Abstract
Cell migration is driven by propulsive forces derived from polymerizing actin that pushes and extends the plasma membrane. The underlying actin network is constantly undergoing adaptation to new mechano-chemical environments and intracellular conditions. As such, mechanisms that regulate actin dynamics inherently contain multiple feedback loops and redundant pathways. Given the highly adaptable nature of such a system, studies that use only perturbation experiments (e.g. knockdowns, overexpression, pharmacological activation/inhibition, etc.) are challenged by the nonlinearity and redundancy of the pathway. In these pathway configurations, perturbation experiments at best describe the function(s) of a molecular component in an adapting (e.g. acutely drug-treated) or fully adapted (e.g. permanent gene silenced) cell system, where the targeted component now resides in a non-native equilibrium. Here, we propose how quantitative live-cell imaging and analysis of constitutive fluctuations of molecular activities can overcome these limitations. We highlight emerging actin filament barbed-end biology as a prime example of a complex, nonlinear molecular process that requires a fluctuation analytic approach, especially in an unperturbed cellular system, to decipher functional interactions of barbed-end regulators, actin polymerization and membrane protrusion.
This article is part of the theme issue ‘Self-organization in cell biology’.
Keywords: actin, system redundancy, real-time, imaging, signalling, fluctuations
1. Introduction
Migrating cells are polarized and extend membrane protrusions, including lamellipodia and filopodia, that are generated through dynamic actin polymerization and remodelling events [1–4]. During this process monomeric, globular actin (G-actin) polymerizes into diverse actin filament (F-actin) networks with distinct sub-functions. Therefore, the architecture, size, location and density of actin networks at a given time in a cell are tightly controlled (figure 1a). The dynamics of formation and turnover of these networks, mostly organized into arrays of linear filaments with various degrees of bundling or branching, are governed by a plethora of actin-binding proteins (ABPs) that individually and cooperatively contribute to the network architecture. Moreover, it is becoming apparent that structurally distinct actin networks compete for a common pool of polymerizable actin [6–9]. However, the molecular mechanism by which cells maintain a distinct network architecture and homeostasis of polymerizable actin pool is not fully understood [10–12]. Acquiring this understanding is important, as the concentration of polymerizable actin determines F-actin growth, which translates into actin-based cellular processes, including cell protrusion and migration [11,12]. Here, we propose how microscopy-based fluctuation analysis of molecular components embedded in highly redundant systems will help elucidate the hierarchy of ABPs in a perturbation-free cellular system.
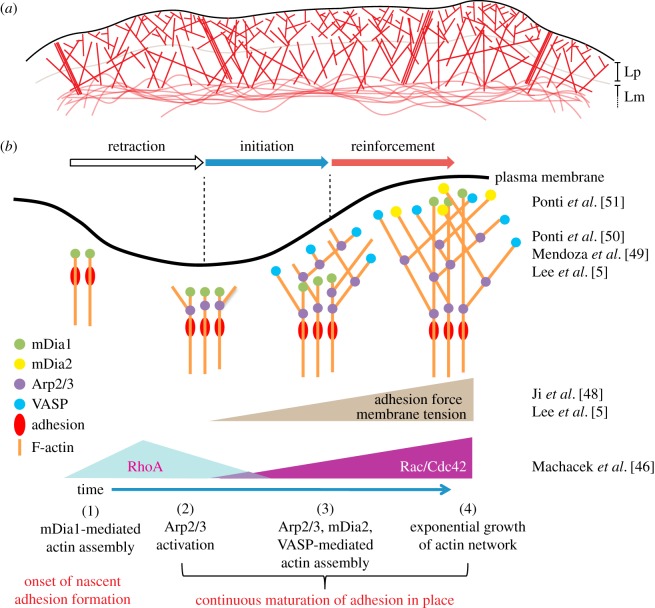
Figure 1.
Actin dynamics at the cell leading edge. (a) Actin network at the cell leading edge. Actin networks at the cell edge are organized into two kinetically, kinematically, molecularly and functionally distinct polymer structures, termed the lamellipodium (Lp) and lamella (Lm). Lp consists of a dense network of branched actin with high polymerization rate and high density of barbed ends. Lm consists of more straight filaments in a less dense array. Thus, the density of barbed ends tends to be lower. Lp and Lm are tightly coordinated. Thus, their filament arrays may not always be clearly delineated. In particular, Lm filaments can reach all the way to the leading edge. (b) Model of initiation and reinforcement of actin assembly during cell protrusions mediated by multiple, functionally overlapping assembly pathways. The pathways are differentially activated by different signalling systems. Key references describing the unified model are inserted to the right. See text for description of the model. Figure is reprinted from Lee et al. [5] with permission of Elsevier.
2. Internetwork crosstalk and actin network (re)arrangements
G-actin polymerizes spontaneously in vitro into polarized filaments with a fast-growing barbed end and a slow-growing pointed end [13,14]. Filaments are generated from an actin seed or nucleus of three G-actin monomers. However, the formation of a nucleus is thermodynamically unstable and rate limiting. Moreover, cellular actin (70–150 µM in non-muscle mammalian cells) is bound to profilin (20–100 µM) or actin-sequestering protein β-thymosin (100–500 µM), which ensures a low steady-state G-actin concentration to prevent spontaneous nucleus formation [9,15–18]. To overcome this rate-limiting nucleation step, cells contain several classes of actin nucleators to initiate and catalyse F-actin polymerization [13,14,19]. Major classes of actin nucleators include the Arp2/3 complex, members of the formin family, and proteins that contain tandem repeats of WASP-Homology 2 domains (WH2). Growth of nucleated filaments is accelerated by processive elongation factors such as Ena/VASP, and again, formins. For an extensive overview of actin nucleators, nucleation-promoting factors, elongation factors and their regulation by extracellular signals, we refer the reader to excellent reviews written by our colleagues [19–22].
In addition to the regulation of filament nucleation and growth, a mechanism is required to constantly exchange or replenish the pool of polymerizable actin. Two models of actin homeostatic mechanisms have been proposed: a global treadmilling model and an internetwork competition model [11,12].
The global treadmilling model proposed by Carlier & Shekhar emphasizes competition between regulators at filament barbed ends as the key factor to dictate network geometry. They further assume that the various networks are in a ‘dynamic’ steady state with coordinated filament turnover and monomer exchange between the filaments. Importantly, they regard profilin-bound actin as the non-exhaustible, steady-state pool of polymerizable actin that is replenished by filament disassembly through mainly the actin depolymerizing factor (ADF)/cofilin family of proteins. Depletion of the pool is blocked by capping of growing barbed ends [3,11,13,14,23–31].
The internetwork competition model of Suarez & Kovar hypothesizes that distinct F-actin networks (i.e. bundled versus branched in a simplified scheme) compete for polymerizable actin from a common finite pool and exchange G-actin between networks [12]. For example, in fission yeast, which has a simplified F-actin network consisting of only branched, bundled or cytokinetic actin rings, inhibition of Arp2/3 complex-mediated branched network polymerization enhanced the bundled network, which was attributed to increased formin activity, and vice versa: inhibition of formin activity and loss of the bundled network enhanced the Arp2/3 complex-mediated branched network [7,32,33]. Suarez & Kovar further highlight that a similar mechanism may be at play in mammalian systems, and refer to studies that show increases in filopodia counts, resulting from inhibition of branched network assembly [12]. However, this may not necessarily be the result of enhanced polymerization in filopodia, but rather collapse of the branched network and exposure of pre-existing filopodia precursors embedded within the lamellipodium [34,35]. Rather, depletion of capping proteins (CPs) was shown to increase global and filopodial F-actin intensity in mammalian systems [36]. In support of the internetwork competition model, we have recently shown a competitive network interaction in control of cell polarization between myosin II-dependent cortical actomyosin bundles and Arp2/3-dependent branched networks [8].
3. Limitations of perturbation-based experiments in cellular process with redundant network circuitries
Both the global treadmilling and internetwork competition models are based on results obtained from cell biological experiments with long-term genetic perturbations and isolated biochemical experiments in vitro. During cellular processes like membrane protrusion, the actin networks constantly adapt to changing mechano-chemical and intracellular signalling environments. How the dynamics between the networks is coordinated in such a scenario has remained largely unknown.
Addressing this question is significantly complicated by the redundancy and feedback between actin regulatory components [1]. Such a pathway system tends to rapidly reconfigure itself under perturbation of a particular pathway component. Long-term genetic perturbation will affect the global steady state of the system, which is significantly shifted from the steady state of the unperturbed native system; and even acute perturbations such as pharmacological interventions or optogenetics change the stoichiometry among regulators with partially overlapping functions, which may also have severe instantaneous adaptation. As a result, the outcomes of perturbation experiments are very difficult to interpret. In particular, they do not allow conclusions as to the function of the perturbed target component in the unperturbed system. For example, at both the functional and structural level, the lamellipodium and the Arp2/3 complex-dependent dendritic actin network therein have always been considered crucial for cell migration. Concomitantly, the Arp2/3 complex has been contemplated as the master regulator of cell migration and lamellipodium formation [37]. However, recent studies have demonstrated that cells lacking the Arp2/3 complex or a lamellipodium are still capable of migration [38–40]. This indicates the redundancy of the migratory system and the existence of Arp2/3 complex-independent F-actin polymerization pathways [41,42]. Similarly, pharmacological inhibition of formins by SMIFH2 revealed that cells exhibited dynamic and alternating cytoskeletal rearrangements and migration velocities over time [43].
While we do not question the power of biochemical experiments and molecular genetics to identify and probe major molecular nodes within a system, we want to point out the limitations of drawing conclusions from perturbed cellular systems that may have undergone adaptation(s). Therefore, we have previously proposed the usage of microscopy-based fluctuation studies in unperturbed systems as a complementary approach to establish causal relations among molecular processes in highly redundant regulatory networks [44]. We have demonstrated how such fluctuation studies in minimally perturbed systems coupled with computer-vision analysis and a rigorous statistical framework provide insight into complex signalling pathways [5,45,46]. Notably, we introduced fluorescent fusions to several ABPs, including actin nucleators mDia1 and Arp3, at very low levels into cells to minimally perturb the system, and followed their dynamic fluctuations over time. After fine-grained registration of the intensity fluctuations relative to local motion events, such as protrusion or retraction onset, or time point of maximal edge advancement, we could extract from very noisy signals the sequential recruitment of ABPs during a protrusion–retraction cycle. We found that with the exception of the formin family member mDia1 all other imaged nucleators, and especially the Arp2/3 complex, were recruited to the growing actin network only after protrusion onset [5]. We interpreted this result as mDia1 being the nucleator of filament polymerization that induces protrusion, whereas the Arp2/3 complex and other nucleators and elongation factors reinforce the polymerization of the growing actin networks against increasing boundary forces of the plasma membrane that are likely resulting from mechanical stretch of the cell edge during protrusion. The proposed functional hierarchy of mDia1-mediated polymerization first and Arp2/3 complex-mediated polymerization second was then validated by co-imaging of mDia1 recruitment and actin assembly. While mDia1 recruitment correlated strongly and with a slightly negative time lag to actin assembly, i.e. mDia1 recruitment precedes actin assembly before protrusion onset, the correlation disappeared during cell edge advancement, indicating that during this phase of the protrusion–retraction cycle other factors drive actin polymerization. A similar cooperation between mDia1 and the Arp2/3 complex was demonstrated [47] using protrusion events in response to stimulation of cells by epidermal growth factor. The study also suggested that the Arp2/3 complex, once activated by mDia1-nucleated filaments, enriches at the tips of expanding lamellipodia, which drives the protrusive actin polymerization. By contrast, mDia1 lagged behind at the base of the lamellipodia, and likely does not contribute to the expansion of the lamellipodia. Thus, both studies independently concluded that mDia1 is an activator of actin filament polymerization prior to Arp2/3 complex recruitment, using completely different experimental approaches: the former study derived the functional hierarchy from signal fluctuation and image registration in a minimally perturbed system, while the latter study used biochemical reconstitution and long-term genetic perturbations in cell lines. We speculate that, in this particular case, the two studies converged in their conclusions, because mDia1-mediated filament polymerization acts upstream of Arp2/3 complex-mediated polymerization without redundancy to another nucleator, and without feedback. Thus, knockdown of mDia1 generated a defect in actin network growth that could unambiguously be interpreted as mDia1-activation residing at the top of the event cascade.
4. Unique insights gained from fluctuation analyses
Based on a series of image fluctuation studies, we have compiled a model of the cascade of molecular events that regulate cell edge protrusion and retraction dynamics (figure 1b). We have shown that nascent adhesions and accumulation of mDia1 precede membrane protrusion, initiating the polymerization and growth of linear actin filaments (figure 1b, initiation phase; [5,48]). This is followed by recruitment and activation of additional actin assembly factors, including the Arp2/3 complex, to reinforce actin polymerization against an increasingly strained plasma membrane (figure 1b, reinforcement phase; [5,48–51]). Intriguingly, we have also demonstrated that RhoA activity precedes Rac1 and Cdc42 activities during membrane protrusion [46]. Collectively, we now envision that RhoA activates mDia1 during the initiation phase of membrane protrusion, and Rac1/Cdc42 activities enhance polymerization in later phases by activating and recruiting the Arp2/3 complex and other actin assembly factors [5,46,49].
Importantly, none of these insights could have been gained by merely perturbation-based approaches. For example, microinjection or expression of dominant negative Rac1 abrogates lamellipodia formation and in many cases protrusion [52,53], showing that Rac1 is an essential activator of pathways that promote assembly of the F-actin network. However, beside concerns of long-term side-effects induced by global inactivation of a signal as central as Rac1, these experiments do not reveal that Rac1 functions only during later phases of the protrusion event. While acute perturbation by, for example, optogenetic activation of Rac1 may eliminate some of these concerns, this experiment still cannot uncover the bona fide role of Rac1 in the protrusion cycle. Forced Rac1 activation does induce (note: uncontrolled) membrane protrusion and ruffling, but this merely shows that hyperactivation of the Arp2/3 complex downstream of Rac1 signalling can be sufficient for protrusion generation. However, with this approach only, the function of Rac1 in the unperturbed and unstimulated protrusion cycle, namely as an enforcer of the Arp2/3 complex-dependent actin assembly pathway in response to the earlier activation of Arp2/3-independent pathways, remains inaccessible.
Genetic perturbations, i.e. knockdown and knockout of genes, besides being global and thus potentially affecting multiple functions, also bear the risk of gene regulatory compensation. To get a sense of the meaning of a phenotype, it would be necessary to monitor the expression changes of every other gene implicated in the same and potentially neighbouring pathways [54,55]. This is not feasible in routine experiments; and even if it were, the interpretation of the significance of any observed expression shifts related to the phenotypic outcome of an experiment would be a daunting task. Indeed, many of the functions assigned to the Arp2/3 complex, for example, have emerged from knockdown and knockout approaches. Quite a few of the studies have generated conflicting results, as discussed by Swaney & Li [56]. While Swaney & Li propose differences in genetic background between cell lines, molecular perturbation methodology, the targeted Arp2/3 complex subunit or experimental conditions as possible reasons for the discrepancies, we surmise here that cells may have adopted divergent compensation mechanisms over the course of genetic manipulation, which usually takes longer than 24 h. It would be very interesting to reproduce some of these controversial data and map out the differences in expression of all other ABPs in the system. Notably, loss of the Arp2/3 complex unambiguously has confirmed the importance of the protein complex in lamellipodia formation, but when and how the complex contributes to lamellipodia formation, and especially its mechanoresponsiveness during lamellipodial dynamics, cannot be unveiled by simply removing the complex from the cells.
The following two sections highlight the emerging complexity of F-actin growth regulation and outline an agenda for a perturbation-free analysis to study the molecular interplay of ABPs to deduce their hierarchy in a complex network.
5. Emerging complexity at filament barbed ends
Elongation of actin filaments in a cell is tightly controlled by an array of barbed-end-binding proteins that either inhibit or promote the addition of G-actin to the barbed end [31]. It has been estimated that essentially all barbed ends (99.0–99.8%) of the lamellipodial network are capped (i.e. inhibited for G-actin addition and thus elongation) by CPs [11,14,24,57,58]. The binding of CPs is extremely tight with a dissociation rate constant of 2.7 × 10−4 s−1 (with an equilibrium dissociation constant Kd of 0.1 nM) [59,60]. The high rates of barbed-end capping may be counterintuitive at first sight since capping terminates filament growth. However, capping a significant portion of filaments may raise the availability of actin monomers for rapid elongation of a select number of uncapped filaments, or enhance Arp2/3 complex-mediated nucleation through a ‘monomer funnelling’ or ‘monomer gating’ model [14,24,57,61–63]. Regardless of which model is at work, the active regulation of barbed-end dynamics by CPs in combination with actin nucleators allows a cell to steer and reorganize its actin network more tightly in response to mechano-chemical cues, rather than merely relying on uninhibited polymerization from uncapped barbed ends. Hence, CPs are essential factors for regulating directed cell protrusion and migration [36,58,63–70].
Capping proteins are also in competition with a host of factors that bind to the barbed ends of actin filaments [31]. These include factors such as Ena/VASP [Kd = 1–9 nM [71,72]], formins [Kd << 0.1 nM [73–75]], and Spire [Kd = 4–8 nM [76,77]]. Ena/VASP binds to uncapped filaments to accelerate actin incorporation to the barbed end for rapid filament extension (up to threefold increase [72]). In addition, Ena/VASP may exhibit anti-capping activities [63,78,79]. The actin nucleator and processive elongator formins directly compete for barbed ends by entering into a ternary complex composed of barbed-end actin, formin and CP [73,80]. When formins outcompete CPs (about 30–40% success rate) [73], they accelerate F-actin elongation up to 10-fold [74,75,81,82]. The WH2 repeat protein Spire on its own inhibits filament elongation, but may cooperate with the formin FMN2 to enhance filament elongation [77].
The protein landscape at the F-actin barbed end is becoming even more complex, as free profilin was recently shown to bind barbed ends in vitro, and with some validation in vivo [83–85] (albeit at three orders of magnitude lower affinity compared to G-actin (Kd = 20 µM versus 0.1 µM, respectively)). Noteworthy, profilin has been shown to inhibit Arp2/3 complex-dependent nucleation and branching both in vitro and in vivo [86,87]. This inhibition may actually occur at F-actin barbed ends, given the recent re-demonstration that the Arp2/3 complex nucleates new branches from the barbed ends with 10 times higher frequency than from the F-actin sides [83]. Thus, an overall scheme where profilin competes with and coordinates the function of barbed-end regulators, including the Arp2/3 complex, formins and elongation factors in vivo may be plausible (figure 2) [20,83,84,88].
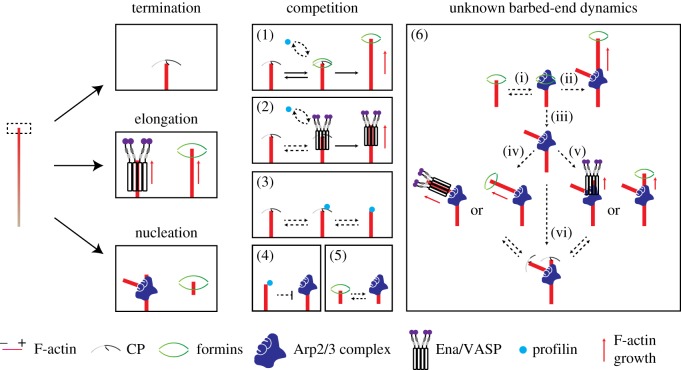
Figure 2.
Open questions on the regulation of growth dynamics of actin filament barbed ends in a complex molecular process. A simplified scheme of the fate of barbed ends at the leading edge. A barbed end can be: capped and terminated for growth (termination), bound by elongation factors for enhanced growth (elongation), or bound by the Arp2/3 complex for branch formation (nucleation). Formins play a dual role in that they promote elongation, but can also lead to de novo nucleation of filaments. Several proteins compete for barbed-end binding (see main text for details): (1) capping proteins (CP) and formins, and (2) CP and Ena/VASP proteins compete for barbed ends. (3) Competition between CP-binding and anti-capping activities by formins and Ena/VASP proteins may be dependent on free profilin. Examples of open questions that await investigation in vivo: (4) Does profilin inhibit the Arp2/3 complex by binding to a barbed end? (5) Do formins and the Arp2/3 complex compete for barbed ends? (6) What is the fate of de novo nucleated filaments: (i) Do formin-polymerized filaments serve as new templates for the Arp2/3 complex? (ii) Does formin-mediated filament elongation and Arp2/3 complex branching occur simultaneously? (iii) Does the Arp2/3 complex compete with formins for barbed ends during branching? (iv) Is there a particular elongation factor (including but not limited to Ena/VASP proteins and formins) preferred for elongation of new branches? (v) What mechanisms drives elongation of the mother filament after Arp2/3 complex-mediated branching? (vi) At what rate do new branches get capped? Is there a difference in this rate between the daughter or the mother filament?
6. An agenda to the quantitative analysis of barbed-end dynamics
With so many proteins competing for actin barbed ends, the recruitment of barbed-end-binding proteins should be tightly controlled to maintain and properly rearrange the actin network. While some data on the hierarchy and interplay between barbed-end-binding proteins is available for single filaments in vitro (figure 2), we have limited knowledge about the precise fate of capped, elongating or de novo nucleated filaments in the lamellipodia in vivo. For example, do formin-nucleated linear filaments recruit and serve as new templates for Arp2/3 complex-nucleation sites [47]? Does the Arp2/3 complex truly operate in an auto-catalytic fashion [20]? Does the Arp2/3 complex compete with formins for the barbed end, or does the Arp2/3 complex prefer side-branching in this case [83]? Does free profilin cooperate with and enhance the anti-capping activity of Ena/VASP and formins in vivo [73,84,89]? If the Arp2/3 complex uses the barbed ends of formin-nucleated (mother) filaments as templates, do formins remain bound to those filaments to maintain elongation? What is the fate of Arp2/3-nucleated branches? Are particular formins or elongation factors recruited to facilitate elongation of the mother, daughter or both filaments? Alternatively, will the filaments elongate passively, or become rapidly capped? How does the interaction of a barbed-end protein affect the global distribution of other ABPs within the lamellipodium? What feedback mechanisms are at play? Are those feedbacks mechanical [90,91]?
These questions will not easily be answered by perturbation approaches, long term or short term. We believe that the dynamic protein kinetic landscape at F-actin barbed ends in the lamellipodium is a prime example of a molecular system requiring fluctuation analysis in an unperturbed or minimally perturbed state. Expression of ABPs (even at low levels) may affect the steady-state concentration of actin and impact the global distribution of other ABPs and their respective actin networks [6,11,12]. Additionally, a change in the G/F-actin ratio has been linked with activation of the SRF/MAL transcriptional program. Overexpression and downregulation of ABPs may thus also trigger the SRF/MAL response, which drastically changes the intracellular environment and behaviour [92,93]. Likewise, compensatory expression of formins including FMN1, FMNL1 and INF1 upon depletion of mDia2 has been reported [94]. Thus, to faithfully study the functional hierarchy of ABPs acting on the barbed ends, we need to not only avoid knockdown or overexpression studies, but analysis of dynamic fluctuations in its native system per se must avoid perturbation associated, for example, with the expression of fluorescent markers of the target proteins. Therefore, truly perturbation-free studies will have to build on recent advances in genome editing to label the endogenous protein and control for potential functional shifts associated with the labelling [95,96].
After establishing a robust kinetic model in an intact cellular system, we can validate the model using acute, local, and reversible perturbation by, for example, increasing or decreasing CP activity through optogenetic trapping of myotrophin and CP interaction motif-containing proteins, or capping protein CAPZB, respectively [31,57,97–99].
7. Limitations of fluctuation analysis
Although conceptually attractive and ultimately required to study complex molecular process, perturbation-free image fluctuation analysis brings significant hurdles, making it a far from standard experimental approach. A major caveat is the need for direct observation of every system component to be included in the model. This means that all key players in a pathway system must be known. Usually, these players are identified in screens using genetic or molecular perturbations. Thus, image fluctuation analysis is not replacing perturbation strategies, but will always be only a complement to define with higher precision the function of key players. Second, as discussed above, to achieve truly minimally invasive experimental conditions, the tagging of target proteins for direct observation must be accomplished at endogenous expression levels and without affecting the function of the target. Moreover, if the parameters relevant to the system behaviour are related to target protein activity rather than concentration, the tagging must rely on activity biosensors. Both the tagging itself and the development of adequate activity probes entail laborious procedures, making image fluctuation analysis definitely not a high-throughput approach. Third, to deduce the hierarchical linkages between system components from spontaneous image fluctuations, it requires (i) sophisticated statistical approaches, and (ii) sufficient repeats of movies that record the system under experimentally similar conditions. On the one hand this enables data pooling, and on the other hand reduces the risk of sampling bias. Meeting this criterion can again be very laborious. Nevertheless, we strongly believe that for a system like F-actin barbed-end regulation, where nearly all molecular players are known, yet their interactions likely are dominated by a high level of redundancy and nonlinearity, a perturbation-free fluctuation analysis will break through major barriers in the current understanding of the system that are caused in large part by the limitations of perturbation-based analysis.
8. Conclusion and perspectives
A wealth of new information and insight is expected to come from the application of fluctuation analysis in highly redundant actin network dynamics. The forthcoming information will aid to better understand thermodynamic properties of ABPs at the level of the entire system. Here, we limited our focus on known ABPs that bind the barbed ends. However, CPs and ADF/cofilin family proteins were recently also shown to cooperate to depolymerize severed filaments, and indirectly contribute to F-actin polymerization through treadmilling of actin [26,37,41,61]. Thus, future analysis can be expanded to other ABPs that function in other actin networks, including but not limited to the lamella [51], and which do not directly interact with the barbed end, such as: F-actin side-binding proteins (e.g. ADF/cofilin, tropomyosins [28,40,100]), F-actin motor proteins (myosins [101,102]), WH2 repeat proteins [103] and actin crosslinkers (e.g. fascin, α-actinin [104,105]). Once again, our proposed approach will permit the determination of functional interactions of these indirect effectors in the assembly of actin into diverse networks. Such analyses will be instrumental to understand how distinct actin networks are organized and maintained by activation of different ABPs, how their assembly and disassembly dynamics crosstalk, and how their partially overlapping (or distinct) functions are balanced in the context of different cellular functions.
Acknowledgements
We thank Kevin M. Dean and Dana Kim Reed for proof-reading and their comments.
Data accessibility
This article has no additional data.
Authors' contributions
T.I. and G.D. wrote the manuscript.
Competing interests
We have no competing interests.
Funding
Work on perturbation-free analysis of actin regulation is funded by the NIH grant no. R01 GM071868 (G.D.).
References
- 1.Krause M, Gautreau A. 2014. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15, 577–590. ( 10.1038/nrm3861) [DOI] [PubMed] [Google Scholar]
- 2.Insall RH, Machesky LM. 2009. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell 17, 310–322. ( 10.1016/j.devcel.2009.08.012) [DOI] [PubMed] [Google Scholar]
- 3.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. 1999. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401, 613–616. ( 10.1038/44183) [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ. 2011. Life at the leading edge. Cell 145, 1012–1022. ( 10.1016/j.cell.2011.06.010) [DOI] [PubMed] [Google Scholar]
- 5.Lee K, Elliott HL, Oak Y, Zee C-T, Groisman A, Tytell JD, Danuser G. 2015. Functional hierarchy of redundant actin assembly factors revealed by fine-grained registration of intrinsic image fluctuations. Cell Syst. 1, 37–50. ( 10.1016/j.cels.2015.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimchev G, Steffen A, Kage F, Dimchev V, Pernier J, Carlier M-F, Rottner K. 2017. Efficiency of lamellipodia protrusion is determined by the extent of cytosolic actin assembly. Mol. Biol. Cell 28, 1311–1325. ( 10.1091/mbc.E16-05-0334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. 2014. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr. Biol. 24, 579–585. ( 10.1016/j.cub.2014.01.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomakin AJ, Lee K-C, Han SJ, Bui DA, Davidson M, Mogilner A, Danuser G. 2015. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat. Cell Biol. 17, 1435–1445. ( 10.1038/ncb3246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. 2003. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 83, 433–473. ( 10.1152/physrev.00026.2002) [DOI] [PubMed] [Google Scholar]
- 10.Chhabra ES, Higgs HN. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110–1121. ( 10.1038/ncb1007-1110) [DOI] [PubMed] [Google Scholar]
- 11.Carlier M-F, Shekhar S. 2017. Global treadmilling coordinates actin turnover and controls the size of actin networks. Nat. Rev. Mol. Cell Biol. 18, 389–401. ( 10.1038/nrm.2016.172) [DOI] [PubMed] [Google Scholar]
- 12.Suarez C, Kovar DR. 2016. Internetwork competition for monomers governs actin cytoskeleton organization. Nat. Rev. Mol. Cell Biol. 17, 799–810. ( 10.1038/nrm.2016.106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard TD. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451–477. ( 10.1146/annurev.biophys.35.040405.101936) [DOI] [PubMed] [Google Scholar]
- 14.Bugyi B, Carlier M-F. 2010. Control of actin filament treadmilling in cell motility. Annu. Rev. Biophys. 39, 449–470. ( 10.1146/annurev-biophys-051309-103849) [DOI] [PubMed] [Google Scholar]
- 15.Weber A, Nachmias VT, Pennise CR, Pring M, Safer D. 1992. Interaction of thymosin beta 4 with muscle and platelet actin: implications for actin sequestration in resting platelets. Biochemistry 31, 6179–6185. ( 10.1021/bi00142a002) [DOI] [PubMed] [Google Scholar]
- 16.Machesky LM, Poland TD. 1993. Profilin as a potential mediator of membrane-cytoskeleton communication. Trends Cell Biol. 3, 381–385. ( 10.1016/0962-8924(93)90087-H) [DOI] [PubMed] [Google Scholar]
- 17.Kang F, Purich DL, Southwick FS. 1999. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 274, 36 963–36 972. ( 10.1074/jbc.274.52.36963) [DOI] [PubMed] [Google Scholar]
- 18.Carlier M-F, Hertzog M, Didry D, Renault L, Cantrelle F-X, van Heijenoort C, Knossow M, Guittet E. 2007. Structure, function, and evolution of the beta-thymosin/WH2 (WASP-Homology2) actin-binding module. Ann. N. Y. Acad. Sci. 1112, 67–75. ( 10.1196/annals.1415.037) [DOI] [PubMed] [Google Scholar]
- 19.Campellone KG, Welch MD. 2010. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251. ( 10.1038/nrm2867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goley ED, Welch MD. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726. ( 10.1038/nrm2026) [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Dominguez R. 2010. Regulation of actin cytoskeleton dynamics in cells. Mol. Cells 29, 311–325. ( 10.1007/s10059-010-0053-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridley AJ. 2015. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 36, 103–112. ( 10.1016/j.ceb.2015.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danuser G, Allard J, Mogilner A. 2013. Mathematical modeling of eukaryotic cell migration: insights beyond experiments. Annu. Rev. Cell Dev. Biol. 29, 501–528. ( 10.1146/annurev-cellbio-101512-122308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlier MF, Pantaloni D. 1997. Control of actin dynamics in cell motility. J. Mol. Biol. 269, 459–467. ( 10.1006/jmbi.1997.1062) [DOI] [PubMed] [Google Scholar]
- 25.Suarez C, et al. 2011. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 21, 862–868. ( 10.1016/j.cub.2011.03.064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wioland H, Guichard B, Senju Y, Myram S, Lappalainen P, Jégou A, Romet-Lemonne G. 2017. ADF/Cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr. Biol. 27, 1956–1967.e7. ( 10.1016/j.cub.2017.05.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De La Cruz EM. 2009. How cofilin severs an actin filament. Biophys. Rev. 1, 51–59. ( 10.1007/s12551-009-0008-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322. ( 10.1083/jcb.136.6.1307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southwick FS. 2000. Gelsolin and ADF/cofilin enhance the actin dynamics of motile cells. Proc. Natl Acad. Sci. USA 97, 6936–6938. ( 10.1073/pnas.97.13.6936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier M-F, Pantaloni D. 2003. A biomimetic motility assay provides insight into the mechanism of actin-based motility. J. Cell Biol. 160, 387–398. ( 10.1083/jcb.200207148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shekhar S, Pernier J, Carlier M-F. 2016. Regulators of actin filament barbed ends at a glance. J. Cell Sci. 129, 1085–1091. ( 10.1242/jcs.179994) [DOI] [PubMed] [Google Scholar]
- 32.Gao L, Bretscher A. 2008. Analysis of unregulated formin activity reveals how yeast can balance F-actin assembly between different microfilament-based organizations. Mol. Biol. Cell 19, 1474–1484. ( 10.1091/mbc.E07-05-0520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu R, Chang F. 2011. Characterization of dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr. Biol. 21, 905–916. ( 10.1016/j.cub.2011.04.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Q, Zhang X-F, Pollard TD, Forscher P. 2012. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J. Cell Biol. 197, 939–956. ( 10.1083/jcb.201111052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korobova F, Svitkina T. 2008. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol. Biol. Cell 19, 1561–1574. ( 10.1091/mbc.E07-09-0964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinnar SA, Antoku S, Saffin J-M, Cooper JA, Halpain S. 2014. Capping protein is essential for cell migration in vivo and for filopodial morphology and dynamics. Mol. Biol. Cell 25, 2152–2160. ( 10.1091/mbc.E13-12-0749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard TD, Borisy GG. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. ( 10.1016/S0092-8674(03)00120-X) [DOI] [PubMed] [Google Scholar]
- 38.Suraneni P, Fogelson B, Rubinstein B, Noguera P, Volkmann N, Hanein D, Mogilner A, Li R. 2015. A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol. Biol. Cell 26, 901–912. ( 10.1091/mbc.E14-07-1250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. 2012. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973–987. ( 10.1016/j.cell.2011.12.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupton SL, et al. 2005. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168, 619–631. ( 10.1083/jcb.200406063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D, Waterman-Storer C, Danuser G, Bokoch GM. 2007. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell 13, 646–662. ( 10.1016/j.devcel.2007.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrianantoandro E, Pollard TD. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23. ( 10.1016/j.molcel.2006.08.006) [DOI] [PubMed] [Google Scholar]
- 43.Isogai T, van der Kammen R, Innocenti M. 2015. SMIFH2 has effects on Formins and p53 that perturb the cell cytoskeleton. Sci. Rep. 5, 9802 ( 10.1038/srep09802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welf ES, Danuser G. 2014. Using fluctuation analysis to establish causal relations between cellular events without experimental perturbation. Biophys. J. 107, 2492–2498. ( 10.1016/j.bpj.2014.10.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilela M, Halidi N, Besson S, Elliott H, Hahn K, Tytell J, Danuser G. 2013. Fluctuation analysis of activity biosensor images for the study of information flow in signaling pathways. Methods Enzymol. 519, 253–276. ( 10.1016/B978-0-12-405539-1.00009-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Machacek M, et al. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103. ( 10.1038/nature08242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isogai T, van der Kammen R, Leyton-Puig D, Kedziora KM, Jalink K, Innocenti M. 2015. Initiation of lamellipodia and ruffles involves cooperation between mDia1 and the Arp2/3 complex. J. Cell Sci. 128, 3796–3810. ( 10.1242/jcs.176768) [DOI] [PubMed] [Google Scholar]
- 48.Ji L, Lim J, Danuser G. 2008. Fluctuations of intracellular forces during cell protrusion. Nat. Cell Biol. 10, 1393 ( 10.1038/ncb1797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendoza MC, Vilela M, Juarez JE, Blenis J, Danuser G. 2015. ERK reinforces actin polymerization to power persistent edge protrusion during motility. Sci. Signal. 8, ra47 ( 10.1126/scisignal.aaa8859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ponti A, Matov A, Adams M, Gupton S, Waterman-Storer CM, Danuser G. 2005. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative fluorescent speckle microscopy. Biophys. J. 89, 3456–3469. ( 10.1529/biophysj.104.058701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786. ( 10.1126/science.1100533) [DOI] [PubMed] [Google Scholar]
- 52.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410. ( 10.1016/0092-8674(92)90164-8) [DOI] [PubMed] [Google Scholar]
- 53.Nobes CD, Hall A. 1999. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144, 1235–1244. ( 10.1083/jcb.144.6.1235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bazellières E, et al. 2015. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 17, 409–420. ( 10.1038/ncb3135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. 2010. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 12, 477–483. ( 10.1038/ncb2049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swaney KF, Li R. 2016. Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr. Opin. Cell Biol. 42, 63–72. ( 10.1016/j.ceb.2016.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. 2014. Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 15, 677–689. ( 10.1038/nrm3869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. 2006. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 175, 947–955. ( 10.1083/jcb.200604176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schafer DA, Jennings PB, Cooper JA. 1996. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179. ( 10.1083/jcb.135.1.169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuhn JR, Pollard TD. 2007. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282, 28 014–28 024. ( 10.1074/jbc.M705287200) [DOI] [PubMed] [Google Scholar]
- 61.Shekhar S, Carlier M-F. 2017. Enhanced depolymerization of actin filaments by ADF/Cofilin and monomer funneling by capping protein cooperate to accelerate barbed-end growth. Curr. Biol. 27, 1990–1998.e5. ( 10.1016/j.cub.2017.05.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akin O, Mullins RD. 2008. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133, 841–851. ( 10.1016/j.cell.2008.04.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. 2004. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 118, 363–373. ( 10.1016/j.cell.2004.07.019) [DOI] [PubMed] [Google Scholar]
- 64.Iwasa JH, Mullins RD. 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17, 395–406. ( 10.1016/j.cub.2007.02.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogienko AA, Karagodin DA, Lashina VV, Baiborodin SI, Omelina ES, Baricheva EM. 2013. Capping protein beta is required for actin cytoskeleton organisation and cell migration during Drosophila oogenesis. Cell Biol. Int. 37, 149–159. ( 10.1002/cbin.10025) [DOI] [PubMed] [Google Scholar]
- 66.Bear JE, et al. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509–521. ( 10.1016/S0092-8674(02)00731-6) [DOI] [PubMed] [Google Scholar]
- 67.Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. 2003. Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160, 951–962. ( 10.1083/jcb.200208059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooper JA, Schafer DA. 2000. Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 12, 97–103. ( 10.1016/S0955-0674(99)00062-9) [DOI] [PubMed] [Google Scholar]
- 69.Cooper JA, Sept D. 2008. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267, 183–206. ( 10.1016/S1937-6448(08)00604-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wear MA, Cooper JA. 2004. Capping protein: new insights into mechanism and regulation. Trends Biochem. Sci. 29, 418–428. ( 10.1016/j.tibs.2004.06.003) [DOI] [PubMed] [Google Scholar]
- 71.Winkelman JD, Bilancia CG, Peifer M, Kovar DR. 2014. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc. Natl Acad. Sci. USA 111, 4121–4126. ( 10.1073/pnas.1322093111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen SD, Mullins RD. 2010. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571–584. ( 10.1083/jcb.201003014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shekhar S, Kerleau M, Kühn S, Pernier J, Romet-Lemonne G, Jégou A, Carlier M-F. 2015. Formin and capping protein together embrace the actin filament in a ménage à trois. Nat. Commun. 6, 8730 ( 10.1038/ncomms9730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier M-F. 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119, 419–429. ( 10.1016/j.cell.2004.09.039) [DOI] [PubMed] [Google Scholar]
- 75.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. 2006. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435. ( 10.1016/j.cell.2005.11.038) [DOI] [PubMed] [Google Scholar]
- 76.Bosch M, Le KHD, Bugyi B, Correia JJ, Renault L, Carlier M-F. 2007. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol. Cell 28, 555–568. ( 10.1016/j.molcel.2007.09.018) [DOI] [PubMed] [Google Scholar]
- 77.Montaville P, Jégou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier M-F. 2014. Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biol. 12, e1001795 ( 10.1371/journal.pbio.1001795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. 2003. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 19, 541–564. ( 10.1146/annurev.cellbio.19.050103.103356) [DOI] [PubMed] [Google Scholar]
- 79.Applewhite DA, Barzik M, Kojima S-I, Svitkina TM, Gertler FB, Borisy GG. 2007. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell 18, 2579–2591. ( 10.1091/mbc.E06-11-0990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bombardier JP, Eskin JA, Jaiswal R, Corrêa IR, Xu M-Q, Goode BL, Gelles J. 2015. Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat. Commun. 6, 8707 ( 10.1038/ncomms9707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. 2003. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13, 1820–1823. ( 10.1016/j.cub.2003.09.057) [DOI] [PubMed] [Google Scholar]
- 82.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. 2005. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–494. ( 10.1038/nature03251) [DOI] [PubMed] [Google Scholar]
- 83.Pernier J, Shekhar S, Jegou A, Guichard B, Carlier M-F. 2016. Profilin interaction with actin filament barbed end controls dynamic instability, capping, branching, and motility. Dev. Cell 36, 201–214. ( 10.1016/j.devcel.2015.12.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE. 2015. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell 32, 54–67. ( 10.1016/j.devcel.2014.10.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kinosian HJ, Selden LA, Gershman LC, Estes JE. 2000. Interdependence of profilin, cation, and nucleotide binding to vertebrate non-muscle actin. Biochemistry 39, 13 176–13 188. ( 10.1021/bi001520%2B) [DOI] [PubMed] [Google Scholar]
- 86.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl Acad. Sci. USA 96, 3739–3744. ( 10.1073/pnas.96.7.3739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. 2000. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007–1011. ( 10.1038/35010008) [DOI] [PubMed] [Google Scholar]
- 88.Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. 2015. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell 32, 43–53. ( 10.1016/j.devcel.2014.10.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siton O, Bernheim-Groswasser A. 2014. Reconstitution of actin-based motility by vasodilator-stimulated phosphoprotein (VASP) depends on the recruitment of F-actin seeds from the solution produced by cofilin. J. Biol. Chem. 289, 31 274–31 286. ( 10.1074/jbc.M114.586958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bieling P, Li T-D, Weichsel J, McGorty R, Jreij P, Huang B, Fletcher DA, Mullins RD. 2016. Force feedback controls motor activity and mechanical properties of self-assembling branched actin networks. Cell 164, 115–127. ( 10.1016/j.cell.2015.11.057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jégou A, Carlier M-F, Romet-Lemonne G. 2013. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat. Commun. 4, 1883 ( 10.1038/ncomms2888) [DOI] [PubMed] [Google Scholar]
- 92.Thurston SF, Kulacz WA, Shaikh S, Lee JM, Copeland JW. 2012. The ability to induce microtubule acetylation is a general feature of formin proteins. PLoS ONE 7, e48041 ( 10.1371/journal.pone.0048041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olson EN, Nordheim A. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365. ( 10.1038/nrm2890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Isogai T, van der Kammen R, Goerdayal SS, Heck AJR, Altelaar AFM, Innocenti M. 2015. Proteomic analyses uncover a new function and mode of action for mouse homolog of Diaphanous 2 (mDia2). Mol. Cell. Proteomics 14, 1064–1078. ( 10.1074/mcp.M114.043885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cong L, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. ( 10.1126/science.1231143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. ( 10.1016/j.cell.2014.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. 2010. Rationally improving LOV domain-based photoswitches. Nat. Methods 7, 623–626. ( 10.1038/nmeth.1473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fujiwara I, Remmert K, Piszczek G, Hammer JA. 2014. Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc. Natl Acad. Sci. USA 111, E1970–E1979. ( 10.1073/pnas.1313738111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jung G, Alexander CJ, Wu XS, Piszczek G, Chen B-C, Betzig E, Hammer JA. 2016. V-1 regulates capping protein activity in vivo. Proc. Natl Acad. Sci. USA 113, E6610–E6619. ( 10.1073/pnas.1605350113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crevenna AH, Arciniega M, Dupont A, Mizuno N, Kowalska K, Lange OF, Wedlich-Söldner R, Lamb DC. 2015. Side-binding proteins modulate actin filament dynamics. eLife 4, e04599 ( 10.7554/eLife.04599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Makowska KA, Hughes RE, White KJ, Wells CM, Peckham M. 2015. Specific myosins control actin organization, cell morphology, and migration in prostate cancer cells. Cell Rep. 13, 2118–2125. ( 10.1016/j.celrep.2015.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kendrick-Jones J, Hodge T, Lister I, Roberts R, Buss F. 2001. Myosin Superfamily. eLS ( 10.1038/npg.els.0000673) [DOI]
- 103.Carlier M-F, Pernier J, Avvaru BS. 2013. Control of actin filament dynamics at barbed ends by WH2 domains: from capping to permissive and processive assembly. Cytoskeleton 70, 540–549. ( 10.1002/cm.21124) [DOI] [PubMed] [Google Scholar]
- 104.Winkelman JD, Suarez C, Hocky GM, Harker AJ, Morganthaler AN, Christensen JR, Voth GA, Bartles JR, Kovar DR. 2016. Fascin- and α-actinin-bundled networks contain intrinsic structural features that drive protein sorting. Curr. Biol. 26, 2697–2706. ( 10.1016/j.cub.2016.07.080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tseng Y, Kole TP, Lee JSH, Fedorov E, Almo SC, Schafer BW, Wirtz D. 2005. How actin crosslinking and bundling proteins cooperate to generate an enhanced cell mechanical response. Biochem. Biophys. Res. Commun. 334, 183–192. ( 10.1016/j.bbrc.2005.05.205) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.