Abstract
Mutations in KRAS exon 2, BRAF and PIK3CA are commonly present in colorectal cancer (CRC) worldwide, but few data about RAS mutations outside KRAS exon 2 are available for Chinese CRCs. We, therefore, determined the mutation frequencies and prognostic values of KRAS exon 2, 3 and 4, NRAS exon 2 and 3, PIK3CA exon 9 and 20, and BRAF exon 15 by PCR and direct sequencing in 353 CRC patients from two Chinese clinical centers. KRAS exon 2, BRAF, PIK3CA mutations were identified in 42.2%, 4.5%, 12.3% of the cases, respectively. We found “rare mutations” in RAS genes in nearly 14% of CRCs-i.e., in almost a quarter (24.0%) of KRAS exon 2 wild type CRCs, including 2.3% in KRAS exon 3, 8.2% in KRAS exon 4 and 3.4% in NRAS. Stage I-III patients with PIK3CA or NRAS mutations developed more distant metastases (3-year risk in PIK3CA mutated and wild type patients: 23.3% vs 11.5%, P = 0.03; multivariate Hazard ratio (HR) = 3.129, P = 0.003; 3-year risk in NRAS mutated and wild type patients: 40.0% vs 12.2%, P = 0.012; multivariate HR = 5.152, P = 0.003). Our data emphasizes the importance of these novel molecular features in CRCs.
Introduction
Colorectal cancer (CRC) is the third most common malignancy in the world. In the last few years, CRC has become the sixth most common malignancy and the fifth leading cause of malignancy-related mortality among the Chinese population1.
Activation of multiple signaling pathways of the Epidermal Growth Factor Receptor (EGFR), the RAS-RAF or the PI3K-PTEN-AKT pathways are considered the most common carcinogenic mechanisms in CRC2. Mutations of RAS, BRAF or PIK3CA cause constitutive activations in these two pathways. Mutations of KRAS exon 2, BRAF and PIK3CA are most common among CRCs with frequencies of 30–50%, 10–15% and 10–20%, respectively. KRAS exon 3 and 4 mutations contribute to a lower degree, only accounting for 1% and 4%3. NRAS mutations are found in about 3–5% and HRAS mutations were rare in previous studies4,5.
The KRAS exon 2 mutation was widely regarded as a predictor for anti-EGFR MoAbs resistance among CRCs6. Nevertheless, the majority of KRAS exon 2 wild-type (wt) patients fail to benefit from anti-EGFR MoAbs, implying the possibility that activating mutations in other KRAS exons or other genes may cause this resistance as well. Recently, a small number of clinical trials have shown that mutations in other RAS exons, such as mutations in KRAS exons 3 and 4, and NRAS can also predict resistance to anti-EGFR MoAbs7–9. Although many clinical data showed that BRAF or PIK3CA mutations were likely to be associated with anti-EGFR MoAbs resistance10, their predictive role is still controversial.
The benefits of individual genetic profiling for the selection of therapy have been proven in clinical use, but studies concerning the mutation frequencies and efficacy of targeted therapies were mostly presented in Western countries and few data are available for China11, especially for mutations in KRAS exons 3 and 4, and NRAS, mainly because the frequency of such mutations was considered low in literature. Moreover, few studies on the prognostic role of these rare mutations are available among Chinese CRC patients, due to lack of follow-up information.
Distant metastasis is a major problem of stage I to III CRC patients after surgery, as it is associated with both high morbidity and mortality. Effective postoperative adjuvant treatment, such as chemotherapy and radiotherapy, can improve patient outcome. Usually such treatment decisions are based on patients’ prognostic features, including traditional clinicopathological features (such as TNM stage, histopathological differentiation grade, invasion to surrounding tissue, and number of lymph-node metastases), microsatellite instability (MSI), and DNA mismatch repair status. According to the National Comprehensive Cancer Network clinical practice guidelines in oncology prior to 2016, stage II and III patients who are assessed to have a poor prognosis, need postoperative chemotherapy, except those who have a high frequency of MSI (MSI-H)12. Although postoperative chemotherapy resulted in a reduction of distant metastasis, patients still had a lower survival rate13. These clinical features, therefore, do not seem to make an exact evaluation of tumor development, so that patients with similar features still reveal difference in survival: quite a few were assessed at low risk and did not receive adjuvant therapy, but developed metastases shortly after surgery; on the other hand, high risk patients endured a decrease in quality of life due to excessive treatment. This discrepancy suggests that patients with similar clinocopathologic characteristics harbor a different genetic biology that regulates their tumor development.
In the last decade, many studies showed that molecular genetic changes can be more accurate markers than clinicopathological features to evaluate the prognosis of cases with early and medium stage CRC. In a study of 450 patients with stage I to III colon cancer, for example, PIK3CA mutations predicted a poorer prognosis, but only among KRAS wild-type CRC patients14. PIK3CA mutation was also identified as an independent biomarker for local recurrence among stage I-III rectal cancers15. Other studies suggested that BRAF mutations confer a poorer prognosis on stage II to III colon cancers, but no conclusive prognostic significance for KRAS mutations could be reached among early and medium stage CRCs16–18. Only one study showed that NRAS mutations predicted a poor outcome for CRC patients with metastases19. Despite the inconsistencies in these studies, they all suggest that biological markers will make a precise assessment of patient outcome in stage I to III CRCs possible and can improve the selection of patients for adjuvant treatment after surgery.
Our previous study of 214 Chinese CRC patients20 reported the mutation status and the prognostic values of KRAS exon 2, BRAF and PIK3CA, respectively. However, we did not analyze the “rare mutation” status at other locations of the KRAS and NRAS genes. Furthermore, the cohort (214) was relatively small. We, therefore, recruited more patients from another clinical center to extend the cohort to 353 CRC patients. We further analyzed additional mutations (including KRAS mutations outside exon 2 and NRAS mutations) and investigated the relationship between mutations and the clinicopathological features. Furthermore, we collected patients’ follow-up information and determined whether a mutation may be used as prognostic biomarker.
Materials and Methods
Samples
Between 2007 and 2012, we consecutively collected 436 CRC patients at Zhongda Hospital (Nanjing, China; the same patients from our previous study20) and 203 CRC patients at Jiangyin People’s Hospital (Jiangyin, China). Patients who did not undergo surgery (n = 35), were lost during the follow-up period (n = 156), had no tissue blocks available, or had poor DNA quality of the tumor sample (n = 95) were excluded. In total, 353 patients were included for genetic detection (Fig. 1). There was no difference in clinicopathological parameters between the in- and excluded patients (see Supplementary Table S1). All cases were diagnosed as CRC by two independent pathologists. No patients had accepted preoperative adjuvant treatment. The patients’ information is listed in Table 1. The collection of tissues and inclusion of patients were approved by the Institutional Ethics Committee of Zhongda Hospital and Jiangyin People’s Hospital. Written informed consent was obtained from all study subjects. The study was conducted according to the institutional guidelines and regulations set by Chinese law for the use of human material for research. The median follow-up for survivors was 33 months.
Figure 1.
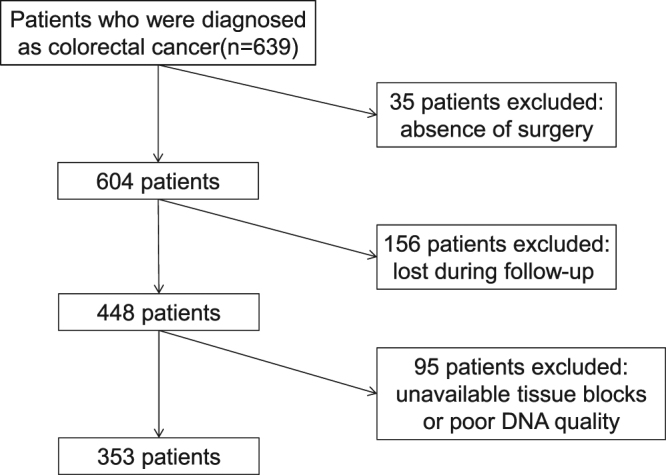
Selection of study population.
Table 1.
Clinicopathological characteristics according to RAS-RAF/PI3K pathway gene mutation status in 353 (350) colorectal cancer patient.
| case | KRAS(exon2/3/4) | NRAS(exon2/3) | BRAF(exon15) | PIK3CA(exon9/20)* | PIK-pathway* | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 353(350) | No, n (%) | Yes, n (%) | p value | No, n (%) | Yes, n (%) | p value | No, n (%) | Yes, n (%) | p value | No, n (%) | Yes, n (%) | p value | No, n (%) | Yes, n (%) | p value | ||
| sex | Male | 204 (202) | 98(58.7) | 106(57.0) | 0.748a | 201(58.9) | 3(25.0) | 0.019 a | 194 (57.6) | 10 (62.5) | 0.696a | 172 (56.6) | 30 (65.2) | 0.269a | 79(59.8) | 124(56.6) | 0.553a |
| Female | 149 (148) | 69 (41.3) | 80(43.0) | 140(41.1) | 9(75.0) | 143 (42.4) | 6 (37.5) | 132 (43.4) | 16 (34.8) | 53 (40.2) | 95(43.4) | ||||||
| Age | 65.04 | 67.02 | 0.137d | 65.96 | 69.5 | 0.335d | 66.03 | 67.06 | 0.748d | 66.4 | 64.37 | 0.298d | 64.44 | 67.21 | 0.040 d | ||
| location | Colon | 210(208) | 106 (63.5) | 104(55.9) | 0.149a | 203(59.5) | 7(58.3) | 1.000b | 197 (58.5) | 13 (81.3) | 0.070a | 173 (56.9) | 35 (76.1) | 0.014 a | 79 (59.8) | 129 (58.9) | 0.862a |
| Rectum | 143(142) | 61(36.5) | 82 (44.1) | 138 (40.5) | 5 (41.7) | 140 (41.5) | 3 (18.8) | 131 (43.1) | 11(23.9) | 53 (40.2) | 90 (41.1) | ||||||
| Differentiation | Well | 44 | 23 (13.8) | 21(11.3) | 0.780c | 42(12.3) | 2(16.7) | 0.484c | 43 (12.8) | 1 (6.3) | 0.030 c | 36 (11.8) | 8 (17.4) | 0.469c | 18 (13.6) | 26 (11.9) | 0.965c |
| moderate | 274(272) | 126 (75.4) | 148(79.6) | 265(77.7) | 9(75.0) | 264 (78.3) | 10 (62.5) | 239 (78.6) | 33 (71.7) | 102 (77.3) | 170 (77.6) | ||||||
| Poor | 12(11) | 9(5.4) | 3 (1.6) | 12 (3.5) | 0 (0) | 9 (2.7) | 3 (18.8) | 9 (3.0) | 2 (4.3) | 6 (4.5) | 6 (2.7) | ||||||
| Missing | 23 | 9 (5.4) | 14(7.5) | 22 (6.5) | 1 (8.3) | 21 (6.2) | 2 (12.5) | 20 (6.6) | 3 (6.5) | 6 (4.5) | 17 (7.8) | ||||||
| Tumor diameter | <5cm | 171(169) | 79 (47.3) | 92 (49.5) | 0.579a | 165 (48.4) | 6(50.0) | 0.936a | 164 (48.7) | 7 (43.8) | 0.676 a | 151 (49.7) | 18 (39.1) | 0.211a | 62 (47.0) | 108 (49.3) | 0.583a |
| >=5cm | 179(178) | 88 (52.7) | 91 (48.9) | 173 (50.7) | 6(50.0) | 170 (50.4) | 9 (56.3) | 151 (49.7) | 27 (58.7) | 70 (53.0) | 108 (49.3) | ||||||
| Missing | 3 | 0 | 3 (1.6) | 3(0.9) | 0(0) | 3 (0.9) | 0 (0) | 2 (0.7) | 1 (2.2) | 0 (2.2) | 3 (1.4) | ||||||
| TNM stage | I | 53(53) | 21(12.6) | 32(17.2) | 0.752c | 52(15.2) | 1(8.3) | 0.692c | 53(15.7) | 0() | 0.001 c | 43(14.1) | 10(21.7) | 0.122c | 17(12.9) | 36(16.4) | 0.405c |
| II | 126(125) | 69(41.3) | 57(30.6) | 121(35.5) | 5(41.7) | 123(36.5) | 3(18.8) | 108(35.5) | 17(37.0) | 56(42.4) | 69(31.5) | ||||||
| III | 126 | 53(31.7) | 73(39.2) | 122(35.8) | 4(33.3) | 120(35.6) | 6(37.5) | 109(35.9) | 16(34.8) | 42(31.8) | 83(37.9) | ||||||
| IV | 45(44) | 22(13.2) | 23(12.4) | 43(12.6) | 2(16.7) | 38(11.3) | 7(43.8) | 41(13.5) | 3(6.5) | 15(11.4) | 30(13.7) | ||||||
| Missing | 3 | 2(1.2) | 1(0.5) | 3(0.9) | 0(0) | 3(0.9) | 0(0) | 3(1.0) | 0(0) | 2(1.5) | 1(0.5) | ||||||
| T | T1 | 5 | 2 (1.2) | 3 (1.6) | 0.197c | 5 (1.5) | 0 | 0.532c | 5 (1.5) | 0 | 0.019 c | 5 (1.6) | 0 | 0.816c | 2 (1.5) | 3 (1.4) | 0.542c |
| T2 | 69 | 28 (16.8) | 41 (22.2) | 68 (19.9) | 1 (8.3) | 69 (20.5) | 0 | 57 (18.8) | 12 (26.1) | 24 (18.2) | 45 (20.5) | ||||||
| T3 | 258(256) | 125 (74.9) | 133 (71.5) | 247 (72.4) | 11 (91.7) | 244 (72.4) | 14 (87.5) | 226 (74.3) | 30 (65.2) | 96 (72.7) | 161 (73.5) | ||||||
| T4 | 19(18) | 10 (6.0) | 9 (4.8) | 19 (5.6) | 0 | 17 (5.0) | 2 (12.5) | 14 (4.6) | 4 (8.7) | 8 (6.1) | 10 (4.6) | ||||||
| Missing | 2 | 2 (1.2) | 0 (0) | 2 (0.6) | 0 | 2 (0.6) | 0 | 2 (0.7) | 0 | 2 (1.5) | 0 | ||||||
| N | N(−) | 189(188) | 97(58.1) | 92 (49.5) | 0.102a | 181 (53.1) | 8 (66.7) | 0.365a | 186 (55.2) | 3(18.8) | 0.004 a | 160 (52.6) | 28(60.9) | 0.317a | 78 (59.1) | 110 (50.2) | 0.099a |
| N(+) | 162(160) | 69 (41.3) | 93 (50.0) | 158(46.3) | 4 (33.3) | 149 (44.2) | 13 (81.3) | 142 (46.7) | 18 ((39.1) | 53 (40.2) | 108 (49.3) | ||||||
| Missing | 2 | 1 (0.6) | 1(0.5) | 2 (0.6) | 0 (0) | 2 (0.6) | 0 | 2(0.7) | 0 (0) | 1 (0.8) | 1 (0.5) | ||||||
| M-synchronous | (−) | 306(304) | 144(86.2) | 162(87.1) | 0.818a | 296(86.8) | 10(83.3) | 0.657b | 297(88.1) | 9(56.3) | 0.002 b | 261(85.9) | 43(93.5) | 0.180a | 116(87.9) | 188(85.8) | 0.533a |
| (+) | 45(44) | 22(13.2) | 23(12.4) | 43(12.6) | 2(16.7) | 38(11.3) | 7(43.8) | 41(13.5) | 3(6.5) | 15(11.4) | 30(13.7) | ||||||
| Missing | 2 | 1(0.6) | 1(0.5) | 2(0.6) | 0(0) | 2(0.6) | 0 | 2(0.6) | 0(0) | 1(0.8) | 1(0.5) | ||||||
| M-metachronous | (−) | 300(297) | 141(84.4) | 159(85.5) | 0.782a | 292(85.6) | 8(66.7) | 0.089b | 290(86.1) | 10(62.5) | 0.021 b | 262(86.2) | 35(76.1) | 0.075a | 117(88.6) | 181(82.6) | 0.129a |
| (+) | 53 | 26(15.6) | 27(14.5) | 49(14.4) | 4(33.3) | 47(13.9) | 6(37.5) | 42(13.8) | 11(23.9) | 15(11.4) | 38(17.4) | ||||||
aChi-square test; bFisher exact test; cMann-Whitney test; dt test. *DNA of three samples was not available for PIK3CA exon 20.
DNA extraction
Genomic DNA was extracted from 5 sections of 10 μm thickness of macro-dissected formalin-fixed paraffin-embedded (FFPE) tumor samples, containing at least 50% tumor epithelium, as determined by two experienced pathologists in H&E-stained paraffin sections. The QIAmp DNA Mini Kits (Qiagen GmbH, Hilden, Germany) were used according to the manufacturer’s instructions.
PCR and Direct sequencing
For each sample, mutations of KRAS exons 2, 3 and 4, NRAS exons 2 and 3, PIK3CA exons 9 and 20, and BRAF exon 15 were amplified by polymerase chain reaction (PCR). Amplification was performed for 30 cycles with the following settings: 95 °C for 4 min (only first cycle); 94 °C for 30 s, 55 °C for 30 s (1 min for the PIK3CA exon 9 and 20 mutations), and 72 °C for 1 min; the final extension cycle was carried out at 72 °C for 7 minutes (10 min for the PIK3CA exon 9 and 20 mutations). The presence of mutations was detected by direct sequencing at Beijing Genomic Institute (BGI, ABI 3730xL Genetic analyzer, Shenzhen, China), using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems). Forward and reverse sequencing were carried out to confirm mutant PCR products. Primer information is listed in Supplementary Table S2.
Statistical analyses
SPSS statistical software (version 18.0 for Windows, SPSS, Inc.) was used for statistical analyses. Categorical variables were compared by the chi-square or Fisher’s exact test; quantitative and ordered variables were compared by the Mann-Whitney test. Normally distributed variables were compared by Student’s t test. Metastasis time was defined as the period between surgery and the detection of a distant metastasis. Overall Survival (OS) was defined as the period between surgery and death of any cause or last follow-up visit. The Kaplan-Meier (KM) method and Log-rank tests were used to evaluate the time to diagnosis of metastases and survival.
To study variables associated with metastasis or survival, we first evaluated the variables (listed in Table 2) in a univariate Cox regression model. Variables with P < 0.1 were taken into a multivariate Cox regression model with stepwise backward elimination. A two-sided P value ≤ 0.05 was considered statistically significant.
Table 2.
Analysis of distant metastasis in 305 CRC patients with TNM stage I to III by univariate and multivariate Cox regression analysis.
| Variables | Univarite analysis | Multivarite analysis | ||
|---|---|---|---|---|
| HR(95% CI) | P | HR(95% CI) | P | |
| Age | 0.578 | |||
| <=65 | 1.0 | |||
| >65 | 1.195(0.638–2.240) | |||
| sex | 0.789 | |||
| Female | 1.0 | |||
| Male | 0.919(0.496–1.704) | |||
| Tumor location | 0.177 | |||
| colon | 1.0 | |||
| rectum | 0.636(0.329–1.228) | |||
| Differention | 0.160 | |||
| well | 1.0 | |||
| moderate | 0.470(0.217–1.019) | 0.056 | ||
| poor | 0.643(0.081–5.080) | 0.675 | ||
| Lymphnode Examined | 0.046 | 0.006 | ||
| >12 | 1.0 | 1.0 | ||
| <=12 | 1.874(1.012–3.471) | 2.500(1.304–4.795) | ||
| Tumor diameter | 0.734 | |||
| <5 cm | 1.0 | |||
| >=5 cm | 1.115(0.595–2.090) | |||
| TNM-stage | 0.101 | |||
| I | 1.0 | |||
| II | 4.137(0.963–17.765) | 0.056 | ||
| III | 4.891(1.142–20.941) | 0.032 | ||
| KRAS status | 0.924 | |||
| wt | 1.0 | |||
| mutant | 1.030(0.558–1.904) | |||
| NRAS status | 0.024 | 0.003 | ||
| wt | 1.0 | 1.0 | ||
| mutant | 3.280(1.167–9.219) | 5.152(1.758–15.101) | ||
| BRAF status | 0.323 | |||
| wt | 1.0 | |||
| mutant | 2.048(0.494–8.499) | |||
| PIK3CA status | 0.038 | 0.003 | ||
| wt | 1.0 | 1.0 | ||
| mutant | 2.131(1.044–4.352) | 3.129(1.463–6.693) | ||
Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding authors.
Results
Mutation frequencies and distributions
KRAS mutations were detected in 186 out of 353 (52.7%) tumor samples, of which 149 (42.2%) had mutations in exon 2, 8 (2.3%) in exon 3, and 29 (8.2%) in exon 4. Among mutations in KRAS exon 2, 109 (30.9%) had single mutations in codon 12 and 38 (10.8%) in codon 13. The main mutant type was 35G>A (G12D, 27.9%), followed by 38G>A (G13D, 18.8%). The most frequently mutation type in exon 3 was 183A>C (Q61H), but 176C>A (A59E), 181C>A (Q61K), 182A>T (Q61L) and 182A>G (Q61R) were also found in our study. In exon 4, the most common mutation was 436G>A (A146T), followed by 351A>T (K117N), 437C>T (A146V), 350A>G (K117R), 436G>C (A146P) and 441G>T (K147N). Moreover, one case harbored both G12V and G12D, while another had both G13D and D54N. NRAS mutations were identified in 12 out of 353 (3.4%) tumor samples, with 5 cases in exon 2 (1.4%) and 7 cases in exon 3 (2.0%). The main mutant types were 35G>A (G12D) in exon 2 and 181C>A (Q61K) in exon 3.
Sixteen (4.5%) patients harbored BRAF exon 15 mutations, with 14 mutations in codon 600 and 2 mutations in codon 601. The most common mutation was 1799T>A (V600E). PIK3CA mutations could not be detected in three samples, 46 out of 350 patients (12.3%) harbored PIK3CA mutations, with 26 mutations in exon 9 (7.4%) and 20 mutations in exon 20 (5.7%). The most frequent mutant types were 1633G>A (E545K) in exon 9 and 3140A>G (H1047R) in exon 20. Mutations are summarized in Table 3.
Table 3.
Frequency and distribution of KRAS, NRAS, BRAF and PIK3CA mutations *DNA of three samples was not available for PIK3CA exon 20.
| Nucleotide | Amino acid | Case(%) | Nucleotide | Amino acid | Case(%) | ||
|---|---|---|---|---|---|---|---|
| KRAS exon 2 | 149(42.2%) | NRAS exon 2 | 5(1.4%) | ||||
| 34G>A | G12S | 5 | 34G>C | G12R | 1 | ||
| 34G>T | G12C | 9 | 35G>C | G12A | 1 | ||
| 34G>C | G12R | 1 | 35G>A | G12D | 2 | ||
| 35G>A | G12D | 52 | 38G>A | G13D | 1 | ||
| 35G>C | G12A | 9 | NRAS exon 3 | 7(2%) | |||
| 35G>T | G12V | 33 | 178G>A | G60R | 1 | ||
| 35G>T&35G>A | G12V&G12D | 1 | 181C>A | Q61K | 6 | ||
| 37G>C | G13R | 1 | BRAF exon 15 | 16(4.5%) | |||
| 37G>T | G13C | 2 | 1799T>A | V600E | 14 | ||
| 38G>A | G13D | 35 | 1801A>G | K601E | 1 | ||
| 38G>A&160G>A | G13D&D54N | 1 | 1803A>C | K601N | 1 | ||
| KRAS exon 3 | 8(2.3%) | PIK3CA exon 9 | 26(7.4%) | ||||
| 176C>A | A59E | 1 | 1624G>A | E542K | 5 | ||
| 181C>A | Q61K | 1 | 1633G>A | E545K | 11 | ||
| 182A>T | Q61L | 1 | 1634A>C | E545A | 2 | ||
| 182A>G | Q61R | 1 | 1635G>C | E545D | 1 | ||
| 183A>C | Q61H | 4 | 1636C>A | Q546K | 5 | ||
| KRAS exon 4 | 29(8.2%) | 1637A>G | Q546R | 1 | |||
| 351A>T | K117N | 3 | 1638G>T | Q546H | 1 | ||
| 350A>G | K117R | 1 | PIK3CA exon 20* | 20(5.7%) | |||
| 436G>A | A146T | 21 | 3062A>T | Y1021F | 2 | ||
| 436G>C | A146P | 1 | 3073A>G | T1025A | 1 | ||
| 437C>T | A146V | 2 | 3129G>A | M1043I | 1 | ||
| 441G>T | K147N | 1 | 3139C>T | H1047Y | 2 | ||
| 3140A>G | H1047R | 11 | |||||
| 3140A>T | H1047L | 1 |
In total, 197 patients (55.8%) had RAS mutations. 49 patients (13.9%) had mutations outside KRAS exon 2. 218 patients (62.3%) carried one or more mutations, of which 178 (50.9%) harbored a single gene mutation, 38 patients (10.9%) two gene mutations, and 2 patients (0.6%) three gene mutations. In patients carrying two mutations, 34 patients had mutations in both KRAS and PIK3CA and 2 patients in both BRAF and PIK3CA. BRAF and KRAS exon 2 mutations were mutually exclusive, but we identified one patient who had concomitant KRAS exon 4 and BRAF mutations and one patient who had both NRAS and BRAF mutations. In addition, one patient suffered from KRAS, BRAF and PIK3CA mutations, while another patient harbored two KRAS and one PIK3CA mutation. The mutation distribution is shown in Fig. 2 and Supplementary Table S3.
Figure 2.
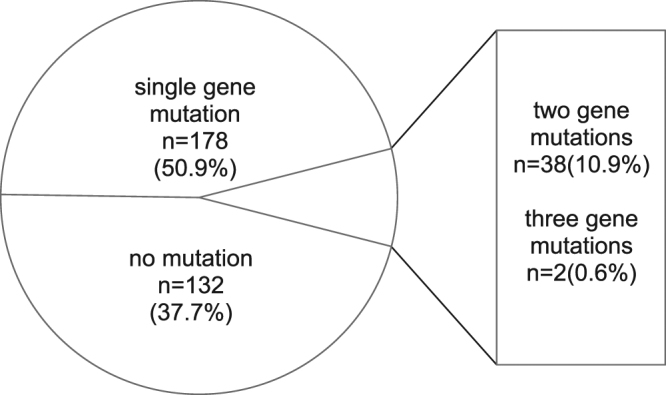
The distribution of mutations is illustrated in a pie chart of 350 colorectal cancer samples.
Clinicopathological characteristics of mutations
We did not find any significant correlation between KRAS (exon 2, 3 and 4) mutations and patients’ clinicopathological characteristics (Table 1). Female patients harbored more NRAS (exon 2 and 3) mutations than male patients (75.0% vs 25.0%; P = 0.019). Compared to BRAF wt patients, patients with BRAF mutations were more likely to exhibit poor differentiation (P = 0.030), advanced TNM stage (P = 0.001), larger/more invasive tumor (P = 0.019), higher lymph node metastasis rate (P = 0.004), and higher synchronous (P = 0.002) and metachronous metastasis rate (P = 0.016). PIK3CA mutations occurred more frequently in colon than in rectal cancers (P = 0.014). Those who had at least one mutation, occurred more frequently among older patients (average age: 67.2 vs 64.4 years old, P = 0.04). There were no significant associations in clinicopathological characteristics between double gene mutant and wt patients (see Supplementary Table S4).
We then investigated the associations between different subtypes of KRAS mutations and patients’ clinicopathological characteristics. KRAS exon 2 mutation appeared more frequent in older patients (average age: 67.7 years old vs 64.9 years old; P = 0.036) and was associated with higher lymph node metastasis rate (52.3% vs 41.6%; P = 0.046). KRAS exon 3 mutation was more likely to appear in lower TNM stage (P = 0.011) and smaller/less invasive tumor (P = 0.001) patients. Data are shown in Supplementary Table S5.
Survival analysis
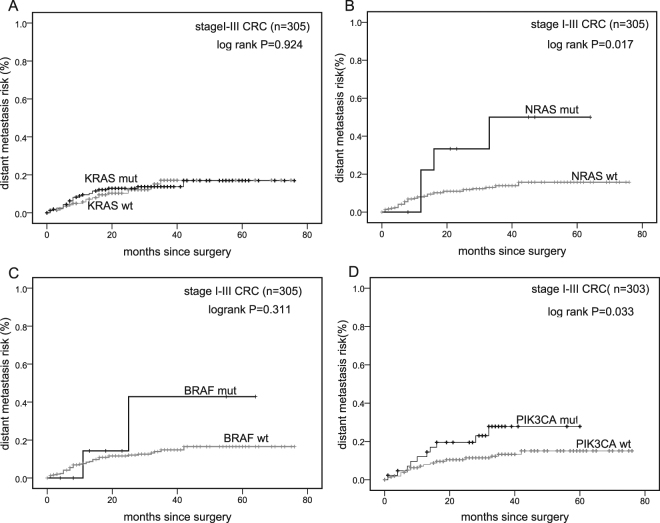
KM analysis showed no difference of OS between KRAS-, NRAS- or PIK3CA-mutant patients and wt patients (P = 0.695; P = 0.847; P = 0.987; Fig. 3A,C,D). Only BRAF mutations had a strong association with poorer OS (3-year OS) in BRAF-mutant vs BRAF-wt patients (14.3% vs 74.6%; P < 0.001; Fig. 3B). In our previous study, BRAF V600E mutations were found only in colon cancers and strongly revealed poorer OS in colon patients. In this extended study, we found BRAF V600E mutations had poorer prognosis both in colon and rectal cancer patients (Fig. 4A,B). Because only two BRAF V600E-mutant patients with rectal cancer were included, the result still needs to be confirmed in the future.
Figure 3.
Kaplan-Meier curves. OS since surgery for patients with (black) and without (gray) mutations in 353 CRC patients. Panel D: DNA of three samples was not available for PIK3CA mutation analysis. wt: Wild-type; mut: Mutant.
Figure 4.
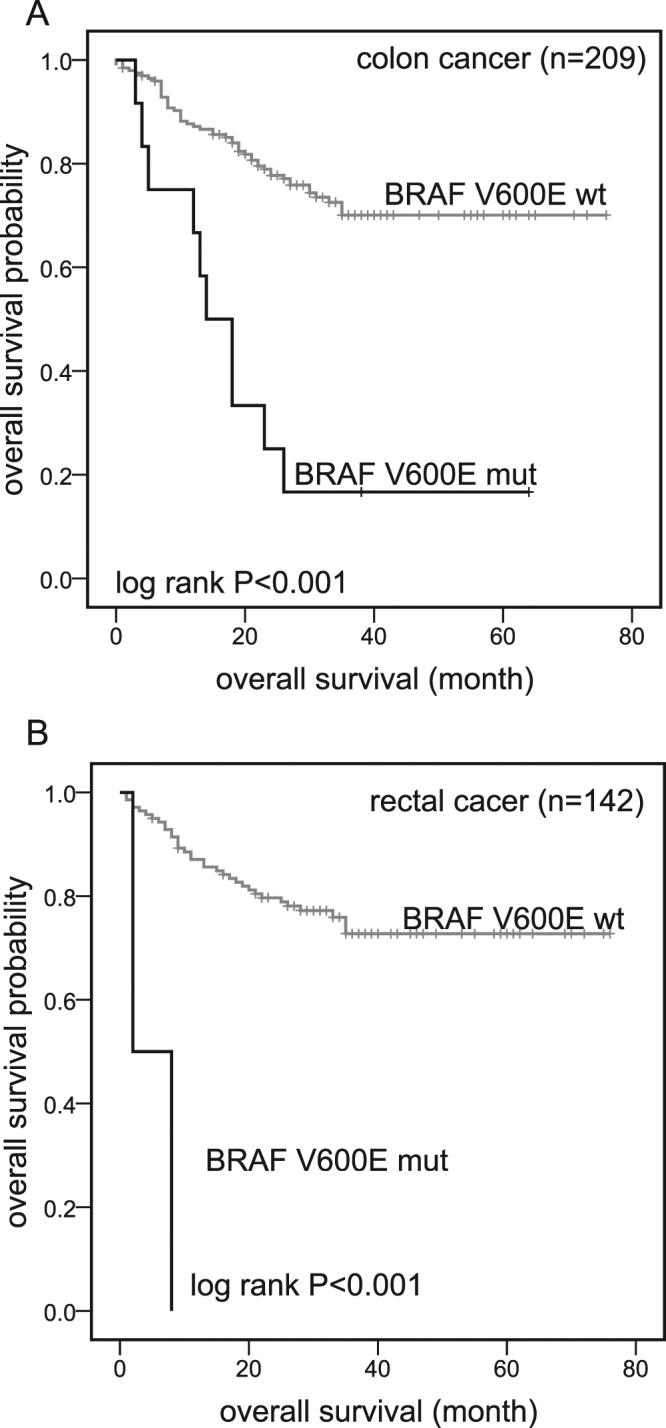
Kaplan-Meier curves. OS since surgery for patients with (black) and without (gray) BRAF V600E mutations in colon or rectal cancer. (A) 3-year OS in BRAF V600E mut versus BRAF wt colon cancer patients: 16.7% versus 74.1%; log-rank P < 0.001. One colon cancer patient harboring a BRAF K601E mutation was excluded in this analysis. (B) 3-year OS in BRAF V600E mut versus BRAF wt rectal cancer patients: 0% versus 75.7%; log-rank P < 0.001. One rectal cancer patient harboring a BRAF K601N mutation was excluded in this analysis. wt: Wild-type; mut: Mutant.
Because BRAF mutations confer a poorer prognosis, several recent studies have suggested to exclude BRAF-mutant patients from KRAS-wt patients when assessing the prognostic value of KRAS17,21,22. After we excluded BRAF-mutant patients, a total of 337 patients (353 patients - 16 BRAF-mutant patients) remained in the analysis (see Supplementary Fig. S1A). OS between KRAS-mutated and wt patients was still not different (P = 0.142). Likewise, there were also no differences in OS between NRAS- (P = 0.524) or PIK3CA- (P = 0.658) mutated and wt patients (data not shown). Finally, we observed no significant correlations between OS and the three subtypes of KRAS mutations.
Previous studies showed that PIK3CA mutations were strongly correlated with a higher local recurrence rate in stage I to III rectal cancer patients, but did not find the correlation with distant metastases15. In our present study, local recurrence analysis was not feasible because of a lack of relevant data. Instead, we analyzed the relationship between different mutations and the occurrence of postoperative distant metastases. In total, 305 patients (353 patients −45 patients with stage IV disease and 3 patients without proper staging) were included in the analysis (see Supplementary Fig. S1B) In this group, 41 patients developed postoperative distant metastases. Patients with PIK3CA mutations had a 2-fold increased distant metastasis rate compared with PIK3CA wt patients (3-year risks, 23.3% vs 11.5%, P = 0.03) and a shorter interval between surgery and the diagnosis of a metastasis (mean metastasis-free intervals: 67.1 months vs 47.6 months; P = 0.033). More intriguingly, patients with NRAS mutations showed a > 3-fold increase in distant metastases and a shorter metastasis-free interval (3-year risks, 40.0% vs 12.2%, P = 0.012; mean metastasis-free intervals: 66.6 months vs 41.9 months, P = 0.017). No correlations were found between other mutations and distant metastases (Fig. 5).
Figure 5.
Kaplan-Meier curves. Distant metastasis rates since surgery for 305 stage I to III patients with (black) and without (gray) mutations. Panel (D) DNA of two samples were not available for PIK3CA mutation analysis. wt: Wild-type; mut: Mutant.
In univariate Cox regression analysis for distant metastases, the variables including age, sex, tumor location, differentiation grade, number of lymph nodes examined, tumor diameter, TNM stage and KRAS/NRAS/BRAF/PIK3CA mutations that are listed in Table 2 were examined. Besides numbers of lymph nodes examined (P = 0.046), PIK3CA (hazard ratio (HR), 2.131; 95% confidence interval (CI), 1.044–4.352; P = 0.038) and NRAS mutations (HR, 3.280; 95%CI, 1.167–9.219; P = 0.024) revealed a higher risk of distant metastases. The three variables were entered into a multivariate analysis with stepwise backward elimination. PIK3CA (HR, 3.129; 95% CI, 1.463–6.693; P = 0.003) and NRAS (HR, 5.152; 95% CI, 1.758–15.101; P = 0.003) mutations both persisted as prognostic markers for distant metastases in stage I to III patients. No significant interactions were observed between the variables.
Discussion
In this study, we identified that 13.9% (49 out of 353) CRC patients carried mutations at RAS exons outside the KRAS exon 2. The mutations were mainly located in exons 3 and 4 of KRAS, and in exons 2 and 3 of NRAS genes. More importantly, we found that stage I to III patients who carried PIK3CA- or NRAS-mutated genes had a higher risk to develop distant metastases after surgery and had shorter metastasis-free intervals.
Mutations of KRAS exon 2 were most common in 353 Chinese CRC patients and were typically located in codons 12 and 13. BRAF mutation frequency was 4.5% in CRCs and substantially lower than in Western countries (10–15%)23, but was consistent with other Asian regions, such as Japan21 and Taiwan24. The PIK3CA mutation occurred in 13.1% of CRCs in our study. All mutation frequencies found were consistent with our previous study20.
Mutations of RAS exons outside KRAS exon 2 occurred in 13.9% of CRC patients in our study, of which 8 (2.5%) had single mutations in KRAS exon 3, 29 (8.2%) in KRAS exon 4, and 12 (3.4%) in NRAS exon 2 or 3. Among the 8 mutations in KRAS exon 3, 7 mutations occurred in codon 61, which is similar to another Chinese study11. Mutations in exon 4 were mainly located in codons 117 and 146. The mutation frequency in codon 146 (6.8%) was higher than the observed rate in Western countries, but similar to the reported frequencies in Hong Kong (5.6%, 7/126)3,25,26. NRAS mutations were identified in 12 (3.4%) out of 353 tumor samples. The mutation frequency of NRAS exon 3 (7, 2.0%) was slightly higher than in NRAS exon 2 (5, 1.4%) and similar to Shen’s study of the Chinese population11.
Among recent Western studies, one showed that 12.1% of 513 KRAS exon 2 wt American CRCs had mutations in either KRAS exon 3 or 4, or NRAS exon 2 or 327. Another study9 in 639 KRAS exon 2 wt European CRCs found that these RAS mutations reached a prevalence of 17%. In our study, mutation frequencies of RAS exons outside KRAS exon 2 reached approximately 14% as well. These frequencies are also close to other Chinese studies published in recent years with the exception of the mutation frequency of KRAS exon 411,28,29. The mutation frequency in KRAS exon 4 (8.2%) in our study was higher than the frequencies in two Western studies mentioned (1.9% and 3.3%), but close to two Chinese reports (one study: 5.6% of 126 Hong Kong CRCs; the other study: 4.1% of 1110 Chinese CRCs)3,28. However, another study reported that only 2.7% of 1506 Hong Kong CRCs had mutation in KRAS exon 4. At present, we have no explanation for this discrepancy. We used tumor samples from two hospitals in different regions and studied only patients without preoperative radiotherapy or chemotherapy. Furthermore, our samples were tested for all RAS mutations regardless of whether they harbored KRAS exon 2 mutations. These boundary conditions enabled us to make a better estimate of mutation frequencies in RAS exons outside KRAS exon 2 in Chinese CRCs.
In western countries a mutation test for KRAS exon 2 is routine clinical practice to qualify for anti-EGFR treatment, but more than half of the KRAS exon 2 wt patients are, nonetheless, resistant to this treatment6. Therefore, it is necessary to further extend the genetic status in KRAS exon 2 wt patients. In agreement, a large proportion of clinical trials has demonstrated mutations at RAS sequences outside KRAS exon 2. These mutations, which are called “rare mutations” in literature, confer a detrimental effect on the response to anti-EGFR MoAbs7,8,23,30. Because most of the data came from Western countries, it was important to establish the mutation distribution and frequencies of these rare mutations in the Chinese population. In our study, we found that up to 14% CRC patients carried these rare mutations, and that these patients represent almost a quarter (24.0%, 48 out of 204 (353 CRCs-149 KRAS exon 2 mutants)) among the KRAS exon 2 wt patients.
More than 60% of cases carried either one or more activating mutations in the RAS-RAF or the PI3K-PTEN-AKT pathways in our study-cohort, so that they may well be resistant to anti-EGFR MoAbs. Irrespective of KRAS exon 2 mutations, it remains controversial whether it is important to investigate these rare mutations in clinical practice. Considering there are a series of rare mutation subtypes in the two pathways, it is hard to test every mutation site. Although the total mutation frequency of these rare mutations in our study was similar to Western countries, there was a difference in the frequency of each mutation subtype, as best shown for KRAS exon 4. Therefore, our findings can contribute to determine frequent mutation sites for priority detection in the Chinese population.
At first, we found no associations between KRAS mutations (including exons 2, 3 and 4) and patients’ clinicopathological features. However, each mutation subtype confers unique conformational or structural alterations, which may confer variable effects on tumor progression31,32. To verify this view, we further analyzed KRAS mutations at different exons. And found KRAS exon 2 mutations occurred more frequently in older patients and patients with lymph-node metastases.
KRAS exon 3 mutations seemed to confer a less aggressive biological behavior, because they were associated with lower TNM stage and smaller/less invasive tumor. Considering there were only 9 patients harboring KRAS exon 3 mutations and those 9 patients contained 5 subtypes, this finding does not yet allow us to draw a firm conclusion.
We found that NRAS mutations occurred more frequently in female patients (75.0% vs 41.1%). Other studies in Chinese populations did not find any correlations between NRAS mutations and clinicopathological characteristics28. Because we observed only 5 cases with mutations in NRAS exon 2 and only 7 cases in exon 3, we could not determine their clinical impact.
Mutations in PIK3CA may be significantly related to relapse-free survival in stage II and III patients, but the sample size was small (n = 96)33. In 450 stage I to III colon cancers, PIK3CA mutation was associated with a significant increase in colon cancer–specific mortality in the KRAS wt patients14. A recent study on Chinese patients showed that among KRAS, BRAF, PIK3CA and NRAS mutations in stage II to III colon cancers (n = 228), only PIK3CA mutations were an independent prognostic biomarker for poor OS among stage III patients34. In agreement, we found that patients with PIK3CA mutations had a 2-fold increased distant metastasis rate and shorter metastasis-free intervals, compared with the PIK3CA wt patients in stages I-III. In the following univariate and multivariate Cox regression analysis, PIK3CA mutations persisted as an independent prognostic marker for distant metastasis among stage I to III patients when mixed with other confounding factors. This observation was consistent with the above mentioned studies, but we had a relative large sample size.
None of the previous studies investigated the prognostic value of the rare mutations among stage I to III patients. In our study, a NRAS mutation acted as an independent prognostic marker for distant metastasis (3-fold increased rate) in stage I to III patients, with shorter metastasis-free intervals than NRAS wt patients. In fact, a large study of patients with in metastatic CRCs (n = 786) showed recently that NRAS mutations were associated with shorter OS19, but the effect of such mutations in stage I to III patients was not discussed. To the best of our knowledge, we are, therefore, the first to illustrate the prognostic role of NRAS mutations on distance metastases in early, mid-stage patients. Nevertheless, this observation requires further confirmation in a larger population of Chinese patients given the relatively low frequencies of NRAS mutations (n = 12) in our data.
KRAS and NRAS proteins are homologous enzymes of the RAS protein family35 and genetic studies have clarified apparent differences between RAS isoforms. Furthermore, KRAS and NRAS mutations have a mutually exclusive mechanism to induce carcinogenesis36. Mutant KRAS induces excessive proliferation and differentiation of human colonic epithelial cell lines, whereas mutant NRAS suppresses apoptosis37. Accordingly, mutant NRAS signals in vivo through the non-canonical RAS-RAF-MEK-STAT3 (Signal transducer and activator of transcription 3) MAPK pathway to regulate cell apoptosis. We speculate that the ability to predict distance metastasis based on NRAS mutations may be responsible for their distinct tumorigenic mechanism.
The limitations of our study are its relatively small sample size, retrospective nature, short follow-up time, and lack of epigenetic or MSI status, which are important for risk assessment of CRC. NRAS mutation frequency was too low to analyze its mutation subgroups. We will, therefore, enlarge our sample size by recruiting more CRC patients from other clinical centers. On the other hand, we collected tumor samples from two hospitals in different regions and included patients who had not received preoperative radiotherapy or chemotherapy. These aspects make our data more representative and prognostic for new patients.
Conclusion
In conclusion, we studied mutations in the RAS-RAF and PI3K-PTEN-AKT pathways, especially the frequencies and distributions of so-called rare mutations, including KRAS exons 3 and 4, and NRAS exons 2 and 3 in 353 Chinese CRC patients. Among the mutations, PIK3CA and NRAS mutations had prognostic value and predicted distant metastasis in stage I to III CRC patients. Our findings show that these new molecular features may be important for better decision making in clinical practice for postoperative chemotherapy, such as the use of anti-EGFR MoAbs and/or radiotherapy as postoperative adjuvant therapy. The NRAS mutation is promising for future studies because of its unique carcinogenic mechanism and biological characteristics.
Electronic supplementary material
Acknowledgements
The authors thank Professor Wouter H Lamers (University of Amsterdam) for careful editing the manuscript. The authors also thank the departments of Pathology and General Surgery of Zhongda Hospital and Jiangyin People’s Hospital affiliated to Southeast University for the preparation of the FFPE sections and the collection of the clinical data.
Author Contributions
J.Z. and Y.H. designed the study. F.G., H.G., J.Z. and Y.H. conducted the experiments and data analysis. F.G., J.Z. and Y.H. interpreted the data and drafted the manuscript. H.Z., J.C., Y.Z., L.Z., X.S. and A.Z. helped with sample preparation, patients’ data collection and interpretation. H.J. helped with statistical analysis. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24306-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianqiong Zhang, Email: zhjq@seu.edu.cn.
Youji He, Email: heyouji@yahoo.com.
References
- 1.Chen W, et al. Report of cancer incidence and mortality in China, 2010. Annals of translational medicine. 2014;2:61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 3.Edkins S, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer biology & therapy. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irahara N, et al. NRAS mutations are rare in colorectal cancer. Diagnostic molecular pathology: the American journal of surgical pathology, part B. 2010;19:157–163. doi: 10.1097/PDM.0b013e3181c93fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos JL. Ras oncogenes in human cancer: a review. Cancer research. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 6.Allegra CJ, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 7.Loupakis F, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. British journal of cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeters M, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:1902–1912. doi: 10.1158/1078-0432.CCR-12-1913. [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. The New England journal of medicine. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 10.Laurent-Puig P, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PloS one. 2013;8:e81628. doi: 10.1371/journal.pone.0081628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Provenzale D, et al. Genetic/Familial High-Risk Assessment: Colorectal Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network: JNCCN. 2016;14:1010–1030. doi: 10.6004/jnccn.2016.0108. [DOI] [PubMed] [Google Scholar]
- 13.Andre T, et al. Adjuvant Fluorouracil, Leucovorin, and Oxaliplatin in Stage II to III Colon Cancer: Updated 10-Year Survival and Outcomes According to BRAF Mutation and Mismatch Repair Status of the MOSAIC Study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33:4176–4187. doi: 10.1200/JCO.2015.63.4238. [DOI] [PubMed] [Google Scholar]
- 14.Ogino S, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:6956–6962. doi: 10.1158/1078-0432.CCR-09-1165. [DOI] [PubMed] [Google Scholar]
- 16.Gavin PG, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:6531–6541. doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farina-Sarasqueta A, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 18.Roth AD, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 19.Schirripa M, et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. International journal of cancer. 2015;136:83–90. doi: 10.1002/ijc.28955. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, et al. BRAF V600E mutation and KRAS codon 13 mutations predict poor survival in Chinese colorectal cancer patients. BMC cancer. 2014;14:802. doi: 10.1186/1471-2407-14-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokota T, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. British journal of cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura Y, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Roock W, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The Lancet. Oncology. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 24.Li HT, Lu YY, An YX, Wang X, Zhao QC. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncology reports. 2011;25:1691–1697. doi: 10.3892/or.2011.1217. [DOI] [PubMed] [Google Scholar]
- 25.Janakiraman M, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer research. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamura Y, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Molecular cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes, chromosomes & cancer. 2011;50:307–312. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Scientific reports. 2015;5:18678. doi: 10.1038/srep18678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong JH, et al. Characterization of rare transforming KRAS mutations in sporadic colorectal cancer. Cancer biology & therapy. 2014;15:768–776. doi: 10.4161/cbt.28550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pentheroudakis G, et al. Biomarkers of benefit from cetuximab-based therapy in metastatic colorectal cancer: interaction of EGFR ligand expression with RAS/RAF, PIK3CA genotypes. BMC cancer. 2013;13:49. doi: 10.1186/1471-2407-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Er TK, Chen CC, Bujanda L, Herreros-Villanueva M. Clinical relevance of KRAS mutations in codon 13: Where are we? Cancer letters. 2014;343:1–5. doi: 10.1016/j.canlet.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert review of molecular diagnostics. 2012;12:621–628. doi: 10.1586/erm.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato S, et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. International journal of cancer. 2007;121:1771–1778. doi: 10.1002/ijc.22890. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, et al. Prognostic impact of mutation profiling in patients with stage II and III colon cancer. Scientific reports. 2016;6:24310. doi: 10.1038/srep24310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nature reviews. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes & cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nature genetics. 2008;40:600–608. doi: 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding authors.