Abstract
Plants utilize energy from sunlight to perform photosynthesis in chloroplast, an organelle that could be damaged by solar UV radiation. The ultraviolet-B (UV-B) photoreceptor UVR8 is required for UV-B perception and signal transduction. However, little is known about how UVR8 influence chloroplast development under UV-B radiation. Here, we characterized tomato UVR8 gene (SlUVR8) and our results indicated that SlUVR8 facilitate plant acclimation to UV-B stress by orchestrating expression of the UVB-responsive genes (HY5 and CHS) and accumulating UV-absorptive compounds. In addition, we also discovered that SlUVR8 promotes fruit chloroplast development through enhancing accumulation of transcription factor GOLDEN2-LIKE2 (SlGLK2) which determines chloroplast and chlorophyll levels. Furthermore, UV-B radiation could increase expression of SlGLK2 and its target genes in fruits and leaves. SlUVR8 is required for UVB-induced SlGLK2 expression. Together, our work not only identified the conserved functions of SlUVR8 gene in response to UV-B stress, but also uncovered a novel role that SlUVR8 could boost chloroplast development by accumulating SlGLK2 proteins.
Introduction
Sunlight provides the energy of photosynthesis in sessile plants and also plays an essential role in regulation of their entire life cycle. However, ultraviolet-B (UV-B) light, as an indispensible component of sunlight, can retard plant growth by causing DNA damage, generating reactive oxygen species, and inhibiting photosynthesis1. To survive in sunlight, plants have to evolve the specific mechanisms perceiving and responding to the UV-B radiation2,3.
Recent studies revealed that UV RESISTANCE LOCUS8 (UVR8) protein was responsible for UV-B perception and signal transduction in Arabidopsis1,4–6. The UVR8 gene was first identified in a mutation screen for UV-sensitive plants and the uvr8 mutant was hypersensitive to UV-B radiation7. The abolished UV acclimation of uvr8 mutant is caused by failure of UV-induced expression of defense genes involved in UV damage repairment and UV protection, such as chalcone synthase (CHS) gene which is the committing enzyme for UV-absorptive flavonoid and anthocyanin biosynthesis7,8. Further investigations revealed that the transcription factor ELONGATED HYPOCOTYL5 (HY5) was a fundamental factor of UV signal pathway9–11 and UVR8 regulated HY5 expression through physical association with chromatin in its promoter region8. Besides, UVR8-mediated signal facilitates HY5 and its homolog HYH binding to a T/G-box cis-acting element in the promoters of the UV-responsive genes12.
UVR8 forms homodimers in cytoplasm and their instant monomerization, which requires two tryptophan residues serving as the UV-B chromophore, can be activated by UV-B radiation2,4,6,13. The monomerized UVR8 proteins are translocated from cytoplasm to the nucleus for fulfilling its function and signal transduction14,15. UVR8 protein interacts with multifunctional E3 ligase CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) protein, a key regulator of light signaling, which is also involved in response to UV-B14,16–19. In the dark, COP1 interacts with DAMAGED DNA BINDING PROTEIN 1 (DDB1) and CULLIN4 (CUL4) to form the super complex of CUL4-DDB1-COP1-SPA E3 ligase which inhibits the photomorphogenesis by targeting HY5 for degradation. Under UV-B radiation, COP1-SPA complex disassociates from CUL4-DDB1 and interacts with monomerized UVR820 to form UVR8-COP1-SPA complex which plays a positive role in stabilizing HY5 protein and its activity21. It appears that UVR8 modulates plant response to UV-B through regulating key transcription factor HY5 at both transcriptional and posttranslational level.
DDB1 was first demonstrated to be involved in damaged DNA repair, since it binds to the UV-induced DNA lesions and mediates nucleotide excision repair processes22. In Arabidopsis, DDB1 is associated with CUL4 and additional substrate receptor proteins including COP1 to form CUL4-RING ubiquitin ligase (CRL4) which is required for many cellular processes23–25. In tomato (Solanum lycopersicum), CUL4-DDB1 complex was proved to participate in plastid development and secondary metabolism26–28, epigenetic regulation29, and stress response30–32. The mutant high pigment -1 (hp1), which is caused by a point mutation in tomato DDB1 (SlDDB1) gene, displays the enhanced fruit nutrient contents resulting from the increased plastid (chloroplast) numbers and sizes in fruit cells26. Genetic suppression of tomato CUL4 (SlCUL4) and SlDDB1 genes resulted in the phenocopy of the hp1 mutant27. These studies suggested that CUL-DDB1 complex plays a crucial role in plastid development in tomato. A transcription factor GOLD2-LIKE (SlGLK2), which determines plastid and chlorophyll levels by enhancing photosynthesis gene expression and chloroplast development33–35, is a target of CRL4 ubiquitin E3 ligase36. The degradation of SlGLK2 protein is impaired in the hp1 mutant and SlCUL4 silencing plants28.
Although the homologous genes of Arabidopsis UVR8 were cloned in several plant species including Arabidopsis, Populus euphratica37, apple38, grape berry39, grapevine40, radish sprouts41, and Chlamydomonas reinhardtii42, little is known about characterization of tomato UVR8 (SlUVR8) gene. In this study, we cloned SlUVR8 gene and confirmed its conserved role in response to UV-B radiation. In addition, our results also revealed that SlUVR8 could mediate fruit plastid development under UV-B radiation, possibly through regulating the accumulation of transcription factor SlGLK2.
Results
Cloning of tomato UVR8 gene
We BLAST in tomato (Solanum lycopersicum) genome sequence database (https://sgn.cornell.edu/organism/Solanum_lycopersicum/genome) with Arabidopsis UVR8 protein sequence and got only one positive hit (Solyc05g018620), termed as SlUVR8, which suggested that the tomato genome contained only one homologous gene of UVR8. We cloned the gene by RT-PCR and the encoding protein shared 79% identities with Arabidopsis UVR8 (Supplemental Fig. 1). SlUVR8 also contains multiple repeated RCC1 domains, similar to UVR8 protein in Arabidopsis. The phylogenetic analysis indicated that SlUVR8 shared the best amino acid identity with Solanum tuberosum UVR8 (StUVR8) (Supplemental Fig. 2).
The SlUVR8 gene was expressed constitutively in all the organs of tomato plants we tested. The expression levels in the leaves and flowers were apparently higher than that in other tissues, as indicated by quantitative RT-PCR (qRT-PCR) analysis (Fig. 1A). To check the sub-cellular localization of SlUVR8 protein, the gene of Green Fluorescent Protein (GFP) was fused with SlUVR8 gene and GFP-SlUVR8 construct was transformed in protoplasts of tobacco leaves. The transformed protoplasts were observed by confocal microscope. As shown in Fig. 1B, GFP-SlUVR8 proteins were localized in the nucleus and cytoplasm simultaneously, similar to the sub-cellular localization of Arabidopsis UVR814,15. Western-blot analysis indicated the both GFP and GFP-SlUVR8 were really expressed in the transformed protoplasts (Fig. 1C).
Figure 1.
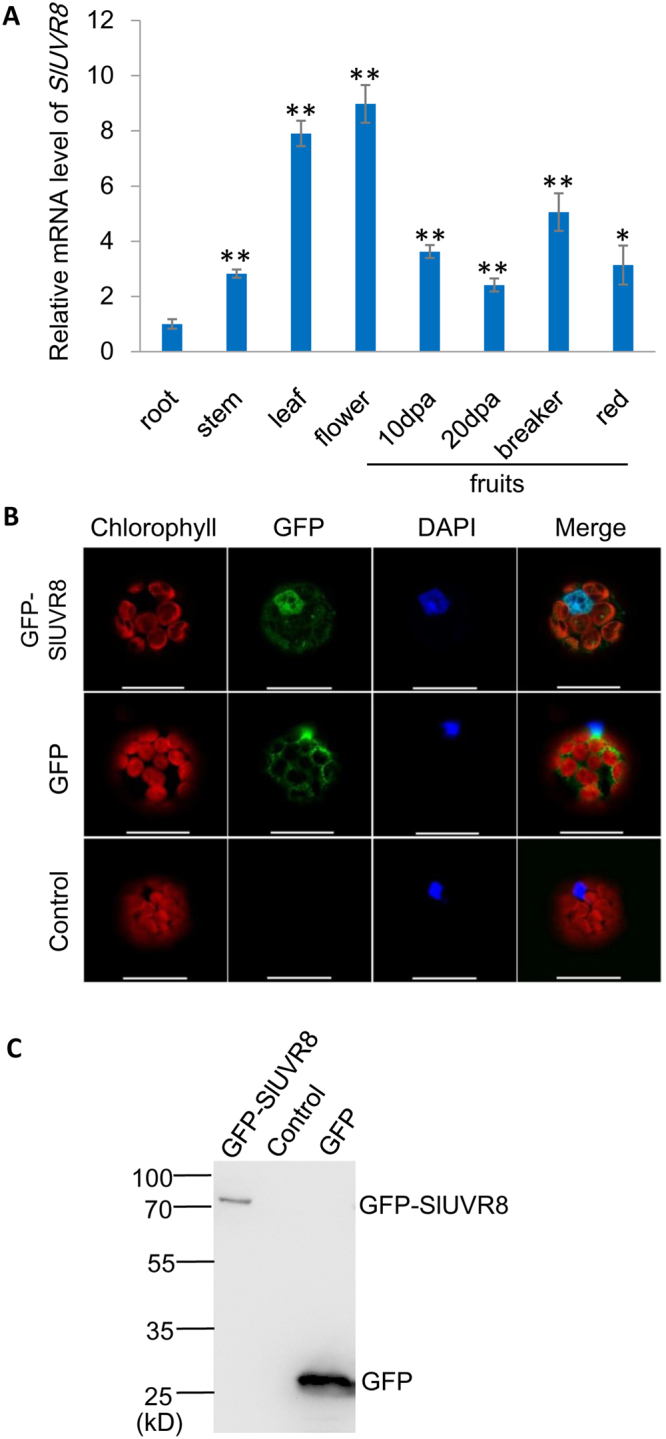
The expression pattern and sub-cellular localization of SlUVR8. (A) Constitutive expression of tomato UVR8 in various tissues. The mRNA levels for tomato UVR8 was analyzed by quantitative RT-PCR. Total RNAs were extracted from roots, stems, leaves, flowers and fruit pericarps at various developmental stages (10, and 20 days post-anthesis, breaker and ripe, respectively). The roots were harvested from plant grown indoor (22–28 °C, 16 h light and 8 h dark) and the rest tissues were harvested from plant grown in the outdoor field. Each bar represents mean value from three biological replicates from each type of tissues (n = 3). Error bars representing standard deviations (SD) are shown in each case. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test). (B) Localization of GFP-UVR8 fusion protein transiently expressed in tobacco protoplasts. Upper panels, GFP-UVR8; middle panels, GFP; bottom panels, an untransformed protoplast as a negative control. Left to right: red, chlorophyll autofluorescence; green, GFP fluorescence; blue, nucleus stained with DAPI; merged, combined fluorescence from GFP, chlorophyll and DAPI. Scale bars = 25 μm. (C) Western-blot analysis of transient expression samples from (B) by using anti-GFP antibody. The positions of protein ladders were marked on the left side of the gel figure.
SlUVR8 is required for tomato acclimation to UV-B radiation
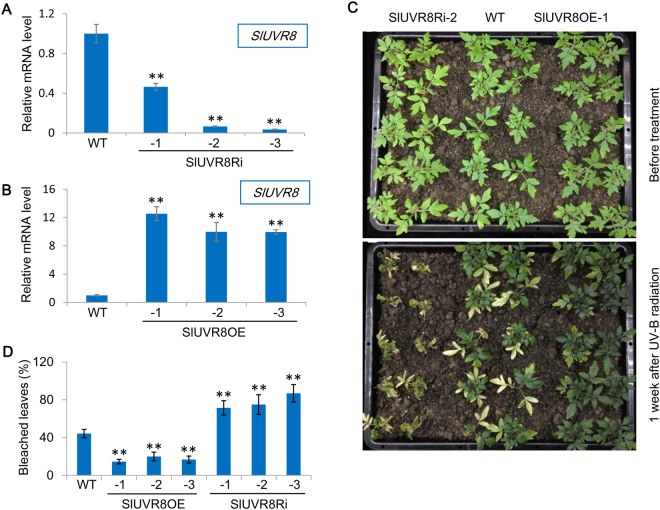
To study the function of SlUVR8-encoded protein and to explore its physiological role in UV-B response, two different kinds of transgenic tomato lines were generated by Agrobacterium tumefaciens-mediated transformation. One was the repression lines by using RNA interference (SlUVR8Ri), expressing SlUVR8-derived inverted-repeat sequences under the direction of the CaMV 35S promoter. The other was over-expression lines of SlUVR8 (SlUVR8OE), also driven by 35S promoter. After screening the T0 generation plants with the quantitative RT-PCR (qRT-PCR) assays of SlUVR8 expression, three independent lines were chosen for SlUVR8Ri (-1, -2, and -3) and SlUVR8OE (-1, -2, and -3), respectively. The homolozygous plants for each line were obtained in T2 generation. As validated by qRT-PCR, SlUVR8 expression were significantly decreased in SlUVR8Ri lines (Fig. 2A) and remarkably increased in SlUVR8OE lines (Fig. 2B) compared to wild type (WT) plants.
Figure 2.
SlUVR8 is required for tomato acclimation to UV radiation. (A) Quantitative RT-PCR analysis of SlUVR8 mRNA levels in young leaves from wild-type Ailsa Craig (WT) and three independent 35S::SlUVR8Ri transgenic lines. Each bar represents mean value from three biological replicates from each line (n = 3). Error bars representing SD are shown in each case. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test). (B) Quantitative RT-PCR analysis of SlUVR8 mRNA levels in young leaves from wild-type Ailsa Craig (WT) and three independent 35S::SlUVR8 transgenic lines. Each bar represents mean value from three biological replicates from each line (n = 3). Error bars representing SD are shown in each case. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test). (C) Upper panels, phenotypes of 4-week-old 35S::SlUVR8Ri-2, Ailsa Craig (WT), and 35S::SlUVR8OE-1 seedlings grown under white light without UV-B. Bottom panels, phenotypes of the seedlings after UV-B radiation for a week. (D) Percentage of bleached leaves of the seedlings after UV-B radiation in (C). For each line, total 50 leaves from 10 different plants were counted (n = 50). “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test).
The growth and development of transgenic plants were indistinguishable from WT plants in absence of UV-B light (Fig. 2C). After additional treatment with UV-B radiation for 1 week, however, SlUVR8Ri plants showed hypersensitive phenotypes including retarded growth, curly and bleached leaves, and even premature cell death (Fig. 2C) while SlUVR8OE plants displayed much less bleached leaves than WT plants (Fig. 2C and D).
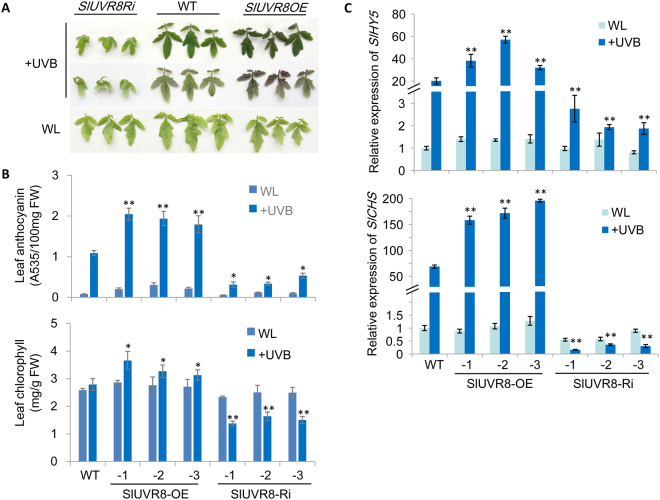
In addition, anthocyanin and chlorophyll accumulation were significiantly reduced in the leaves of SlUVR8Ri plants after UV-B treatment (Fig. 3A and B). However, SlUVR8OE plants enhanced accumulation of anthocyanin and chlorophyll in leaves (Fig. 3A and B). Moreover, qRT-PCR assays indicated that expression of tomato HY5 (SlHY5), as well as CHS (SlCHS) gene, was down-regulated in SlUVR8Ri plants and up-regulated SlUVR8OE plants (Fig. 3C) compared with WT. Together, these results demonstrated that SlUVR8 was required for tomato acclimation to UV-B radiation. Over-expression of SlUVR8 could increase plant tolerance to UV-B stress by up-regulating SlHY5 expression and enhancing anthocyanin accumulation.
Figure 3.
SlUVR8 mediates UV-induced gene expression and anthocyanin accumulation. (A) Leaves detached from 30-d-old 35S::SlUVR8Ri, Ailsa Craig (WT), and 35S::SlUVR8OE seedlings grown under white light (photoperiod: 16 h light and 8 h dark) with or without 3-days UV-B radiation (12 h/day). (B) Quantitative measurement of anthocyanin and chlorophyll content in the fully expanded leaves in (A). Values were shown as “means ± SD”; Error bars represent SD of ten biological replicates. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test). (C) The quantitative RT-PCR analysis of UVB-induced genes SlHY5 and SlCHS of the seedlings in (A). Values were shown as “means ± SD”; Each bar represents mean value from three biological replicates from each line (n = 3). “**” means P < 0.001 (Student’s t test).
SlUVR8 affects chloroplast development and nutrient quality of tomato fruits
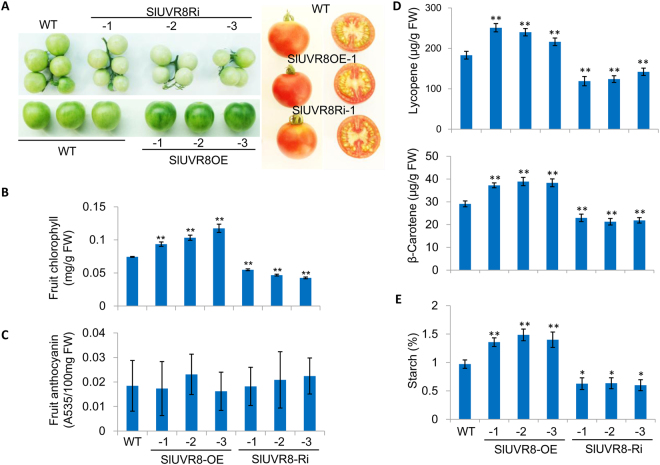
We grew the transgenic lines and WT plants in the outdoor fields, as described in “Materials and Methods”, with exposure to the natural sunlight. The SlUVR8Ri plants displayed normal growth without any phenotypes of hypersensitivity to UV radiation (data not shown). However, the SlUVR8Ri plants had pale green immature fruits (Fig. 4A). In contrast, SlUVR8OE plants had darker green fruits compared to WT plants (Fig. 4A). Accordingly, ripe fruits expressing SlUVR8 had a 20% increase in chlorophyll content (Fig. 4B) and a 25% increase in carotenoids (Lycopene and β-Carotene) contents (Fig. 4D), as compared with WT fruits. In addition, starch accumulation were boosted in fruits of SlUVR8OE plants but reduced in SlUVR8Ri plants (Fig. 4E). However, the anthocyanin contents in fruits were very low and indistinguishable between WT and transgenic fruits (Fig. 4C). The previous studies revealed that low accumulation of flavonoids (including anthocyanins) in tomato fruits was caused by low expression of chalcone isomerase (CHI) gene43 and other genes44 required for anthocyanin biosynthesis. Our results indicated that overexpression of SlUVR8 had no apparent effect on anthocyanin accumulation in tomato fruits, although SlHY5 expression was also up-regulated in fruits of SlUVR8OE plants (Supplemental Fig. 3).
Figure 4.
SlUVR8 affects tomato fruit pigment and nutrient quality. (A) Phenotypes of immature fruits (25 DPA) and red ripe fruits from field-grown plants of wild-type Ailsa Craig (WT), 35S::SlUVR8Ri lines and 35S::SlUVR8OE lines. (B) Total chlorophyll levels from immature green fruit in (A). Values were shown as “means ± SD”; Error bars represent SD of 10 biological replicates. “**” means P < 0.001 (Student’s t test). (C) Lycopene, β-carotene, and starch levels in Ailsa Craig (WT), 35S::SlUVR8Ri and 35S::SlUVR8OE fruit. Lycopene and β-carotene levels were measured in red ripe fruits. Starch was measured in immature green fruits. Values were shown as “means ± SD”; Error bars represent SD of 10 biological replicates. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test).
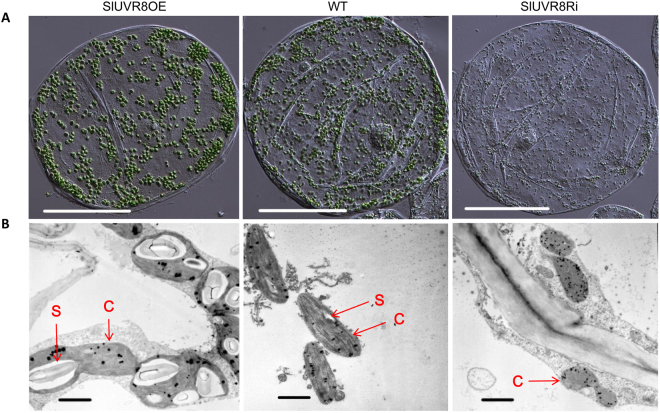
Meanwhile, we investigated chloroplast compartment sizes in pericarp cells of mature green fruits from WT and transgenic plants. The results indicated that plastid number per cell showed no substantial difference between WT and transgenic plants (Table 1), but plastid size (plastid plan area) was altered significantly (Table 1 and Fig. 5A). Larger chloroplasts were developed in fruit pericarp cells of SlUVR8OE plants while smaller chloroplast were displayed in SlUVR8Ri plants, as observed by optical microscope (Fig. 5A) and transmission electron microscope (Fig. 5B). The quantitative measurements confirmed our observations (Table 1). These results revealed that SlUVR8 could influence chloroplast size but not chloroplast number in tomato fruits. In addition, bigger starch grains were observed in chloroplasts from SlUVR8OE fruits while SlUVR8Ri fruits showed on obvious starch grains, as indicated by note “S” in Fig. 5B, in chloroplasts. This observation was consistent with our previous quantitative measurements of total starch content (Fig. 4D). It was noteworthy that we observed much less thylakoid stacks (grana) in the chloroplasts of SlUVR8Ri plants than in WT and SlUVR8OE plants (Fig. 5B), which implied that thylakoid membrane or photosynthesis apparatus in SlUVR8Ri plants might be damaged by solar UV radiation.
Table 1.
Cell and plastid characteristics of fruit pericarp cells from tomato cv. Ailsa Craig (WT), SlUVR8OE and SlUVR8Ri transgenic plants. Values are means ± SE. Cell index is calculated as total plastid area per cell plan area. Immature green fruit were studied at 25d post-anthesis.
| Cell plan area (μm2, n = 30) | Plastid number per cell (n = 30) |
Total plastid area per cell | Plastid density (per μm2 cell plan area, n = 30) |
Plastid plan area (μm2, n > 1000) |
Cell index |
|
|---|---|---|---|---|---|---|
| WT | 57579 ± 3707 | 817 ± 58 | 10348 ± 811 | 0.014 | 12.7 ± 0.8 | 0.180 |
| UVR8-OE1 | 56494 ± 5344 | 859 ± 65 | 13590 ± 799 | 0.015 | 15.8 ± 1.1 | 0.241 |
| UVR8-OE2 | 58665 ± 4740 | 928 ± 67 | 13668 ± 785 | 0.016 | 14.8 ± 1.3 | 0.234 |
| UVR8-OE3 | 55966 ± 4265 | 846 ± 50 | 12390 ± 829 | 0.015 | 14.7 ± 0.8 | 0.222 |
| UVR8-Ri1 | 54370 ± 4505 | 942 ± 75 | 8368 ± 690 | 0.017 | 8.9 ± 0.6 | 0.154 |
| UVR8-Ri2 | 56956 ± 5406 | 885 ± 83 | 7459 ± 789 | 0.016 | 8.4 ± 0.9 | 0.131 |
| UVR8-Ri3 | 59592 ± 4893 | 872 ± 68 | 8782 ± 671 | 0.015 | 10.1 ± 0.9 | 0.148 |
Figure 5.
SlUVR8 affects chloroplast development in fruit cells. (A) Isolated pericarp cells from immature green tomato fruits (25 DPA) of SlUVR8OE, wild-type (WT) and SlUVR8Ri plants grown in the outdoor field. Bars = 50 μm. (B) Transmission electron microscopy images of immature green fruit (25 DPA) chloroplasts from SlUVR8OE, wild-type (WT) and SlUVR8Ri plants grown in the outdoor field. Bars = 1 μm. Red “C” indicated Chloroplasts, Red “S” indicated Starch in chloroplast.
SlUVR8 enhances the accumulation of transcriptional factor SlGLK2 under UV-B radiation
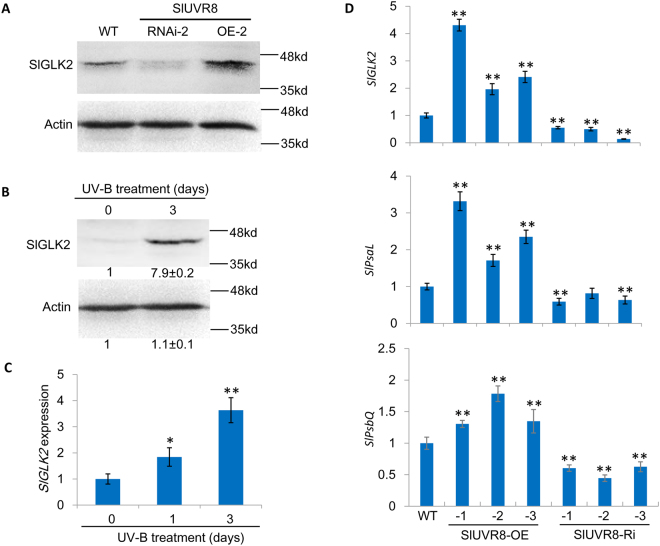
Tomato GOLDEN2-LIKE (GLK) transcription factor SlGLK2 determines chlorophyll accumulation and chloroplast development in fruits through enhancing photosynthesis gene expression28,33,34. Therefore, we checked the SlGLK2 protein abundance in the fruits of transgenic plants grown under sunlight in open fields by using a SlGLK2-specific antibody which doesn’t recognize SlGLK128. As shown in Fig. 6A, SlGLK2 proteins were over-accumulated in SlUVR8OE plants but less accumulated in SlUVR8Ri plants grown under sunlight. In addition, UV-B radiation could enhance the accumulation of SlGLK2 in fruits of WT plants grown in chamber with white light (Fig. 6B). These results indicated that SlUVR8 might affect chloroplast development through targeting transcription factor SlGLK2. Next, the qRT-PCR assays indicated that UV-B could increase the mRNA levels of SlGLK2 gene in WT fruits (Fig. 6C). It was noteworthy that UV-B radiation could increase SlGLK2 protein abundance by almost 7 times while mRNA level of SlGLK2 gene was increased by only 3 times (Fig. 6B and C). These results suggested that there possibly was a post-translational regulation in UV-B enhanced SlGLK2 accumulation. Also, SlGLK2 expression was increased in fruits of SlUVR8OE plants but decreased in fruits of SlUVR8Ri plants (Fig. 6D). We also randomly selected two SlGLK2-targeted genes (SlPsaL and SlPsbQ), which encode photosystem I subunit L and photosystem II subunit Q respectively, and checked their expression in fruits of transgenic plants (Fig. 6D). The results indicated that over-expression of SlUVR8 could increase the expression of transcriptional factor SlGLK2 and its target genes, while silencing of SlUVR8 led to suppression of SlGLK2 and its target genes in fruits, when plants were exposed to the sunlight.
Figure 6.
SlUVR8 enhances SlGLK2 accumulation under UV radiation. (A) Immunoblot analysis of SlGLK2 levels among Ailsa Craig (WT), 35S::SlUVR8Ri and 35S::SlUVR8OE transgenic lines. The proteins extracted from immature fruits (25 DPA) from field-grown plants was resolved by SDS-PAGE, then probed with anti-SlGLK2 antibody and anti-β-actin antibodies. (B) Immunoblot analysis of SlGLK2 levels between Micro Tom (WT) plant grown in white light and plant grown in white light with added 3 days of UV-B irradiation. The proteins extracted from immature fruits (15 DPA) was resolved by SDS-PAGE, then probed with anti-SlGLK2 antibody and anti-β-actin antibodies. Blots were quantitatively analyzed by software ImageJ 1.46r. Values were shown as “means ± SD” from results of three independent experiments. (C) Quantitative RT-PCR analysis of SlGLK2 mRNA levels in immature fruits (15 DPA) from Micro Tom (WT) with different UV-B irradiation time (0 d, 1 d and 3 d respectively). Values were shown as “means ± SD”; Error bars represent SD of 3 biological replicates. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test). (D) Quantitative RT-PCR analysis of chloroplast development related genes SlGLK2, SlPsaL and SlPsbQ in Ailsa Craig (WT), 35S::SlUVR8Ri and 35S::SlUVR8OE transgenic lines. Total RNAs were extracted from fruit pericarps of immature fruits (25 DPA) from field-grown plants. Values were shown as “means ± SD”; Error bars represent SD of 3 biological replicates. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test).
Since SlGLK2 was also expressed in leaves33,34, we also checked its expression in leaves of transgenic plants with or without UV-B treatment. The qRT-PCR assays indicated that, when plants were cultured in indoor chamber with white light, SlUVR8 had no apparent effect on expression of SlGLK2 and its target genes (Fig. 7A). However, after UV-B treatment, SlGLK2 expression was significantly increased in WT and SlUVR8OE leaves but not in SlUVR8Ri leaves. In addition, the expression of the target gene SlPsaL and SlPsbQ showed no obvious difference under white light but was suppressed severely after UV-B treatment in the leaves of SlUVR8Ri plants compared with WT and SlUVR8OE plants (Fig. 7A). These results confirmed that SlUVR8 was required for UVB-enhanced expression of SlGLK2 and its target genes. We also checked the expression of SlGLK1 gene which was expressed only in leaves but not in fruits33,34. The qRT-PCR analysis indicated that UV-B radiation, as well as SlUVR8 expression, had no significant effect SlGLK1 expression in leaves (Fig. 7B).
Figure 7.
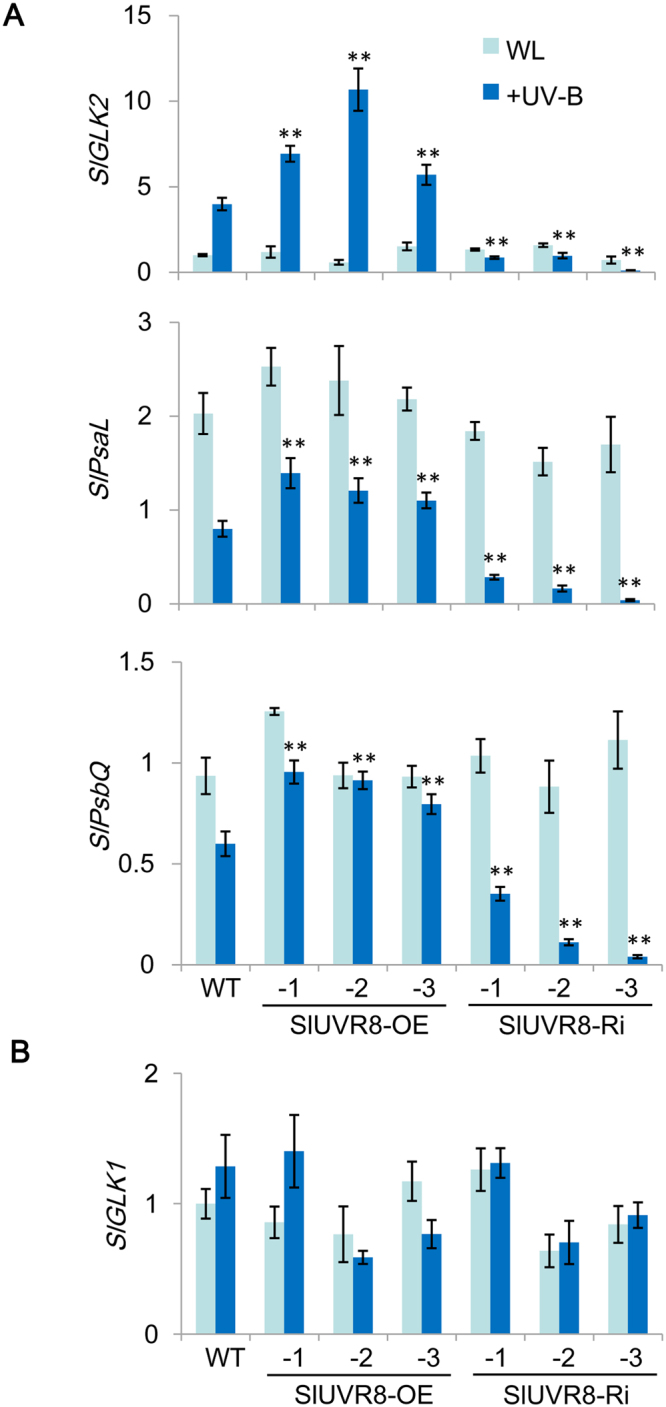
SlUVR8 is required for UV-induced SlGLK2 expression. (A) Quantitative RT-PCR analysis of chloroplast development related genes SlGLK2 and its target genes (SlPsaL and SlPsbQ) in Ailsa Craig (WT), 35S::SlUVR8OE and 35S::SlUVR8Ri transgenic lines with or without 3-d UV-B irradiation. Total RNAs were extracted from totally expanded leaves of 30-d old seedlings grown in white light. Values were shown as “means ± SD”; Error bars represent SD of 3 biological replicates. “*” and “**” means P < 0.05 and P < 0.001 respectively (Student’s t test). (B) Quantitative RT-PCR analysis of SlGLK1 gene in the same samples as described in (A). “**” means P < 0.001 (Student’s t test).
Discussion
As UV-B photoreceptor, UVR8 gene has been characterized in several plant species including Arabidopsis, Populus euphratica37, apple38, grape berry39, grapevine40 and radish sprouts41, and in Chlamydomonas reinhardtii42. Here, we isolated tomato SlUVR8 gene and identified its functions in plant’s adaptation to UV-B radiation. Transgenic evidences suggested overexpression of SlUVR8 increased plant tolerance whereas silencing SlUVR8 gene led to the hypersensitivity to UV-B stress (Fig. 2). In accordance with the previous studies in Arabidopsis, UV-B-induced HY5 and CHS expression, as well as anthocyanin accumulation, were impaired in SlUVR8Ri plants but were enhanced in SlUVR8OE plants compared to WT (Fig. 3). These results demonstrated that SlUVR8 played an essential role of orchestrating expression of key UV-responsive genes (such as SlHY5 and SlCHS) and accumulating UV-absorptive compounds with sunscreen functions of protecting plants from UV damage.
Sunlight provides energy for photosynthesis of chloroplast. However, solar UV radiation could damage photosynthesis and chloroplasts as well45. Numerous studies revealed that photosynthetic apparatus was susceptible to damage caused by UV-B46–50. The adverse effects from enhanced UV-B radiation on chloroplasts include but not limited to loss in integrity of the thylakoid membranes51, impaired activity of photosystem II (PSII)52,53, enhanced protein degradation50, inhibited carbon fixation54 and reduced content of starch and chlorophyll53,55. Therefore, plants must employ some strategies or mechanisms, some of which remain to be elucidated yet, to respond to these problems and to maintain photosynthesis and chloroplast functions. A few studies suggested that UVR8 is required for maintaining the photosynthesis efficiency under enhanced UV-B radiation in Arabidopsis49,50. The underlying mechanism, however, has not been revealed yet. In our work, we proved that SlUVR8 promote chloroplast development in tomato fruits when plants were grown in outdoor fields and exposed to natural sunlight (Fig. 4). Silencing SlUVR8 caused some developmental issues in fruits including reduced size of chloroplasts and decreased contents of chlorophyll and starch, while overexpresssion of SlUVR8 led to larger chloroplasts and increased contents of chlorophyll and starch (Fig. 5). The previous studies revealed that low accumulation of flavonoids (including anthocyanins) in tomato fruits resulted from low expression of chalcone isomerase (CHI) gene43 and other genes44 required for anthocyanin biosynthesis. Our results also showed that the anthocyanin contents in fruits were very low and indistinguishable between WT and transgenic fruits (Fig. 4C). It is conceivable that deficiency of anthocyanins was not responsible for chloroplast abnormality in fruit cells from SlUVR8Ri plants. In our case, SlUVR8 might play another role in affecting chloroplast development of tomato fruits rather than its role in accumulating UV-absorbing compounds. Therefore, we speculated that SlUVR8 could enhance chloroplast development, facilitate the recycling of damaged chloroplasts, and thereby contribute to maintain photosynthesis under UV-B stress.
In tomato, silencing SlHY5 also caused abnormalities in both organization and abundance of thylakoids26. However, no reports yet showed that overexpression of SlHY5 could lead to larger chloroplasts and high pigment of fruits in tomato. Therefore, we surmise that other genes are also involved in the UVR8-mediated chloroplast development. Our results disclosed that UV-B could enhance accumulation of SlGLK2, which was a key regulator of fruit chloroplast development33,34, and the UVB-enhanced SlGLK2 accumulation was dependent on SlUVR8 gene. Several studies demonstrated that SlGLK2 influenced chloroplast development and chlorophyll level by increasing fruit photosynthesis and chloroplast gene expression33,34. The accumulation of carbohydrates and carotenoids were elevated in fruits of SlGLK2 overexpression plants. Our results showed that more chlorophyll, starch, and carotenoids were accumulated in fruits of SlUVR8OE plants (Fig. 4), similar to the phenotypes of fruits expressing SlGLK2. In addition, our results indicated that SlGLK2 proteins were over-accumulated in SlUVR8OE fruits but less-accumulated in SlUVR8Ri fruits under sunlight (Fig. 6). Further studies revealed that expression of SlGLK2 and its randomly-selected target genes were elevated in SlUVR8OE plants under UV-B radiation (Figs 6 and 7). Silencing SlUVR8 abolish UVB-induced expression of SlGLK2 and its target gene (Figs 6 and 7). These results suggested that SlUVR8 gene is required for UVB-enhanced SlGLK2 expression. Therefore, we proposed an unknown pathway that photoreceptor UVR8 promote chloroplast development in tomato under UV-B stress by enhancing accumulation of SlGLK2. Of course, we can’t exclude the role of SlHY5 in UV-mediated chloroplast development26. A study about the root greening in Arabidopsis56 showed that GLK2 could induce the accumulation of HY5 and combination of GLK2 and HY5 mediated the coordinated expression of many key genes of chloroplast biogenesis. This study might shed some light on the mechanism that both SlHY5 and SlGLK2 are possibly required for UVB-mediated chloroplast development. The previous study showed that expression of GLK2 was also responsive to photooxidative chloroplast damage and retrograde signals from plastid in Arabidopsis57, indicating that GLK2 might play an additional role in adjusting photosynthetic capacity in changing environmental conditions. It will be interesting to check whether silencing SlGLK2 alone could affect UVB-mediated chloroplast development and plant response to UV-B stress in further investigations.
In Arabidopsis, transcription factor HY5 is regulated by UVR8 at both transcriptional and post-translational levels8,17,21. Here, our immunoblot analysis showed that UV-B radiation could increase SlGLK2 protein abundance by about 7 times while mRNA level of SlGLK2 gene was increased by only 3 times, as compared with untreated condition (Fig. 6B and C). These results suggested that a post-translational modification, which requires further studies, might exist in this case. In Arabidopsis, UVR8 were associated with chromatin in the region of the HY5 promoter and regulate HY5 transcription8,58,59. However, a recent study60 indicated that UVR8 didn’t bind to chromatin in vivo or in vitro, possibly because of lacking critical histone- and DNA-interaction residues. Therefore, the exact mechanism of UVR8 regulating transcription remains controversial and elusive. In our study, there is no evidence yet that SlUVR8 interacts with the promoter of SlGLK2 gene directly. Therefore, we can’t exclude the possibility that SlUVR8 might regulate SlGLK2 gene expression through another transcription factor or proteins. Further investigations are needed to validate our speculations and figure out the mysteries.
Together, we revealed that SlUVR8 played a conserved role in plant acclimate to UV-B stress. Besides, our results indicated that SlUVR8 mediated the fruit chloroplast development by enhancing accumulation of transcription factor SlGLK2. Moreover, manipulation of SlUVR8 level represents a new option to enhance both tolerance to UV-B stress and nutrient values of tomato fruits, especially when plants were grown in outdoor fields with intense sunlight.
Materials and Methods
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum) cv Ailsa Craig (LA2838A) were acquired from the Tomato Genetics Resource Center (UC Davis, USA). Tomato plants were grown under artificial conditions (22–28 °C, 16 h light and 8 h dark) for several weeks and then transplanted into the outdoor field (located at the suburbs of Chengdu, China) for 4 months (from May to August). During this time, the maximum value of UV-B irradiance at noon (1 p.m.) is 9.5 μmol/m2/s and the minimum value is 4.3 μmol/m2/s, as measured by TBQ-ZW-2 UV-B (280–320 nm) light meter (Shanghai Minyin Electrics co., Ltd, China). For photomorphogenic UV-B treatment, seedlings were grown under white light (photoperiod: 16 h light and 8 h dark) supplemented with Philips TL20W/01RS narrowband UV-B tubes (23 μmol/m2/s) for 12 hours per day.
Generation of transgenic tomato
The Agrobacterium EHA105 strains containing vectors pBI121-SlUVR8 or pBI121-SlUVR8-RNAi were used to transform tomato cv Ailsa Craig by using Agrobacterium tumefaciens-mediated transformation61. Primary transformants (T0) and their offspring were cultivated under the artificial conditions for 4 weeks and then transplanted into the outdoor field (located at the suburbs of Chengdu, China). The T-DNA insertions of transgenic plants were identified by PCR using NPTII-specific primers and the expression of SlUVR8 gene was validated by real-time RT-PCR using SlUVR8-specific primers. Three independent transgenic lines were used for phenotypic and molecular analysis.
Quantitative RT-PCR analysis
Total RNAs were isolated using Trizol reagent (Invitrogen). The genomic DNAs in total RNAs were erased and first-strand cDNAs were synthesized with oligo-dT(18T) primers by using cDNA Synthesis kit, according to the manufacturer’s protocol (TransGen, Beijing, China). Quantitative PCR was conducted on the ABI StepOnePlus PCR System by using the TransStart Green qPCR SuperMix (TransGen). Gene expression was normalized by the expression of UBI3 gene. All the primers used are listed in Table S1.
Subcellular localization analysis
The coding sequence of the SlUVR8 gene was cloned into the expression vector pART27-mcs:GFP and generated the pART27- GFP-SlUVR8 construct. Plasmid vectors pART27-GFP-SlUVR8 and pART27-GFP were introduced into protoplasts of tobacco (Nicotiana benthamiana) according to the methods described previously62,63. After incubation at 25 °C for 12–16 h, protoplasts were observed under a Leica TCS SPII confocal microscope using 488 and 633 nm excitation wavelengths and three-channel measurement of emission: 435 nm (blue/DAPI), 522 nm (green/GFP) and 680 nm (red/chlorophyll).
Protein extraction and immunoblot analysis
Tissue was ground in liquid N2 and was treated in boil water for 5 min with SDS-PAGE loading buffer to extract total proteins. Immunoblot analysis was performed as described previously64. The SlGLK2-specific antibodies recognized the SlGLK2 N-terminal unique sequence (synthetic peptide SSSLSYKNERENYD, 5–18), but not SlGLK1 protein, as described in the previous study28. The secondary antibody Goat-anti-rabbit IgG conjugated to horseradish peroxidase (HRP) were purchased from Hangzhou HuaAn Biotechnology Co., Ltd.
Plastid Analysis and Transmission Electron Microscopy
The fruit tissues used for plastid analysis were harvested from plants grown in outside open fields and processed as previously reported65. In brief, the fruit pericarp tissues were immersed in 3.5% glutaraldehyde for 1 hour at least, then incubated in 0.1 M Na2-EDTA for 30 min at 60 °C. The sliced and smashed samples were imaged with a Leica DM2500 microscope, and cell plan area was measured by Image-Pro Plus. For transmission electron microscopy, outer pericarp tissues were prefixed in 3% glutaraldehyde in 0.1 M phosphate buffer at 4 °C. Tissues were post fixed in 1% osmium tetroxide, dehydrated in series acetone, infiltrated in Epox 812 for a longer, and embedded. The semithin sections were stained with methylene blue and ultrathin sections were cut with diamond knife, stained with uranyl acetate and lead citrate. Sections were examined with a Transmission Electron Microscope (TEM; HITACHI, H-600IV, Japan).
Anthocyanin, chlorophyll and carotenoid assays
Anthocyanin was assayed according to the procedures described previously66. chlorophyll and green fruits’ total carotenoid was extracted into 80% acetone, and their content was calculated by using Lichtenthaler’s formulas, in which chlorophyll a equals 12.21A663-2.81A645, chlorophyll b equals 20.13A645-5.03A663, total carotenoids equals (1000 A470-3.27Ca-104Cb)/229. The red fruits’ carotenoid contents were measured by HPLC as previously described27, with analytical reagent lycopene and β-carotene as standards.
Starch Analysis
Starch quantification was determined using a starch assay kit (STA20; Sigma-Aldrich) following the manufacturer’s protocol34.
Electronic supplementary material
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 31400265), the Hundred Talents Program (Chinese Academy of Sciences), the National Natural Science Foundation of China (Grant Nos. 31701922, 31671259, 31171179 and 90717110), National Science Fund for Distinguished Young Scholars (Grant No. 30825030), the 973 Program of China (Grant No. 2011CB100401), and Advanced Program of Doctoral Fund of Ministry of Education of China (20110181130009).
Author Contributions
H.L., S.W. and Y.L. designed the experiments. H.L., Y.L., H.D., X.S., A.W., X.T., Y.G., N.Z., L.W. and S.Y. conducted the experiments. H.L. and S.W. analyzed the data. S.W., Y.L. and H.L. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24309-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongsheng Liu, Email: liu_ys@scu.edu.cn.
Songhu Wang, Email: wangsh1@cib.ac.cn.
References
- 1.Tilbrook K, et al. The Arabidopsis book. 2013. The UVR8 UV-B Photoreceptor: Perception, Signaling and Response; p. e0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenkins GI. Structure and function of the UV-B photoreceptor UVR8. Current opinion in structural biology. 2014;29:52–57. doi: 10.1016/j.sbi.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins GI. The UV-B photoreceptor UVR8: from structure to physiology. The Plant cell. 2014;26:21–37. doi: 10.1105/tpc.113.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 5.Heijde M, Ulm R. UV-B photoreceptor-mediated signalling in plants. Trends in plant science. 2012;17:230–237. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant physiology. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown BA, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binkert M, et al. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. The Plant cell. 2014;26:4200–4213. doi: 10.1105/tpc.114.130716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stracke R, et al. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant, cell & environment. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- 12.Binkert M, et al. UV-B-Responsive Association of the Arabidopsis bZIP Transcription Factor ELONGATED HYPOCOTYL5 with Target Genes, Including Its Own Promoter. The Plant cell. 2014;26:4200–4213. doi: 10.1105/tpc.114.130716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 14.Yin R, Skvortsova MY, Loubery S, Ulm R. COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E4415–4422. doi: 10.1073/pnas.1607074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiserli E, Jenkins GIUV-B. promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. The Plant cell. 2007;19:2662–2673. doi: 10.1105/tpc.107.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oravecz A, et al. Constitutively Photomorphogenic1 is required for the UV-B response in Arabidopsis. The Plant cell. 2006;18:1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. The EMBO journal. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safrany J, et al. Identification of a novel cis-regulatory element for UV-B-induced transcription in Arabidopsis. The Plant journal: for cell and molecular biology. 2008;54:402–414. doi: 10.1111/j.1365-313X.2008.03435.x. [DOI] [PubMed] [Google Scholar]
- 19.Gruber H, et al. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20132–20137. doi: 10.1073/pnas.0914532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin R, Arongaus AB, Binkert M, Ulm R. Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. The Plant cell. 2015;27:202–213. doi: 10.1105/tpc.114.133868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, et al. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16669–16674. doi: 10.1073/pnas.1316622110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu G, Chang E. Xeroderma Pigmentosum Group-E Cells Lack a Nuclear Factor That Binds to Damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 23.Seo KI, et al. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. The Plant cell. 2014;26:695–711. doi: 10.1105/tpc.113.119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. The Plant cell. 2008;20:1437–1455. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernhardt A, et al. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 2006;47:591–603. doi: 10.1111/j.1365-313X.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- 26.Liu YS, et al. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9897–9902. doi: 10.1073/pnas.0400935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, et al. Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. The Plant journal: for cell and molecular biology. 2008;55:89–103. doi: 10.1111/j.1365-313X.2008.03489.x. [DOI] [PubMed] [Google Scholar]
- 28.Tang X, et al. Ubiquitin-conjugated degradation of golden 2-like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato. The New phytologist. 2016;209:1028–1039. doi: 10.1111/nph.13635. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, et al. A role of tomato UV-damaged DNA binding protein 1 (DDB1) in organ size control via an epigenetic manner. PloS one. 2012;7:e42621. doi: 10.1371/journal.pone.0042621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao L, et al. The tomato DDI2, a PCNA ortholog, associating with DDB1-CUL4 complex is required for UV-damaged DNA repair and plant tolerance to UV stress. Plant science: an international journal of experimental plant biology. 2015;235:101–110. doi: 10.1016/j.plantsci.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Miao M, et al. The tomato DWD motif-containing protein DDI1 interacts with the CUL4-DDB1-based ubiquitin ligase and plays a pivotal role in abiotic stress responses. Biochemical and biophysical research communications. 2014;450:1439–1445. doi: 10.1016/j.bbrc.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, et al. The tomato UV-damaged DNA-binding protein-1 (DDB1) is implicated in pathogenesis-related (PR) gene expression and resistance to Agrobacterium tumefaciens. Molecular plant pathology. 2012;13:123–134. doi: 10.1111/j.1364-3703.2011.00735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell AL, et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science. 2012;336:1711–1715. doi: 10.1126/science.1222218. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen CV, et al. Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. The Plant cell. 2014;26:585–601. doi: 10.1105/tpc.113.118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. The Plant journal: for cell and molecular biology. 2002;31:713–727. doi: 10.1046/j.1365-313X.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- 36.Tang X, et al. Ubiquitin-conjugated degradation of golden 2-like transcription factor is mediated by CUL4-DDB1-based E3 ligase complex in tomato. The New phytologist. 2015;209:1028–1039. doi: 10.1111/nph.13635. [DOI] [PubMed] [Google Scholar]
- 37.Mao, K., Wang, L., Li, Y. Y. & Wu, R. L. Molecular Cloning and Functional Analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus euphratica. PloS one 10, 10.1371/journal.pone.0132390 (2015). [DOI] [PMC free article] [PubMed]
- 38.Zhao C, et al. Molecular cloning and functional analysis of a UV-B photoreceptor gene, MdUVR8 (UV Resistance Locus 8), from apple. Plant science: an international journal of experimental plant biology. 2016;247:115–126. doi: 10.1016/j.plantsci.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Liu LL, Gregan S, Winefield C, Jordan B. From UVR8 to flavonol synthase: UV-B-induced gene expression in Sauvignon blanc grape berry. Plant Cell and Environment. 2015;38:905–919. doi: 10.1111/pce.12349. [DOI] [PubMed] [Google Scholar]
- 40.Loyola R, et al. The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. Journal of experimental botany. 2016;67:5429–5445. doi: 10.1093/jxb/erw307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q, et al. Hydrogen peroxide, nitric oxide and UV RESISTANCE LOCUS8 interact to mediate UV-B-induced anthocyanin biosynthesis in radish sprouts. Scientific reports. 2016;6:29164. doi: 10.1038/srep29164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilbrook K, et al. UV-B Perception and Acclimation in Chlamydomonas reinhardtii. The Plant cell. 2016;28:966–983. doi: 10.1105/tpc.15.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muir SR, et al. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nature biotechnology. 2001;19:470–474. doi: 10.1038/88150. [DOI] [PubMed] [Google Scholar]
- 44.Butelli E, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nature biotechnology. 2008;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- 45.Kataria S, Jajoo A, Guruprasad KN. Impact of increasing Ultraviolet-B (UV-B) radiation on photosynthetic processes. Journal of photochemistry and photobiology. B, Biology. 2014;137:55–66. doi: 10.1016/j.jphotobiol.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Waring J, Underwood GJ, Baker NR. Impact of elevated UV-B radiation on photosynthetic electron transport, primary productivity and carbon allocation in estuarine epipelic diatoms. Plant, cell & environment. 2006;29:521–534. doi: 10.1111/j.1365-3040.2005.01429.x. [DOI] [PubMed] [Google Scholar]
- 47.Joshi P, Gartia S, Pradhan MK, Biswal B. Photosynthetic response of clusterbean chloroplasts to UV-B radiation: energy imbalance and loss in redox homeostasis between Q(A) and Q(B) of photosystem II. Plant science: an international journal of experimental plant biology. 2011;181:90–95. doi: 10.1016/j.plantsci.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Lidon FC, Ramalho JC. Impact of UV-B irradiation on photosynthetic performance and chloroplast membrane components in Oryza sativa L. Journal of photochemistry and photobiology. B, Biology. 2011;104:457–466. doi: 10.1016/j.jphotobiol.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Singh S, Agrawal SB, Agrawal M. UVR8 mediated plant protective responses under low UV-B radiation leading to photosynthetic acclimation. Journal of photochemistry and photobiology. B, Biology. 2014;137:67–76. doi: 10.1016/j.jphotobiol.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 50.Davey MP, et al. The UV-B photoreceptor UVR8 promotes photosynthetic efficiency in Arabidopsis thaliana exposed to elevated levels of UV-B. Photosynthesis research. 2012;114:121–131. doi: 10.1007/s11120-012-9785-y. [DOI] [PubMed] [Google Scholar]
- 51.Gupta R, Bhadauriya P, Chauhan VS, Bisen PS. Impact of UV-B radiation on thylakoid membrane and fatty acid profile of Spirulina platensis. Current microbiology. 2008;56:156–161. doi: 10.1007/s00284-007-9049-9. [DOI] [PubMed] [Google Scholar]
- 52.Dobrikova AG, Krasteva V, Apostolova EL. Damage and protection of the photosynthetic apparatus from UV-B radiation. I. Effect of ascorbate. Journal of plant physiology. 2013;170:251–257. doi: 10.1016/j.jplph.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 53.van Rensen JJ, Vredenberg WJ, Rodrigues GC. Time sequence of the damage to the acceptor and donor sides of photosystem II by UV-B radiation as evaluated by chlorophyll a fluorescence. Photosynthesis research. 2007;94:291–297. doi: 10.1007/s11120-007-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Luscher J, et al. Climate change conditions (elevated CO2 and temperature) and UV-B radiation affect grapevine (Vitis vinifera cv. Tempranillo) leaf carbon assimilation, altering fruit ripening rates. Plant science: an international journal of experimental plant biology. 2015;236:168–176. doi: 10.1016/j.plantsci.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Fagerberg WR. Below-ambient levels of UV induce chloroplast structural change and alter starch metabolism. Protoplasma. 2007;230:51–59. doi: 10.1007/s00709-006-0221-z. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi K, et al. Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. The Plant cell. 2012;24:1081–1095. doi: 10.1105/tpc.111.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waters MT, et al. GLK transcription factors coordinate expression of the Photosynthetic Apparatus in Arabidopsis. The Plant cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velanis CN, Herzyk P, Jenkins GI. Regulation of transcription by the Arabidopsis UVR8 photoreceptor involves a specific histone modification. Plant molecular biology. 2016;92:425–443. doi: 10.1007/s11103-016-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cloix C, Jenkins GI. Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Molecular plant. 2008;1:118–128. doi: 10.1093/mp/ssm012. [DOI] [PubMed] [Google Scholar]
- 60.Binkert M, et al. Revisiting chromatin binding of the Arabidopsis UV-B photoreceptor UVR8. BMC plant biology. 2016;16:42. doi: 10.1186/s12870-016-0732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fillatti JJ, Kiser J, Rose R, Comai L. Efficient Transfer of a Glyphosate Tolerance Gene into Tomato Using a Binary Agrobacterium-Tumefaciens Vector. Bio-Technol. 1987;5:726–730. [Google Scholar]
- 62.Nguyen HP, et al. Methods to Study PAMP-Triggered Immunity Using Tomato and Nicotiana benthamiana. Mol Plant Microbe In. 2010;23:991–999. doi: 10.1094/MPMI-23-8-0991. [DOI] [PubMed] [Google Scholar]
- 63.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 64.Li YX, et al. Tomato MBD5, a methyl CpG binding domain protein, physically interacting with UV-damaged DNA binding protein-1, functions in multiple processes. New Phytologist. 2016;210:208–226. doi: 10.1111/nph.13745. [DOI] [PubMed] [Google Scholar]
- 65.Cookson PJ, et al. Increases in cell elongation, plastid compartment size and phytoene synthase activity underlie the phenotype of the high pigment-1 mutant of tomato. Planta. 2003;217:896–903. doi: 10.1007/s00425-003-1065-9. [DOI] [PubMed] [Google Scholar]
- 66.Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C. Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. The Plant cell. 1999;11:145–157. doi: 10.1105/tpc.11.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.