2.9 | 1.2 ± 0.09 |
All enzymes were assayed with their preferred substrate; AtACX1 and AtACX3 were assayed with C12:0-CoA and AtSACX was assayed with C6:0-CoA. In all cases substrate concentrations ranged from 0 to 27.5 mm.
Expression Pattern of AtACX3
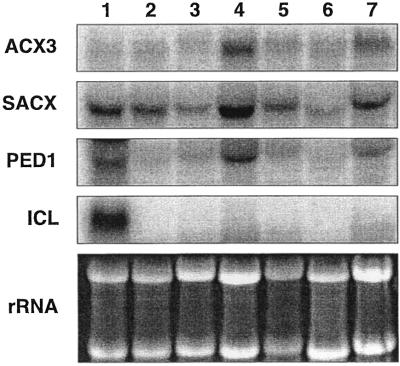
Northern-blot analysis of total RNA isolated from various Arabidopsis tissues (etiolated and green cotyledons, young and senescing rosette leaves, cauline leaves, stems, and flowers) were employed to determine the temporal and spatial expression patterns of AtACX3 as compared with other β-oxidation genes, SACX, PED1, and isocitrate lyase (ICL). The full-length cDNAs representing these genes were used as probes (Fig. 4). The ethidium bromide-stained rRNA bands were included as a loading control. Based on these data, the steady-state AtACX3 transcripts were most abundant in senescing rosette leaves and flowers, and least abundant in stems and young rosette leaves, cotyledons, and cauline leaves. The expression pattern of SACX was very similar to that of AtACX3. The expression pattern and levels of PED1 were very similar to those of AtACX3 and AtSACX, except that PED1 steady-state transcript levels were higher in etiolated cotyledons than green cotyledons. Expression patterns of ICL were distinct from AtACX3, AtSACX, and PED1 in that ICL transcripts were detected in etiolated cotyledons only.
Figure 4.
Northern-blot analyses of AtACX3, AtSACX, PED1, and ICL in different Arabidopsis tissues. Each lane was loaded with 10 μg of total RNA isolated from the following Arabidopsis tissues. Lane 1, 6-d-old etiolated cotyledons; lane 2, 6-d-old light-grown green cotyledons; lane 3, rosette leaves; lane 4, senescing rosette leaves; lane 5, cauline leaves; lane 6, stem; lane 7, flowers. Blots were probed with: AtACX3, Arabidopsis ACX3; AtSACX, Arabidopsis SACX (Hayashi et al., 1999); PED1, Arabidopsis thiolase (Hayashi et al., 1998b); ICL, B. napus ICL (accession no. Y13356). The ethidium bromide-stained ribosomal RNA bands are shown as a loading control.
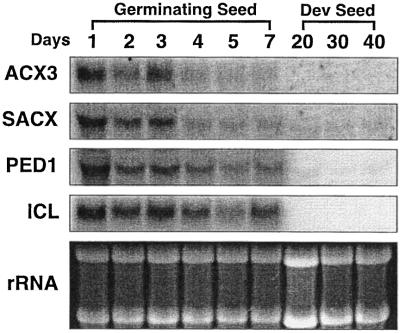
Engineering B. napus seeds to produce high levels of medium-chain fatty acids (MCFAs) has been one of our long standing goals. Several investigators (Eccleston and Ohlrogge, 1998; Mittendorf et al., 1999) have shown that accumulation of unusual fatty acids such as MCFA leads to increased expression of β-oxidation enzymes. As the first step toward metabolic regulation of β-oxidation, we have employed northern-blot analysis to examine the presence of an AtACX3 homolog in developing and germinating B. napus seeds (Fig. 5). In addition to the AtACX3 probe, we also included Arabidopsis SACX, Arabidopsis thiolase (PED1), and B. napus ICL cDNAs probes as a point of reference to other genes involved in fatty acid catabolism. The steady-state transcript levels for AtACX3 were highest 1 d after germination followed by a gradual decline through d 7. AtACX3 transcripts were not detected in developing seeds at 20, 30, or 40 DPA. Similar patterns of expression were observed for SACX, PED1, and ICL transcripts. Additional bands were not detected upon longer exposure of the northern blot, except for a low intensity signal for the PED1 transcript (data not shown). The panel with ethidium bromide-stained rRNA bands shows equal loading.
Figure 5.
Northern-blot analyses of ACX3, SACX, PED1, and ICL in developing and germinating B. napus seeds. Each lane was loaded with 10 μg of total RNA isolated from either B. napus cotyledons (except for d 1, which included the entire seedling) or developing seeds. The developmental stage are shown as day after imbibition for the germinating seed or DPA for the developing seed. Blots were probed with: AtACX3, Arabidopsis ACX3; AtSACX, Arabidopsis SACX (Hayashi et al., 1999); PED1, Arabidopsis thiolase (Hayashi et al., 1998b); ICL, B. napus ICL (accession no. Y13356). The ethidium bromide-stained ribosomal RNA bands are show as a loading control.
Chromosomal Location of ACX Genes
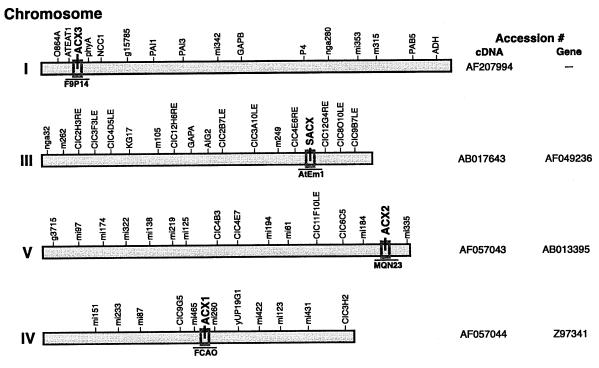
A search of the currently available Arabidopsis databases (covering approximately 85% of the genome) with all the published ACX clones enabled us to identify the chromosomal location of each corresponding gene (Fig. 6). Based on these data AtACX3 is located at the top of the chromosome I. The Southern-blot analysis (data not shown) that encompasses the entire genome supports the result of the database search suggesting that AtACX3 is a single gene. The database search also revealed that AtACX2, AtSACX, and AtACX1 are located on the chromosome V, III, and IV, respectively. To date, we have not been able to find other homologs for either AtACX2 or AtSACX, suggesting that they may also be present as single gene copies. We however, have isolated a cDNA clone encoding an enzyme of unknown function that shares 86% identity with AtACX1 (data not shown). Based on these data one may assume that AtACX1 is not a single gene copy.
Figure 6.
Chromosomal location of AtACX genes. Identification of chromosomal location of AtACX1, 2, 3, and AtSACX genes by a BLAST search of Arabidopsis databases using corresponding cDNAs. The designated clone containing each ACX gene is displayed under the appropriate chromosomal location. Genomic and cDNA GenBank accession numbers are shown to the right of the their respective chromosome.
DISCUSSION
The initial step of β-oxidation is catalyzed by a small family of ACX enzymes that differ in many properties including the preference for different length acyl-CoA substrates. A plant long-chain preferring ACX enzyme was first identified from cucumber (Kirsch et al., 1986). Later the presence of at least three ACXs that differed in a number of biochemical properties, including subunit composition and substrate preference was demonstrated (Hooks et al., 1996). These authors reported that ACXs in maize have a preference for short- (C4:0- to C8:0-CoA), medium- (C10:0- to C14:0-CoA), and long-chain (C16:0- to C18:0-CoA) substrates. In recent years, a number of ACX cDNAs have been cloned including a pumpkin long-chain ACX (Hayashi et al., 1998a), an Arabidopsis SACX (Hayashi et al., 1999), and two long-chain ACXs designated as AtACX1 and AtACX2 (Hooks et al., 1999). In this study, we have isolated and characterized a cDNA encoding a polypeptide distinct, in both amino acid sequence and enzymatic activity, from other previously cloned ACXs. Hence, we have designated this cDNA AtACX3. A search of the currently available Arabidopsis databases (Fig. 6) supported by Southern-blot analysis (data not shown) suggest that AtACX3 is present as a single copy gene.
Sequence analysis data showed that although AtACX3 is distinct from the other three Arabidopsis ACXs, it is most similar to AtACX2. AtACX3 is 28% identical to AtACX2 and both contain the conserved PTS2 sequence (R/K)-(L/V/I)-X5-(H/Q)(L/A) present at the N-terminal domain (Subramani, 1996). In contrast, AtACX1 and the AtSACX both contain the consensus PTS1 (C/A/S/P)-(K/R)-(I/L/M) sequence located at the C terminus (Hayashi et al., 1997). Functional analysis of the PTS2 sequence in MDH and CS from pumpkin identified two regions that were required for proper import and processing (Kato et al., 1996a, 1998). The first region, required for proper import into glyoxysomes has the conserved sequence R(I/Q/L)-X5-HL as underlined in Figure 1. The second region was required for proper processing of the immature protein once it was imported into the glyoxysome and is represented by the conserved Cys shown at position 34 of AtACX3. This processing site has been determined by N-terminal sequence analysis of the mature proteins of long-chain ACX, MDH, CS, and thiolase from pumpkin, and a long-chain ACX from Phalaenopsis sp. (Kato et al., 1995, 1996b, 1998; Do and Huang, 1997; Hayashi et al., 1998a). In addition, two thiolases (PED1 and PKT2) from Arabidopsis also shared a number of amino acids within this region (Ferreira da Rocha et al., 1996; Hayashi et al., 1998b). The role of the other conserved amino acids within the PTS2 region of plant glyoxysomal proteins is yet to be determined, specifically the conserved amino acid substitution of a Val for Ala at position 6, and Ile for a Leu at position 13, and a Val or Ile substituting for a Leu at position 29 of AtACX3 (Fig. 2). Future import studies will be required to determine the role of these residues.
Recombinant E. coli expressed protein fused to an N-terminal His6 tag was used to determine the substrate profile of AtACX3 (Fig. 3). AtACX3 activity ranged from C8:0-CoA to C14:0-CoA with maximal activity on C12:0-CoA substrate. Comparison of AtACX3 activity with other ACXs showed that these enzymes differed not only in their substrate specificity but also in their specific activities. AtSACX had a narrow range of activity (C4:0- to C8:0-CoA), the AtACX3 activity range was slightly broader (C8:0- to C14:0-CoA), and AtACX1 had the broadest range (C6:0- to C20:0-CoA). The specific activity measurements together with the apparent Vmax values demonstrate that AtACX3 and AtSACX have a higher intrinsic specific activity with their preferred substrate than AtACX1 with its respective acyl-CoA substrate. Several lines of evidence suggest that plants contain a medium-chain ACX (Hayashi et al., 1998a; Hooks et al., 1999). These authors reported that in crude seedling extracts the level of medium-chain ACX activity, as measured with C10:0-CoA, were approximately 5-fold higher than long-chain ACX activity (C16:0-CoA). Both AtSACX and AtACX1 lack the proper substrate specificity to account for the 5-fold differences between medium- and long-chain acyl-CoA enzyme activities. The AtSACX had no detectable activity in the medium-chain acyl-CoA range and AtACX1 had similar levels of activity on both C10:0-CoA and C16:0-CoA. Therefore, the activity profile of these two enzymes could not account for the 5-fold higher activity on C10:0-CoA as compared to C16:0-CoA, measured in germinating seeds. In contrast, AtACX3 had the appropriate profile and level of activity to account for the ACX activities observed in these tissues.
The study reported by Eccleston and Ohlrogge (1998), demonstrated that transgenic B. napus plants engineered to produce high levels of MCFAs show increased expression of both β-oxidation and glyoxylate cycle enzymes. Production of MCFAs in an established oil crop such as B. napus has been the goal of several research programs. One strategy within this effort has been metabolic regulation of β-oxidation for production of high levels of MCFAs. We have, therefore, employed northern-blot analysis to examine not only the expression patterns of AtACx3 in different Arabidopsis tissues, but also the presence of its homolog in developing and germinating B. napus seeds. The presence of cross-hybridizing bands in the northern blots after stringent wash conditions indicated that ACX3 is found in Arabidopsis and B. napus (Figs. 4 and 5) and hence one may predict that ACX3 is present in other plant species. Furthermore, these data also showed that the patterns and levels of AtACX3 expression were similar to those of other β-oxidation genes such as SACX and PED1. All the examined transcripts showed highest levels of expression 1 d after germination and reducing levels during post-germinative growth. This high level of expression during early development followed by a steady decline in transcript levels is consistent with that of the pumpkin long-chain ACX (Hayashi et al., 1998a) and Arabidopsis ACX1 and ACX2 (Hooks et al., 1999). Even the glyoxylate gene, ICL, showed a similar trend in expression pattern during germination. This was not surprising since transcript levels are generally declining at a time when protein levels are increasing following germination of B. napus seeds (Bewley and Black, 1994). As expected, all of the β-oxidation transcripts were high in senescing rosette leaves, as they are involved in the remobilization of lipids from leaves. In contrast, ICL transcripts were only detected in etiolated cotyledons, suggesting a separate regulatory mechanism to that of the coordinately regulated genes ACX and PED1. Similar to the other three Arabidopsis ACXs (SACX, ACX1, and ACX2) (Hayashi et al., 1999; Hooks et al., 1999), AtACX3 transcripts were expressed at moderately high levels in flowers. The requirement of β-oxidation in flower development has been demonstrated in Arabidopsis (Richmond and Bleecker, 1999). A mutation in one of the genes encoding a multifunction protein (aim 1, abnormal inflorescence meristem 1), an enzyme that catalyzes the second step in β-oxidation, resulted in a range of phenotypes. In the most severe cases the inflorescence meristem did not produce any recognizable floral structures, but rather produced only a small mass of undifferentiated tissues that subsequently ceased to develop. It is interesting that this mutation did not affect the growth of germinating seeds as was observed for the ped1 mutant described by Hayashi et al. (1998b). This suggests that various β-oxidation enzymes perform distinct roles at different temporal and spatial developmental stages, such as the mobilization of lipids during early germination, development of flowers, and/or the redistribution of lipids during senescence.
In conclusion, a random genomic sequence was used to isolate and characterize AtACX3 cDNA encoding a medium-chain ACX, which is distinct from AtSACX (short-chain), AtACX1 (medium- and long-chain), and AtACX2 (long-chain) enzymes. This addition to the existing array of ACX enzymes will enhance the range of tools available to study the mechanism of fatty acid breakdown during plant growth and development. Furthermore, our demonstration that B. napus seeds express ACX3 establishes that, in addition to ACX1 and SACX, this gene is a suitable target to engineer the β-oxidation pathway for production of high levels of MCFAs in this oil crop.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (ecotype No-O) was employed in these studies. To obtain light-grown cotyledons, seeds were routinely surface sterilized and sown onto 0.7% (w/v) agar plates containing 1× Murashige-Skoog medium and vernalized in the dark for 3 d at 4°C, followed by the transfer of the plates to a growth chamber under a cycle of 16 h of light and 8 h of dark at a temperature of 22°C for 6 d. For dark-grown cotyledons, surface-sterilized seeds were imbibed on filter discs saturated with sterile water and grown in the dark at 22°C for 6 d. For all other tissues, seeds were sown directly onto soil and grown for 6 weeks under a cycle of 16 h of light and 8 h of dark at 19°C. All of the tissues, including young and senescing rosette leaves, stem, and mature open flowers, were collected from individual plants of approximately the same size and chronological age. Cauline leaves were removed from secondary bolts at least 8 to 10 cm from the bottom.
Brassica napus (Quantum) seeds were all imbibed on filter discs saturated with sterile water and grown in the dark at 22°C for 1 to 7 d, prior to collection of the cotyledons. Developing seeds were harvested at the indicated time points from plants grown in a greenhouse and stored at −70°C before RNA isolation.
RNA Isolation
Total RNA was extracted from Arabidopsis tissues using the TRI Reagent (Sigma-Aldrich, St. Louis) method. Total RNA from B. napus cotyledons and developing seeds was isolated as described by Jones et al. (1995).
Cloning of AtACX3 cDNA
Total RNA from 4-d-old etiolated cotyledons was used as template for PCR amplification of AtACX3 cDNA employing the Marathon cDNA Amplification Kit (CLONTECH Laboratories, Palo Alto, CA). The Marathon Adaptor Primer 1 and gene-specific primers 5′-CCATCCTAATACGACTCACTATAGGGC-3′ and 5′-GCTGCTTTGCAATGACTGAGCTAGGCCATG-3′ were employed for 5′ and 3′ RACE using the touchdown thermocycling procedure as described by the manufacturer (CLONTECH Laboratories). The final full-length cDNA as determined by the presence of a stop codon 5′ of the first in-frame Met, was obtained by PCR employing 5′-ACTTTCTTCCCGGATAATTGAAATCGGTGA-3′ as the gene-specific primer and Marathon Adaptor Primer 1 as the antisense primer. Cloning of the PCR product into pCR2.1, using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), generated the construct pCGN9897.
DNA Sequencing and Sequence Analysis
The cDNA along with all the other PCR-amplified clones (see below) were sequenced completely in both directions using an automated ABI PRISM sequencer (Applied Biosystems, Foster City, CA). DNA sequences were analyzed with OMIGA 2.0 software (Oxford Molecular Ltd., Oxford). The BLAST server was utilized for the analysis of homologies between DNA and protein sequences. Alignment of several ACXs was performed using the CLUSTAL V program included in the Megalign software package using the PAM 250 residue weight table (DNASTAR, Madison, WI).
Bacterial Expression and Affinity Purification of Recombinant ACX
PCR-amplified products of Arabidopsis ACX1, SACX, and ACX3 cDNAs were cloned into pQE30 (QIAexpress, Qiagen USA, Valencia, CA) generating in-frame N-terminal His6 fusion proteins. The mature portion of the ACX cDNAs was PCR amplified using the following procedure and oligonucleotide primers.
AtACX3
pCGN9897 was employed as the template for PCR amplification of the insert using the sense primer 5′-GATATCGGATCCTTGCAGTACTCTCCACCGGAG-3′ and the antisense primer 5′-GATATCGGTACCCTAAACTGAAGACCAAGCATTGG-3′ containing a BamHI and KpnI restriction site at the 5′ ends, respectively. The cloning of the generated insert into the appropriate sites of pQE30 vector resulted in the construction of pCGN10409.
AtSACX
An Arabidopsis etiolated cotyledon cDNA library (Marathon, CLONTECH Laboratories) was used as a template to PCR amplify a DNA fragment using the sense primer 5′-GGATCCGCGGTGCTTTCATCTGCAGAT-3′ and antisense primer 5′-GTCGACTTAGAGACGGCTACGTGTAGC-3′ containing a BamHI and SalI restriction site at the 5′ ends, respectively. The cloning of the generated product into appropriate sites of pQE30 resulted in the construction of pCGN10438.
AtACX1
The sense primer 5′-CCCGGGAGATCTATGGAAGGAATTGATCACCTCGC-3′ and the antisense primer 5′-CCCGGGGTCGACTCAGAGCCTAGCGGTACGAAG-3′ containing a BglII and SalI restriction site at the 5′ ends, respectively were used to construct pCGN10410 as described for AtSACX.
These plasmids were subsequently sequenced and their authenticity was verified. Escherichia coli strain M15[pREP4] was transformed with each plasmid and the transformed bacteria were grown to late log phase at 30°C and induced with 2 mm isopropyl β-d-thiogalactopyranoside for approximately 18 h. The cells were harvested by centrifugation at 14,000g for 10 min at 4°C and the resultant pellets were suspended in buffer containing 25 mm Tris-HCl, pH 8.0, 30 mm imidazole, 300 mm NaCl, 10% (v/v) glycerol, 0.5% (v/v) Triton X-100, 10 mm β-mercaptoethanol, and 10 μm FAD, and lysed by sonication. Debris was sedimented by a 15-min centrifugation at 14,000g and the supernatant fraction was used for affinity purification of the recombinant protein over Ni-nitrilotriacetic acid resin according to the manufacturer's instruction (Qiagen USA) with the addition of 10 μm FAD to the wash and elution buffers. Eluates from each chromatographic step were analyzed by SDS-PAGE to verify ACX expression levels. Protein concentrations were determined with a protein assay mix (Bio-Rad Laboratories, Hercules, CA).
ACX Enzyme Assay
ACX activity assays were carried out according to the method of Gerhardt (1987), except that the reaction was measured at an A490 and the concentration of the acyl-CoA substrates was reduced to 50 μm. Enzyme assays were carried out in the presence and absence of 100 μg mL−1 BSA (Sigma-Aldrich), and activities were measured in 200-μL reaction volumes, at 30°C, with an Ultramark Microplate Imaging System (Bio-Rad Laboratories). The kinetic constants were calculated by nonlinear regression analysis.
Northern-Blot Hybridization
Northern blots were performed with 10 μg of total RNA per lane, electrophoretically fractionated through a 1.2% (w/v) agarose gel containing 0.66 m formaldehyde and 1× MOPS (3-[N-morpholino]propanesulfonic acid) buffer. The RNA was transferred onto Hybond-N membrane (Amersham-Pharmacia Biotech, Uppsala) in 10× SSC. Probes were generated by labeling PCR-amplified cDNA with [α-32P]dCTP using a Prime-It II random primer labeling kit (Stratagene, La Jolla, CA). Hybridizations were carried out in 40% (v/v) formamide, 6× SSPE, 5× Denhardt's, 0.5% (w/v) SDS, and 0.1 mg mL−1 herring sperm DNA with 5 × 106 to 1 × 107 cpm mL−1 of radiolabeled DNA, for 16 h at 42°C. The membranes were washed in 2× SSC buffer and 0.1% (w/v) SDS at 60°C for 15 min followed by an additional wash in 0.2× SSC buffer, 0.1% (w/v) SDS at 60°C for 15 min. A phosphor screen was exposed to the membrane for 16 h and the radioactive signal was visualized using a Storm 860 optical scanner with ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
ACKNOWLEDGMENTS
We would like to thank Jason Fenner at Calgene for sequencing the AtACX3 PCR products and Eugene Losev for his help constructing the expression plasmids. We would also like to thank Stan Noteboom for his assistance in preparation of digital artwork and John Harada in the Plant Biology Department at the University of California at Davis for providing the B. napus ICL cDNA clone.
LITERATURE CITED
- Bairoch A, Bucher P, Hofmann K. The PROSITE database: its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M. Seeds: Physiology of Development and Germination. New York: Plenum Press; 1994. pp. 311–342. [Google Scholar]
- Do YY, Huang PL. Characterization of a pollination-related cDNA from Phalaenopsis encoding a protein which is homologous to human peroxisomal acyl-CoA oxidase. Arch Biochem Biophys. 1997;344:295–300. doi: 10.1006/abbi.1997.0212. [DOI] [PubMed] [Google Scholar]
- Dubourdieu M, Fox JL. Amino acid sequence of Desulfovibrio vulgaris flavodoxin. J Biol Chem. 1977;252:1453–1463. [PubMed] [Google Scholar]
- Eccleston VS, Ohlrogge JB. Expression of lauroyl-acyl carrier protein thioesterase in Brassica napus seeds induces pathways for both fatty acid oxidation and biosynthesis and implies a set point for triacylglycerol accumulation. Plant Cell. 1998;10:613–621. doi: 10.1105/tpc.10.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira da Rocha PSC, Topping JF, Lindsey K. Promoter trapping in Arabidopsis-One T-DNA tag, three transcripts and two thiolase genes. J Exp Bot. 1996;47:ss24–ss24. [Google Scholar]
- Gerhardt B. Peroxisomes and fatty acid degradation. Methods Enzymol. 1987;148:516–525. [Google Scholar]
- Grossi M, Gulli M, Stanca AM, Cattivelli L. Characterization of two barley genes that respond rapidly to dehydration stress. Plant Sci. 1995;105:71–80. [Google Scholar]
- Hayashi H, De Bellis L, Ciurli A, Kondo M, Hayashi M, Nishimura M. A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J Biol Chem. 1999;274:12715–12721. doi: 10.1074/jbc.274.18.12715. [DOI] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Yamaguchi K, Kato A, Hayashi M, Nishimura M. Molecular characterization of a glyoxysomal long chain acyl-CoA oxidase that is synthesized as a precursor of higher molecular mass in pumpkin. J Biol Chem. 1998a;273:8301–8307. doi: 10.1074/jbc.273.14.8301. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Aoki M, Kondo M, Nishimura M. Changes in targeting efficiencies of proteins to plant microbodies caused by amino acid substitutions in the carboxy-terminal tripeptide. Plant Cell Physiol. 1997;38:759–768. doi: 10.1093/oxfordjournals.pcp.a029233. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell. 1998b;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks MA, Bode K, Couee I. Higher-plant medium- and short-chain acyl-CoA oxidases: identification, purification and characterization of two novel enzymes of eukaryotic peroxisomal beta-oxidation. Biochem J. 1996;320:607–614. doi: 10.1042/bj3200607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks MA, Kellas F, Graham IA. Long-chain acyl-CoA oxidases of Arabidopsis. Plant J. 1999;20:1–13. doi: 10.1046/j.1365-313x.1999.00559.x. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Okamura-Ikeda K, Tanaka K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. J Biol Chem. 1985;260:1311–1325. [PubMed] [Google Scholar]
- Jones A, Davies HM, Voelker TA. Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Kondo M, Nishimura M. Targeting and processing of a chimeric protein with the N-terminal pre-sequence of the precursor to glyoxysomal citrate synthase. Plant Cell. 1996a;8:1601–1611. doi: 10.1105/tpc.8.9.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Mori H, Nishimura M. Molecular characterization of a glyoxysomal citrate synthase that is synthesized as a precursor of higher molecular mass in pumpkin. Plant Mol Biol. 1995;27:377–390. doi: 10.1007/BF00020191. [DOI] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Takeuchi Y, Nishimura M. cDNA cloning and expression of a gene for 3-ketoacyl-CoA thiolase in pumpkin cotyledons. Plant Mol Biol. 1996b;31:843–852. doi: 10.1007/BF00019471. [DOI] [PubMed] [Google Scholar]
- Kato A, Takeda-Yoshikawa Y, Hayashi M, Kondo M, Hara-Nishimura I, Nishimura M. Glyoxysomal malate dehydrogenase in pumpkin: cloning of a cDNA and functional analysis of its pre-sequence. Plant Cell Physiol. 1998;39:186–195. doi: 10.1093/oxfordjournals.pcp.a029356. [DOI] [PubMed] [Google Scholar]
- Kindl H. β-Oxidization of fatty acids by specific organelles. In: Stumpf PK, editor. The Biochemistry of Plants. Vol. 9. New York: Academic Press; 1987. pp. 31–50. [Google Scholar]
- Kirsch T, Loffler HG, Kindl H. Plant acyl-CoA oxidase: purification, characterization, and monomeric apoprotein. J Biol Chem. 1986;261:8570–8575. [PubMed] [Google Scholar]
- Meinke D, Koorneef M. Community standards for Arabidopsis genetics. Plant J. 1997;12:247–253. [Google Scholar]
- Mittendorf V, Bongcam V, Allenbach L, Coullerez G, Martini N, Poirier Y. Polyhydroxyalkanoate synthesis in transgenic plants as a new tool to study carbon flow through β-oxidation. Plant Physiol. 1999;20:45–55. doi: 10.1046/j.1365-313x.1999.00572.x. [DOI] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell. 1999;11:1911–1923. doi: 10.1105/tpc.11.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Protein translocation into peroxisomes. J Biol Chem. 1996;271:32483–32496. doi: 10.1074/jbc.271.51.32483. [DOI] [PubMed] [Google Scholar]