Abstract Abstract
Cylindrocladiella spp. are widely distributed especially in tropical and sub-tropical regions, where they are mainly known as saprobes although some species are plant pathogens. Very little is known about these fungi in South-East Asia. The aim of this study was to identify a collection of Cylindrocladiella isolates from soils collected in forest nurseries and plantations in Vietnam and Malaysia. This was achieved using DNA sequence comparisons and morphological observations. The study revealed two previously described species, Cy. lageniformis and Cy. peruviana as well as five novel taxa, described here as Cy. arbusta sp. nov., Cy. malesiana sp. nov., Cy. obpyriformis sp. nov., Cy. parvispora sp. nov. and Cy. solicola sp. nov. A relatively small collection of isolates from a limited geographic sampling revealed an unexpectedly high level of Cylindrocladiella diversity suggesting that many more species in this genus await discovery in South-East Asia.
Keywords: multigene phylogeny, plantation forestry, taxonomy
Introduction
Cylindrocladiella (Hypocreales, Nectriaceae) are soil-borne fungi that have commonly been confused with the asexual morph of the closely related genus Calonectria (Crous 2002). Species of Cylindrocladiella can be distinguished from Calonectria spp. by their aseptate stipe extensions, distinctive conidiophore branching patterns and their small 1-septate conidia. In addition, they have sexual morphs in Nectricladiella that are very different to those in Calonectria (Boesewinkel 1982, Crous and Wingfield 1993, Schoch et al. 2000, Crous 2002). Multigene phylogenetic inference has led to the description of a relatively large number of novel species and to the delimitation of cryptic species (Schoch et al. 2000, van Coller et al. 2005, Lombard et al. 2012, 2017). Currently, Cylindrocladiella accommodates 35 species (Crous 2002, van Coller et al. 2005, Inderbitzin et al. 2012, Lombard et al. 2012, 2017, Crous et al. 2017).
Species of Cylindrocladiella are distributed globally, especially in the tropical, sub-tropical and temperate regions of the world (Crous 2002, Lombard et al. 2012). These fungi are not typically considered primary pathogens although their role in causing plant disease is likely underestimated. The fact that they are isolated using baiting with living plant tissue similar to the approach for Calonectria spp. (Crous 2002), suggests some level of pathogenicity. Disease symptoms that have been associated with Cylindrocladiella include leaf spot (Mohanan and Sharma 1985, Crous et al. 1991, Crous and Wingfield 1993), damping off (Sharma and Mohanan 1982, Scattolin and Montecchio 2007) and shoot die-back (Brielmaier-Liebetanz et al. 2013). Cylindrocladiella spp. are, however, most frequently associated with root diseases (Crous et al. 1991, Crous and Wingfield 1993, Crous 2002). They have, for example, been reported causing root rot on Eucalyptus spp. (Mohanan and Sharma 1985, Crous and Wingfield 1993) and Pinus sp. (Boesewinkel 1982) in forestry nurseries. They have also been associated with root rot of peanut (Crous and Wingfield 1993), tea (Peerally 1974), kiwi fruit (Erper et al. 2013) and black-foot disease of grapevines (Agustí-Brisach and Armengol 2013, Armengol and Gramaje 2016, Carlucci et al. 2017).
Thirteen species of Cylindrocladiella have been reported from South-East Asia from Indonesia and Thailand (Crous 2002, Lombard et al. 2012, 2017). Of these, only four species (Cy. camelliae, Cy. infestans, Cy. microcylindrica and Cy. viticola), have been isolated from plant tissues, with the other nine species having been isolated from soil (Crous 2002, Lombard et al. 2012, 2017). However, nothing is known regarding their role as plant pathogens in this region.
In order to provide a better understanding about the diversity of Cylindrocladiella species in South-East Asia, this study aimed at identifying a collection of Cylindrocladiella isolates obtained from soils collected in plantations and nurseries in Malaysia and Vietnam. This was achieved using multigene sequence comparisons and morphological observations.
Materials and methods
Isolates
Soil samples were collected from various plantations and nurseries in Malaysia and Vietnam and baited with germinating alfalfa (Medicago sativa) seeds as described by Crous (2002). Direct isolations from fungal structures were made on to malt extract agar (MEA; 2 % w/v; Biolab, Midrand, South Africa). Cultures were incubated for 3–7 d at 25 °C and purified by transferring single hyphal tips from primary isolations to fresh MEA plates. Cultures were deposited in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa with representative isolates in the collection of the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands. Dried specimens were deposited in the National Collection of Fungi (PREM), Pretoria, South Africa.
DNA sequencing and phylogenetic analyses
Seven-day-old fungal cultures grown on MEA at 25 °C were used for DNA extraction using Prepman® Ultra Sample Preparation Reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the protocols provided by the manufacturer. Four loci were amplified and sequenced including the internal transcribed spacer (ITS) region using primers ITS1F (Gardes and Bruns 1993) and ITS4 (White et al. 1990); partial fragments of the translation elongation factor 1-alpha (tef1) gene region using primers EF1-728F (Carbone and Kohn 1999) and EF-2 (O’Donnell et al. 1998); partial fragments of the β-tubulin (tub2) gene region using primers T1 (O’Donnell and Cigelnik 1997) and CYLTUB1R (Crous et al. 2004a) and part of the Histone H3 (his3) gene region using primers CYLH3F and CYLH3R (Crous et al. 2004a).
The PCR reactions were conducted as described by Pham (2018). Amplified fragments were purified using ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The products were sequenced in both directions with the same primers used for amplification, using the BigDye terminator sequencing kit v. 3.1 (Applied Biosystems, USA) on an ABI PRISM 3100 DNA sequencer (Applied Biosystems, USA).
Raw sequences were assembled and edited using Geneious v. 7.0 (Kearse et al. 2012). Sequence data were compared with other closely related Cylindrocladiella spp. available on the GenBank database. Sequences were aligned using MAFFT v. 7 (Katoh and Standley 2013), then edited manually in MEGA v. 7 (Kumar et al. 2016).
Maximum Parsimony (MP) and Maximum Likelihood (ML) analyses were performed on data sets for each gene region and the combined data set. For MP, analyses were conducted using PAUP v. 4.0b10 (Swofford 2003) with phylogenic relationships estimated by heuristic searches with 1000 random stepwise addition sequences and tree bisection and reconstruction (TBR) branch-swapping. Alignment gaps were treated as missing data and all characters were weighted equally. Measures calculated for parsimony included tree length (TL), retention index (RI), consistency index (CI), rescaled consistency index (RC) and homoplasy index (HI). Statistical support for branch nodes in the most parsimonious trees was obtained by performing 1000 bootstrap replicates. For ML, the appropriate substitution model was obtained using the software package jModeltest v. 2.1.5 (Posada 2008). The ML phylogenetic trees were generated using PhyML v. 3.0 (Guindon and Gascuel 2003). Confidence levels for the nodes were determined using 1000 replication bootstrap analyses. For both MP and ML, Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5638) were used as the outgroup taxa. All resulting trees were viewed using MEGA v. 7 (Kumar et al. 2016).
Taxonomy
Morphological characteristics were assessed using single hyphal tip cultures on synthetic low-nutrient agar (SNA; Nirenburg 1981) and incubated at 25 °C for 3–7 d. In some cases, pieces of carnation leaf were added to the media to induce sporulation. Fungal structures were studied by mounting in 80 % lactic acid on glass sides and examined using a Nikon H550L microscope (Nikon, Japan). Thirty to fifty measurements were made for all taxonomically informative characters depending on their availability. The 95 % confidence levels were determined and extremes of conidial measurements are given in parentheses. For all other fungal structures, only extremes are presented. Colony colour and morphology were assessed using 7-d-old cultures on MEA grown at 25 °C using the colour charts of Rayner (1970). To determine the optimal temperature for growth, cultures were transferred to MEA and incubated at temperatures ranging from 5 to 35 °C at 5 °C intervals. Fungal descriptions and associated metadata were deposited in MycoBank (Crous et al. 2004b).
Results
Isolates
Nineteen isolates in total were obtained from soil baits. Of these, 15 were from Vietnam (nine from Tuyen Quang, four from Nghe An, one from Vinh Phuc and one from Hanoi) and four were from Sabah, Malaysia. The majority (16) of the isolates were from soils collected from Acacia plantations (Table 1).
Table 1.
Collection details and GenBank accessions of Cylindrocladiella isolates included in the phylogenetic analysis.
| Species | Isolate number1,3 | Substrate | Locality | Genbank accession2 | References | |||
|---|---|---|---|---|---|---|---|---|
| tub2 | his3 | tef1 | ITS | |||||
| Cy. arbusta | CMW 47295T; CBS 143546 | soil in Acacia mangium plantation | Tan Ky, Nghe An, Vietnam | MH016958 | MH016996 | MH016977 | MH017015 | This study |
| CMW 47296; CBS 143547 | soil in A. mangium plantation | Tan Ky, Nghe An, Vietnam | MH016959 | MH016997 | MH016978 | MH017016 | This study | |
| Cy. camelliae | CPC 234; PPRI 3990; IMI 346845 | Eucalyptus grandis | South Africa | AY793471 | AY793509 | JN099087 | AF220952 | Boesewinkel 1982 |
| CPC 237 | E. grandis | South Africa | JN098749 | JN098839 | JN099090 | JN100573 | Boesewinkel 1982 | |
| Cy. clavata | CBS 129563; CPC 17591 | soil | Australia | JN098751 | JN098859 | JN098975 | JN099096 | Lombard et al. 2012 |
| CBS 129564T; CPC 17592 | soil | Australia | JN098752 | JN098858 | JN098974 | JN099095 | Lombard et al. 2012 | |
| Cy. cymbiformis | CBS 129553T; CPC 17393 | soil | Australia | JN098753 | JN098866 | JN098988 | JN099103 | Lombard et al. 2012 |
| Cy. elegans | CBS 338.92T; PPRI 4050; IMI 346847 | leaf litter | South Africa | AY793474 | AY793512 | JN099039 | AY793444 | Crous and Wingfield 1993 |
| CBS 110801; CPC 525 | leaf litter | South Africa | JN098755 | JN098916 | JN099044 | JN100609 | Crous and Wingfield 1993 | |
| Cy. lageniformis | CBS 340.92T; PPRI 4449; UFV 115 | Eucalyptus sp. | Brazil | AY793481 | AY793520 | JN099003 | AF220959 | Crous and Wingfield 1993 |
| CBS 111060; CPC 1240 | Eucalyptus sp. | South Africa | JN098770 | JN098918 | JN099046 | JN100611 | Crous and Wingfield 1993 | |
| CMW 47419 | soil in E. camaldulensis plantation | Hoang Mai, Nghe An, Vietnam | MH016972 | MH017010 | MH016991 | MH017029 | This study | |
| Cy. lanceolata | CBS 129565; CPC 17566 | soil | Australia | JN098788 | JN098939 | JN099069 | JN100632 | Lombard et al. 2012 |
| CBS 129566T; CPC 17567 | soil | Australia | JN098789 | JN098862 | JN098978 | JN099099 | Lombard et al. 2012 | |
| Cy. longiphialidica | CBS 129557T; CPC 18839 | soil | Thailand | JN098790 | JN098851 | JN098966 | JN100585 | Lombard et al. 2012 |
| CBS 129558 | soil | Thailand | JN098791 | JN098852 | JN098967 | JN100586 | Lombard et al. 2012 | |
| Cy. malesiana | CMW 48276; CBS 143549 | soil in A. mangium plantation | Tawau, Sabah, Malaysia | MH016960 | MH016998 | MH016979 | MH017017 | This study |
| CMW 48277; CBS 143550 | soil in A. mangium plantation | Tawau, Sabah, Malaysia | MH016961 | MH016999 | MH016980 | MH017018 | This study | |
| CMW 48278T; CBS 143548 | soil in A. mangium plantation | Tawau, Sabah, Malaysia | MH016962 | MH017000 | MH016981 | MH017019 | This study | |
| Cy. malesiana | CMW 48279 | soil in A. mangium plantation | Tawau, Sabah, Malaysia | MH016963 | MH017001 | MH016982 | MH017020 | This study |
| Cy. microcylindrica | CBS 111794T; ATCC 38571; CPC 2375 | Echeveria elegans | Indonesia | AY793483 | AY793523 | JN099041 | AY793452 | Schoch et al. 2000 |
| Cy. natalensis | CBS 110800; CPC 529 | soil | South Africa | JN098793 | JN098915 | JN099043 | JN100608 | Lombard et al. 2012 |
| CBS 114943T; CPC 456 | Arachis hypogaea | South Africa | JN098794 | JN098895 | JN099016 | JN100588 | Lombard et al. 2012 | |
| Cy. nederlandica | CBS 143.95; PD94/1353 | Kalanchoe sp. | The Netherlands | JN098798 | JN098891 | JN099013 | JN099129 | Lombard et al. 2012 |
| CBS 152.91T; PD90/2015 | Pelargonium sp. | The Netherlands | JN098800 | JN098910 | JN099033 | JN100603 | Lombard et al. 2012 | |
| Cy. novaezelandica | CBS 486.77T; ATCC 44815; CPC 2397 | Rhododendron indicum | New Zealand | AY793485 | AY793525 | JN099050 | AF220963 | Boesewinkel 1982 |
| Cy. obpyriformis | CMW 47194T; CBS 143552 | soil in Acacia hybrid plantation | Tuyen Quang, Vietnam | MH016965 | MH017003 | MH016984 | MH017022 | This study |
| CMW 49940; CBS 143553 | soil in Camellia chrysantha nursery | Tam Dao, Vinh Phuc, Vietnam | MH016966 | MH017004 | MH016985 | MH017023 | This study | |
| Cy. parvispora | CMW 47193 | soil in Acacia hybrid plantation | Tuyen Quang, Vietnam | MH016967 | MH017005 | MH016986 | MH017024 | This study |
| CMW 47197T; CBS 143554 | soil in Acacia hybrid plantation | Tuyen Quang, Vietnam | MH016968 | MH017006 | MH016987 | MH017025 | This study | |
| CMW 47207; CBS 143555 | soil in Acacia hybrid plantation | Tuyen Quang, Vietnam | MH016969 | MH017007 | MH016988 | MH017026 | This study | |
| CMW 47208; CBS 143556 | soil in Acacia hybrid plantation | Tuyen Quang, Vietnam | MH016970 | MH017008 | MH016989 | MH017027 | This study | |
| CMW 47315 | soil in A. mangium plantation | Son Duong, Tuyen Quang, Vietnam | MH016971 | MH017009 | MH016990 | MH017028 | This study | |
| Cy. peruviana | CBS 113022; CPC 4291 | Eucalyptus sp. | South Africa | JN098801 | JN098906 | JN099029 | JN100599 | Boesewinkel 1982 |
| CPC 2404T; IMUR 1843 | ants | Peru | AY793500 | AY793540 | JN098968 | AF220966 | Boesewinkel 1982 | |
| CMW 47297 | soil in A. mangium plantation | Tan Ky, Nghe An, Vietnam | MH016973 | MH017011 | MH016992 | MH017030 | This study | |
| CMW 47304 | soil in A. mangium plantation | Son Duong, Tuyen Quang, Vietnam | MH016974 | MH017012 | MH016993 | MH017031 | This study | |
| CMW 47333 | soil in A. mangium plantation | Son Duong, Tuyen Quang, Vietnam | MH016975 | MH017013 | MH016994 | MH017032 | This study | |
| Cy. peruviana | CMW 47416 | soil | Bac Tu Liem, Hanoi, Vietnam | MH016976 | MH017014 | MH016995 | MH017033 | This study |
| Cy. pseudocamelliae | CBS 129555T; CPC 18825 | soil | Thailand | JN098814 | JN098843 | JN098958 | JN100577 | Lombard et al. 2012 |
| CBS 129556; CPC 18832 | soil | Thailand | JN098815 | JN098846 | JN098961 | JN100580 | Lombard et al. 2012 | |
| Cy. solicola | CMW 47198T; CBS 143551 | soil in Acacia hybrid plantation | Tuyen Quang, Vietnam | MH016964 | MH017002 | MH016983 | MH017021 | This study |
| Cy. variabilis | CBS 375.93; IMI 317057 | Mangifera indica | India | JN098836 | JN098881 | JN099000 | JN099119 | Lombard et al. 2012 |
|
CBS 129561T; CPC 17505 |
soil | Australia | JN098719 | JN098950 | JN099080 | JN100643 | Lombard et al. 2012 | |
1 CBS: Culture collection of Westerdijk Fungal Biodiversity Institute (WI), Utrecht, the Netherlands; CMW: Culture collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa; CPC: Pedro Crous working collection housed at WI; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, UK; IMUR: Institute of Mycology, University of Recife, Recife, Brazil; ATCC: American Type Culture Collection, Virginia, U.S.A; PPRI: Plant Protection Research Institute, Agricultural Research Council, Pretoria, South Africa; UFV: Universidade Federal de Viçosa, Viçosa, Brazil.
2tub2 = β-tubulin;
his3 = histone H3;
tef1 = translation elongation factor 1-alpha; ITS = Internal transcribed spacer regions 1 and 2 and the 5.8S gene of the ribosomal RNA.
T Ex-type cultures.
3 Isolates obtained during the survey in this study are indicated in bold.
Phylogenetic analyses
Approximately 500–570 bases were obtained for each of the his3, tef1, tub2 and ITS loci. For the ML analyses of each individual data sets, the TIM2+G model was selected for his3; GTR+G model for tef1; TrN+I+G for tub2 and the K80+I+G for ITS. The ML tree of each individual gene region with bootstrap support values of both the ML and MP analyses are presented in Suppl. materials 1–4.
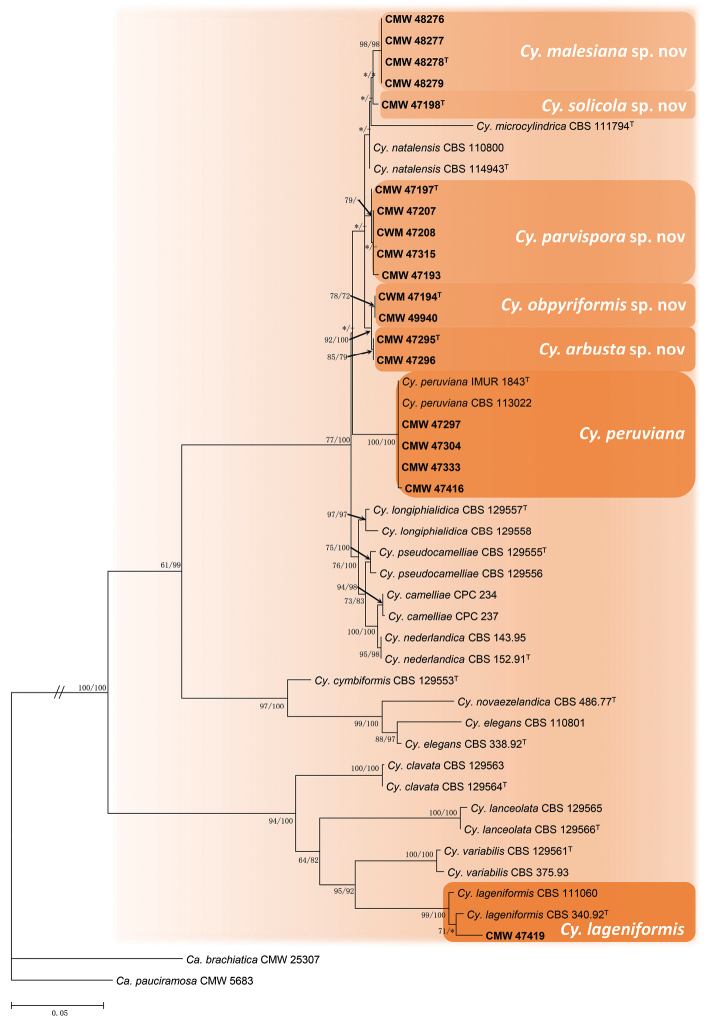
The combined data set of his3, tef1, tub2 and ITS, included 44 ingroup taxa and two outgroup taxa. The data set consisted of 2054 characters, of which 640 were parsimony-informative and 1414 characters were excluded. The MP analysis yielded 1000 trees (TL = 1414; CI = 0.691; RI = 0.880; RC = 0.608; HI = 0.309). The TIM2+I+G model was selected for the combined data set for the ML analyses. The ML tree with bootstrap support values of both the ML and MP analyses is presented in Figure 1.
Figure 1.
Phylogenetic tree based on maximum likelihood (ML) analysis of a combined data set of his3, tef1, tub2 and ITS sequence alignments. Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60 % are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
In the phylogenetic tree (Figure 1), four isolates (CMW 47297, CMW 47304, CMW 47333, CMW 47416) clustered in the clade representing Cy. peruviana (ex-type IMUR 1843). Cylindrocladiella lageniformis (ex-type CBS 340.92) was represented by CMW 47419. The remaining isolates resided in five distinct clades representing novel taxa, accommodating four isolates (CMW 48276, CMW 48277, CMW 48278, CMW 48279), one isolate (CMW 47198), five isolates (CMW 47193, CMW 47197, CMW 47207, CMW 47208, CMW 47315), two isolates (CMW 47194, CMW 49940) and two isolates (CMW 47295, CMW 47296) respectively.
Taxonomy
Morphological comparisons and phylogenetic inference showed that 19 Cylindrocladiella isolates represented five novel species along with two previously described species, Cy. lageniformis (CMW 47419) and Cy. peruviana (CMW 47297, CMW 47304, CMW 47333, CMW 47416). The novel taxa are provided with names in Cylindrocladiella and their important morphological characteristics are compared in Table 2.
Table 2.
Comparisons of morphological characteristics of Cylindrocladiella spp. included in this study.
| Species | Stipe extension | Vesicle | Macroconidia | Subverticillate conidiophores | References | ||
|---|---|---|---|---|---|---|---|
| Length (µm) | Diam (µm) | Shape | Size (µm) | Average (µm) | |||
| Cy. arbusta | 93–139 | 4–5.5 | obpyriform to lanceolate | (8.5–)10–12 (–13.5) × 2–3 | 11 × 2.5 | moderate | This study |
| Cy. malesiana | 114.5–144.5 | 4.5–6 | fusoid to lanceolate | (10–)11–13(–13.5) × (1.5–)2–2.5 | 12 × 2 | abundant | This study |
| Cy. microcylindrica | 70–130 | 3–4 | cylindrical to lanceolate | (10–)12–14 (–15) × 2(–3) | 12.5 × 2 | abundant | Schoch et al. 2000 |
| Cy. natalensis | 82–127 | 6–8 | ellipsoidal to fusoid | (12–)14–16 (–17) × 2–3 | 15× 3 | moderate | Lombard et al. 2012 |
| Cy. obpyriformis | 86.5–150 | 4–7 | obpyriform | (9–)11–13(–15) × 2–3(–3.5) | 12 × 2.5 | abundant | This study |
| Cy. parvispora | 112.5–141 | 4.5–6.5 | fusoid to cylindrical | (8–)10– 12 (–13) × 2–2.5 | 11 × 2 | moderate | This study |
| Cy. solicola | 93.5–170 | 3.5–6.5 | broadly clavate to lanceolate to fusiform | (10.5–)12.5–14.5(–15.5) × 2–3 | 13.5 × 2.5 | abundant | This study |
Cylindrocladiella arbusta
N.Q. Pham, T.Q. Pham & M.J. Wingf. sp. nov.
MB824550
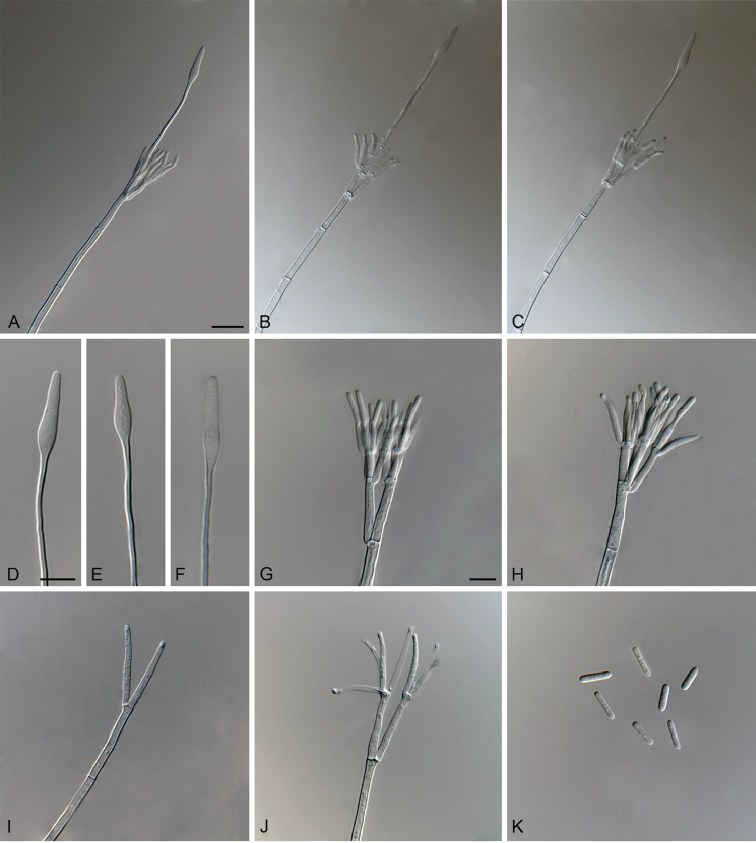
Figure 2.
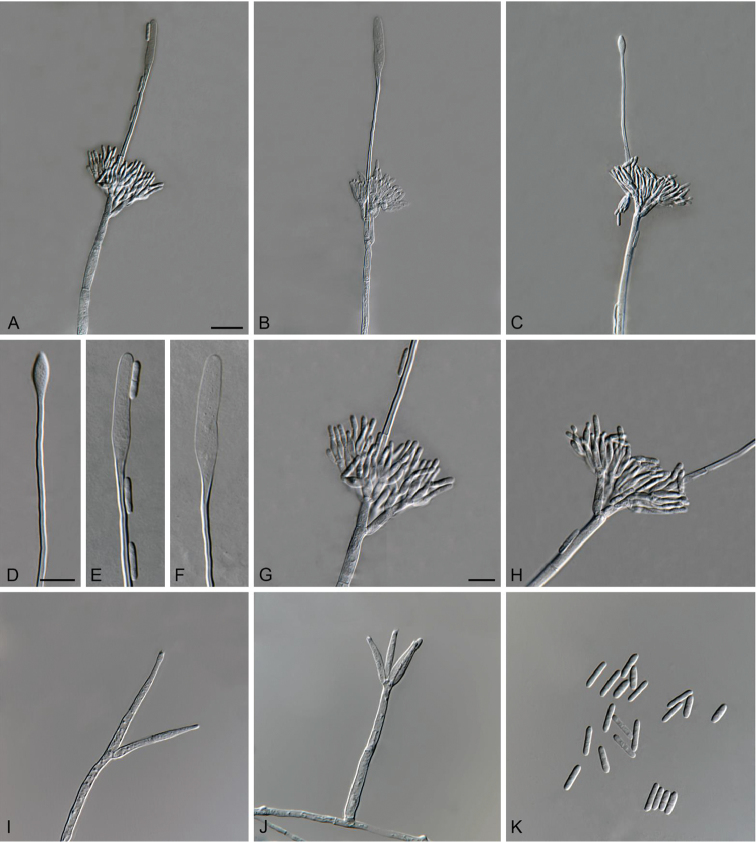
Cylindrocladiella arbusta (ex-type CMW 47295). A–C Penicillate conidiophores D–F Obpyriform to lanceolate vesicles G–H Penicillate conidiogenous apparatus I–J Subverticillate conidiophores K Conidia. Scale bars: A = 20 µm (apply to B–C); D = 10 µm (apply to E–F); G = 10 µm (apply to H–K).
Etymology.
Name refers to a plantation and the environment where this fungus was isolated.
Type material.
VIETNAM. Nghe An Province: Tan Ky, from soil in Acacia mangium plantation, Nov. 2013, N.Q. Pham & T.Q. Pham, herbarium specimen of dried culture, PREM 62159 (holotype), CMW 47295 = CBS 143546 (ex-type culture).
Description.
Sexual morph not observed. Conidiophores dimorphic, penicillate and subverticillate, mononematous and hyaline. Penicillate conidiophores comprising a stipe, a penicillate arrangement of fertile branches, a stipe extension and a terminal vesicle; stipe septate, hyaline, smooth, 116–166.5 × 4–5 µm; stipe extension aseptate, straight, 93–139 µm long, thick-walled with one basal septum, terminating in thin-walled, obpyriform to lanceolate vesicles, 4–5.5 µm wide. Penicillate conidiogenous apparatus with primary branches aseptate, 15–28.5 × 2.5–5 µm, secondary branches aseptate, 12–22.5 × 2.5–3.5 µm, each terminal branch producing 2–4 phialides; phialides doliiform to reniform to cymbiform, hyaline, aseptate, 10–18 × 2–3 µm, apex with minute periclinal thickening and collarette. Subverticillate conidiophores in moderate numbers, comprising of a septate stipe and primary branches terminating in 2–4 phialides; primary branches straight, hyaline, 0–1-septate, 25–31 × 2.5–3.5 µm; phialides cymbiform to cylindrical, hyaline, aseptate, 16.5–30.5 × 2–3.5 µm, apex with minute periclinal thickening and collarette. Conidia cylindrical, rounded at both ends, straight, 1-septate, (8.5–)10–12(–13.5) × 2–3 µm (av. = 11 × 2.5 µm), frequently slightly flattened at the base, held in asymmetrical clusters by colourless slime.
Culture characteristics.
Colonies white to buff on the surface and salmon to sienna in reverse on MEA after 7 d; smooth margins; extensive aerial mycelium in the middle and the margins; chlamydospores moderate, arranged in chains. Optimal growth temperature at 25 °C, no growth at 5 °C and 35 °C; after 7 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C and 30 °C reached 3.5 mm, 27.7 mm, 49.2 mm, 67.9 mm and 52.7 mm, respectively.
Additional material examined.
VIETNAM, Nghe An Province: Tan Ky, from soil in Acacia mangium nursery, Nov. 2013, N.Q. Pham & T.Q. Pham, PREM 62160, culture CMW 47296 = CBS 143547.
Distribution.
Nghe An, Vietnam.
Notes.
Cylindrocladiella arbusta is phylogenetically closely related to Cy. natalensis, Cy. obpyriformis and Cy. parvispora. The stipe extensions of Cy. arbusta are longer than those of Cy. natalensis and shorter than those of Cy. obpyriformis and Cy. parvispora. Conidia of Cy. arbusta are shorter than those of Cy. natalensis and Cy. obpyriformis (Table 2).
Cylindrocladiella malesiana
N.Q. Pham & M.J. Wingf. sp. nov.
MB824551
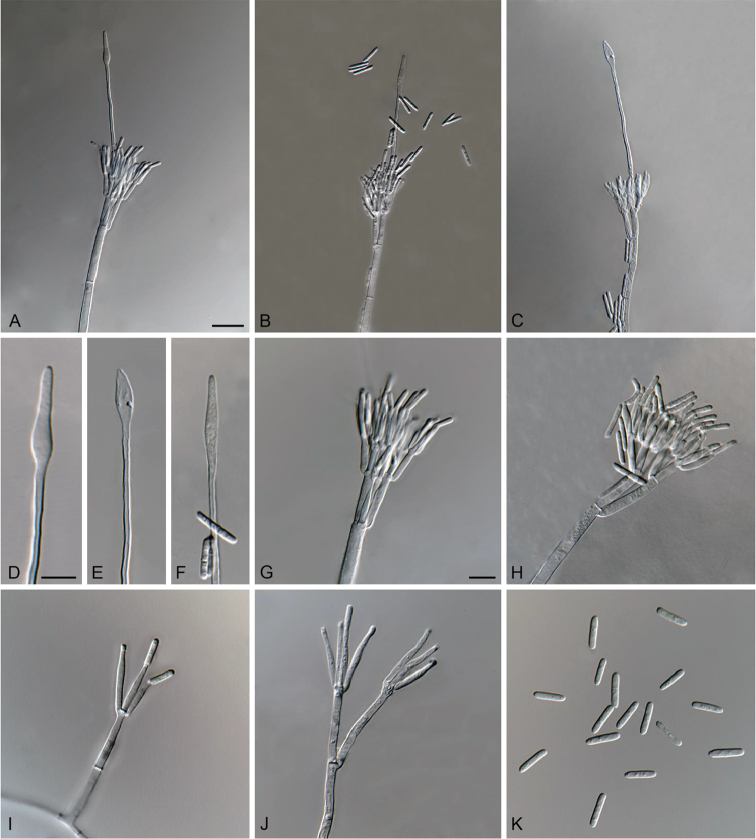
Figure 3.
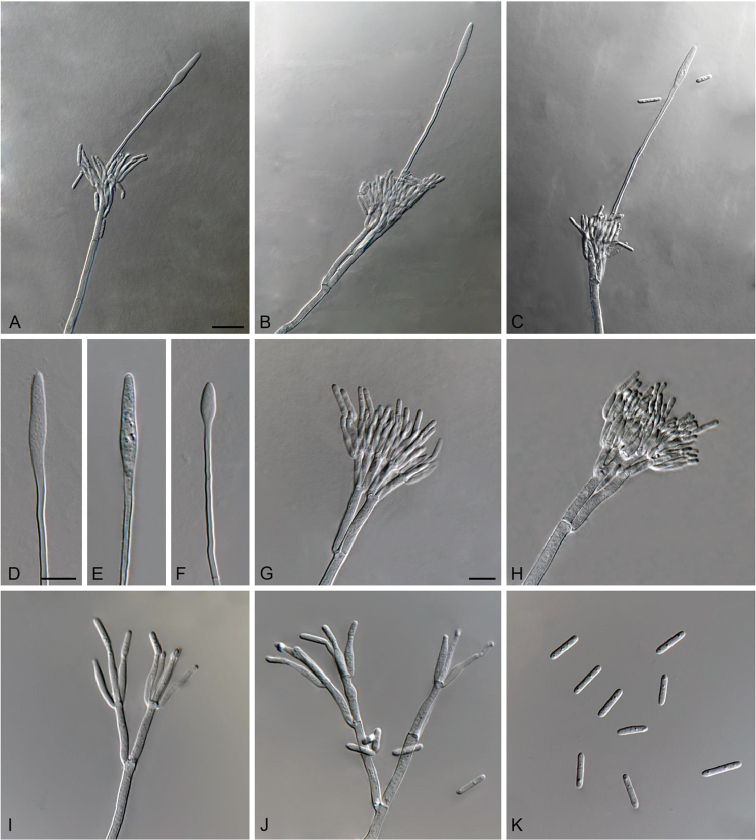
Cylindrocladiella malesiana (ex-type CMW 48278). A–C Penicillate conidiophores D–F Fusoid to lanceolate vesicles G–H Penicillate conidiogenous apparatus I–J Subverticillate conidiophores K Conidia. Scale bars: A = 20 µm (apply to B–C); D = 10 µm (apply to E–F); G = 10 µm (apply to H–K).
Etymology.
Name refers to Malaysia, the country where this species was first collected.
Type material.
MALAYSIA. Sabah State: Tawau, Brumas, from soil in Acacia mangium plantation, Mar. 2013, M.J. Wingfield, herbarium specimen of dried culture, PREM 62161 (holotype), CMW 48278 = CBS 143548 (ex-type culture).
Description.
Sexual morph not observed. Conidiophores dimorphic, penicillate and subverticillate, mononematous and hyaline. Penicillate conidiophores comprising a stipe, a penicillate arrangement of fertile branches, a stipe extension and a terminal vesicle; stipe septate, hyaline, smooth, 76.5–126 × 3.5–5 µm; stipe extension aseptate, straight, 114.5–144.5 µm long, thick-walled with one basal septum, terminating in thin-walled, fusoid to lanceolate vesicles, 4.5–6 µm wide. Penicillate conidiogenous apparatus with primary branches aseptate, 16.5–24 × 3–4.5 µm, secondary branches aseptate, 10.5–15 × 2–3.5 µm, each terminal branch producing 2–4 phialides; phialides cymbiform to cylindrical, hyaline, aseptate, 9–15.5 × 2–3.5 µm, apex with minute periclinal thickening and collarette. Subverticillate conidiophores abundant, comprising of a septate stipe and primary branches terminating in 2–4 phialides; primary branches straight, hyaline, 0–1-septate, 13.5–35 × 2.5–4 µm; phialides cymbiform to cylindrical, hyaline, aseptate, 14.5–27 × 2–3.5 µm, apex with minute periclinal thickening and collarette. Conidia cylindrical, rounded at both ends, straight, 1-septate, (10–)11–13(–13.5) × (1.5–)2–2.5 µm (av. = 12 × 2 µm), frequently slightly flattened at the base, held in asymmetrical clusters by colourless slime.
Culture characteristics.
Colonies buff to hazel on the surface and dark brick to brown vinaceous in reverse on MEA after 7 d; smooth to undulate margins; moderate aerial mycelium; chlamydospores moderate, arranged in chains. Optimal growth temperature at 25 °C, no growth at 5 °C and 35 °C; after 7 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C and 30 °C reached 3.8 mm, 24.3 mm, 45.2 mm, 74.4 mm and 48.8 mm, respectively.
Distribution.
Sabah, Malaysia
Additional material examined.
MALAYSIA. Sabah state: Tawau, Brumas, from soil in Acacia mangium plantation, Mar. 2013, M.J. Wingfield, PREM 62162, culture CMW 48276 = CBS 143549; ibid., PREM 62163, culture CMW 48277 = CBS 143550.
Notes.
Cylindrocladiella malesiana is phylogenetically closely related to Cy. microcylindrica, Cy. natalensis and Cy. solicola. Conidia of Cy. malesiana are shorter than those of Cy. microcylindrica, Cy. natalensis and Cy. solicola (Table 2).
Cylindrocladiella obpyriformis
N.Q. Pham, T.Q. Pham & M.J. Wingf. sp. nov.
MB824552
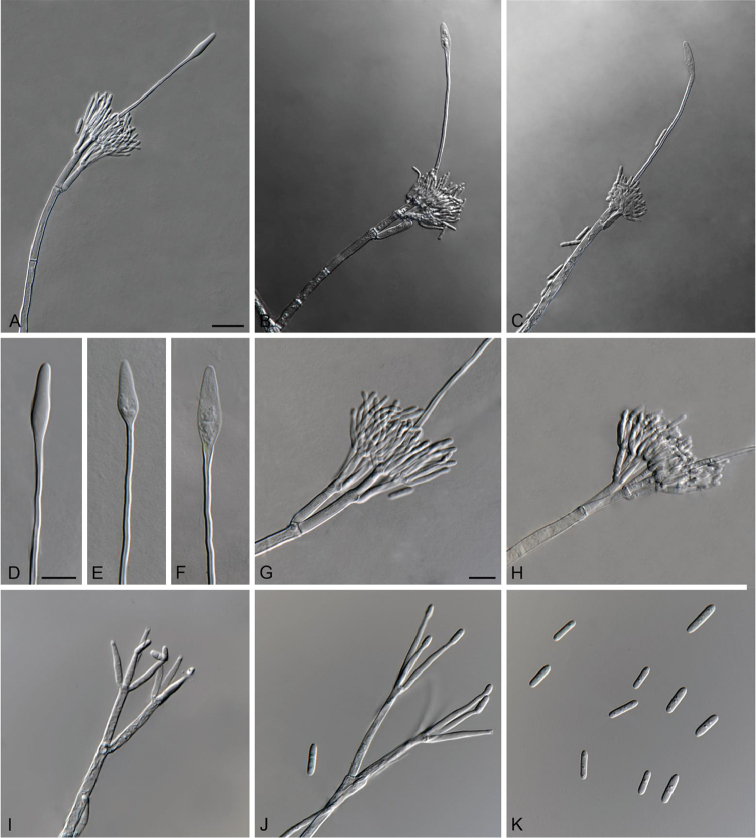
Figure 4.
Cylindrocladiella obpyriformis (ex-type CMW 47194). A–C Penicillate conidiophores D–F Obpyriform vesicles G–H Penicillate conidiogenous apparatus I–J Subverticillate conidiophores K Conidia. Scale bars: A = 20 µm (apply to B–C); D = 10 µm (apply to E–F); G = 10 µm (apply to H–K).
Etymology.
Name refers to the obpyriform terminating vesicles in this species.
Type material.
VIETNAM. Tuyen Quang Province, from soil in Acacia hybrid plantation, Nov. 2013, N.Q. Pham & T.Q. Pham, herbarium specimen of dried culture, PREM 62165 (holotype), CMW 47194 = CBS 143552 (ex-type culture).
Description.
Sexual morph not observed. Conidiophores dimorphic, penicillate and subverticillate, mononematous and hyaline. Penicillate conidiophores comprising a stipe, a penicillate arrangement of fertile branches, a stipe extension and a terminal vesicle; stipe septate, hyaline, smooth, 58.5–148 × 4–6 µm; stipe extension aseptate, straight, 86.5–150 µm long, thick-walled with one basal septum, terminating in thin-walled, obpyriform vesicles, 4–7 µm wide. Penicillate conidiogenous apparatus with primary branches aseptate, 17.5–31.5 × 3–5 µm, secondary branches aseptate, 10–19 × 2–4 µm, each terminal branch producing 2–4 phialides; phialides cymbiform to cylindrical, hyaline, aseptate, 10.5–18 × 2–3 µm, apex with minute periclinal thickening and collarette. Subverticillate conidiophores abundant, comprising of a septate stipe and primary branches terminating in 2–4 phialides; primary branches straight, hyaline, 0–1-septate, 15–38.5 × 2–4 µm; phialides cymbiform to cylindrical, hyaline, aseptate, 13–30.5 × 2–3 µm, apex with minute periclinal thickening and collarette. Conidia cylindrical, rounded at both ends, straight, 1-septate, (9–)11–13(–15) × 2–3(–3.5) µm (av. = 12 × 2.5 µm), frequently slightly flattened at the base, held in asymmetrical clusters by colourless slime.
Culture characteristics.
Colonies buff to isabelline on the surface and dark brick to sepia in reverse on MEA after 7 d; smooth to undulate margins; extensive aerial mycelium especially in the middle; chlamydospores moderate, arranged in chains. Optimal growth temperature at 25 °C, no growth at 5 °C and 35 °C; after 7 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C and 30 °C reached 5.4 mm, 25.5 mm, 47.2 mm, 74.0 mm and 50.8 mm, respectively.
Distribution.
Tuyen Quang & Vinh Phuc, Vietnam
Additional material examined.
VIETNAM. Vinh Phuc Province: Tam Dao, from soil in Camellia chrysantha nursery, Sept. 2013, N.Q. Pham, Q.N. Dang & T.Q. Pham, PREM 62166, culture CMW 49940 = CBS 143553.
Notes.
Cylindrocladiella obpyriformis is phylogenetically closely related to Cy. arbusta, Cy. natalensis and Cy. parvispora. The stipe extensions of Cy. obpyriformis are longer than those of Cy. arbusta, Cy. natalensis and Cy. parvispora (Table 2).
Cylindrocladiella parvispora
N.Q. Pham, T.Q. Pham & M.J. Wingfield sp. nov.
MB824553
Figure 5.
Cylindrocladiella parvispora (ex-type CMW 47197). A–C Penicillate conidiophores D–F Fusoid to cylindrical vesicles G–H Penicillate conidiogenous apparatus I–J Subverticillate conidiophores K Conidia. Scale bars: A = 20 µm (apply to B–C); D = 10 µm (apply to E–F); G = 10 µm (apply to H–K).
Etymology.
Name refers to the small conidia produced by this species.
Type material.
VIETNAM. Tuyen Quang Province, from soil in Acacia hybrid plantation, Nov. 2013, N.Q. Pham & T.Q. Pham, herbarium specimen of dried culture, PREM 62167 (holotype), CMW 47197 = CBS 143554 (ex-type culture).
Description.
Sexual morph not observed. Conidiophores dimorphic, penicillate and subverticillate, mononematous and hyaline. Penicillate conidiophores comprising a stipe, a penicillate arrangement of fertile branches, a stipe extension and a terminal vesicle; stipe septate, hyaline, smooth, 67–107 × 3–6.5 µm; stipe extension aseptate, straight, 112.5–141 µm long, thick-walled with one basal septum, terminating in thin-walled, fusoid to cylindrical vesicles, 4.5–6.5 µm wide. Penicillate conidiogenous apparatus with primary branches aseptate, 10.5–25 × 2–4 µm, secondary branches aseptate, 7.5–17 × 2–3 µm, each terminal branch producing 2–4 phialides; phialides doliiform to reniform to cymbiform, hyaline, aseptate, 7.5–13 × 2–3 µm, apex with minute periclinal thickening and collarette. Subverticillate conidiophores in moderate numbers, comprising of a septate stipe and primary branches terminating in 2–4 phialides; primary branches straight, hyaline, 0–1-septate, 15.5–27 × 2.5–4 µm; phialides cymbiform to cylindrical, hyaline, aseptate, 13.5–41 × 2.5–6 µm, apex with minute periclinal thickening and collarette. Conidia cylindrical, rounded at both ends, straight, 1-septate, (8–)10– 12(–13) × 2–2.5 µm (av. = 11 × 2 µm), frequently slightly flattened at the base, held in asymmetrical clusters by colourless slime.
Culture characteristics.
Colonies buff to honey to isabelline on the surface and umber to sepia in reverse on MEA after 7 d; smooth to undulate margin; abundant aerial mycelium especially in the middle; chlamydospores moderate, arranged in chains. Optimal growth temperature at 25 °C, no growth at 5 °C and 35 °C; after 7 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C and 30°C reached 5.5 mm, 23.4 mm, 43.8 mm, 63.6 mm and 49.2 mm, respectively.
Distribution.
Tuyen Quang, Vietnam
Additional material examined.
VIETNAM. Tuyen Quang Province, from soil in Acacia hybrid plantation, Nov. 2013, N.Q. Pham & T.Q. Pham, PREM 62168, culture CMW 47207 = CBS 143555; ibid., PREM 62169, culture CMW 47208 = CBS 143556.
Notes.
Cylindrocladiella parvispora is phylogenetically closely related to Cy. arbusta, Cy. natalensis and Cy. obpyriformis. Conidia of Cy. parvispora are slightly smaller than those of Cy. arbusta, Cy. natalensis and Cy. obpyriformis (Table 2).
Cylindrocladiella solicola
N.Q. Pham, T.Q. Pham & M.J. Wingf. sp. nov.
MB824554
Figure 6.
Cylindrocladiella solicola (ex-type CMW 47198). A–C Penicillate conidiophores D–F Broadly clavate to lanceolate to fusiform vesicles G–H Penicillate conidiogenous apparatus I–J Subverticillate conidiophores K Conidia. Scale bars: A = 20 µm (apply to B–C); D = 10 µm (apply to E–F); G = 10 µm (apply to H–K).
Etymology.
Name refers to soil, the substrate from which this fungus was first isolated.
Type material.
VIETNAM. Tuyen Quang Province, from soil in Acacia hybrid plantation, Nov. 2013, N.Q. Pham & T.Q. Pham, herbarium specimen of dried culture, PREM 62164 (holotype), CMW 47198 = CBS 143551 (ex-type culture).
Description.
Sexual morph not observed. Conidiophores dimorphic, penicillate and subverticillate, mononematous and hyaline. Penicillate conidiophores comprising a stipe, a penicillate arrangement of fertile branches, a stipe extension and a terminal vesicle; stipe septate, hyaline, smooth, 58.5–120 × 2.5–5 µm; stipe extension aseptate, straight, 93.5–170 µm long, thick-walled with one basal septum, terminating in thin-walled, broadly clavate to lanceolate to fusiform vesicles, 3.5–6.5 µm wide. Penicillate conidiogenous apparatus with primary branches aseptate, 16–36.5 × 3–4.5 µm, secondary branches aseptate, 10–16 × 2.5–3.5 µm, each terminal branch producing 2–4 phialides; phialides cymbiform to cylindrical, hyaline, aseptate, 9–15.5 × 2–3 µm, apex with minute periclinal thickening and collarette. Subverticillate conidiophores abundant, comprising of a septate stipe and primary branches terminating in 2–4 phialides; primary branches straight, hyaline, 0–1-septate, 16.5–25 × 2.5–5 µm; phialides cymbiform to cylindrical, hyaline, aseptate, 12–28 × 2.5–4 µm, apex with minute periclinal thickening and collarette. Conidia cylindrical, rounded at both ends, straight, 1-septate, (10.5–)12.5–14.5(–15.5) × 2–2.5(–3) µm (av. = 13.5 × 2.5 µm), frequently slightly flattened at the base, held in asymmetrical clusters by colourless slime.
Culture characteristics.
Colonies honey to isabelline on the surface and sepia to brown vinaceous in reverse on MEA after 7 d; undulate margins; extensive aerial mycelium especially in the middle; chlamydospores moderate, arranged in chains. Optimal growth temperature at 25 °C, no growth at 5 °C and 35 °C; after 7 d, colonies at 10 °C, 15 °C, 20 °C, 25 °C and 30 °C reached 5.2 mm, 20.4 mm, 37.8 mm, 61.2 mm and 37.1 mm, respectively.
Distribution.
Tuyen Quang, Vietnam
Notes.
Cylindrocladiella solicola is phylogenetically closely related to Cy. malesiana, Cy. microcylindrica and Cy. natalensis. The stipe extensions of Cy. solicola are longer than those of Cy. malesiana, Cy. microcylindrica and Cy. natalensis (Table 2).
Discussion
Application of multigene phylogenetic inference made it possible to identify five novel and two known species of Cylindrocladiella in this study. The seven species found bring the number of Cylindrocladiella known from South-East Asia to 20 (Crous 2002, Lombard et al. 2012, 2017), thus suggesting that this geographical region could be a possible centre of diversity for the genus Cylindrocladiella. A relatively small collection of isolates was shown to represent a high diversity of Cylindrocladiella spp. This indicates that more Cylindrocladiella spp. remain to be discovered in South-East Asia.
The his3 gene region provided the best resolution for species delineation amongst the four gene regions applied. This was the only gene region that could distinguish between all five novel species in the study. The ITS could not resolve any single lineage and the tef1 gene region failed to distinguish between Cy. arbusta and Cy. parvispora. The phylogenetic relationship between Cy. arbusta, Cy. malesiana and Cy. obpyriformis could not be resolved using the tub2 gene region (Suppl. materials 1–4). In the most recent study of species of Cylindrocladiella (Lombard et al. 2017), the his3 gene region was not used in the analyses because it provided limited information compared with tef1 and tub2 gene sequences that were more informative. However, the results of the present study suggest that his3 sequence data should be included in future studies as they provide valuable additional information on the relationships amongst some groups of species.
Five novel species, described as Cy. arbusta, Cy. malesiana, Cy. obpyriformis, Cy. parvispora and Cy. solicola, were all isolated from soil samples associated with Acacia plantations across Malaysia and Vietnam. In comparison with a previous study on Calonectria spp. from South-East Asia (Pham 2018), even though they share similar ecological niches, Cylindrocladiella spp. seemed to have a relatively narrow distribution and host association. This suggests that there is some substrate specialisation for these species of Cylindrocladiella. It is possible that they are mild pathogens of roots but no evidence of disease was observed.
This study includes the first report of Cy. lageniformis and Cy. peruviana in Vietnam. These two species have been reported as causal agents of black-foot disease, one of the most economically important fungal disease and a major constraint to wine and grape production (van Coller et al. 2005, Koike et al. 2016). The detection of these species from plantations soils in Vietnam might suggest that they infect the roots of Acacia spp. but this would require further investigation. These species have also been reported to cause leaf spots as well as root and cutting rot of Eucalyptus in Brazil (Crous et al. 1991, Crous and Wingfield 1993) and they clearly deserve further study in South-East Asia.
Supplementary Material
Acknowledgements
We acknowledge financial support from the Tree Protection and Cooperation Programme (TPCP) and the DST/NRF Centre of Excellence in Tree Health Biotechnology (CTHB), South Africa. We thank the members of the Forest Protection Research Centre (FPRC), Vietnam, especially Quynh N. Dang for the valuable assistance with the cultures.
Citation
Pham NQ, Barnes I, Chen S, Pham TQ, Lombard L, Crous PW, Wingfield MJ (2018) New species of Cylindrocladiella from plantation soils in South-East Asia. MycoKeys 32: 1–24. https://doi.org/10.3897/mycokeys.32.23754
Supplementary materials
Figure S1. Phylogenetic tree based on maximum likelihood (ML) analysis of his3 sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60 % are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
Figure S2. Phylogenetic tree based on maximum likelihood (ML) analysis of tef1 sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Phylogenetic tree based on maximum likelihood (ML) analysis of tef1 sequence alignments. Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60% are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
Figure S3. Phylogenetic tree based on maximum likelihood (ML) analysis of tub2 sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60 % are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
Figure S4. Phylogenetic tree based on maximum likelihood (ML) analysis of ITS sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60 % are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
References
- Agustí-Brisach C, Armengol J. (2013) Black-foot disease of grapevine: an update on taxonomy, epidemiology and management strategies. Phytopathologia Mediterranea 52: 245–261. http://dx.doi.org/10.14601/Phytopathol_Mediterr-12662 [Google Scholar]
- Armengol J, Gramaje D. (2016) Soilborne fungal pathogens affecting grapevine rootstocks: current status and future prospects. Acta Horticulturae 1136: 235–328. http://doi.org/10.17660/ActaHortic.2016.1136.32 [Google Scholar]
- Boesewinkel HJ. (1982) Cylindrocladiella, a new genus to accommodate Cylindrocladium parvum and other small-spored species of Cylindrocladium. Canadian Journal of Botany 60: 2288–2294. https://doi.org/10.1139/b82-280 [Google Scholar]
- Brielmaier-Liebetanz U, Wagner S, Werres S. (2013) First report of dieback on Euonymus fortunei caused by Cylindrocladiella parva in Germany. Plant Disease 97: 1120. https://doi.org/10.1094/PDIS-02-13-0162-PDN [DOI] [PubMed]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. http://doi.org/10.2307/3761358 [Google Scholar]
- Carlucci A, Lops F, Mostert L, Halleen F, Raimondo ML. (2017) Occurrence fungi causing black foot on young grapevines and nursery rootstock plants in Italy. Phytopathologia Mediterranea 56: 10–39. http://dx.doi.org/10.14601/Phytopathol_Mediterr-18769 [Google Scholar]
- Crous PW. (2002) Taxonomy and pathology of Cylindrocladium (Calonectria) and allied genera. American Phytopathological Society, Minnesota, USA.
- Crous PW, Groenewald JZ, Risède J-M, Simoneau P, Hywel-Jones NL. (2004a) Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004b) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Phillips AJL, Wingfield MJ. (1991) The genera Cylindrocladium and Cylindrocladiella in South Africa, with special reference to forest nurseries. South African Forestry Journal 157: 69–85. https://doi.org/10.1080/00382167.1991.9629103 [Google Scholar]
- Crous PW, Wingfield MJ. (1993) A re-evaluation of Cylindrocladiella, and a comparison with morphologically similar genera. Mycological Research 97: 433–448. https://doi.org/10.1016/S0953-7562(09)80131-7 [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GEStJ, Barber PA et al. (2017) Fungal Planet description sheets: 558–624. Persoonia 38: 240–384. https://doi.org/10.3767/003158517X698941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erper I, Agustí-Brisach C, Tunali B, Armengol J. (2013) Characterization of root rot disease of kiwifruit in the Black Sea region of Turkey. European Journal of Plant Pathology 136: 291–300. https://doi.org/10.1007/s10658-012-0163-6 [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes ‐ application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 55: 696–704. [DOI] [PubMed] [Google Scholar]
- Inderbitzin P, Bostock RM, Subbarao KV. (2012) Cylindrocladiella hahajimaensis, a new species of Cylindrocladiella transferred from Verticillium. MycoKeys 4: 1–8. https://doi.org/10.3897/mycokeys.4.2619 [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. https://doi.org/10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike ST, Bettiga LJ, Nguyen TT, Gubler WD. (2016) First report of Cylindrocladiella lageniformis and C. peruviana as grapevine pathogens in California. Plant Disease 100: 1783. https://doi.org/10.1094/PDIS-02-16-0157-PDN
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Cheewangkoon R, Crous PW. (2017) New Cylindrocladiella spp. from Thailand soils. Mycosphere 8: 1088–1104. https://doi.org/10.5943/mycosphere/8/8/14 [Google Scholar]
- Lombard L, Shivas RG, To-Anun C, Crous PW. (2012) Phylogeny and taxonomy of the genus Cylindrocladiella. Mycological Progress 11: 835–868. https://doi.org/10.1007/s11557-011-0799-1 [Google Scholar]
- Mohanan C, Sharma JK. (1985) Cylindrocladium causing seedling diseases of Eucalyptus in Kerala, India. Transactions of the British Mycological Society 84: 538–539. https://doi.org/10.1016/S0007-1536(85)80019-X [Google Scholar]
- Nirenberg HI. (1981) A simplified method for identifying Fusarium spp. occurring on wheat. Canadian Journal of Botany 59: 1599–1609. https://doi.org/10.1139/b81-217 [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences 95: 2044–2049. https://doi.org/10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. https://doi.org/10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Peerally MA. (1974) Cylindrocladium camelliae CMI Descriptions of Pathogenic Fungi and Bacteria 428.
- Pham NQ. (2018) New Calonectria and Cylindrocladiella species from Vietnam, Malaysia and Indonesia. MSc Thesis. University of Pretoria, South Africa.
- Posada D. (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. http://doi.org/10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Rayner RW. (1970) A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society. Kew, Surrey, UK.
- Scattolin L, Montecchio L. (2007) First report of damping-off of common oak plantlets caused by Cylindrocladiella parva in Italy. Plant Disease 91: 771. https://doi.org/10.1094/PDIS-91-6-0771B [DOI] [PubMed]
- Schoch CL, Crous PW, Wingfield MJ, Wingfield BD. (2000) Phylogeny of Calonectria and selected hypocrealean genera with cylindrical macroconidia. Studies in Mycology 45: 45–62. [Google Scholar]
- Sharma J, Mohanan C. (1982) Cylindrocladium spp. associated with various diseases of Eucalyptus in Kerala. European Journal of Forest Pathology 12: 129–136. https://doi.org/10.1111/j.1439-0329.1982.tb01385.x [Google Scholar]
- Swofford DL. (2003) PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts, USA.
- van Coller GJ, Denman S, Groenewald JZ, Lamprecht SC, Crous PW. (2005) Characterisation and pathogenicity of Cylindrocladiella spp. associated with root and cutting rot symptoms of grapevines in nurseries. Australasian Plant Pathology 34: 489–498. https://doi.org/10.1071/AP05058 [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic tree based on maximum likelihood (ML) analysis of his3 sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60 % are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
Figure S2. Phylogenetic tree based on maximum likelihood (ML) analysis of tef1 sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Phylogenetic tree based on maximum likelihood (ML) analysis of tef1 sequence alignments. Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60% are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
Figure S3. Phylogenetic tree based on maximum likelihood (ML) analysis of tub2 sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60 % are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.
Figure S4. Phylogenetic tree based on maximum likelihood (ML) analysis of ITS sequence alignments
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Nam Q. Pham, Irene Barnes, ShuaiFei Chen, Thu Q. Pham, Lorenzo Lombard, Pedro W. Crous, Michael J. Wingfield
Data type: molecular data
Explanation note: Bootstrap value ≥ 60 % for maximum parsimony (MP) and ML analyses are indicated at the nodes. Bootstrap values lower than 60 % are marked with “*” and absent are marked with “–”. Isolates representing ex–type material are marked with “T” and isolates collected in this study are highlighted in bold. Calonectria brachiatica (CMW 25307) and Calonectria pauciramosa (CMW 5683) represent the outgroups.