Abstract
A dynamic self-organized morphology is the hallmark of network-shaped organisms like slime moulds and fungi. Organisms continuously reorganize their flexible, undifferentiated body plans to forage for food. Among these organisms the slime mould Physarum polycephalum has emerged as a model to investigate how an organism can self-organize their extensive networks and act as a coordinated whole. Cytoplasmic fluid flows flowing through the tubular networks have been identified as the key driver of morphological dynamics. Inquiring how fluid flows can shape living matter from small to large scales opens up many new avenues for research. This article is part of the theme issue ‘Self-organization in cell biology’.
Keywords: morphogenesis, fluid flows, transport
1. Introduction
Many organisms, including a broad range of species of slime moulds and fungi, grow and forage as a single large network. Networks change their morphology over and over again during growth and migration to locate food in a patchy environment [1,2] (see also figure 1a). Moreover, network morphology adapts in response to newly acquired food sources, even connecting food sources in an efficient and robust manner [3]. The striking similarity of the morphological dynamics in foraging fungi and slime moulds is even more surprising if one takes into account that slime moulds and fungi are genetically distinct, with slime moulds being genetically closer to animals than fungi. It is, therefore, likely that it is not biological make-up but the physics of fluid flows within the tubular networks that are critical to the self-organization of network morphology across an individual. Both kinds of living, adaptive networks exhibit oscillatory, long-ranged fluid flows [4–6]. Here, the syncytial plasmodia of the slime mould Physarum polycephalum emerged as a model system to understand the role of flows in coordinating morphology. Fluid flows in this organism are highly coordinated, driving intracellular transport on short time scales but also migration and likely morphological self-organization at long time scales.
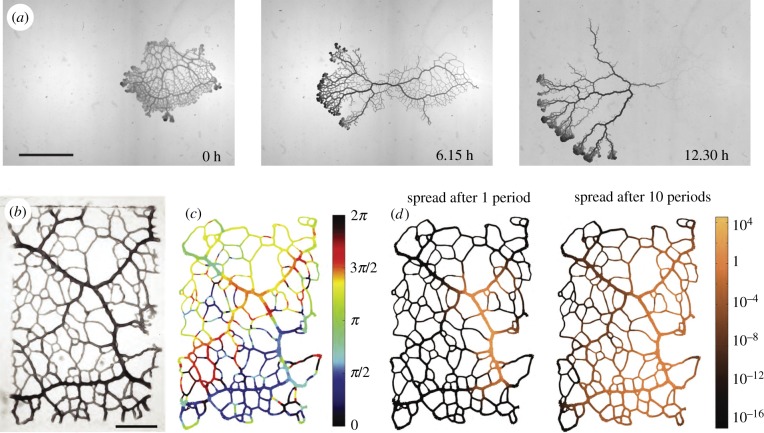
Figure 1.
Network morphology and peristaltic wave driving long-ranged cytoplasmic flows in the plasmodial slime mould P. polycephalum. (a) Self-organization of the network morphology over the course of 12.5 h. An initially well-reticulated network migrates to the left thereby retracting connections and altering tube radius hierarchy. Scale bar, 5 mm. (b) Bright-field microscopy image of P. polycephalum network trimmed to a rectangular shape. Scale bar, 2 mm. (c) Analysis of actin-cytoskeleton-driven contractions of tube walls in (b) reveal phases of contractions to be patterned in a roughly linear gradient. The wavelength of this peristaltic phase pattern on a network matches the organism size. (d) Simulation results of the spread of particles (copper colour) due to the streaming of fluid flow driven by the peristaltic contractions.
2. Cytoplasmic flows organized in a peristaltic wave
In P. polycephalum the fluid cytoplasm within the tubular network streams forth and back in a shuttle flow [6,7]. Network sizes are macroscopic ranging from about 500 μm to 0.5 m—experiments are typically conducted on specimens of up to a few centimetres in size. Flows generally exhibit a Poiseuille profile [6,8] with deviations likely in smaller tubes [9]. Flow is dominated by small Reynolds number Re = 2UR/ν ≈ 0.002 and small Womersley number 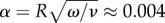 , based on a representative tube radius of R = 50 μm, a flow velocity reaching up to U = 1 mm s−1 [6], a kinematic viscosity of cytoplasm ν = 6.4 × 10−6 m2 s−1 [10] and an oscillation frequency of ω = 0.05 Hz. The cytoplasm is enclosed by gel-like walls that are lined with an actin cortex [11,12]. Actin organizes in circumferential fibrils that contract periodically [13] and drive the cytoplasms' shuttle flow. Contrary to long-lasting speculations about localized pumps driving pressure difference, the common understanding now is that flows arise through network-wide, self-organized contractions of the actin cortex [14,15].
, based on a representative tube radius of R = 50 μm, a flow velocity reaching up to U = 1 mm s−1 [6], a kinematic viscosity of cytoplasm ν = 6.4 × 10−6 m2 s−1 [10] and an oscillation frequency of ω = 0.05 Hz. The cytoplasm is enclosed by gel-like walls that are lined with an actin cortex [11,12]. Actin organizes in circumferential fibrils that contract periodically [13] and drive the cytoplasms' shuttle flow. Contrary to long-lasting speculations about localized pumps driving pressure difference, the common understanding now is that flows arise through network-wide, self-organized contractions of the actin cortex [14,15].
The shuttling cytoplasm itself is very rich in actin. In 1 mm-sized cytoplasm extract droplets, so-called protoplasmic droplets, the actin cortex and the contractions self-organize over time, showing first irregular contraction patterns and later highly coordinated spatio-temporal patterns including standing, travelling and spiral waves [16–19]. Similar dynamics and patterns can be reproduced in models by describing the protoplasmic droplets by two phases of a viscoelastic solid phase representing the cytoskeleton, interpenetrated by a second fluid phase representing the cytosol and coupling both phases by a soluble molecule that activates tension in the solid phase [20,21]. Even on the much larger scale of an entire tubular network contractions are also highly coordinated, forming a peristaltic wave spanning specimens of at least 2 cm in size [14,22] (figure 1b–c). While the contraction period only increases moderately with network size [23,24], the wavelength of the peristaltic wave matches the organism size spanning two orders of magnitude [14]. It is fascinating to investigate how this scaling can arise. Given the success of mechanochemical models for protoplasmic droplets it is likely that the organism-spanning peristaltic wave is also a result of mechanochemical patterning, which could nicely complement our emerging understanding of this novel patterning mechanism widespread in biological systems [25,26]. From a fluid mechanics point a view, a peristaltic wave matching organism size induces the highest flow velocities throughout a network. As flows change with organism morphology, they are likely not only important for organism homoeostasis but also for the coordination of morphological adaptation itself.
3. Effective intracellular transport by oscillatory flow
Peristalsis is a common mechanism in biological systems, creating oscillatory flow and pumping fluid along a tube [27–30]. In P. polycephalum, the tubular network can be considered to be of fixed volume on the time scales of tens of contraction periods. Therefore, net fluid transport is not relevant on these short time scales [14,31]. Yet, creating shuttle flows by a peristaltic wave of contractions is a simple but powerful mechanism to increase the spread of any particles, like metabolites or signalling molecules, within this closed network. Rare measurements show organism-wide, approximately 2 cm, transport of particles within half a contraction period [22] out-competing diffusive spread that would have travelled only 0.25 mm in that time frame. Because of the oscillatory nature of flow, particles flow mainly back to their initial site after a whole period. But the peristaltic flow also increases the effective diffusion 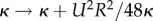 according to Taylor dispersion in long slender tubes [32,33], also applicable to contractile tubes [34,35]. Here, κ denotes the bare molecular diffusivity. Rapid diffusion across the tiny tube cross-sections allows particles to transition between fast and slow streamlines of the Poiseuille profile rapidly increasing their dispersion along the tube by an order of magnitude (figure 1d). The adjacency of large and small tubes and therefore different flow velocities in a slime mould network make it a hard task to theoretically map out how far particles can spread [36]. One successful strategy is to map out an effective dispersion [37,38], which unveils, for example, that particle spread is increased by pruning/coarsening of the network when P. polycephalum is left to starve [38]. This already provides a glimpse of the challenging question on how network morphology impacts network-wide transport [22]. Is morphology geared to optimizing dispersion? If so, how does it evolve towards an optimized morphology? Transport by fluid flow seems to lie at the basis of network self-organization since advected signalling molecules may propagate information about the acquisition of a food source throughout the network by hijacking fluid flows [39]. Here, signalling molecules advected by fluid flow directly increase contraction activity. Further, flow is necessary to synchronize and coordinate contractions [40–42]. It is, therefore, likely that flows play a crucial role in coordinating contractions over space and time.
according to Taylor dispersion in long slender tubes [32,33], also applicable to contractile tubes [34,35]. Here, κ denotes the bare molecular diffusivity. Rapid diffusion across the tiny tube cross-sections allows particles to transition between fast and slow streamlines of the Poiseuille profile rapidly increasing their dispersion along the tube by an order of magnitude (figure 1d). The adjacency of large and small tubes and therefore different flow velocities in a slime mould network make it a hard task to theoretically map out how far particles can spread [36]. One successful strategy is to map out an effective dispersion [37,38], which unveils, for example, that particle spread is increased by pruning/coarsening of the network when P. polycephalum is left to starve [38]. This already provides a glimpse of the challenging question on how network morphology impacts network-wide transport [22]. Is morphology geared to optimizing dispersion? If so, how does it evolve towards an optimized morphology? Transport by fluid flow seems to lie at the basis of network self-organization since advected signalling molecules may propagate information about the acquisition of a food source throughout the network by hijacking fluid flows [39]. Here, signalling molecules advected by fluid flow directly increase contraction activity. Further, flow is necessary to synchronize and coordinate contractions [40–42]. It is, therefore, likely that flows play a crucial role in coordinating contractions over space and time.
4. Cell migration by pumping of peristaltic wave
Cytoplasmic flows form the basis of fast locomotion of very different kinds of amoeba [43–46]. Flows arise by local expansion of the actin cortex and subsequent myosin-dependent contraction [44]. Flows and contractility underlying cell migration are well accessible in the amoeboid plasmodia of P. polycephalum. Plasmodia of 100–500 μm length adopt an amoeboid shape migrating rapidly [47,48] by net fluid transport generated by a contractile peristaltic wave and cortex expansion at the front [49]. Changes in contraction pattern affect locomotion velocity [50]. Plasmodia that exceed ∼500 μm form a full network structure often including multiple migration fronts. Over the peristaltic cycle, a front advances and retracts asymmetrically leading to net advancement at long time scales [51]. As networks grow they move faster [23]. When confined to lanes, migration velocity scales linearly with maximal plasmodium height, reaching locomotion speeds of up to v = 0.4 mm s−1 [23]. Reorientation of migration direction, for example, towards a food source, is associated with a redirection of the peristaltic wave direction to that site [39] further substantiating that migration is governed by fluid flows on long time scales. Given that signalling molecules are advected by fluid flows affecting actin cortex dynamics [39], it seems possible that flows play an important role in the navigation of organisms, acting, for example, during chemotaxis.
5. Morphological changes triggered by cytoplasmic flows
Fluid flows not only transport particles and fluid mass but also exert forces themselves that may induce long-term changes to morphology. Forces may directly feed back onto biochemical reactions triggering complex spatio-temporal dynamics due to this mechanochemical coupling as currently more and more observed in morphogenetic processes [25,26]. Even without the ability to pin down a specific feedback on chemical reactions the influence of forces generated by flow can be investigated on a coarse-grained level. For example, in animal vasculature, it is observed that tube diameters grow with increased flow rate regulating shear force to a balanced level [52]. This observation inspired the idea that fluid shear force induces morphological changes in vasculature, a concept also successfully used in models of P. polycephalum dynamics [22]. Further support is the success of Murray's Law, particularly in plant and animal vasculature [53] which predicts the ratio of tube diameters at a network node under the assumption of conserved shear force. Murray's Law is also consistent with minimizing dissipation, inspiring theoretical work on optimal network architectures [54–56]. Given that P. polycephalum grows its almost transparent tubes in a planar network, testing principles such as Murray's Law [57] and, in general, relating morphological dynamics to flow properties is very feasible. In particular, the adaptability of the network morphology makes it a very suitable system to explore how well certain properties like dissipation, robustness [3] or transport capabilities [58,59] are optimized by living organisms. Equally, as flows are globally coupled throughout the network, there is considerable additional complexity in this system. Indeed, it might well be that precisely this added complexity due to the coupling is the key to have simple mechanisms based on fluid flow give rise to the complex dynamics of self-organization of morphology we observe.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
This work has been supported by the Max Planck Society.
References
- 1.Boddy L, Hynes J, Bebber DP, Fricker MD. 2009. Saprotrophic cord systems: dispersal mechanisms in space and time. Mycoscience 50, 9–19. ( 10.1007/S10267-008-0450-4) [DOI] [Google Scholar]
- 2.Stephenson SL, Stempen H. 1994. Myxomycetes: a handbook of slime molds. Portland, OR: Timber Press, Inc. [Google Scholar]
- 3.Tero A, Takagi S, Saigusa T, Ito K, Bebber DP, Fricker MD, Yumiki K, Kobayashi R, Nakagaki T. 2010. Rules for biologically inspired adaptive network design. Science 327, 439–42. ( 10.1126/science.1177894) [DOI] [PubMed] [Google Scholar]
- 4.Tlalka M, Watkinson SC, Darrah PR, Fricker MD. 2002. Continuous imaging of amino-acid translocation in intact mycelia of Phanerochaete velutina reveals rapid, pulsatile fluxes. New Phytol. 153, 173–184. ( 10.1046/j.0028-646X.2001.00288.x) [DOI] [Google Scholar]
- 5.Tlalka M, Bebber DP, Darrah PR, Watkinson SC, Fricker MD. 2007. Emergence of self-organised oscillatory domains in fungal mycelia. Fungal Genet. Biol. 44, 1085–1095. ( 10.1016/j.fgb.2007.02.013) [DOI] [PubMed] [Google Scholar]
- 6.Kamiya N. 1950. The rate of the protoplasmic flow in the myxomycete plasmodium. I. Cytologia 15, 183–193. ( 10.1508/cytologia.15.183) [DOI] [Google Scholar]
- 7.Stewart PA, Stewart BT. 1959. Protoplasmic movement in slime mold plasmodia: the diffusion drag force hypothesis. Exp. Cell Res. 17, 44–58. ( 10.1016/0014-4827(59)90151-X) [DOI] [PubMed] [Google Scholar]
- 8.Bykov AV, Priezzhev AV, Lauri J, Myllylä R. 2009. Doppler OCT imaging of cytoplasm shuttle flow in Physarum polycephalum. J. Biophotonics 2, 540–547. ( 10.1002/jbio.200910057) [DOI] [PubMed] [Google Scholar]
- 9.Romanovskii Y, Teplov VA. 1995. The physical bases of cell movement. The mechanisms of self-organisation of amoeboid motility. Phys.-Usp. 38, 521–542. ( 10.1070/PU1995v038n05ABEH000086) [DOI] [Google Scholar]
- 10.Swaminathan R, Hoang CP, Verkman AS. 1997. Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys. J. 72, 1900–1907. ( 10.1016/S0006-3495(97)78835-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isenberg G, Wohlfarth-Bottermann KE. 1976. Transformation of cytoplasmic actin. Importance for the organization of the contractile gel reticulum and the contraction–relaxation cycle of cytoplasmic actomyosin. Cell Tissue Res. 173, 495–528. ( 10.1007/BF00224311) [DOI] [PubMed] [Google Scholar]
- 12.Naib-Majani W, Stockem W, Weber K, Wehland J, Wohlfarth-Bottermann KE. 1983. Cytoplasmic actin patterns in Physarum as revealed by NBD-phallacidin staining. Cell Biol. Int. 7, 637–640. ( 10.1016/0309-1651(83)90118-2) [DOI] [PubMed] [Google Scholar]
- 13.Rieu J-P, Delanoë-Ayari H, Takagi S, Tanaka Y, Nakagaki T. 2015. Periodic traction in migrating large amoeba of Physarum polycephalum. J. R. Soc. Interface 12, 20150099 ( 10.1098/rsif.2015.0099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alim K, Amselem G, Peaudecerf F, Brenner MP, Pringle A. 2013. Random network peristalsis in Physarum polycephalum organizes fluid flows across an individual. Proc. Natl Acad. Sci. USA 110, 13 306–13 311. ( 10.1073/pnas.1305049110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teplov VA. 2017. Role of mechanics in the appearance of oscillatory instability and standing waves of the mechanochemical activity in the Physarum polycephalum plasmodium. J. Phys. D: Appl. Phys. 50, 213 002–213 022. ( 10.1088/1361-6463/aa6727) [DOI] [Google Scholar]
- 16.Matsumoto K, Ueda T, Kobatake Y. 1988. Reversal of thermotaxis with oscillatory stimulation in the plasmodium of Physarum polycephalum. J. Theor. Biol. 131, 175–182. ( 10.1016/S0022-5193(88)80235-2) [DOI] [Google Scholar]
- 17.Ueda T. 2005. An intelligent slime mold: a self-organizing system of cell shape and information. In World scientific lecture notes in complex systems: networks of interacting machines (eds Armbruster D, Kaneko K, Mikhailov AS), pp. 221–255. Singapore: World Scientific Publishing Co; ( 10.1142/9789812703248_0009) [DOI] [Google Scholar]
- 18.Takagi S, Ueda T. 2008. Emergence and transitions of dynamic patterns of thickness oscillation of the plasmodium of the true slime mold Physarum polycephalum. Physica D 237, 420–427. ( 10.1016/j.physd.2007.09.012) [DOI] [Google Scholar]
- 19.Takagi S, Ueda T. 2010. Annihilation and creation of rotating waves by a local light pulse in a protoplasmic droplet of the Physarum plasmodium. Physica D 239, 873–878. ( 10.1016/j.physd.2009.06.011) [DOI] [Google Scholar]
- 20.Radszuweit M, Alonso S, Engel H, Bär M. 2013. Intracellular mechanochemical waves in an active poroelastic model. Phys. Rev. Lett. 110, 138102 ( 10.1103/PhysRevLett.110.138102) [DOI] [PubMed] [Google Scholar]
- 21.Alonso S, Strachauer U, Radszuweit M, Bär M, Hauser MJB. 2016. Oscillations and uniaxial mechanochemical waves in a model of an active poroelastic medium: application to deformation patterns in protoplasmic droplets of Physarum polycephalum. Physica D 318–319, 58–69. ( 10.1016/j.physd.2015.09.017) [DOI] [Google Scholar]
- 22.Nakagaki T, Yamada H, Ueda T. 2000. Interaction between cell shape and contraction pattern in the Physarum plasmodium. Biophys. Chem. 84, 195–204. ( 10.1016/S0301-4622(00)00108-3) [DOI] [PubMed] [Google Scholar]
- 23.Kuroda S, Takagi S, Nakagaki T, Ueda T. 2015. Allometry in Physarum plasmodium during free locomotion: size versus shape, speed and rhythm. J. Exp. Biol. 218, 3729–3738. ( 10.1242/jeb.124354) [DOI] [PubMed] [Google Scholar]
- 24.Ueda T, Kobatake Y. 1982. Initiation, development and termination of contraction rhythm in plasmodia of myxomycete Physarum polycephalum. J. Theor. Biol. 97, 87–93. ( 10.1016/0022-5193(82)90280-6) [DOI] [Google Scholar]
- 25.Howard J, Grill SW, Bois JS. 2011. Turing's next steps: the mechanochemical basis of morphogenesis. Nat. Rev. Mol. Cell Biol. 12, 392–398. ( 10.1038/nrm3120) [DOI] [PubMed] [Google Scholar]
- 26.Gross P, Kumar KV, Grill SW. 2017. How active mechanics and regulatory biochemistry combine to form patterns in development. Annu. Rev. Biophys. 46, 337–356. ( 10.1146/annurev-biophys-070816-033602) [DOI] [PubMed] [Google Scholar]
- 27.Shapiro AH, Jaffrin MY, Weinberg SL. 1969. Peristaltic pumping with long wavelengths at low Reynolds number. J. Fluid Mech. 37, 799–825. ( 10.1017/S0022112069000899) [DOI] [Google Scholar]
- 28.Shapiro AH, Jaffrin MY. 1971. Reflux in peristaltic pumping—is it determined by Eulerian or Lagrangian mean velocity. J. Appl. Mech. 38, 1060–1062. ( 10.1115/1.3408917) [DOI] [Google Scholar]
- 29.Selverov KP, Stone HA. 2001. Peristaltically driven channel flows with applications toward micromixing. Phys. Fluids 13, 1837–1859. ( 10.1063/1.1377616) [DOI] [Google Scholar]
- 30.Li M, Brasseur JG. 1993. Non-steady peristaltic transport in finite-length tubes. J. Fluid Mech. 248, 129–151. ( 10.1017/S0022112093000710) [DOI] [Google Scholar]
- 31.Iima M, Nakagaki T. 2012. Peristaltic transport and mixing of cytosol through the whole body of Physarum plasmodium. Math. Med. Biol. 29, 263–281. ( 10.1093/imammb/dqr010) [DOI] [PubMed] [Google Scholar]
- 32.Aris R. 1956. On the dispersion of a solute in a fluid flowing through a tube. Proc. R. Soc. Lond. A 235, 67–77. ( 10.1098/rspa.1956.0065) [DOI] [Google Scholar]
- 33.Taylor G. 1953. Dispersion of soluble matter in solvent flowing slowly through a tube. Proc. R. Soc. Lond. A 219, 186–203. ( 10.1098/rspa.1953.0139) [DOI] [Google Scholar]
- 34.Mercer GN, Roberts AJ. 1990. A center manifold description of contaminant dispersion in channels with varying flow properties. SIAM J. Appl. Math. 50, 1547–1565. ( 10.1137/0150091) [DOI] [Google Scholar]
- 35.Mercer GN, Roberts AJ. 1994. A complete model of shear dispersion in pipes. Japan J. Ind. Appl. Math. 11, 499–521. ( 10.1007/BF03167234) [DOI] [Google Scholar]
- 36.Heaton LL, López E, Maini PK, Fricker MD, Jones NS. 2012. Advection, diffusion, and delivery over a network. Phys. Rev. E 86, 021905 ( 10.1103/PhysRevE.86.021905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saffman PG. 1959. A theory of dispersion in a porous medium. J. Fluid Mech. 6, 321–349. ( 10.1017/S0022112059000672) [DOI] [Google Scholar]
- 38.Marbach S, Alim K, Andrew N, Pringle A, Brenner MP. 2016. Pruning to increase Taylor dispersion in Physarum polycephalum networks. Phys. Rev. Lett. 117, 178103 ( 10.1103/PhysRevLett.117.178103) [DOI] [PubMed] [Google Scholar]
- 39.Alim K, Andrew N, Pringle A, Brenner MP. 2017. Mechanism of signal propagation in Physarum polycephalum. Proc. Natl Acad. Sci. USA 114, 5136–5141. ( 10.1073/pnas.1618114114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimoto Y, Kamiya N. 1978. Studies on contraction rhythm of plasmodial strand: III. Role of endoplasmic streaming in synchronization of local rhythms. Protoplasma 95, 111–121. ( 10.1007/BF01279699) [DOI] [Google Scholar]
- 41.Samans KE, Hinz I, Hejnowicz Z, Wohlfarth-Bottermann KE. 1984. Phase relation of oscillatory contraction cycles in Physarum plasmodia: I. A serial infrared registration device and its application to different plasmodial stages. J. Interdiscip. Cycle Res. 15, 241–250. ( 10.1080/09291018409359857) [DOI] [Google Scholar]
- 42.Achenbach U, Wohlfarth-Bottermann KE. 1981. Synchronization and signal transmission in protoplasmic strands of Physarum. Planta 151, 574–583. ( 10.1007/BF00387437) [DOI] [PubMed] [Google Scholar]
- 43.Taylor DL, Condeelis JS, Moore PL, Allen RD. 1973. The contractile basis of amoeboid movement: I. The chemical control of motility in isolated cytoplasm. J. Cell Biol. 59, 378–394. ( 10.1083/jcb.59.2.378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lämmermann T. et al. 2008. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453, 51–55. ( 10.1038/nature06887) [DOI] [PubMed] [Google Scholar]
- 45.Charras G, Paluch E. 2008. Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 9, 730–736. ( 10.1038/nrm2453) [DOI] [PubMed] [Google Scholar]
- 46.Yoshida K, Soldati T. 2006. Dissection of amoeboid movement into two mechanically distinct modes. J. Cell Sci. 119, 3833–3844. ( 10.1242/jcs.03152) [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto K, Takagi S, Nakagaki T. 2008. Locomotive mechanism of Physarum plasmodia based on spatiotemporal analysis of protoplasmic streaming. Biophys. J. 94, 2492–504. ( 10.1529/biophysj.107.113050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S, Guy RD, Lasheras JC, del Álamo JC. 2017. Self-organized mechano-chemical dynamics in amoeboid locomotion of Physarum fragments. J. Phys. D: Appl. Phys. 50, 204004 ( 10.1088/1361-6463/aa68be) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis OL, Zhang S, Guy RD, delAlamo JC. 2015. Coordination of contractility, adhesion and flow in migrating Physarum amoebae. J. R. Soc. Interface 12, 20141359 ( 10.1098/rsif.2014.1359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodiek B, Takagi S, Ueda T, Hauser MJB. 2015. Patterns of cell thickness oscillations during directional migration of Physarum polycephalum. Eur. Biophys. J. 44, 349–358. ( 10.1007/s00249-015-1028-7) [DOI] [PubMed] [Google Scholar]
- 51.Baumgarten W, Hauser MJB. 2014. Dynamics of frontal extension of an amoeboid cell. Europhys. Lett. 108, 50010 ( 10.1209/0295-5075/108/50010) [DOI] [Google Scholar]
- 52.Kamiya A, Togawa T. 1980. Adaptive regulation of wall shear-stress to flow change in the canine carotid-artery. Am. J. Physiol. 239, H14–H21. ( 10.1152/ajpheart.1980.239.1.H14) [DOI] [PubMed] [Google Scholar]
- 53.Murray CD. 1926. The physiological principle of minimum work: I. The vascular system and the cost of blood volume. Proc. Natl Acad. Sci. USA 12, 207–214. ( 10.1073/pnas.12.3.207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corson F. 2010. Fluctuations and redundancy in optimal transport networks. Phys. Rev. Lett. 104, 048703 ( 10.1103/PhysRevLett.104.048703) [DOI] [PubMed] [Google Scholar]
- 55.Katifori E, Szöllosi GJ, Magnasco MO. 2010. Damage and fluctuations induce loops in optimal transport networks. Phys. Rev. Lett. 104, 048704 ( 10.1103/PhysRevLett.104.048704) [DOI] [PubMed] [Google Scholar]
- 56.Durand M. 2007. Structure of optimal transport networks subject to a global constraint. Phys. Rev. Lett. 98, 088701 ( 10.1103/PhysRevLett.98.088701) [DOI] [PubMed] [Google Scholar]
- 57.Akita D, Kunita I, Fricker MD, Kuroda S, Sato KA, Nakagaki T. 2016. Experimental models for Murray's law. J. Phys. D: Appl. Phys. 50, 024001 ( 10.1088/1361-6463/50/2/024001) [DOI] [Google Scholar]
- 58.Meigel FJ, Alim K. 2017. Flow rate of transport network controls uniform metabolite supply to tissue. (http://arxiv.org/abs/1707.05616[physics.bio-ph]). [Google Scholar]
- 59.Chang SS, Tu S, Baek KI, Pietersen A, Liu Y-H, Savage VM, Hwang S-PL, Hsiai TK, Roper M. 2017. Optimal occlusion uniformly partitions red blood cells fluxes within a microvascular network. PLoS Comput. Biol. 13, e1005892 ( 10.1371/journal.pcbi.1005892) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.