Abstract
Stylo has a great potential for Al3+ resistance in acidic soils through secretion of citrate from the roots. To get insight into the molecular mechanisms responsible, transcriptomic changes were investigated in the roots after treatment with T01 (−Al3+, pH6.0), T02 (−Al3+, pH4.3) and T03 (50 µM AlCl3, pH4.3). In total, 83,197 unigenes generated from 130,933 contigs were obtained. Of them, 282, 148 and 816 differentially expressed unigenes (DEGs) were revealed in T01_vs_T02, T02_vs_T03 and T01_vs_T03 comparison, respectively (FDR < 0.001, log2FC > 2). DEGs by Al3+ were related to G-proteins, diacyglycerol and inositol metabolism, calcium-signaling, transcription regulation, protein modification and transporters for detoxification of Al3+. Additionally, Al3+ facilitates citrate synthesis via modifying gene expression of pathways responsible for citrate metabolism. Overall, Al3+ resistance in stylo involves multiple strategies and enhancement of citrate anabolism. The Al3+ signal transmits through heterotrimeric G-proteins, phospholipase C, inositol triphosphate, diacylglycerol, Ca2+ and protein kinases, thereby activating transcription and anion channels in plasma membrane, and resulting in citrate secretion from stylo roots.
Introduction
Trivalent aluminum (Al3+) toxicity is a major constraint for root growth and crop yields of plants in acidic soils, which constitute 50% of the potentially arable lands worldwide1. At soil pH below 5.0, the soluble Al3+ species damages cells at the root apex, and subsequently inhibits the uptake of water and nutrient2,3. Although Al3+ toxicity can be ameliorated through application of lime which raises the soil pH, this amendment does not reduce acidity in the subsoil layer. Moreover, liming is not a cost effective method, thus may not always be practical. Alternatively, one of the most appropriate strategies to increase crop productivity on acidic soils is to cultivate Al3+ resistant varieties. A solid understanding of Al3+-resistance mechanisms will facilitate development of improved crops well suitable for acidic soils.
Plants thriving in acidic soils have evolved both external avoidance and internal tolerance mechanisms. The external avoidance mechanisms prevent Al3+ from entering root cells by secretion of organic acid anions (OAs), such as citrate, malate and oxalate, from the root apex, resulting in the formation of stable nonphytotoxic OA-Al chelates2,3. The internal tolerance mechanisms enable plants to uptake and sequester Al3+ in the root vacuole once it enters the cell in the cytosol, or to allow the plant to tolerate the Al3+ in the cell wall.
Over the past decades, great efforts have been made to identify the genes for external and internal detoxification of Al3+ in plants. The first class of resistance genes encoding malate and citrate efflux transporters (ALMTs) has been identified in wheat, Arabidopsis, rape and rye2. Another protein family involved in Al3+ tolerance is from multidrug and toxic compound extrusion proteins (MATEs) that are associated with Al3+-activated citrate exudation in plant species2. Internally, Nramp aluminum transporter 1 (Nrat1), operates in concert with a tonoplast-localized half-size ABC transporter, ALS1, to remove Al3+ from the cell wall, and to sequester it in the root-cell vacuole in rice3,4. Subsequent studies have identified sensitive to proton rhizotoxicity 1 (STOP1) in Arabidopsis and Al3+ resistance transcription factor 1 (ART1) in rice as transcription factors that regulate expression of a suite of Al3+-resistance genes5,6. Despite of these findings, the mechanisms that modulate gene expression responsible for Al3+-induced secretion of citrate remain unclear.
Stylo (Stylosanthes) has a great potential for Al3+ tolerance on highly acidic soils prevalent in the tropics and sub-tropics. We documented a cultivar genotype Reyan 2 (R2) of stylo exhibiting high Al3+ resistance via Al3+-induced release of citrate from root apex7. In addition, citrate efflux upon Al3+ exposure was delayed by several hours, suggesting that Al3+ induces the expression of responsive genes in this species. Yet, to date, research on mechanisms of stylo responses and tolerance to Al3+ have been hampered because of the lack of the genomic sequence and transcriptomic data under Al3+ stress. Here, the RNA-Seq was applied to analyze the transcriptomic changes in stylo roots after Al3+ exposure. The aim was to characterize the pathways that orchestrate expression of genes responsible for Al3+-induced secretion of citrate in stylo.
Results
Optimal concentration and duration of Al3+ treatment
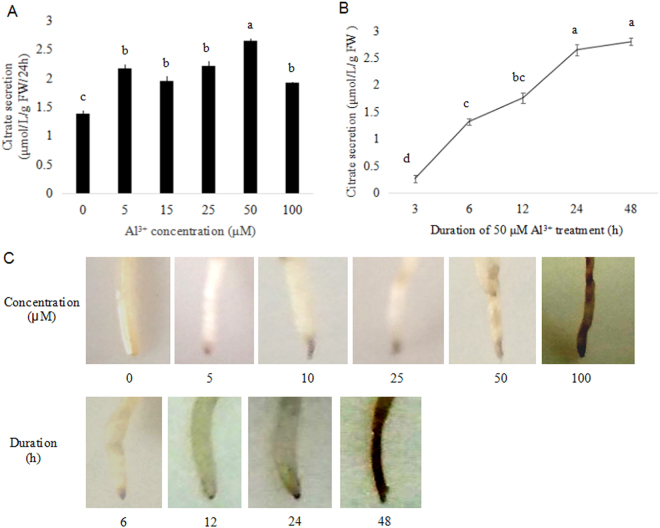
We previously reported that in R2 roots the amount of citrate efflux was Al3+ concentration and time dependent7. To further optimize Al3+ treatment for RNA-Seq analysis, measurement of citrate secretion was performed at different dosage and treatment time of Al3+. The largest quantity of citrate exudation was detected at 24 h after 50 μM AlCl3 (pH4.3) treatment with no more than a 25% inhibition of relative root growth (RRG) compared to the control (−Al3+) (Fig. 1A,B). Although Chrome Azurol S staining deepened with increasing of Al3+ concentration and treatment time, the 24 h group demonstrated much less damage in the roots than that of the 48 h group (Fig. 1C).
Figure 1.
Effect of AlCl3 concentration for 24 h (A) and duration of 50 µM AlCl3 treatment (B) on citrate secretion in R2 roots and Chrome Azurol S staining (C). Values are represent as Mean ± SE (n = 5). Different letters indicate significance at P < 0.05.
Transcripts de novo assembly and annotation
RNA-Seq of T01 (control, −Al3+, pH6.0), T02 (acid treatment, −Al3+, pH4.3) and T03 (+Al3+, 50 µM AlCl3, pH4.3) treatments generated a range of 20.59–34.36 million clean reads per sample (Table 1), which represented an range of 91.3% to 93% of the raw reads. In total, there were 61.91 Gb clean paired-end reads from all samples with a GC content of about 45% and with equal distribution of G and C (or A and T) in the reads. De novo assembly of the reads produced 130,933 transcripts corresponding to 83,197 unigenes (Table S1). The assembled unigenes had a length distribution from 203 to 4,017 bp with an N50 length of 1,307 bp and an average length of 959 bp (Table S1), and 54,976 unigenes were annotated (Table S2). Saturation simulation for transcriptomic data showed that the number of novel genes decreased with total read number increasing in each sample (Fig. S1), and a good correlation between the replicates was observed (R2 > 0.847, Fig. S2A).
Table 1.
Summary of short-read data of RNA-Seq.
| Sample | Treatment | Clean Read | GC (%) | ≥Q30 (%) | Mapped Read | Expressed Unigene |
|---|---|---|---|---|---|---|
| T01–1 | T01(−Al3+, pH6.0) | 23,593,254 | 45.41 | 92.50 | 19,028,551 | 52,176 |
| T01–2 | 20,594,687 | 45.26 | 91.93 | 14,329,659 | 65,806 | |
| T01–3 | 23,598,964 | 45.25 | 92.01 | 17,737,584 | 62,682 | |
| T02–1 | T02(−Al3+, pH4.3) | 23,359,649 | 45.09 | 92.71 | 19,325,346 | 48,568 |
| T02–2 | 34,364,939 | 44.58 | 92.20 | 26,577,280 | 60,741 | |
| T02–2 | 21,974,723 | 45.21 | 92.32 | 16,229,807 | 60,414 | |
| T03–1 | T03(+Al3+, pH4.3) | 24,804,328 | 45.17 | 92.79 | 20,331,769 | 49,133 |
| T03–2 | 22,992,711 | 44.85 | 92.60 | 16,830,708 | 61,540 | |
| T03–3 | 23,474,652 | 44.97 | 92.01 | 17,067,386 | 60,030 |
Global transcriptomic analysis of Al3+-affected genes
Differentially expressed unigenes (DEGs) in R2 roots between treatments were further analyzed. A total of 281 DEGs were discovered in the comparison of T01_vs_T02 (2 up-regulated and 279 down-regulated), whereas there were 148 DEGs in T02_vs_T03 with 49 increasing and 99 decreasing in expression (FDR < 0.001, log2FC (fold change) > 2) (Table 2). The number of DEGs in T01_vs_T03 was 819, of which 40 unigenes were induced and 799 unigenes were inhibited. A hierarchical clustering and heatmap analysis showed that the expression patterns observed in plants of acid and Al3+ treatment clustered together, which showed clear difference to the control plants (Fig. S2B).
Table 2.
Differentially expressed unigenes between treatments.
| Group | Total | Up-regulated | Down-regulated |
|---|---|---|---|
| T01_vs_T02 | 281 | 2 | 279 |
| T02_vs_T03 | 148 | 49 | 99 |
| T01_vs_T03 | 819 | 40 | 799 |
Functional classification of DEGs
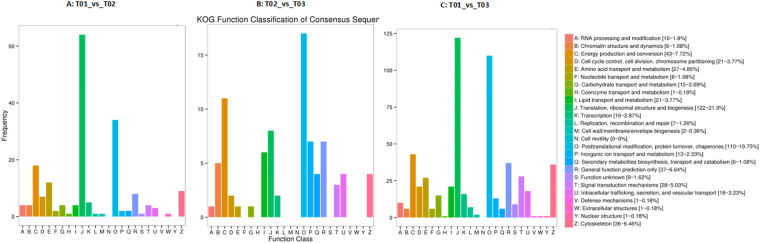
The annotated DEGs were compared with Eukaryotic Orthologous Groups (KOG) database for functional prediction and the putative proteins were classified into 25 KOG groups. In T01_vs_T02, the larger groups included “translation, ribosomal structure and biogenesis”, “posttranslational modification, protein turnover, chaperones”, “energy production and conversion”, “amino acid transport and metabolism” and “cytoskeleton” (Fig. 2). This result was supported by T02_vs_T03 and T01_vs_T03 except “inorganic ion transport and metabolism” and “signal transduction mechanisms” instead of “cytoskeleton”, respectively. GO (Gene Ontology) enrichment on the basis of the biological process (BP), cellular component (CC) and molecular function (MF) showed that most affected GO terms common in the three groups included “translation”, “regulation of transcription, DNA-templated” and “small GTPase mediated signal transduction” in BP, “ribosome” and “small ribosomal subunit” in CC, and “structural constituent of ribosome”, “beta-galactosidase activity” and “phospholipase activity” in MF (Table S3). GO terms only represent in T01_vs_T03 included “regulation of response to biotic stimulus”, “regulation of response to external stimulus” and “xylan metabolic process” in BP, “trans-Golgi network”, “organelle” and “extracellular region” in CC, and “transition metal ion binding” in MF (Table S3). For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, DEGs related to “ribosome”, “nitrogen metabolism” and “citrate cycle” were significantly enriched in both T02_vs_T03 and T01_vs_T03 groups, but “aminoacyl-tRNA biosynthesis”, “fatty acid degradation” and “tryptophan metabolism” were only over-represented in T01_vs_T03 group (P < 0.05, rich factor > 2, Fig. S3).
Figure 2.
Histogram of KOG classification for differentially expressed genes in R2 roots.
Al3+-affected transporter genes in R2 roots
Transporters are required for Al3+-induced secretion of OAs and for Al3+ uptake, sequestration and distribution8. Therefore, we extracted transporter genes whose expression levels were altered by Al3+. In total, 36 transporter genes were found (Table 3, Table S4). Among them, 16 unigenes were up-regulated and 20 unigenes were down-regulated. The upregulated genes belonged to ATP-binding cassette (ABC) transporter family, multidrug and toxic compound extrusion (MATE) family, major facilitator superfamily (MFS), inorganic ion transporters, H+-ATPase, lipid transport and metabolism and others.
Table 3.
Summary of unigenes affected by Al3+.
| Gene category | Total No. | Up-regulated | Down-regulated |
|---|---|---|---|
| Transporter | 36 | 16 | 20 |
| Transcription | 16 | 2 | 14 |
| Translation and posttranslational modification | 4 | 4 | 0 |
| Signal transduction | 37 | 6 | 31 |
Influence of Al3+ on the genes involved in citrate metabolism
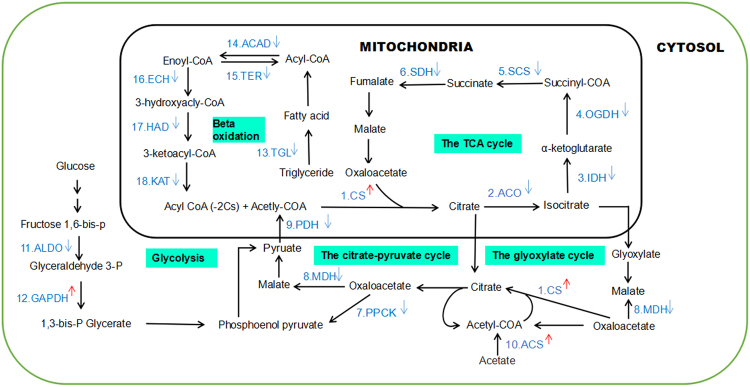
As citrate play an important role in Al3+ tolerance in R2 roots7, we examined the genes associated with citrate metabolism. For genes in tricarboxylic cycle (TCA), CS experienced a more than 4-fold increased expression by Al3+, but transcription levels of ACO, IDH, OGDH, DLST, SCS and SDH were decreased (Fig. 3, Table S5). No change of gene expression in the glyoxylate cycle was observed except elevated transcripts of ACS and CS, which catalyze acetate to acetyl-COA, then to citrate. As regards the genes in glycolysis pathway, GAPDH was up-regulated though ALDO and PPCK were down-regulated. Of interest, decreased expression was observed for genes related to triglyceride metabolism (TGL) and fatty acid beta-oxidation (HCAD, TER, ECH, HAD and KAT), and down-regulation of MDH and PDH in citrate-pyruvate cycle was also detected.
Figure 3.
Expression changes of genes involved in citrate metabolism under Al3+ treatment. 1. CS, citrate synthase; 2. ACO, aconitate hydratase; 3. IDH, isocitrate dehydrogenase; 4. OGDH, 2-oxoglutarate dehydrogenase; 5. SCS, succinyl- CoA synthetase; 6. SDH, succinate dehydrogenase; 7. PPCK, phosphoenolpyruvate carboxykinase; 8. MDH, malate dehydrogenase, cytoplasmic; 9. PDH, pyruvate dehydrogenase; 10. ACS, acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal; 11. ALDO, fructose-bisphosphate aldolase; 12. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; 13. TGL, triacylglycerol lipase; 14. ACAD, acyl-CoA dehydrogenase; 15. TER, trans-2-enoyl-CoA reductase, mitochondrial; 16. ECH, enoyl-CoA hydratase; 17. HAD, 3-hydroxyacyl-CoA dehydrogenase; 18. KAT2, 3-ketoacyl-CoA thiolase 2. Significant difference between treatments is indicated with ↑ (increase in expression) and ↓ (decrease in expression).
Expression changes of genes associated with transcription and translation
Twenty unigenes, mainly encoding transcription factors (9) and translation and posttranslational modification proteins (4), were affected with Al3+ treatment (Table 3, Table S6). Particularly, Al3+ enhanced the expression of transcription factor STOP1 and unigenes involved in translation and posttranslational modification. Previous reports have determined that STOP1/ART1 regulates 32 genes in rice and 43 genes in Arabidopsis2,5. In R2 roots, Al3+ altered 6 STOP1-regulated genes in expression, of which 3 unigenes were down-regulated, and 4 unigenes, which belong to ALS1 homolog, MATE family and DUF642 family, were up-regulated (Table S7).
Expression changes of genes associated with signal transduction
Six unigenes involved in cell signal were increased while 31 unigenes were decreased in expression by Al3+ (Table 3, Table S8). These unigenes were associated with ras family and G-proteins (4), diacyglycerol and inositol metabolism (2), calcium-binding proteins (4), CBL-interacting protein kinases (CIPK) (3), serine/threonine kinases (10), Carboxylesterase (3), phosphatases (4) and others (6).
Validation of RNA-Seq data by qRT-PCR
To validate the results of RNA-Seq, real-time RT-PCRs were performed for 7 DEGs (PIPK5D6, GPA2, PLC, INPS1, DGAT, STOP1, ABCC9) involving the G-protein signaling pathways and citrate secretion (Table S9). A significant correlation (R2 = 0.89, P < 0.05) was observed between the two data sets.
Pharmacological disclosure of Al3+-signaling pathways
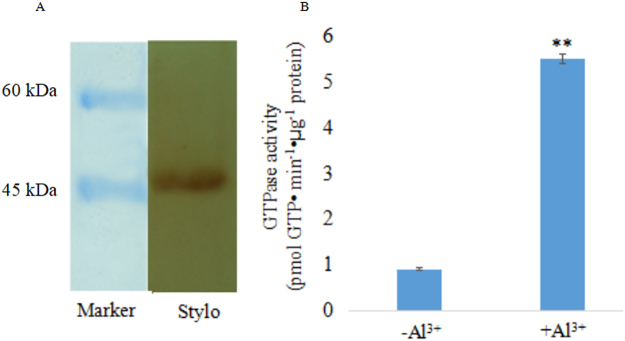
Our early investigation detected the activation of GTPase in response to Al3+ in root plasma membrane of Arabidopsis and rye9. To test the role of heterotrimeric G-proteins in Al3+-induced secretion of citrate in stylo roots, presence of the proteins in the plasma membrane of R2 roots was evaluated (Fig. 4). After exposure to 50 μM AlCl3 (+Al3+) for 3 h, GTPase activity in root plasma membrane was elevated about 5 times compared to that of the control (−Al3+) (P < 0.01, Fig. 4). Citrate secretion from R2 roots was significantly stimulated by G-proteins agonist cholera toxin (CTX, 5–50 ng/mL), whereas the secretion was significantly inhibited by the antagonist pertussis toxin (PTX, 50–200 ng/mL) in a dose-dependent manner (P < 0.05) (Fig. S4A). In the absence of Al3+, citrate secretion from roots was not detectable, regardless of whether the roots were exposed to CTX or not. To support the involvement of G-proteins in Al3+ enhancement of citrate excretion, the downstream effectors were also analyzed pharmacologically. Citrate exudation was significantly induced (P < 0.05) by the specific agonist of IP3 receptor thapsigargin (TSG, 2–10 µM), but was inhibited by the specific antagonsits of phosphoinositide phospholipase C (PLC) (neomycin, 5–20 µM and U73122, 2–8 µM), inositol trisphosphate (IP3) receptor (2-aminoeyhyl diphenylborinate (2-APB), 5–20 µM), diacylglycerol (DAG) (R599499, 2–8 µM) and protein kinase (K-252a, 1–10 µM) (Fig. S4B–S4E). Also, CTX stimulated exudation of citrate under Al3+ treatment was arrested by anion channel antagonsits phenylglyoxal (PG) and anthracene-9-carboxylic acid (A-9-C). After co-treated CTX (20 ng·mL−1) with PG and A-9-C (30 µmol·L−1), secretion rate of citrate dropped to 71.4% (CTX + PG) and 72.0% (CTX + A-9-C), respectively (P < 0.05, Fig. S4F).
Figure 4.
Effect of Al3+ on the GTPase activity in the plasma membrane of R2 roots. (A) Immunoblot analyses for heterotrimeric G-proteins probed with antibody raised against the Arabidopsis Gα. (B) R2 seedlings were exposed to 0.5 mM CaCl2 solution containing 0 (−Al3+) or 50 μM AlCl3 (+Al3+) at pH 4.3 for 3 h. Vertical bars represent Mean ± SEM (n = 3). Pairwise Student’s t test was used to compare values to the control (−Al3+). **Significant at P < 0.01.
Discussion
In the present work, the optimal concentration and duration of Al3+ treatment, at which citrate secretion reached its highest level with slight damage in the roots, was determined for RNA-Seq analysis. This makes it possible to analyse Al3+-tolerance genes in stylo. The high Q-score and no GC and AT separation in clean reads indicate the high accuracy of base calling in RNA-Seq data (Table 1, Fig. S1). Especially, the N50 length of transcripts and unigenes (Table S1) and the number of genes detected with mapped reads tending to stabilize in each sample (Fig. S1) suggest the high integrity of assembly. Thus, the transcriptome assembly provides a rich source for investigating the genes and pathways related to Al3+-tolerance in stylo.
The differential expression analysis identified that most of DEGs were down-regulated in acidic Al3+ treatment (Table 2). These results are consistent with the fact that stylo thrives in acidic soil but not in neutral environment in practice, reflecting the adaptation of stylo to acidic Al3+ soil but being stressed under neutral environment. KOG and GO classification and KEGG pathways of DEGs enriched in acid and Al3+ treatment were involved in regulation of transcription, ribosome, translation and posttranslational modification (Figs 2 and S3). Especially, Al3+ enhanced the expression of transcription factor STOP1, ribosomal protein S30 and 3 other unigenes functioning in posttranslational modification (Table S6). These results lend supports to the observation that the protein-synthesis inhibitor, cycloheximide, significantly inhibits Al3+-induced secretion of citrate7, and emphasize the importance of transcription and translation regulation and protein modification during the activation of Al3+resistance responses in stylo.
Of particular interest, DEGs associated with transporters were over-represented. The importance of membrane-localized MATE transporters and H+-ATPase in mediating OA exudation has been well documented10. We also established the important role of anion-channel or citrate transporters in Al3+-enhancement of citrate secretion in stylo roots using pharmacological methods7. In this work, although only 2 unigenes induced by Al3+ were annotated as MATE transporter family (Table S4), high expression levels of MATE efflux family protein FRD3 and NRAMP family Nramp5 were detected. Al3+-induced expression of another ABC transporter ABCF4, plasma membrane ATPase and acetolactate synthase ALS1 were also observed (Table S4). FRD3 transports citrate into the xylem in Arabidopsis and over-expression of the gene in barley confers citrate efflux in the roots11. The H+-ATPase extrudes protons from cells to create a gradient of electrochemical proton across the plasma membrane. The electrochemical gradient promotes the activity of OA transporters and a passive efflux of organic anions from root tips10. Increasing evidence indicates a significant and positive correlation of plasma membrane H+-ATPase with the rates of citrate exudation under conditions of Al3+ stress and P-deficiency. Internally, a member of NRAMP family, Nrat1, was implicated in cooperating with ALS1 for Al3+ sequestration into vacuole in rice3,4, and internal Al3+ detoxification was mainly responsible for the 72 h Al3+ tolerance of TPRC2001–1 genotype in stylo12. Besides, STAR1, an ABC transporter in rice and Arabidopsis, form a complex with STAR2 to facilitate the export of UDP-glucose for cell wall modification and to mask Al3+-binding sites4. Thus, the expression patterns of above important transporters suggest that stylo harbors multiple strategies for Al3+-detoxification via Al3+ avoidance and tolerance mechanisms as well as modifications in the carbohydrate composition of the root cell wall.
Evidence indicates that Al3+ resistance in common bean depends on the capacity to sustain citrate synthesis for maintaining the cytosolic citrate pool enabling exudation13. Plants over-expressing genes related to citrate biosynthesis, such as CS and MDH, also showed more citrate production and efflux when compared to the untransformed control2. In an Al3+-toxicity environment, the substitution of Fe3+ by Al3+ in organisms leads to a block of TCA and oxidative phosphorylation due to dysfunctional proteins/enzymes that are dependent on Fe3+ to function14. In this study, expression of genes coding for Fe3+-dependent enzymes in TCA cycle, including ACO, IDH, SCS and SDH, were down-regulated (Fig. 3). Researchers documented a modified TCA cycle via oxidation of glyoxylate enabling the soil microbe P. fluorescens and rice exposed to Al3+ to generate ATP15,16. This novel ATP-producing module works in tandem with SCS, isocitrate lyase, acylating glyoxylate dehydrogenase and oxalate CoA-transferase to generate oxalate, a dicarboxylic acid involved in Al3+ immobilization14. By contrast, our data demonstrated no significant difference in expression for the above genes related to glyoxylate oxidation except down-regulation of SCS and up-regulation of CS. Given the decreased expression of MDH and increased expression of ACS, expression patterns of genes in TCA and in glyoxysome lead to an increased synthesis of citrate. The increased citrate anabolism is further supported by decreased expression of genes involved in the citrate-pyruvate cycle, glycolysis and beta oxidation. In consistence with these findings, intracellular citrate concentration increases with Al3+ dosage increasing in stylo roots as detected by our group previously7. Therefore, expression of genes involving citrate metabolism pathways has to be modulated to facilitate citrate synthesis for Al3+ detoxification in stylo roots.
OAs secretion from roots under Al3+ stress is achieved by the combined action of multiple transcription factors that regulate expression levels and tissue specificity17. Multiple transcription factors are required for STOP1/ART1-dependent secretion of malate in Arabidopsis and rice17. Similarly, our work revealed Al3+ activated expression of 2 transcription factor genes including STOP1 homolog (Table S6) and 3 STOP1-regulated genes including MATE and ALS1 (Table S7). In contrast to the finding in Arabidopsis2, the others of STOP1/ART1-regulated genes were not induced by Al3+. These results suggest that transcription regulation of citrate secretion differs from that of malate exudation.
According to the current reports, the signal cascades from Al3+ sensing to transcription activation remain to be resolved. To our knowledge, G-proteins relay the information from G-protein coupled receptors (GPCRs) on the plasma membrane by forming functional subunits of Gα and Gβγ18. Gα signaling is terminated by the intrinsic GTPase activity of Gα, in which ras-related small GTPase superfamily function as switch molecules by cycling between GTP-bound activated state to a GDP-bound inactivated state19,20. In consistence with our previous data in Arabidopsis and rye9, the involvement of G-proteins in Al3+ enhancement of citrate excretion in stylo roots was supported by pharmacological analysis and enhancement of GTPase activity revealed in this work. Importantly, our recent data demonstrated Al3+-induced secretion of oxalate in buckwheat and exudation of malate in wheat without elevation of GTPase activity in the roots of both species (unpublished data). Together with Al3+ decreased 4 of DEGs classed as ras family and G-proteins (Table S8), pharmacological observations point to a pivotal for heterotrimeric G-proteins specific in mediating Al3+-induced release of citrate at translation level.
It has been recognized that second messengers are important components in G-protein-mediated signaling pathways18. When a ligand binds to a GPCR, Gα can bind to and induce activity of PLC, which results in the cleavage of phosphatidylinositol 4, 5-bisphosphate (PIP2) into IP3 and DAG. IP3 then binds to its receptor and initiates intracellular calcium (as Ca2+) release from the ER. In agreement with previous findings in Arabidopsis and rye9, Al3+-enhancement of citrate secretion from stylo roots was significantly reduced by antagonists of PLC, IP3 receptor and DAG, whereas treatment with TSG, a specific activator of PLC, resulted in an increase in citrate release (Fig. S4). The expression patterns of unigenes participating in DAG and IP3 metabolism (Table S8), lend additional supports to functional involvement of PLC, IP3 and DAG in G-protein-medicated Al3+-signaling pathways also at translation level.
Ca2+ is another important component in G-protein-mediated signal in organisms21. In plants, Ca2+-decoding involves calcineurin B-like proteins (CBLs) that bind Ca2+ and activate CIPKs. The CBL-CIPK network targets to the cell membranes where CIPKs phosphorylate the substrate proteins and activate the transport activity21. In these processes, calcium-dependent protein kinases (CPKs) are activated by Ca2+ and lead to the phosphorylation of transcription factors, thereby regulating expression of responding genes including OA transporter genes22. Our previous data established cytoplasmic Ca2+ mediation of Al3+-induced citrate secretion in rye roots23. In this work, Al3+ disrupted expression of 21 Ca2+ signal members (Table S8). Although only 3 unigenes (At1g34300, FERONIA, PTP1) of DEGs were increased (Table S8), Al3+ has been reported to induce either increases or decreases in cytosolic Ca2+ depending on the experimental system studied24, and Al3+ triggers Ca2+ signaling activity at post-transcription, translation and posttranslational levels via protein phosphorylation and dephosphorylation as exemplified by recent reports25,26. Furthermore, the involvement of 16 up-regulated unigenes functional unkown (Table S10) in Ca2+ signal remains to be determined. Collectively, these results indicate that multiple Ca2+ signaling pathways function synergistically in resistance to Al3+ in stylo.
In conclusion, Al3+ tolerance in stylo involves multiple strategies and enhancement of citrate anabolism. Importantly, the Al3+-signaling cascades compose G-proteins, PLC, IP3, DAG, Ca2+ and protein kinases, which lead to the activation of transcription and anion channels in plasma membrane, and result in root secretion of citrate (Fig. 5). However, the Al3+-receptors and the Al3+-activated genes responsible for citrate exudation remain to be determined.
Figure 5.
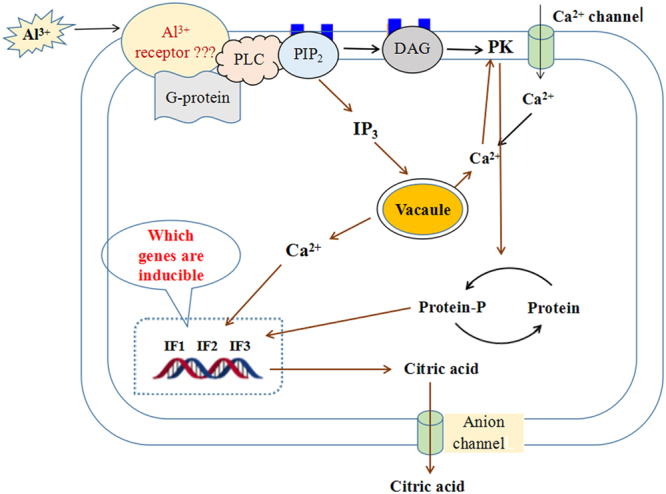
Putative model illustrating signaling cascades of Al3+- induced exudation of citrate from stylo roots. Al3+ interacts with a receptor on the plasma membrane to initiate a signaling pathway. Al3+ signal is transmitted via heterotrimeric G-proteins, inositol trisphosphate (IP3), diacylglycerol (DAG), Ca2+ and protein kinases (PK), leading to the activation of transcription and anion channels in plasma membrane, and resulting in citrate secretion from stylo roots.
Methods
Plant cultivation and measurements of Al3+ treatments
R2 seeds were obtained from Yunnan Academy of Agricultural Sciences (Kunming city, Yunnan province, Southwest China). Cultivation of seeds was performed according to Li et al.7. Seedlings after growing in the dark for 4 d at 25 °C were grown on a floating mesh in plastic containers with full nutrient solution. The nutrient solution described by Li et al.7 was renewed every other day. For Al3+ treatment, the 30-day-old uniform seedlings were used. Seedlings pre-grown overnight in a 0.5-mM CaCl2 solution (pH 4.3) were transferred into a 0.5-mM CaCl2 solution containing 0 (control), 5, 15, 25, 50 and 100 µM AlCl3 (pH 4.3) for 24 h, respectively. Each experiment was repeated for three times. RRG and Chrome Azurol S staining methods were used to measure the effects of Al3+ treatment, and citrate exudation was determined using HPLC as described previously7.
RNA extraction, library construction and sequencing
R2 seedings cultivated as above were subject to treatments of T01, T02 and T03 with three replicates for each treatment and 30 seedlings for each replicate. After 2, 6 and 24 h, root apex (0–1 cm) of 10 seedlings chosen randomly from the same treatment were cut and collected into centrifuge tube, and frozen in liquid nitrogen immediately. Total RNA was extracted using an RNeasy Plant Mini Kit (QIAGEN) and was digested with DNase I (TAKARA). RNA purity, concentration and integrity were checked using Nanodrop 1000 (Thermo Fisher Scientific, CA, USA), Qubit® 2.0 (Life Technologies, CA, USA) and Aglient 2100 (Aglient Technologies, CA, USA), respectively. The quality of the total RNA ranged from RIN 9.6 to 9.9 over the suggested RIN 8, proving to be excellent and stable for RNA-Seq analysis. Total RNA from the three time points of the same treatment were pooled, and a total amount of 3 μg RNA was used mRNA sample preparations. Ploy(A) mRNAs were purified separately from the 3 RNA pools using Oligo (dT), and fragmented. The first strand cDNA was reverse transcribed with random hexamers, followed by second strand cDNAs synthesis with DNA polymerase I (New England BioLabs) and RNase H (Invitrogen). After purification using AMPure XP beads (New England Biolabs), adaptor-ligation and size-selection, the double-stranded cDNA was subjected to PCR-enrichment for 14–16 cycles. The quality of the libraries was checked using Qubit2.0, Agilent 2100 Bioanalyzer and Q-PCR. The cDNA library was subject to paired-end sequencing in HiSeq X Ten system with PE125 by Biomarker Technologies (Beijing, China). The raw sequence data obtained have been deposited at the NCBI in the Short Read Archive (SRA) database under the accession number SRP131721.
Transcriptome assembly and annotation
Raw reads were quality-checked, and adaptor sequences and low quality reads were removed according to the method of Chen et al.10. The obtained clean reads were assembled into unigenes using the Trinity program27, which partitions the sequence data into many individual de Bruijn graphs, and processes each graph independently to extract full-length splicing isoforms and to tease apart transcripts derived from paralogous genes. For annotation, all unigenes, which proved to be over 200 bp, were subjected to BLAST search (E-value < 1e-5) against the databases of NCBI non-redundant protein (NR), Swissprot, GO, KOG, eggNOG, KEGG and Pfam.
Differential expression and enrichment analyses
The clean reads per sample were mapped back to assembled contigs by Bowtie28. Transcripts abundance was calculated and represented by reads per kilobase of transcript per million mapped reads (RPKM) value using the RSEM package29. The assembled transcripts with RPKM ≥ 0.1 were defined as expression.
The assembled transcripts with more than 10 reads mapped were subjected to differential expression analysis between treatments by DESeq30. DEGs were defined when FDR (false discovery rate) < 0.001 and log2FC > 2, and were extracted for GO and KEGG enrichment analysis, which was tested using Fisher’s exact test at a significance cutoff of FDR < 0.05.
Real-time RT-PCR
Total RNA was isloated from R2 seedlings for the RNA-Seq library construction. After DNase I treatment, the RNA samples were subjected to the first-strand cDNA synthesis using HiScript® 1st Strand cDNA Synthesis Kit (Vazyme). One twentieth of the cDNA products and the SYBR® Green Master Mix kit (Vazyme) were used for qRT-PCR analysis with the CFX96 Real-Time PCR Detection System (Bio-Rad) with primers listed in Table S9. Reactions were performed in triplicate for each sample with SgEF-1a as an endogenous control. Relative expression was calculated with the 2−∆∆Ct method.
GTPase activity and pharmacological assays
R2 seedlings aforementioned were exposed to a 0.5-mM CaCl2 solution (pH4.3) containing 50 μM AlCl3 (+Al3+) or not (−Al3+) for 3 h. The details for isolation of plasma membrane by two-phase partitioning protocol (6.2% dextran T-500 and 6.2% polyethylene glycol 3350), measurement of GTPase activity with a spectrofluorometer (LS-55, Perkin Eimer Inc., Liantrisant, UK) and pharmacological analyses referred to Li et al.7.
Data availability
Raw sequence reads can be found in the SRA database under BioProject PRJNA431518. This Transcriptome Assembly project has been deposited SRA database under the accession SRP131721.
Electronic supplementary material
Acknowledgements
The reported work was supported by the National 973 Project of China (2014CB138701), People’s Livelihood Special innovation Projects of CQ CSTC (cstc2017shms - kjfp0114 and cstc2017shms - zdyfx0045), Fundamental Research Funds for the Central University (XDJK2017A003) and the Innovation Team Building Program in Chongqing Universities.
Author Contributions
J.C. analyzed the results and wrote the paper. L.X., L.L., H.R., and W.Y. conducted the experiments and participated in sample assay and data collection. Y.Y. designed the study. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24536-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Caode Jiang, Email: jcdpjx@swu.edu.cn.
Yongxiong Yu, Email: yuyongxiong8@126.com.
References
- 1.Kochian LV, Piñeros MA, Liu J, Magalhaes JV. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015;66:571–598. doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- 2.Sade H, et al. Toxicity and tolerance of aluminum in plants: tailoring plants to suit to acid soils. Biometals. 2016;29:187–210. doi: 10.1007/s10534-016-9910-z. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Piñeros MA, Kochian LV. The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 2014;56:221–230. doi: 10.1111/jipb.12162. [DOI] [PubMed] [Google Scholar]
- 4.Huang CF, Yamaji N, Chen ZC, Ma JF. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012;69:857–867. doi: 10.1111/j.1365-313X.2011.04837.x. [DOI] [PubMed] [Google Scholar]
- 5.Sawaki Y, et al. STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009;150:281–294. doi: 10.1104/pp.108.134700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaji N, et al. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell. 2009;21:3339–3349. doi: 10.1105/tpc.109.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li XF, et al. Secretion of citrate from roots in response to aluminum and low phosphorus stresses in Stylosanthes. Plant Soil. 2009;325:219. doi: 10.1007/s11104-009-9971-7. [DOI] [Google Scholar]
- 8.Ma JF, Ryan PR, Delhaize E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001;6:273–278. doi: 10.1016/S1360-1385(01)01961-6. [DOI] [PubMed] [Google Scholar]
- 9.Li YY, Tang XL, Yang LG, Yu YX, Li XF. Possible involvement of heterotrimeric G-protein signaling in Al-induced secretion of organic acid anions in Arabidopsis and rye. Plant Soil. 2015;388:55–63. doi: 10.1007/s11104-014-2325-0. [DOI] [Google Scholar]
- 10.Zhou G, et al. Enhancing the aluminium tolerance of barley by expressing the citrate transporter genes SbMATE and FRD3. J. Exp. Bot. 2014;5:2381–2390. doi: 10.1093/jxb/eru121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu W, Kan Q, Zhang J, Zeng B, Chen Q. Role of the plasma membrane H(+)-ATPase in the regulation of organic acid exudation under aluminum toxicity and phosphorus deficiency. Plant Signal Behav. 2016;11:e1106660. doi: 10.1080/15592324.2015.1106660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, et al. Superior aluminium (Al) tolerance of Stylosanthes is achieved mainly by malate synthesis through an Al-enhanced malic enzyme, SgME1. New Phytol. 2014;202:209–219. doi: 10.1111/nph.12629. [DOI] [PubMed] [Google Scholar]
- 13.Eticha D, et al. Transcriptomic analysis reveals differential gene expression in response to aluminium in common bean (Phaseolus vulgaris) genotypes. Ann. Bot. 2010;105:1119–1128. doi: 10.1093/aob/mcq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemire J, Mailloux R, Auger C, Whalen D, Appanna VD. Pseudomonas fluorescens orchestrates a fine metabolic-balancing act to counter aluminium toxicity. Environ. Microbiol. 2010;12:1384–90. doi: 10.1111/j.1462-2920.2010.02200.x. [DOI] [PubMed] [Google Scholar]
- 15.Yu L, et al. Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J. Exp. Bot. 2010;61:1625–1634. doi: 10.1093/jxb/erq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, et al. An ATP and oxalate generating variant tricarboxylic acid cycle counters aluminum toxicity in Pseudomonas fluorescens. PLoS One. 2009;4:e7344. doi: 10.1371/journal.pone.0007344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokizawa M, et al. Sensitive to proton rhizotoxicity1, calmodulin binding transcription activator2, and other transcription factors are involved in aluminum-activated malate transporter1 expression. Plant Physiol. 2015;167:991–1003. doi: 10.1104/pp.114.256552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syrovatkina V, Alegre KO, Dey R, Huang XY. Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 2016;428:3850–68. doi: 10.1016/j.jmb.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishida ME, Oguchi M, Fukuda M. Multiple types of guanine nucleotide exchange factors (GEFs) for Rab small GTPases. Cell Struct. Funct. 2016;41:61–79. doi: 10.1247/csf.16008. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, et al. Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell. 2010;22:4031–4044. doi: 10.1105/tpc.110.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang RJ, et al. Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2015;112:3134–3139. doi: 10.1073/pnas.1420944112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto K, et al. Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 2012;287:7956–7968. doi: 10.1074/jbc.M111.279331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang XL, Li XF, Ling GZ, Gu MH, Yu YX. The involvement of Ca2+ signal in the regulation of Al-induced secretion of organic acids in rye. Sci. Agri. Sin. 2008;41:2279–2285. [Google Scholar]
- 24.Liu J, Piñeros MA, Kochian LV. The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant. Biol. 2014;56:221–230. doi: 10.1111/jipb.12162. [DOI] [PubMed] [Google Scholar]
- 25.Chen Q, et al. Phosphorylation and interaction with the 14-3-3 protein of the plasma membrane H+ -ATPase are involved in the regulation of magnesium-mediated increases in aluminum-induced citrate exudation in broad bean (Vicia faba. L) Plant Cell Physiol. 2015;56:1144–1153. doi: 10.1093/pcp/pcv038. [DOI] [PubMed] [Google Scholar]
- 26.Huang W, et al. Hyperforin attenuates aluminum-induced Aβ production and Tau phosphorylation via regulating Akt/GSK-3β signaling pathway in PC12 cells. Biomed Pharmacother. 2017;96:1–6. doi: 10.1016/j.biopha.2017.09.114. [DOI] [PubMed] [Google Scholar]
- 27.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:33. doi: 10.1186/1471-2105-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads can be found in the SRA database under BioProject PRJNA431518. This Transcriptome Assembly project has been deposited SRA database under the accession SRP131721.