Abstract
Bats represent one of the largest and most striking nocturnal mammalian radiations, exhibiting many visual system specializations for performance in light-limited environments. Despite representing the greatest ecological diversity and species richness in Chiroptera, Neotropical lineages have been undersampled in molecular studies, limiting the potential for identifying signatures of selection on visual genes associated with differences in bat ecology. Here, we investigated how diverse ecological pressures mediate long-term shifts in selection upon long-wavelength (Lws) and short-wavelength (Sws1) opsins, photosensitive cone pigments that form the basis of colour vision in most mammals, including bats. We used codon-based likelihood clade models to test whether ecological variables associated with reliance on visual information (e.g. echolocation ability and diet) or exposure to varying light environments (e.g. roosting behaviour and foraging habitat) mediated shifts in evolutionary rates in bat cone opsin genes. Using additional cone opsin sequences from newly sequenced eye transcriptomes of six Neotropical bat species, we found significant evidence for different ecological pressures influencing the evolution of the cone opsins. While Lws is evolving under significantly lower constraint in highly specialized high-duty cycle echolocating lineages, which have enhanced sonar ability to detect and track targets, variation in Sws1 constraint was significantly associated with foraging habitat, exhibiting elevated rates of evolution in species that forage among vegetation. This suggests that increased reliance on echolocation as well as the spectral environment experienced by foraging bats may differentially influence the evolution of different cone opsins. Our study demonstrates that different ecological variables may underlie contrasting evolutionary patterns in bat visual opsins, and highlights the suitability of clade models for testing ecological hypotheses of visual evolution.
Keywords: visual opsins, chiroptera, niche specialization, likelihood-based codon models, clade models of molecular evolution, evolution of bat vision
1. Introduction
Bats represent the only mammalian radiation to have occupied the aerial nocturnal environment and diversified into a variety of ecological niches [1]. Niche specialization was accompanied by a suite of molecular modifications leading to major shifts in mechanisms underlying sensory perception. The diverse ecological niches bats occupy have been associated with many adaptations, including variation in their chemosensory receptors [2,3], along with contrasting signatures of selection in auditory genes (reviewed in [1]). Although impossible in complete darkness, vision is important for bats and may complement other sensory modalities, such as echolocation, under typical nocturnal light conditions [4].
Vision initiates through the activation of visual pigments, light-sensitive complexes expressed in photoreceptor cells in the retina. These molecules mediate the first steps in the visual transduction cascade and act as an interface between organisms and their environment, having been shown to evolve in response to changes in ecology and light environment, such as spectral composition and light intensity (reviewed in [5]). Because of their functional properties, visual pigments are particularly amenable to molecular evolutionary studies and may greatly contribute to a better understanding of how ecology mediates visual evolution.
Similar to other therian mammals, most bats have two classes of cone visual pigments in their retinae (figure 1) [12,15,16], which are tuned to maximally absorb at very different ranges of the light spectrum: Lws is maximally sensitive to long wavelengths, whereas Sws1 responds to short wavelengths, with peak sensitivity in the UV range in bats [16]. Typically, these cone opsins contribute to colour vision under daylight conditions in most mammals, but their effectiveness is reduced drastically under lower light levels [17]. Because nocturnal light environments are much dimmer than daylight environments, genes associated with daylight vision would be expected to undergo relaxation in bats. However, bat cones operate optimally under lower light levels, such as the intermediate (mesopic) light intensities available at twilight and full moonlight [16], which may have contributed to maintaining bat cone opsin genes under strong evolutionary constraint [12].
Figure 1.
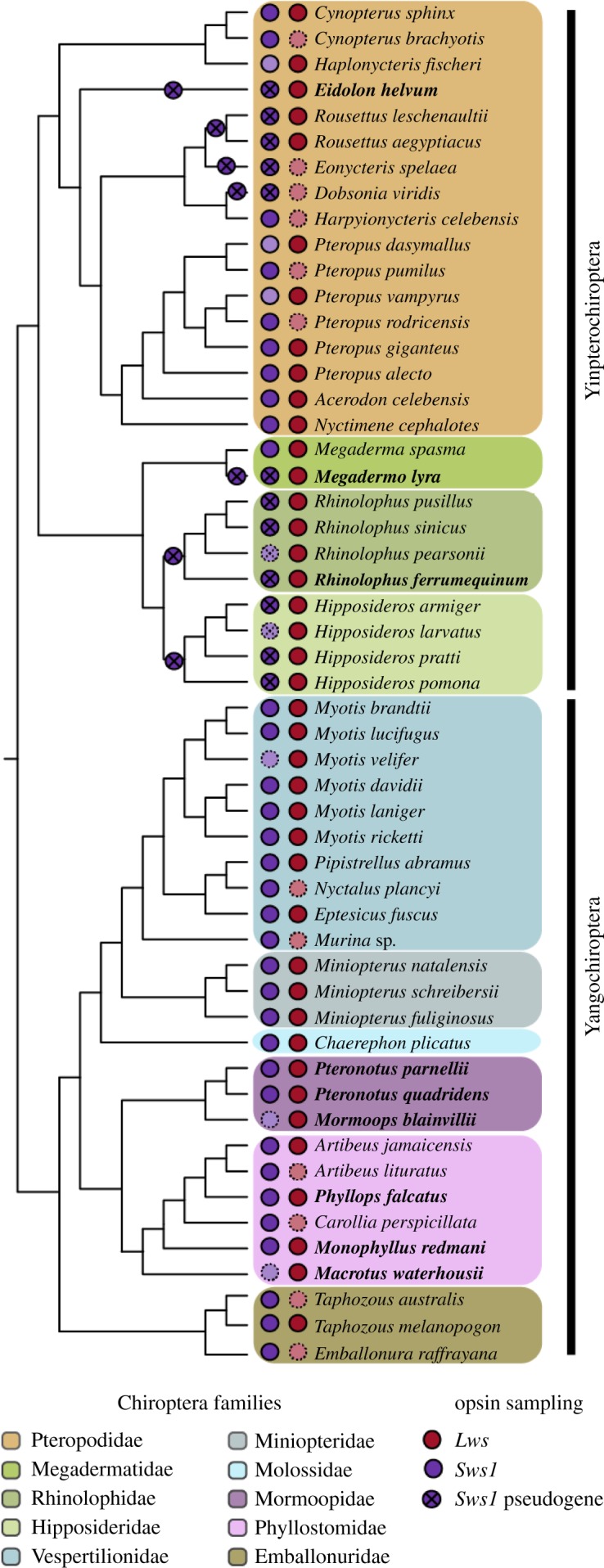
Chiroptera tree depicting species relationships of all bat representatives included in this study, along with respective families and suborders, according to recent literature [6–11]. Symbols preceding species names illustrate Sws1 and Lws gene sequence sampling of each species. Faded symbols with dashed outlines indicate predicted cone opsin set of each bat species for which sequence information is currently unavailable. Species in bold represent new sequences obtained in this study through RNA-seq or genome mining. Symbols above branches denote losses of Sws1 function, according to [12–14]. (Online version in colour.)
While most bats have two classes of cone opsins (figure 1), many exhibit a reduced repertoire resulting from loss of a functional Sws1 via pseudogenization [12], leading to the absence of S cones [18] and loss of sensitivity to short wavelengths of light [19], a pattern that is also observed in several mammalian lineages that inhabit light-limited environments (reviewed in [20]). These losses have occurred in Old World lineages that use sophisticated high-duty cycle (HDC) echolocation (Rhinolophidae and Hipposideridae), which employ constant frequency calls with Doppler shift compensation to navigate and also to detect and classify food items [21]. In these HDC lineages, pseudogenization and subsequent loss of Sws1 function is proposed to represent a sensory trade-off between vision and echolocation [12]. However, Sws1 has also undergone pseudogenization in several cave-roosting Old World fruit bats, which are unable to generate laryngeal echolocation calls, whereas closely related tree-roosting species have maintained a putatively functional Sws1 gene. In this group, the light availability at the roosting site has been proposed to act as a key selective pressure influencing the maintenance of short-wavelength sensitivity [12]. This suggests that not only the degree of reliance on vision but also exposure to light may mediate bat visual evolution.
Echolocation ability and roosting behaviour are not the only ecological variables that may shape bat visual gene evolution. Diet may play an important role, as frugivorous bats generally have proportionally larger projections from retina to brain and cortical areas associated with vision compared with their omnivorous and insectivorous counterparts, which may suggest a greater reliance on visual information [22,23]. Differences in ganglion cell distribution in the retina are also observed among insect-eating bats according to the foraging strategy adopted, such as between aerial insectivores (AI) (seize prey in flight) and insect gleaners (capture prey from substrate), highlighting potential differences in visual ecology [24,25]. Species that forage in distinct habitats (among, near or outside vegetation) [26] are exposed to varying vegetation coverage and light spectra at night, which may also influence how visual pigments evolve, as observed in other nocturnal mammalian groups [27,28]. However, understanding how the impressively diverse ecological niches bats occupy influence their visual evolution remains to be satisfactorily investigated, particularly from the perspective of cone opsin genes.
Despite comprising the greatest ecological diversity and global species richness in Chiroptera [6], Neotropical lineages have been remarkably undersampled in visual opsin studies, limiting the potential for identifying complex signatures of selection associated with differences in bat ecology. Among those lineages, families Phyllostomidae and Mormoopidae account for most of the species diversity in the Neotropics, which may still be greatly underestimated due to cryptic diversity in the region [29,30]. Phyllostomids (New World leaf-nosed bats) arguably represent the most ecologically diverse radiation among bats, comprising species with a variety of roosting behaviours and also highly specialized feeding habits and diets, including convergence of plant-based diets with Old World fruit bats [31]. By contrast, mormoopids (ghost-faced and moustached bats) exclusively comprise insect-eating and rock- or cave-roosting representatives, but include the only species complex (Pteronotus parnellii) in the New World to have independently evolved HDC echolocation [21], which may have had important effects on their visual ecologies.
To capture a wide range of niche specializations of bats, we first sequenced whole-eye transcriptomes of six species of Neotropical bats of families Phyllostomidae and Mormoopidae, from which we obtained new Sws1 and Lws sequences, and combined it with publicly available bat opsin sequences. We hypothesized that certain ecological aspects either associated with the use of visual information (e.g. echolocation ability and diet) or exposure to varying light environment (e.g. roosting behaviour and foraging habitat) may mediate shifts in evolutionary pressure in bat visual opsins and tested this hypothesis using codon-based likelihood clade models. These models have been increasingly applied to evaluate whether complex ecological aspects (e.g. foraging depth and habitat) and evolutionary transitions may have influenced divergent evolutionary rates in distinct lineages in a wide range of vertebrate groups [32–39]. We predicted that ecological specializations associated with diminished reliance on visual cues (e.g. sophisticated echolocation ability and aerial insectivory) and with decreased exposure to light (cave-roosting and foraging among vegetation) may lead to weaker constraint in opsin gene evolution in certain bat lineages. By contrast, variables associated with greater dependence on vision (e.g. non-laryngeal echolocation and frugivory) and increased exposure to ambient light (tree-roosting and foraging outside of vegetation) were expected to mediate greater evolutionary constraint in bat cone opsins.
2. Results
(a). New neotropical bat opsin gene sequences
Nearly complete Lws sequences were obtained from whole-eye transcriptomes of six bat species of families Phyllostomidae and Mormoopidae, not only expanding sampling of Neotropical bat opsin sequences but also improving representation of species with diverse ecological specializations that allowed us to refine hypotheses tested using molecular evolutionary analyses. Putatively functional Sws1 sequences were obtained from the transcriptomes of Phyllops falcatus and Monophyllus redmani (Phyllostomidae) and of Pteronotus parnellii and P. quadridens (Mormoopidae). However, only partial Sws1 fragments were detected for the phyllostomid Macrotus waterhousii and the mormoopid Mormoops blainvillii, and were too short to be included in further analyses. We also identified a number of substitutions in the translated amino acid sequence of M. waterhousii and M. blainvilli Sws1 in regions that are otherwise conserved in bats, hinting at possible pseudogenization (electronic supplementary material, figure S1). In addition to our transcriptomic data, we were also able to obtain Lws sequences from three publicly available chiropteran draft genomes [40], and to confirm the presence of Sws1 pseudogenes in those species as previously reported [12,13]. All new bat visual opsin sequences were deposited on GenBank (accession IDs in electronic supplementary material, table S1).
(b). Bat cone opsin genes are under strong purifying selection
Random sites models [41] were used to estimate selective constraints acting upon each chiropteran cone opsin gene in PAML [42]. Results were consistent on both the species and gene tree topology (electronic supplementary material, figures S2 and S3 and tables S2 and S3), and revealed that overall substitution rate ratios are very low in Lws and Sws1, in agreement with previous analyses in smaller datasets [12]. Significant among-site rate variation in ω was observed for both cone opsin genes (likelihood ratio test (LRT) M3 versus M0, p < 0.001; electronic supplementary material, tables S5 and S6), which is characteristic of functional protein-coding sequences. We also tested whether bat opsin genes may be experiencing positive selection in a subset of sites. Previous studies have hypothesized that bat cone opsin genes would experience relaxation in response to their nocturnal lifestyle, limiting the use of the photopic visual system [12], whereas the lower threshold for cone activation in bats [16] may suggest the acquisition of an enhanced function in cone opsins to operate under lower light levels. However, we found no evidence for relaxed or positive selection in any opsin dataset using random sites models in PAML (LRT M2a versus M1a, M8 versus M8a, p > 0.05; electronic supplementary material, tables S5 and S6) or FUBAR in HyPhy [43] (electronic supplementary material, figure S4), which agreed with previous molecular evolutionary analyses in bat opsin genes [12].
(c). High-duty cycle echolocation underlies significant evolutionary divergence in bat Lws
PAML's Clade model C (CmC) [44], which allows ω to vary along different lineages, was implemented to test whether distinct ecological variables underlie long-term shifts in selective constraint in bat visual opsin genes (hereafter referred to as divergent selection [36]). This hypothesis was tested by partitioning the species tree of each opsin gene according to variables associated with reliance on vision (echolocation ability and diet) or with exposure to light (roosting behaviour and foraging habitat) (electronic supplementary material, figure S5 and table S3) and comparing it with the null model M2a_rel, which does not permit a different ω to be estimated along the phylogeny [45]. Partitioning the Lws species tree according to echolocation ability and diet, and according to roosting behaviour yielded a significantly better fit over the null model (figure 2a). Allowing a separate ω to be estimated in HDC echolocating lineages supported significant shifts in constraint when compared with the null model (p < 0.001, figure 2a; electronic supplementary material, table S4). Significant variation in ω was also observed in AI (p < 0.001) and in cave-roosting species (p < 0.01, figure 2a; electronic supplementary material, table S4).
Figure 2.
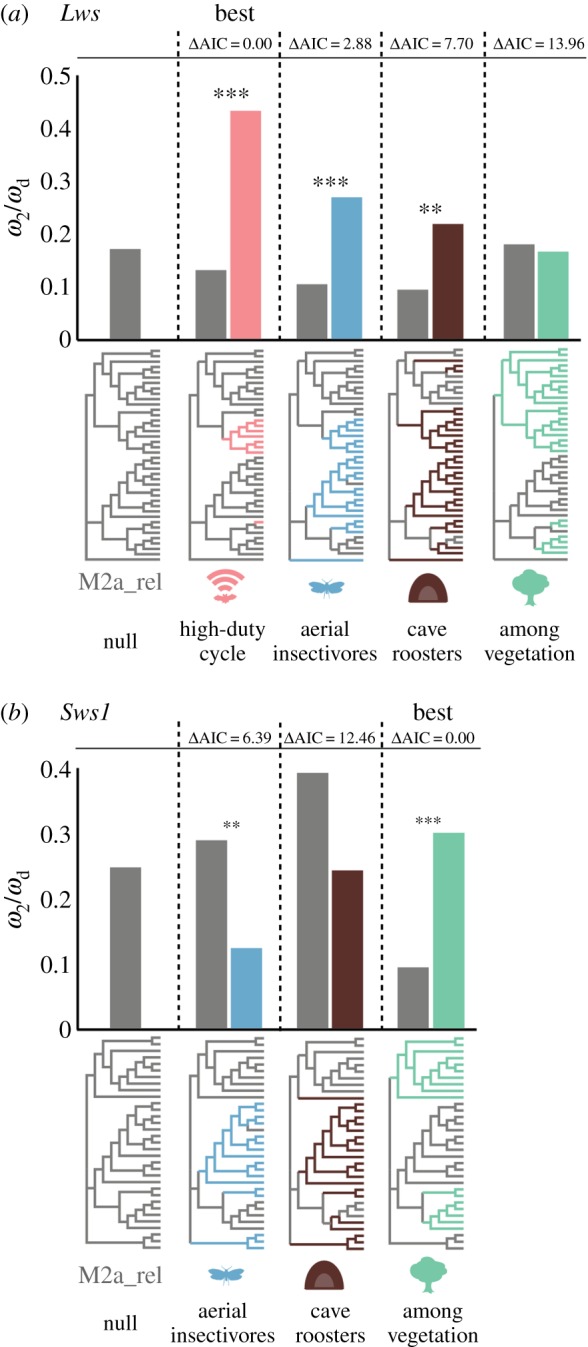
Comparison of divergent omega classes ωd among non-nested partitions obtained in PAML's CmC analysis of bat (a) Lws and (b) Sws1, along with respective ω2 from the null model M2a_rel. Partitions illustrating ecological hypotheses associated with reliance on visual information (HDC echolocation and aerial insectivory) and exposure to varying ambient light (cave roosting and among vegetation foraging) are represented. HDC partition is not depicted in (b) due to widespread losses of Sws1 function in HDC lineages. Topologies below x-axis represent species relationships and partitions used in CmC analyses. Lineages highlighted on each tree were included in the foreground clade. Lineages in grey represent the background clade. LRTs with χ2 distribution were performed to compare CmC with the null model M2a_rel. Statistical significance is indicated by **p < 0.01 and ***p < 0.001. The likelihood of each of non-nested partition models was compared using differences in AIC. Partition that best fits the data is highlighted. (Online version in colour.)
Although these three partitions provided a superior fit to data, overlap among the species tested as foreground clades made it difficult to discriminate which ecological force may have shaped Lws evolution in bats. To discern the effect of distinct ecologies underlying divergent Lws evolution, we compared significant model partitions according to their likelihood using Akaike's information criterion (AIC). We observed that the HDC partition best fitted the data in comparison with AI (ΔAIC = 2.88) as well as cave roosters (ΔAIC = 7.70), suggesting greater influence of sophisticated HDC echolocation on bat Lws evolution (figure 2a; electronic supplementary material, table S4). In this partition, we observed that about 30% of sites in Lws were estimated to be evolving under much lower constraint in HDC bats (ωd = 0.433) compared to the background lineages, which included LDC and non-laryngeal echolocating bats (ω2 = 0.132; electronic supplementary material, table S4). Relaxation of constraint in Lws in HDC lineages was also supported using HyPhy's RELAX model, which estimates a selection intensity parameter k that modulates the magnitude to which ω deviates from neutrality (ωk) [46]. Testing the HDC partition in RELAX resulted in a significantly better fit compared to the null model (p < 0.01) with an estimated k = 0.52, driving ω classes closer to 1 and supporting relaxation in Lws in response to HDC echolocation (electronic supplementary material, table S5).
(d). Foraging habitat mediates significant shifts in evolutionary constraint in bat Sws1
Unlike Lws, our HDC echolocation partition did not perform better than the null model for Sws1 (electronic supplementary material, table S9). This partition was limited to a single branch leading to the Neotropical mormoopid P. parnellii, as a result of widespread losses of Sws1 function in most HDC lineages (figure 1). Instead, our CmC analysis of bat Sws1 yielded a superior fit over the null model when the species tree was partitioned according to diet and foraging habitat (figure 2b). Significant variation in constraint was detected in Sws1 when AI were separated in the foreground clade and compared with a background clade that included bats with other dietary requirements, supporting intensified purifying selection in AI lineages (ωd = 0.13, p < 0.01; figure 2b; electronic supplementary material, table S6). Significant shifts in selection were also detected in species that forage among vegetation relative to species that forage outside or near vegetation coverage, though indicating elevated rates in among vegetation foragers (ωd = 0.30, p < 0.001; figure 2b; electronic supplementary material, table S9). To examine these contrasting results, we compared the likelihood of the two significant partition models using AIC and observed that the among vegetation partition fit the data significantly better compared with AI (ΔAIC = 6.39). In this partition, about 27% of sites in Sws1 were estimated to have evolved under much lower constraint in bats that forage among vegetation (ωd = 0.30) compared with background species (ω2 = 0.09; electronic supplementary material, table S6). We also tested this partition using RELAX and observed significant support for relaxation in Sws1 in among vegetation foragers. Estimating a selection intensity parameter k ≠ 1 fitted the data significantly better compared to the null model (p < 0.001), supporting relaxation of selective forces in Sws1 in response to foraging habitat (k = 0.73; electronic supplementary material, table S7).
3. Discussion
Here, we have used codon-based likelihood models to estimate overall selective pressures acting upon bat cone opsin genes to test whether ecologies associated with reliance on vision or exposure to varying ambient light mediate long-term shifts in constraint in chiropteran Lws and Sws1. We have used eye transcriptome sequencing to obtain new opsin gene sequences from six Neotropical species, which exhibit great ecological diversity and have been poorly sampled in opsin sequencing studies. Along with publicly available sequences, we found that bat cone opsins are experiencing overall strong evolutionary constraint, but we also provide significant evidence for shifts in selection in Lws and Sws1 associated with distinct ecological pressures. Using clade models, we detected significant divergent selection in bat Lws occurring in response to echolocation ability, resulting in elevated rates of evolution in HDC lineages. By contrast, foraging habitat underlies significant shifts in selection pressure in Sws1, leading to relaxation of constraint in species that forage among vegetation.
Our random sites analysis on an expanded bat Lws and Sws1 dataset supported strong purifying selection acting upon the majority of codon sites in bat cone opsin sequences. Although bats have long occupied the nocturnal environment, our work and that of others [12] indicate high levels of sequence conservation in bat cone opsin genes associated with maintenance of protein-coding gene function, suggesting that cone-based vision may play an underappreciated role in chiropteran biology. Although Lws and Sws1 were maintained in most mammalian lineages throughout the nocturnal bottleneck and most extant species still have two functionally distinct classes of cones [47], cone opsins and other genes involved in the bright light visual pathway have been observed to undergo relaxed selection in organisms that either inhabit [48] or have occupied photic-limited environments during their evolutionary history [49]. However, bat cones are predominantly active under intermediate (mesopic) light intensities, suggesting significant cone opsin contribution to bat vision at twilight and full moonlight [16]. Interestingly, several species of bats emerge from the day roost to forage at dusk, when ambient light is enriched with short wavelengths [50], thus probably benefitting from capturing photons with a functional Sws1 pigment that is maximally sensitive in UV range [16]. Similarly, the Lws opsin in bats, with expected peak sensitivity between 530 and 560 nm [16], as with other nocturnal mammals, is presumably tuned to absorb in a range similar to the long-wavelength dominated light available in forested areas, possibly contributing to maximizing capture of scarce photons available at night [27]. Finally, although cone opsins may make individual contributions to maximizing photon capture, the interaction between Sws1 and Lws may support more intricate aspects of vision under intermediate light intensities. Having two different classes of cone opsins in the retina may provide the genetic basis for dichromatic colour vision, though its utility for species that are predominantly nocturnal is still debated [20]. By contrast, maintaining two distinct cone opsin classes may also improve luminance perception. Signal summation from cones expressing Lws and Sws1 may occur in bipolar cells that signal to ganglion cells of the magnocellular pathway, which are responsible for encoding the perception of brightness [51]. The ability to detect and discriminate brightness cues may be particularly advantageous for bats that use vision to identify objects in flight, especially when those are beyond the range of echolocation [4].
Although cone opsin genes experience great sequence conservation in bats, our clade model analyses identified significant long-term shifts in selective constraint intensity in Lws and Sws1 associated with ecological differences in bats. Here, we detected substantial divergent and elevated selection in lineages that use HDC echolocation compared to LDC and non-echolocating groups, suggesting weakened constraint in Lws in response to echolocation ability. The evolution of sophisticated HDC echolocation is hypothesized to have resulted in greater reliance on the auditory system for navigation and foraging behaviour, and was probably accompanied by relaxation of constraint on the visual system, resulting in a sensory trade-off [1]. This hypothesis is supported at the morphological and physiological levels by a reduction in visual brain structures and eye size in HDC bats relative to auditory-associated structures [23,52]. At the molecular level, loss of function of several vision-associated genes has been reported in several HDC species [14,53], including Sws1 [12,14]. In Old World HDC lineages, pseudogenization of vision-associated genes also coincided with faster rates of evolution in the Slc26a5 gene, which encodes the voltage-sensitive membrane protein Prestin in auditory outer hair cells, mediating high-frequency sensitivity in echolocating bats [54]. The hypothesis of a sensory trade-off between hearing and vision is further supported by our analysis in RELAX, which indicates that Lws is undergoing significant relaxation of constraint in HDC bats. This may suggest that maintaining a functional Lws opsin may not be as crucial in species that are more reliant on auditory information, leading to relaxation of selective pressures that eventually result in loss of gene function.
In contrast with Lws, we found no evidence for shifts in selection constraint intensity in response to echolocation ability in species predicted to have a functional Sws1 opsin. Instead, our clade model analyses indicated significant and elevated divergent selection in species that forage among vegetation relative to those that look for food in open space or near the edge of vegetation. This suggests that the environment where bats forage may have a greater contribution for bat visual evolution than previously hypothesized. In forested areas, light attenuation and absorption by leaf photosynthetic pigments influence the intensity and spectral composition of environmental light [55]. As a result, the nocturnal light environment in forests is dim, dominated by middle-to-long wavelengths, and poor in shorter wavelengths, particularly in areas with dense canopy coverage [27]. As such, reduced light intensity and narrower light spectra may result in insufficient photons for Sws1 activation in bats that forage among vegetation, potentially leading to relaxation of constraint upon this visual pigment, which is consistent with our analysis in RELAX. Relaxation of constraint in Sws1 in response to habitat light has also been observed in nocturnal lemurs, in which species that inhabit areas with dense vegetation coverage exhibit either weaker selection upon or even loss of Sws1 function [56]. Interestingly, several species of bats in which a Sws1 pseudogene was identified also forage among vegetation [12,14], which may further support our hypothesis that light spectrum at the foraging site may have greater implications for Sws1 evolution than previously appreciated.
Our study is the first to explicitly test how diverse aspects of bat ecology hypothesized to underlie differences in visual ability may influence cone opsin evolution. We expanded opsin sequence sampling of ecologically diverse Neotropical lineages and provided significant statistical evidence for long-term shifts in selection constraint intensity in Lws and Sws1 in response to ecological variables associated with decreased reliance on visual information and limited exposure to ambient light spectrum. While hypotheses for cone opsin evolution in bats have mostly focused on patterns of loss of gene function associated with ecological differences [12], our analyses of predictably functional Lws and Sws1 sequences highlight the power of codon-based likelihood models combined with knowledge of species ecology to discern the role of ecological factors in shaping bat visual gene evolution. As new bat cone opsin sequences become available and sampling along Chiroptera phylogenetic and ecological diversity is improved, the associations observed in our study can be further tested and refined.
4. Material and methods
(a). Transcriptome sequencing, de novo assembly and opsin gene sequence extraction
Total RNA was extracted from eyes of six Neotropical bat species (the mormoopids M. blainvillii, P. parnellii and P. quadridens, and the phyllostomids M. waterhousii, M. redmani and P. falcatus) collected in the Dominican Republic and deposited at the ROM (electronic supplementary material, table S9). The phyllostomid species represent a wide range of diet specializations, including the frugivorous P. falcatus and the insect gleaner M. waterhousii, whereas the mormoopid P. parnellii is the only lineage in the Neotropics capable of HDC echolocation. RNA extraction was performed using a combined Trizol/RNeasy (Qiagen) protocol according to the manufacturer's instructions, followed by library construction and sequencing on the Illumina HiSeq pipeline at The Centre for Applied Genomics, the Hospital for Sick Children (Toronto). After quality control, transcriptomes were assembled de novo using various assemblers (electronic supplementary material, table S10). Opsin sequences were extracted from assembled transcriptomes using blastn with full-length opsin sequences obtained from available bat genomes as references, and included in its specific dataset for further phylogenetic and molecular evolutionary analyses. A detailed description of tissue acquisition, transcriptome sequencing and de novo assembly can be found in electronic supplementary material.
(b). Dataset preparation, sequence alignment and phylogenetic analysis
Along with new Neotropical bat cone opsin sequences, bat Lws and Sws1 coding sequences were also obtained from GenBank and identified from three recently published bat draft genomes [40] through standalone BLAST. Datasets spanning representatives of ≅ 10 chiropteran families (40 and 33 species for Lws and Sws1, respectively) and outgroup sequences (figure 1; electronic supplementary material, table S3) were aligned by codons in MEGA v.6.0 [57] and used to estimate maximum-likelihood gene trees in PhyML v.3.0 [58]. Estimated gene trees recovered monophyletic relationships of Chiroptera and Phyllostomidae/Mormoopidae lineages, but exhibited contrasting topologies with accepted species relationships at both deep and recent nodes. Because of these discrepancies, we constrained tree topologies to best represent accepted species relationships [6–11], with a basal trichotomy formed by Yinpterochiroptera, Emballonuridae and all remaining Yangochiroptera, as required to run in PAML.
(c). Molecular evolutionary analysis
To investigate the strength and form of selection acting on bat Lws and Sws1, alignments and trees of each opsin gene were analysed with the codeml program in PAML 4.9a [42]. Random sites models were used to estimate variation in the ratio of non-synonymous to synonymous substitution rate (dN/dS or ω) among sites [41]. LRTs with χ2 distribution were carried out to compare random-sites models in order to test for among-site variation in ω (M3 versus M0) and positive selection (M2a versus M1a, and M8 versus M8a) in bat Lws, and Sws1. Datasets were also analysed in HyPhy [59] using the FUBAR model, which performs similarly to PAML's random sites models, but estimates dN and dS independently [43].
PAML's CmC [44] was used to statistically examine whether divergent evolutionary rates (ωd) in bat cone opsin genes occur in response to diverse ecologies that may influence either reliance on visual information (e.g. echolocation and diet) or exposure to ambient light (e.g. roosting behaviour and foraging habitat). A complete description of the various clade model partitions performed on each opsin tree can be found in electronic supplementary material, figure S4 and table S3. Significant divergent selection was tested for using an LRT to compare CmC with the null model M2a_rel, which does not allow divergence of ω in the third class of sites [45]. For each ecological hypothesis, nested models were compared through LRT. For non-nested partitions, AIC values were compared to determine which ecological variable had the greatest effect on the divergence of each visual opsin in bats. All molecular evolutionary analyses were run with starting branch lengths estimated in and repeated at least three times with varying initial starting values for ω and κ (transition to transversion ratio) to avoid local optima. The best-fitting partition in each opsin gene dataset was also analysed using HyPhy's RELAX model [46] to test for evidence of relaxed selection. A detailed description of models used to test for divergent (CmC) and relaxed selection (RELAX) can be found in the electronic supplementary material.
Supplementary Material
Supplementary Material
Supplementary Material
Ethics
Fieldwork to capture and obtain genetic samples followed procedures in accordance with the animal care and use guidelines of the American Society of Mammalogists and was approved by the Animal Use Committee of the Royal Ontario Museum (ROM), Toronto, Ontario, Canada. Collecting permits in the Dominican Republic were issued by Ministerio de Medio Ambiente y Recursos Naturales (Permiso de Exportacion No.: VAPB-01095).
Data accessibility
New sequence data are available from GenBank (accession nos MG775055–MG775064) and listed in electronic supplementary material, table S1.
Authors' contributions
E.A.G. and B.S.W.C. conceived and designed the study. B.K.L. and L.O.L. collected specimens in the field. E.A.G. acquired and analysed experimental data, conducted computational and statistical analyses with help from R.K.S. and M.W.P., and drafted the manuscript. B.S.W.C. and E.A.G. interpreted the results and revised the manuscript, with assistance from R.K.S. and B.K.L.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by a Natural Sciences and Engineering Research Council (NSERC) Discovery grant to B.S.W.C. and a Coordination for Higher Education Personnel (CAPES) Science without Borders fellowship to E.A.G.
References
- 1.Jones G, Teeling EC, Rossiter SJ. 2013. From the ultrasonic to the infrared: molecular evolution and the sensory biology of bats. Front. Physiol. 4, 117 ( 10.3389/fphys.2013.00117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden S, Bekaert M, Goodbla A, Murphy WJ, Dávalos LM, Teeling EC. 2014. A cluster of olfactory receptor genes linked to frugivory in bats. Mol. Biol. Evol. 31, 917–927. ( 10.1093/molbev/msu043) [DOI] [PubMed] [Google Scholar]
- 3.Hong W, Zhao H. 2014. Vampire bats exhibit evolutionary reduction of bitter taste receptor genes common to other bats. Proc. R. Soc. B 281, 20141079 ( 10.1098/rspb.2014.1079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonman A, Bar-On Y, Cvikel N, Yovel Y. 2013. It's not black or white-on the range of vision and echolocation in echolocating bats. Front. Physiol. 4, 248 ( 10.3389/fphys.2013.00248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies WIL, Collin SP, Hunt DM. 2012. Molecular ecology and adaptation of visual photopigments in craniates. Mol. Ecol. 21, 3121–3158. ( 10.1111/j.1365-294X.2012.05617.x) [DOI] [PubMed] [Google Scholar]
- 6.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307, 580–584. ( 10.2307/3840006) [DOI] [PubMed] [Google Scholar]
- 7.Miller-Butterworth CM, Murphy WJ, O'Brien SJ, Jacobs DS, Springer MS, Teeling EC. 2007. A family matter: conclusive resolution of the taxonomic position of the long-fingered bats, Miniopterus. Mol. Biol. Evol. 24, 1553–1561. ( 10.1093/molbev/msm076) [DOI] [PubMed] [Google Scholar]
- 8.Stadelmann B, Lin LK, Kunz TH, Ruedi M. 2007. Molecular phylogeny of new world Myotis (Chiroptera, Vespertilionidae) inferred from mitochondrial and nuclear DNA genes. Mol. Phylogenet. Evol. 43, 32–48. ( 10.1016/j.ympev.2006.06.019) [DOI] [PubMed] [Google Scholar]
- 9.Stoffberg S, Jacobs DS, Mackie IJ, Matthee CA. 2010. Molecular phylogenetics and historical biogeography of Rhinolophus bats. Mol. Phylogenet. Evol. 54, 1–9. ( 10.1016/j.ympev.2009.09.021) [DOI] [PubMed] [Google Scholar]
- 10.Almeida FC, Giannini NP, DeSalle R, Simmons NB. 2011. Evolutionary relationships of the old world fruit bats (Chiroptera, Pteropodidae): another star phylogeny? BMC Evol. Biol. 11, 281 ( 10.1186/1471-2148-11-281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojas D, Warsi OM, Dávalos LM. 2016. Bats (Chiroptera: Noctilionoidea) challenge a recent origin of extant Neotropical diversity. Syst. Biol. 65, 432–448. ( 10.1093/sysbio/syw011) [DOI] [PubMed] [Google Scholar]
- 12.Zhao H, Rossiter SJ, Teeling EC, Li C, Cotton JA, Zhang S. 2009. The evolution of color vision in nocturnal mammals. Proc. Natl Acad. Sci. USA 106, 8980–8985. ( 10.1073/pnas.0813201106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerling CA, Huynh HT, Nguyen MA, Meredith RW, Springer MS. 2015. Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proc. R. Soc. B 282, 20151817 ( 10.1098/rspb.2015.1817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong D, Lei M, Hua P, Pan Y-H, Mu S, Zheng G, Pang E, Lin K, Zhang S. 2017. The genomes of two bat species with long constant frequency echolocation calls. Mol. Biol. Evol. 34, 20–34. ( 10.1093/molbev/msw231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feller KD, Lagerholm S, Clubwala R, Silver MT, Haughey D, Ryan JM, Loew ER, Deutschlander ME, Kenyon KL. 2009. Characterization of photoreceptor cell types in the little brown bat Myotis lucifugus (Vespertilionidae). Comp. Biochem. Physiol. B 154, 412–418. ( 10.1016/j.cbpb.2009.08.006) [DOI] [PubMed] [Google Scholar]
- 16.Müller B, Glösmann M, Peichl L, Knop GC, Hagemann C, Ammermüller J. 2009. Bat eyes have ultraviolet-sensitive cone photoreceptors. PLoS ONE 4, e6390–e6397. ( 10.1371/journal.pone.0006390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb TD. 2010. Phototransduction: adaptation in cones. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 18.Müller B, Goodman SM, Peichl L. 2007. Cone photoreceptor diversity in the retinas of fruit bats (Megachiroptera). Brain Behav. Evol. 70, 90–104. ( 10.1159/000102971) [DOI] [PubMed] [Google Scholar]
- 19.Xuan F, Hu K, Zhu T, Racey P, Wang X, Zhang S, Sun Y. 2012. Immunohistochemical evidence of cone-based ultraviolet vision in divergent bat species and implications for its evolution. Comp. Biochem. Physiol. B 161, 398–403. ( 10.1016/j.cbpb.2012.01.005) [DOI] [PubMed] [Google Scholar]
- 20.Jacobs GH. 2013. Losses of functional opsin genes, short-wavelength cone photopigments, and color vision—a significant trend in the evolution of mammalian vision. Vis. Neurosci. 30, 39–53. ( 10.1017/S0952523812000429) [DOI] [PubMed] [Google Scholar]
- 21.Fenton MB, Faure PA, Ratcliffe JM. 2012. Evolution of high duty cycle echolocation in bats. J. Exp. Biol. 215, 2935–2944. ( 10.1242/jeb.073171) [DOI] [PubMed] [Google Scholar]
- 22.Pettigrew JD. 1986. Flying primates? Megabats have the advanced pathway from eye to midbrain. Science 231, 1304–1306. ( 10.2307/1696160) [DOI] [PubMed] [Google Scholar]
- 23.Baron G, Stephan H, Frahm HD. 1996. Comparative neurobiology in Chiroptera: brain characteristics in functional systems, ecoethological adaptation, adaptive radiation, and evolution. Basel, Switzerland: Birkhauser. [Google Scholar]
- 24.Bell GP, Fenton MB. 1986. V Visual acuity, sensitivity and binocularity in a gleaning insectivorous bat, Macrotus californicus (Chiroptera: Phyllostomidae). Anim. Behav. 34, 409–414. ( 10.1016/S0003-3472(86)80110-5) [DOI] [Google Scholar]
- 25.Pettigrew JD, Dreher B, Hopkins CS. 1988. Peak density and distribution of ganglion cells in the retinae of microchiropteran bats: implications for visual acuity (part 1 of 2). Brain Behav. Evol. 32, 39–56. ( 10.1159/000116531) [DOI] [PubMed] [Google Scholar]
- 26.Denzinger A, Schnitzler H-U. 2013. Bat guilds, a concept to classify the highly diverse foraging and echolocation behaviors of microchiropteran bats. Front. Physiol. 4, 164 ( 10.3389/fphys.2013.00164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veilleux CC, Cummings ME. 2012. Nocturnal light environments and species ecology: implications for nocturnal color vision in forests. J. Exp. Biol. 215, 4085–4096. ( 10.1242/jeb.071415) [DOI] [PubMed] [Google Scholar]
- 28.Moritz GL, Ong PS, Perry GH, Dominy NJ. 2017. Functional preservation and variation in the cone opsin genes of nocturnal tarsiers. Phil. Trans. R. Soc. B 372, 20160075 ( 10.1098/rstb.2016.0075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clare EL, Adams AM, Maya-Simões AZ, Eger JL, Hebert PD, Fenton MB. 2013. Diversification and reproductive isolation: cryptic species in the only New World high-duty cycle bat, Pteronotus parnellii. BMC Evol. Biol. 13, 1 ( 10.1186/1471-2148-13-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim BK, Loureiro LO, Upham NS, Brocca JL. 2017. Phylogeography of dominican republic bats and implications for systematic relationships in the Neotropics. J. Mammal. 98, 986–993. ( 10.1093/jmammal/gyw147) [DOI] [Google Scholar]
- 31.Dumont ER, Davalos LM, Goldberg A, Santana SE, Rex K, Voigt CC. 2012. Morphological innovation, diversification and invasion of a new adaptive zone. Proc. R. Soc. B 279, 1797–1805. ( 10.1098/rspb.2011.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schott RK, Refvik SP, Hauser FE, López-Fernández H, Chang BSW. 2014. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol. Biol. Evol. 31, 1149–1165. ( 10.1093/molbev/msu064) [DOI] [PubMed] [Google Scholar]
- 33.Torres-Dowdall J, Henning F, Elmer KR, Meyer A. 2015. Ecological and lineage-specific factors drive the molecular evolution of rhodopsin in cichlid fishes. Mol. Biol. Evol. 32, 2876–2882. ( 10.1093/molbev/msv159) [DOI] [PubMed] [Google Scholar]
- 34.Van Nynatten A, Bloom D, Chang BSW, Lovejoy NR. 2015. Out of the blue: adaptive visual pigment evolution accompanies Amazon invasion. Biol. Lett. 11, 20150349 ( 10.1098/rsbl.2015.0349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dungan SZ, Kosyakov A, Chang BSW. 2016. S Spectral tuning of killer whale (Orcinus orca) rhodopsin: evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol. Biol. Evol. 33, 323–336. ( 10.1093/molbev/msv217) [DOI] [PubMed] [Google Scholar]
- 36.Baker JL, Dunn KA, Mingrone J, Wood BA, Karpinski BA, Sherwood CC, Wildman DE, Maynard TM, Bielawski JP. 2016. Functional divergence of the nuclear receptor NR2C1 as a modulator of pluripotentiality during hominid evolution. Genetics 203, 905–922. ( 10.1534/genetics.115.183889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castiglione GM, et al. 2017. Evolution of nonspectral rhodopsin function at high altitudes. Proc. Natl Acad. Sci. USA 114, 7385–7390. ( 10.1073/pnas.1705765114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castiglione GM, Schott RK, Hauser FE, Chang BSW. 2017. Convergent selection pressures drive the evolution of rhodopsin kinetics at high altitudes via non-parallel mechanisms. Evolution 72, 170–186. ( 10.1111/evo.13396) [DOI] [PubMed] [Google Scholar]
- 39.Hauser FE, Ilves KL, Schott RK, Castiglione GM, López-Fernández H, Chang BSW. 2017. Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol. Biol. Evol. 34, 2650–2664. ( 10.1093/molbev/msx192) [DOI] [PubMed] [Google Scholar]
- 40.Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ. 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502, 228–231. ( 10.1038/nature12511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z, Nielsen R, Goldman N, Pedersen AM. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 43.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 30, 1196–1205. ( 10.1093/molbev/mst030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bielawski JP, Yang Z. 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 59, 1–12. ( 10.1007/s00239-004-2597-8) [DOI] [PubMed] [Google Scholar]
- 45.Weadick CJ, Chang BSW. 2012. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol. Biol. Evol. 29, 1297–1300. ( 10.1093/molbev/msr311) [DOI] [PubMed] [Google Scholar]
- 46.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2014. RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 32, 820–832. ( 10.1093/molbev/msu400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerkema MP, Davies WIL, Foster RG, Menaker M, Hut RA. 2013. The nocturnal bottleneck and the evolution of activity patterns in mammals. Proc. R. Soc. B 280, 20130508 ( 10.1098/rspb.2013.0508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emerling CA, Springer MS. 2014. Eyes underground: regression of visual protein networks in subterranean mammals. Mol. Biol. Evol. 78, 260–270. ( 10.1016/j.ympev.2014.05.016) [DOI] [PubMed] [Google Scholar]
- 49.Emerling CA, Springer MS. 2014. Genomic evidence for rod monochromacy in sloths and armadillos suggests early subterranean history for Xenarthra. Proc. R. Soc. B 282, 20142192 ( 10.1098/rspb.2014.2192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnsen S, Kelber A, Warrant E, Sweeney AM, Widder EA, Lee RL, Hernández-Andrés J. 2006. Crepuscular and nocturnal illumination and its effects on color perception by the nocturnal hawkmoth Deilephila elpenor. J. Exp. Biol. 209, 789–800. ( 10.1242/jeb.02053) [DOI] [PubMed] [Google Scholar]
- 51.Li W, DeVries SH. 2006. Bipolar cell pathways for color and luminance vision in a dichromatic mammalian retina. Nat. Neurosci. 9, 669–675. ( 10.1038/nn1686) [DOI] [PubMed] [Google Scholar]
- 52.Thiagavel J, Cechetto C, Santana SE, Jakobsen L, Warrant EJ, Ratcliffe JM. 2017. Auditory opportunity and visual constraint enabled the evolution of echolocation in bats. Nat. Commun. 9, 1–10. ( 10.1038/s41467-017-02532-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen B, Fang T, Dai M, Jones G, Zhang S. 2013. Independent losses of visual perception genes Gja10 and Rbp3 in echolocating bats (Order: Chiroptera). PLoS ONE 8, e68867 ( 10.1371/journal.pone.0068867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z, Qi F-Y, Zhou X, Ren H-Q, Shi P. 2014. Parallel sites implicate functional convergence of the hearing gene prestin among echolocating mammals. Mol. Biol. Evol. 31, 2415–2424. ( 10.1093/molbev/msu194) [DOI] [PubMed] [Google Scholar]
- 55.Endler JA. 1993. The color of light in forests and its implications. Ecol. Monograph 63, 1–27. ( 10.2307/2937121) [DOI] [Google Scholar]
- 56.Veilleux CC, Louis EE, Bolnick DA. 2013. Nocturnal light environments influence color vision and signatures of selection on the OPN1SW opsin gene in nocturnal lemurs. Mol. Biol. Evol. 30, 1420–1437. ( 10.1093/molbev/mst058) [DOI] [PubMed] [Google Scholar]
- 57.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 59.Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. ( 10.1093/bioinformatics/bti079) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New sequence data are available from GenBank (accession nos MG775055–MG775064) and listed in electronic supplementary material, table S1.


