Highlights
-
•
The Trigger Loop is one of the major determinants of transcription fidelity.
-
•
Intrinsic proofreading occurs via transcript-assisted cleavage.
-
•
Factor-assisted proofreading takes place via exchange of RNAP active centres.
-
•
Misincorporation is a major source of transcription pausing.
-
•
Another role of fidelity is the prevention of conflicts with other cellular processes.
Abstract
Accuracy of transcription is essential for productive gene expression, and the past decade has brought new understanding of the mechanisms ensuring transcription fidelity. The discovery of a new catalytic domain, the Trigger Loop, revealed that RNA polymerase can actively choose the correct substrates. Also, the intrinsic proofreading activity was found to proceed via a ribozyme-like mechanism, whereby the erroneous nucleoside triphosphate (NTP) helps its own excision. Factor-assisted proofreading was shown to proceed through an exchange of active centres, a unique phenomenon among proteinaceous enzymes. Furthermore, most recent in vivo studies have revised the roles of transcription accuracy and proofreading factors, as not only required for production of errorless RNAs, but also for prevention of frequent misincorporation-induced pausing that may cause conflicts with fellow RNA polymerases and the replication machinery.
Current Opinion in Microbiology 2018, 42:13–18
This review comes from a themed issue on Cell regulation
Edited by Rita Tamayo and Jan-Willem Veening
For a complete overview see the Issue and the Editorial
Available online 29th September 2017
http://dx.doi.org/10.1016/j.mib.2017.08.004
1369-5274/© 2017 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/)..
Introduction
Gene expression relies on the accurate copy of genetic information. The fidelity of RNA synthesis results from the accuracy of correct NTP selection (versus non-complementary NTPs and complementary 2'-deoxy NTPs), the proofreading of misincorporation events, and the efficiency of extension of the misincorporated nucleotide. In this review, we summarize the structural and biochemical determinants of transcription fidelity that have been uncovered in the last decade, and we describe very recent insights on the consequences that stalled misincorporated complexes may have on cellular functions and gene expression.
Determinants of the accuracy of NTP choice
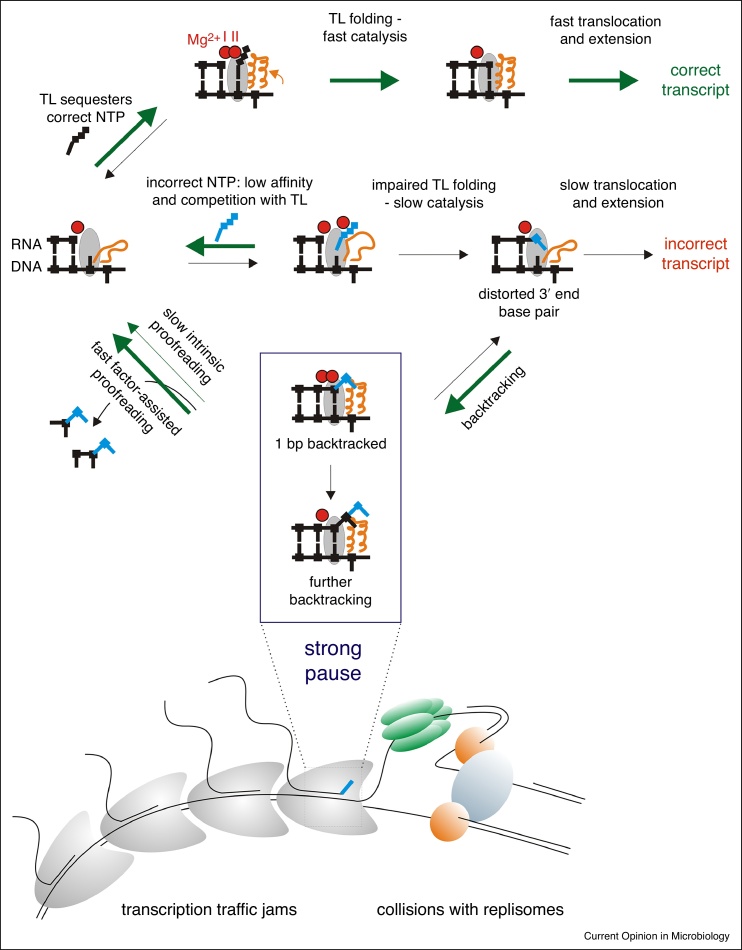
For a long time, the catalysis of phosphodiester bond formation by RNA polymerase (RNAP) was thought to be performed solely via a two metal ion (Mg2+) mechanism within a relatively rigid active centre. However, at saturating NTPs concentrations (close to cellular levels), such a ‘motionless’ active site would provide as low as ~10-fold kinetic discrimination against some non-complementary NTPs (though 103 for certain misincorporations), and would not discriminate at all against complementary 2'-deoxy NTPs [1••]. The discovery of a flexible domain of the active site, the Trigger Loop (TL) [2••], revealed that the active centre of RNAP actively participates in choosing NTPs via an induced fit mechanism [1••, 3]. TL is essential for the catalysis of phosphodiester bond formation, and it acts by stabilising the transition state of the reaction [1••, 4]. The key property of the TL for the accuracy of transcription is its ability to accommodate catalytically active (folded) and inactive (open) structural states. The correct NTP binding in the i + 1 site (grey in Figure 1) induces folding of the TL (orange in Figure 1), which, in turn, participates in the catalysis of nucleoside monophosphate (NMP) incorporation into the transcript. Binding of a non-cognate NTP in the i + 1 site cannot induce productive folding of the TL because of the wrong geometry of base pairing with the template (in case of non-complementary NTPs) or the lack of critical contacts of the NTP's sugar moiety with the TL (in case of complementary deoxy NTPs) [1••]. Such an induced fit mechanism of selection provides 1–3 extra orders of magnitude of kinetic discrimination against non-complementary NTPs, and 3 orders of magnitude against complementary dNTPs [1••].
Figure 1.
Multistep processes ensuring transcription fidelity. A schematic representation of the active centre of RNAP is given for different transcription intermediates, and shows template DNA and RNA (black lines), metal ions (red circles), the i + 1 site (grey oval) and the Trigger Loop (orange ribbon). Correct and incorrect incoming NTPs are coloured in black and blue, respectively. Green arrows show the direction of reactions leading to a correct transcript. The different thickness of the arrows serves only as a qualitative indication of the rates of reactions or conformational changes. At the bottom of the figure, a cartoon depicts a stalled misincorporated elongation complex, which may potentially cause transcription traffic jams with trailing RNAPs (left), and conflicts with replication forks (right).
The affinity discrimination against non-complementary NTPs takes place due to their weaker base pairing with the template, and may increase discrimination by more than an order of magnitude. Furthermore, the TL competes with non-cognate NTPs in the i + 1 site [1••], while sequestering the correct NTPs bound there [5]. Such ‘active’ expulsion of only wrong substrates adds another order of magnitude to the discrimination against non-complementary NTPs. Notably, TL-mediated expulsion is the only ‘affinity’ component for discrimination against dNTPs because the affinity of their binding in the active site is the same as for ribonucleotides [1••].
It must be noted that, while the above-mentioned mechanisms are general and conserved, their efficiencies may vary greatly depending on the identity of incoming NTP, the acceptor base in the template DNA as well as surrounding sequences [1••, 6, 7]. For example, overall kinetic discrimination in the active centre fluctuates from 103 to 105 fold, depending on the particular misincorporation [1••]. The lower affinity of non-complementary NTPs may improve discrimination to 105–107 fold, although this may differentially drop according to the concentrations of NTPs in the cell. It should also be noted that some accessory factor may influence RNAP accuracy, such as, in E. coli, the global transcription regulator DksA, that binds close to the RNAP active centre and slows down the incorporation of erroneous nucleotides [8•].
The fate of misincorporated complexes
Misincorporation does happen occasionally. Because of the absence of Watson-Crick base pairing with the template, the RNA 3' end becomes misaligned in various ways, relatively to the rest of RNAP active centre. This impairs the catalysis of the subsequent NMP addition, though to various extents depending on the mismatched pair at the 3' end of RNA [6], the incoming NTP and surrounding sequences. Thermodynamically, however, misincorporated complexes are likely to accommodate a 1 base pair (bp) backtracked state [9••]. In this conformation, the erroneous NMP of the 3' end loses contacts with the template and flips out of the active site, thus shifting the elongation complex by 1 bp backwards (Figure 1). Backtracking of these complexes may continue even further, depending on the thermodynamics of surrounding sequences (Figure 1). Backtracked complexes are inactive in transcript elongation because the 3' end of RNA is away from the active site. Only an occasional reversion of backtracking, followed by the slow extension of the incorrect 3' end, would result in the retention of the misincorporated nucleotide in the transcript. These delays are one of the major contributors to the overall fidelity of synthesis of the final RNA products as they provide time for resolution of misincorporated complexes via proofreading mechanisms. However, at the same time, they also constitute a major source of paused complexes in the cell, as we discuss below.
Intrinsic proofreading of transcription
RNAP active centre is able to hydrolyse the phosphodiester bonds of the transcript [10•]. This reaction is used by RNAP to proofread the mistakes in RNA, as the new 3' end of RNA generated as a result of hydrolysis becomes available for extension (Figure 1). The reaction is catalysed by the same two metal ions mentioned earlier and the TL [11], though the extent of the TL involvement may differ in different organisms [4, 11, 12, 13] In the 1 bp backtracked state, adopted after misincorporation, it is the second phosphodiester bond that is positioned in the active site for hydrolysis (Figure 1). Interestingly, in this conformation the erroneous 3' end NMP of the transcript directly participates in the hydrolysis, thus facilitating its own removal in the form of a dinucleotide [9••]. The 3' end NMP provides coordination bonds for the second catalytic metal ion, as well as stabilises and activates the attacking water molecule [9••]. Though it is difficult to assess the contribution of this transcript-assisted proofreading to the overall fidelity of transcription, in vitro it was shown to proofread most misincorporation events before the wrong transcript is extended, even in high concentrations of substrates [9••].
Factor-assisted proofreading of transcription
Most organisms possess factors that strongly stimulate hydrolysis of the phosphodiester bonds in the transcript and thus proofreading of transcription. In bacteria these are the Gre factors, while archaea and eukaryotes employ homologues of RNA polymerase II factor TFIIS. These accessory factors stabilise the second catalytic metal ion and activate the attacking water molecule [14•, 15]. To do that, they physically displace and substitute for the TL in the RNAP active centre, thus changing the catalytic properties of RNAP from slow intrinsic hydrolysis (catalysed by TL) to fast factor-assisted hydrolysis [16••, 17••]. In vitro, Thermus aquaticus GreA stays bound to the elongation complex, but is inactive during correct synthesis, and substitutes for the TL only upon misincorporation or occasional backtracking [16••]. E. coli GreB, however, was shown to dissociate quickly from the elongation complex, reflecting possible different modes of regulation of the activities of different Gre factors [18]. In vitro, GreA proofreads almost all misincorporation events before their extension [9••], but the general contribution of Gre to prevent retention of mistakes in the final transcripts could be moderate [7, 19••, 20•].
Visualizing transcription errors in vivo
In bacteria, the study of transcriptional fidelity in vivo relied for a long time on lacZ reporter genes carrying a nonsense codon in the open reading frame [21, 22, 23]. Such constructs allowed to estimate transcriptional error rates of ~10-5–10-4 [21, 22], and were used to identify RNAP mutants with reduced accuracy of chain elongation [23]. More recently, similar constructs have detected an increase in error rate in a greA mutant of Streptococcus pneumoniae [20•], and in a dksA mutant of E. coli [8•, 24]. Comparable approaches in Saccharomyces cerevisiae gave contradictory results on the role of TFIIS [25, 26, 27].
A new reporter assay, based on the suppression of a missense mutation in the active site of Cre recombinase, has recently been developed for the detection of G?A (misincorporation of A instead of G) errors [28, 29]. In E. coli, a greA mutant strain showed over 100-fold increase in error rate, similarly to a double greA greB mutant, while deletion of greB alone did not have any effect, revealing a major role for GreA in transcription proofreading [28]. Overexpression of GreB could however complement deletion of greA [28]. In yeast, the same approach successfully detected an increase in G?A errors in strains lacking TFIIS or the RNA polymerase II subunit Rpb9, with the former inducing 3 times more errors than the latter, and was used to identify new fidelity mutants of Pol II, which mapped in the Trigger Loop, the bridge helix, and in the sites involved in binding to TFIIS [29].
In recent years, next-generation sequencing technologies have allowed the study of transcription fidelity in greater detail [7, 19••, 20•, 30••, 31]. Nascent elongating transcript sequencing (NET-seq) selectively captures the 3' end of transcripts that are being actively elongated by the RNAP, and has revealed sequence-dependent transcriptional pausing with nucleotide resolution [32, 33••]. When applied to the analysis of errors in the actively transcribing complexes, it revealed that misincorporated complexes are 1–3% of all elongation complexes in wild-type cells of Saccharomyces cerevisiae and E. coli, respectively [19••], a much higher proportion than expected from the overall error rate of RNA synthesis. In the absence of cleavage factors (TFIIS or Gre), the fraction became 7% and 5%, respectively [19••]. A somewhat lower proportion of misincorporated complexes was observed in another study [30••], though the native RNA preparation protocol used in that case may have favoured the intrinsic proofreading activity of RNAP, as we have discussed previously [19••].
The misincorporation pattern showed a strong bias towards G?A misincorporation [19••, 30••], in line with previous in vitro observations [1••, 6, 7], and data suggested that CG motifs increase G?A misincorporation [30••]. This bias however seems to be apparent only at positions of very frequent misincorporation (hotspots), which are a minor fraction of the total events [19••]. Interestingly, in E. coli these hotspots are ~8 times more abundant in untranslated regions compared to protein coding sequences, while no difference was observed in S. cerevisiae [19••].
Phenotypic consequences of transcription infidelity
The study of transcription fidelity in vivo remains challenging, but several reports have linked transcription errors to detrimental cellular phenotypes in eukaryotes [34, 35, 36, 37, 38].
In bacteria, transcriptional infidelity was shown to be a significant source of molecular noise, which could lead to heritable phenotypic changes via activation of a bistable switch [39••, 40]. Bistable feedback loops regulate important pathways in bacteria, including cellular differentiation, virulence and expression of metabolic genes, and are particularly sensitive to noise in gene expression [41]. In E. coli, deletion of both greA and greB, but not single deletions alone, considerably increased the switching frequency of the lac operon [39••, 40], and the error-prone ack-1 mutation of RNAP also promoted the switching [39••].
It seems now questionable whether transcription infidelity influences cellular phenotypes via the actual production of erroneous proteins. Misincorporation events cause long-lived pauses in vitro because of backtracking [9••, 42]. Backtracked pauses were shown to cause conflicts with replication forks in vivo, leading to detrimental consequences such as double strand brakes and genome instability [43••, 44•]. It was also suggested that queues of RNAPs forming behind the stalled one might actually be the main obstacle to replication fork progression and/or cause changes in gene expression [20•]. The substantial proportion of misincorporated complexes detected by NET-seq indicates that such stalled complexes are slowly resolved in vivo, and therefore may be a major source of conflicts with fellow RNAPs and replication complexes [19••]. In this context, the physical block of transcription of regulatory genes is likely to have a greater impact on molecular noise than the rare mistakes in final RNA products. Also, accumulation of misincorporated complexes may exacerbate the conflicts between RNAP and other cellular machineries, which could be responsible for the deleterious phenotypes that have been linked to infidelity.
Consequently, the most relevant role of cleavage factors Gre and TFIIS (and its homologues) in vivo may be the resolution of stalled misincorporated complexes [19••, 20•]. Gre factors and DksA were previously shown to be important to resolve conflicts between DNA replication and transcription under certain conditions [44•, 45]. For instance, viability of E. coli strains lacking greA and dksA is reduced when DNA repair is compromised [44•]. Also, DksA was shown to ensure replication completion upon amino acid starvation by removing transcription roadblocks [45]. Furthermore, a triple mutant greA greB dksA grows extremely slowly and with a high degree of filamentation [46, 47] and showed a significant decrease in replication fork progression [45]. Severe growth and morphological defects, including aberrant nucleoid morphology, were also observed in a greA mutant of S. pneumoniae, which does not encode other cleavage factors nor DksA homologues [20•].
Conclusions
Recent biochemical, genetic and next-generation sequencing advances have revised and improved our view of the mechanisms and the roles of transcription fidelity in both bacteria and eukaryotes. However, a number of questions remain unanswered. For example, the exact structural basis for the differences in discrimination against various misincorporation events remains only hypothetical. Also, the involvement of transcription factors such as DksA, or RNA polymerase II subunits such as Rbp9, in transcription accuracy is still unclear. Most interestingly, the mechanisms by which cells resolve the apparently detrimental misincorporated complexes in the absence of proofreading factors remain elusive.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by grants from the UK Biotechnology and Biological Sciences Research Council [BB/L010003/1], Wellcome Trust [102851], and Leverhulme Trust [PLP-2014-229], awarded to N.Z.
Contributor Information
Pamela Gamba, Email: pamela.gamba2@ncl.ac.uk.
Nikolay Zenkin, Email: nikolay.zenkin@ncl.ac.uk.
References
- 1••.Yuzenkova Y., Bochkareva A., Tadigotla V.R., Roghanian M., Zorov S., Severinov K., Zenkin N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive study of misincorporation events, and of the involvement of the Trigger Loop in discrimination against them, as well as in catalysis, in general.
- 2••.Temiakov D., Zenkin N., Vassylyeva M.N., Perederina A., Tahirov T.H., Kashkina E., Savkina M., Zorov S., Nikiforov V., Igarashi N. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]; The discovery of the Trigger Loop as a catalytic domain of the RNAP active centre.
- 3.Zhang J., Palangat M., Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishanina T.V., Palo M.Z., Nayak D., Mooney R.A., Landick R. Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading. Proc Natl Acad Sci U S A. 2017;114:E5103–E5112. doi: 10.1073/pnas.1702383114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kireeva M.L., Nedialkov Y.A., Cremona G.H., Purtov Y.A., Lubkowska L., Malagon F., Burton Z.F., Strathern J.N., Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sydow J.F., Brueckner F., Cheung A.C., Damsma G.E., Dengl S., Lehmann E., Vassylyev D., Cramer P. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Imashimizu M., Oshima T., Lubkowska L., Kashlev M. Direct assessment of transcription fidelity by high-resolution RNA sequencing. Nucleic Acids Res. 2013;41:9090–9104. doi: 10.1093/nar/gkt698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Roghanian M., Zenkin N., Yuzenkova Y. Bacterial global regulators DksA/ppGpp increase fidelity of transcription. Nucleic Acids Res. 2015;43:1529–1536. doi: 10.1093/nar/gkv003. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that the global transcription regulator DksA and the alarmone ppGpp directly participate in increasing the accuracy of RNA synthesis by slowing down misincorporation (see also Ref. [24]).
- 9••.Zenkin N., Yuzenkova Y., Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]; The discovery of the transcript-assisted transcriptional proofreading, whereby the erroneously incorporated nucleotide participates in its own removal from the transcript.
- 10•.Orlova M., Newlands J., Das A., Goldfarb A., Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci U S A. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work identified the RNA hydrolytic activity of RNAP, which was later shown to participate in proofreading (see annotation to Ref. [9••]).
- 11.Yuzenkova Y., Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc Natl Acad Sci U S A. 2010;107:10878–10883. doi: 10.1073/pnas.0914424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen S., Zenkin N. Transcript assisted phosphodiester bond hydrolysis by eukaryotic RNA polymerase II. Transcription. 2013;4:209–212. doi: 10.4161/trns.27062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esyunina D., Turtola M., Pupov D., Bass I., Klimasauskas S., Belogurov G., Kulbachinskiy A. Lineage-specific variations in the trigger loop modulate RNA proofreading by bacterial RNA polymerases. Nucleic Acids Res. 2016;44:1298–1308. doi: 10.1093/nar/gkv1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Borukhov S., Sagitov V., Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]; The discovery of bacterial proofreading factors Gre.
- 15.Jeon C., Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci U S A. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Roghanian M., Yuzenkova Y., Zenkin N. Controlled interplay between trigger loop and Gre factor in the RNA polymerase active centre. Nucleic Acids Res. 2011;39:4352–4359. doi: 10.1093/nar/gkq1359. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that proofreading factors Gre act by substituting for the Trigger Loop in the active centre, thus changing the catalytic properties of RNAP in a controlled manner.
- 17••.Sekine S., Murayama Y., Svetlov V., Nudler E., Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57:408–421. doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]; The structural evaluation of the substitution of the Trigger Loop by an elongation factor, shown biochemically in Ref. [16••].
- 18.Tetone L.E., Friedman L.J., Osborne M.L., Ravi H., Kyzer S., Stumper S.K., Mooney R.A., Landick R., Gelles J. Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue. Proc Natl Acad Sci U S A. 2017;114:E1081–E1090. doi: 10.1073/pnas.1616525114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.James K., Gamba P., Cockell S.J., Zenkin N. Misincorporation by RNA polymerase is a major source of transcription pausing in vivo. Nucleic Acids Res. 2017;45:1105–1113. doi: 10.1093/nar/gkw969. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reveals an unexpectedly high proportion of misincorporated elongation complexes in the cell, suggesting that misincorporation might be a major cause of transcription pausing and, as a result, of conflicts with other cellular processes (see also annotation to Ref. [30••]).
- 20•.Yuzenkova Y., Gamba P., Herber M., Attaiech L., Shafeeq S., Kuipers O.P., Klumpp S., Zenkin N., Veening J.W. Control of transcription elongation by GreA determines rate of gene expression in Streptococcus pneumoniae. Nucleic Acids Res. 2014;42:10987–10999. doi: 10.1093/nar/gku790. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work proposes that one of the major roles of the proofreading factor Gre is to prevent transcription traffic jams by resolving backtracked and misincorporated complexes.
- 21.Rosenberger R.F., Foskett G. An estimate of the frequency of in vivo transcriptional errors at a nonsense codon in Escherichia coli. Mol Gen Genet. 1981;183:561–563. doi: 10.1007/BF00268784. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberger R.F., Hilton J. The frequency of transcriptional and translational errors at nonsense codons in the lacZ gene of Escherichia coli. Mol Gen Genet. 1983;191:207–212. doi: 10.1007/BF00334815. [DOI] [PubMed] [Google Scholar]
- 23.Blank A., Gallant J.A., Burgess R.R., Loeb L.A. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 24.Satory D., Gordon A.J., Wang M., Halliday J.A., Golding I., Herman C. DksA involvement in transcription fidelity buffers stochastic epigenetic change. Nucleic Acids Res. 2015;43:10190–10199. doi: 10.1093/nar/gkv839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw R.J., Bonawitz N.D., Reines D. Use of an in vivo reporter assay to test for transcriptional and translational fidelity in yeast. J Biol Chem. 2002;277:24420–24426. doi: 10.1074/jbc.M202059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesser N.K., Peterson D.O., Hawley D.K. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci U S A. 2006;103:3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama H., Ito T., Nakanishi T., Kawamura N., Sekimizu K. Transcription elongation factor S-II maintains transcriptional fidelity and confers oxidative stress resistance. Genes Cells. 2003;8:779–788. doi: 10.1046/j.1365-2443.2003.00677.x. [DOI] [PubMed] [Google Scholar]
- 28.Bubunenko M.G., Court C.B., Rattray A.J., Gotte D.R., Kireeva M.L., Irizarry-Caro J.A., Li X., Jin D.J., Court D.L., Strathern J.N. A Cre transcription fidelity reporter identifies GreA as a major RNA proofreading factor in Escherichia coli. Genetics. 2017;206:179–187. doi: 10.1534/genetics.116.198960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irvin J.D., Kireeva M.L., Gotte D.R., Shafer B.K., Huang I., Kashlev M., Strathern J.N. A genetic assay for transcription errors reveals multilayer control of RNA polymerase II fidelity. PLoS Genet. 2014;10:e1004532. doi: 10.1371/journal.pgen.1004532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Imashimizu M., Takahashi H., Oshima T., McIntosh C., Bubunenko M., Court D.L., Kashlev M. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16:98. doi: 10.1186/s13059-015-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nascent elongating transcript sequencing coupled to RNase footprinting reveals ubiquitous pauses of transcription and an unusually high proportion of stalled misincorporated complexes (see also annotation to Ref. [19••]).
- 31.Gout J.F., Thomas W.K., Smith Z., Okamoto K., Lynch M. Large-scale detection of in vivo transcription errors. Proc Natl Acad Sci U S A. 2013;110:18584–18589. doi: 10.1073/pnas.1309843110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Churchman L.S., Weissman J.S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nascent elongating transcript sequencing revealed ubiquitous transcription pausing in bacteria, and the high proportion of misincorporated complexes found in a later analysis (Ref. [19••]).
- 34.Vermulst M., Denney A.S., Lang M.J., Hung C.W., Moore S., Moseley M.A., Thompson J.W., Madden V., Gauer J., Wolfe K.J. Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat Commun. 2015;6:8065. doi: 10.1038/ncomms9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carey L.B. RNA polymerase errors cause splicing defects and can be regulated by differential expression of RNA polymerase subunits. Elife. 2015:4. doi: 10.7554/eLife.09945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brulliard M., Lorphelin D., Collignon O., Lorphelin W., Thouvenot B., Gothie E., Jacquenet S., Ogier V., Roitel O., Monnez J.M. Nonrandom variations in human cancer ESTs indicate that mRNA heterogeneity increases during carcinogenesis. Proc Natl Acad Sci U S A. 2007;104:7522–7527. doi: 10.1073/pnas.0611076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxowsky T.T., Meadows K.L., Klungland A., Doetsch P.W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Leeuwen F.W., de Kleijn D.P., van den Hurk H.H., Neubauer A., Sonnemans M.A., Sluijs J.A., Koycu S., Ramdjielal R.D., Salehi A., Martens G.J. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 39••.Gordon A.J., Halliday J.A., Blankschien M.D., Burns P.A., Yatagai F., Herman C. Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 2009;7:e44. doi: 10.1371/journal.pbio.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first evidence that transcription fidelity may have heritable phenotypic consequences by affecting bistable switches.
- 40.Gordon A.J., Satory D., Halliday J.A., Herman C. Heritable change caused by transient transcription errors. PLoS Genet. 2013;9:e1003595. doi: 10.1371/journal.pgen.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norman T.M., Lord N.D., Paulsson J., Losick R. Stochastic switching of cell fate in microbes. Annu Rev Microbiol. 2015;69:381–403. doi: 10.1146/annurev-micro-091213-112852. [DOI] [PubMed] [Google Scholar]
- 42.Erie D.A., Hajiseyedjavadi O., Young M.C., von Hippel P.H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 43••.Dutta D., Shatalin K., Epshtein V., Gottesman M.E., Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reveals that backtracked elongation complexes, and possibly misincorporated ones, present a direct obstacle for replication forks.
- 44•.Trautinger B.W., Jaktaji R.P., Rusakova E., Lloyd R.G. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]; This work reveals the role of transcription proofreading factors in the resolution of conflicts between transcription and replication.
- 45.Tehranchi A.K., Blankschien M.D., Zhang Y., Halliday J.A., Srivatsan A., Peng J., Herman C., Wang J.D. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutherford S.T., Lemke J.J., Vrentas C.E., Gaal T., Ross W., Gourse R.L. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamba P., James K., Zenkin N. A link between transcription fidelity and pausing in vivo. Transcription. 2017;8:99–105. doi: 10.1080/21541264.2016.1274812. [DOI] [PMC free article] [PubMed] [Google Scholar]